Abstract
Peroxidatic activation of the anti-tuberculosis pro-drug isoniazid by Mycobacterium tuberculosis catalase-peroxidase (KatG) is regulated by gating residues of a heme access channel. The steric restriction at the bottleneck of this channel is alleviated by replacement of residue Asp137 with Ser, according to crystallographic and kinetic studies.
Several antibiotics are currently in use to treat tuberculosis (TB) infections including one of the first anti-TB agents, isoniazid (INH, isonicotinic acid hydrazide). Our goals include explaining the catalytic function of Mycobacterium tuberculosis (M.tb) catalase-peroxidase (KatG) in formation of a bactericidal molecule through oxidation of this pro-drug and how mutations in KatG found in INH-resistant strains of the TB pathogen interfere with the process. INH resistance is a global problem in TB treatment.
Catalase-peroxidases protect aerobic microorganisms from oxidative damage through their high catalase activity (2H2O2 → 2H2O + O2). M.tb lacks monofunctional catalase genes, and KatG therefore is crucial for virulence in macrophage infection by this human pathogen.1 On the other hand, KatG’s peroxidatic activity is central to INH action as it converts the pro-drug into an active bactericidal molecule in vivo.2
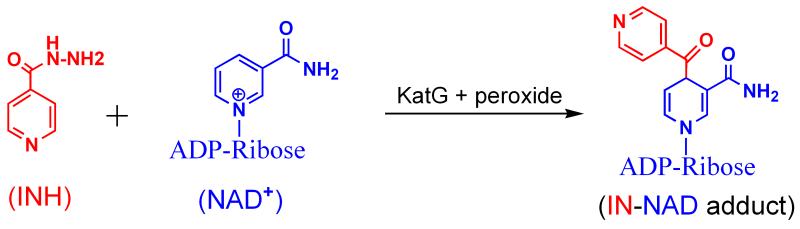
Scheme 1 summarizes the process of INH activation. Oxidation of the hydrazide group by KatG, followed by nonenzymatic acylation of NAD+ by the isonicotinoyl fragment of INH formed after nitrogen release, produces the IN-NAD adduct molecule shown.3 IN-NAD is a potent inhibitor of the vital InhA, an enoyl reductase of Fatty Acid Synthase II in M.tb responsible for biosynthesis of cell wall components.4, 5 Thus, non-lethal mutations either in the katg gene or associated with InhA are responsible for nearly all INH resistance in clinical isolates of M.tb.6
Scheme 1.
Activation of the pro-drug INH by KatG
Some of the KatG mutations linked to INH resistance (for example, Ser315Thr or Asn, and Asn138Ser) are consistent with these replacements interfering with enzyme-pro-drug interactions near the heme edge. Other loci associated with resistance, such as Asp735Asn,7 are remote from the heme. The most common INH-resistant M. tb. strain has the [Ser315Thr] replacement in KatG. We reported that a steric block within a substrate access channel, created by the methyl group of Thr, explained poor activation kinetics for this mutant.8 There is no X-ray crystal structure evidence, however, for INH being bound to KatG near the heme edge analogous to the cytochrome c peroxidase-INH complex.9 In fact, and contradicting conclusions about Ser315 mutants, a crystal structure of B. pseudomallei KatG in complex with INH shows the drug may be bound in any of three sites remote from the heme.10 One site is near Trp139 (Trp135 in M.tb KatG), a residue proposed to be responsible for INH oxidation by a radical formed during turnover of KatG with alkyl peroxide.10 These issues impede formulating a coherent view of INH activation by KatG, prompting our structural and kinetic studies to document the effects of amino acid replacement near the heme edge.
The side chain of Asp137 in WT KatG is opposite residue Ser315 at the base of a substrate access channel. We reasoned that replacing this Asp with Ser could enhance INH oxidation rates and thereby enhance IN-NAD formation, the converse of the effect produced by the Ser315Thr mutation causing resistance. Here, the 3-dimensional crystal stucture of M.tb KatG[Asp137Ser] was solved and revealed an enlarged access channel. This mutant also exhibited greatly improved INH-activation catalysis compared to WT KatG. Another mutant, [Arg418Leu] was examined because like the Asp137Ser mutant, it lacks catalase activity, but does not exhibit altered INH activation. Other mutants were used in the activation kinetics study to probe structural issues as described below.
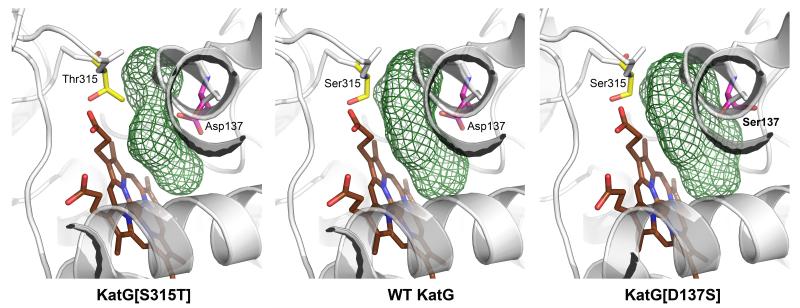
KatG[Asp137Ser] and KatG[Arg418Leu] were crystallized under conditions similar to those previously reported.8 The structures of these mutants were refined to 2.5 Å and 3.1 Å respectively (ESI,† Table S1 and Fig. S2), and there are no significant differences in the overall structures compared to WT KatG (2CCA.pdb) (Cα RMSD values = 0.3-0.4 Å). The region of most interest is the bottom of a substrate access channel where a bottleneck is formed by the carboxyl of Asp137, which is evident in WT KatG and is narrower in the Ser315Thr mutant (Fig. 1).8 In the Asp137Ser mutant, the Ser137 side-chain points away from the channel (while the Ser315 side chain is unchanged), forming a hydrogen bond to the carbonyl of Gly226 (ESI,† Fig. S2B). Thus, in contrast with the restricted access in KatG[Ser315Thr] causing INH KatG 13, 14 as the primary event in activation, and that drug radicals are generated.15, 16 INH may also react with amino acid radicals generated in KatG by internal electron transfers also mediated by hypervalent heme.14, 17 The above mentioned reports relied upon using excess alkyl peroxide for enzymatic turnover. A more physiologically relevant route to initiate catalysis by KatG involves using glucose oxidase to generate a slow flux of H2O2. Such a biomimetic approach, while it does not allow direct observation of enzyme intermediates, leads to formation of the IN-NAD adduct in the presence of INH and NAD+. We have relied on this method (notably without addition of Mn ions used elsewhere 10, 18 to enhance IN-NAD formation) to provide an apparent KM for INH in the peroxidatic activation of the drug by KatG. The steps proposed for activation include the following (irreversible) reactions:
| (1) |
| (2) |
| (3) |
Dilute H2O2 was generated in situ using aerobic glucose/glucose oxidase.8 Reaction of resting KatG with peroxide is rapid (though only a small fraction of KatG will be turning over) and generates an intermediate (KatG) at the oxidation level of peroxidase Compound I,19 which is the species assumed to oxidize INH to a hydrazyl radical.15, 16 resistance, the Asp137Ser mutant exhibits an expansion of this site. The diameter at the bottleneck is calculated to be 2.7 Å in the Ser315Thr mutant, 3.6 Å in WT KatG, and 4.6 Å in the Asp137Ser mutant (Fig. 1). The substrate access channel in the Arg418Leu mutant is very similar to that in WT KatG. No alterations occur at the Trp135 site (ESI† Fig. S3) proposed as a residue responsible for INH activation by a radical.10.
Fig. 1.
Side view of the substrate access channel in M.tb KatG and in mutants S315T and D137S. This figure was generated using PyMol 11 based on crystal structures of M.tb KatG (2CCA.pdb), KatG[S315T] (2CCD.pdb), and KatG[D137S] (4C50.pdb). The substrate access channels (green mesh) were generated with HOLLOW 12 using a 1.4 Å probe radius.
Prior reports by us and other laboratories presented evidence that INH reacts with high-valent (ferryl) heme in This radical is subject to rapid non-enzymatic rearrangements and nitrogen release, leading to an acyl radical that acylates NAD+ also non-enzymatically, to give IN-NAD.20, 21
The rates of IN-NAD generation were determined spectrophotometrically8 as a function of INH concentration and were fitted to a hyperbola from which the INH concentration at 1/2Vmax was calculated as in a Michaelis-Menten kinetics model (See ESI† Fig. S4). Since a large excess of NAD+ was supplied, the rate of IN-NAD formation is directly dependent upon the concentration of INH-derived radical, which reflects the efficiency of drug turnover (oxidation) by the active enzyme intermediate.
The apparent KM for IN-NAD formation is 192 μM for WT KatG and 17.5 μM for KatG[Asp137Ser] (Table 1). This difference reflects a higher apparent “affinity” making the mutant about 8 times more efficient than WT KatG at low INH concentrations. On the other hand, the KM for the Ser315Thr mutant is around 40-fold higher than for WT enzyme, which is in agreement with other reports showing that INH affinity in this mutant is significantly reduced.22, 23 One complication here is that the Asp137Ser replacement causes KatG to lose catalase activity,24, 25 raising the possibility that enhanced activation arises from greater availability of H2O2 and a higher rate of Rxn. 1. While a change in KM value may not be expected as a result, we documented the impact of the lack of catalase turnover by testing two other catalase-deficient KatG mutants, Tyr229Phe and Arg418Leu,25-27 having replacements close to and far from the heme (ESI,† Fig. S1). Both mutants exhibited apparent KM values close to that for WT KatG (Table 1). These findings argue against effects due to disruption of catalase activity (and also eliminate a role in activation for the covalent Met-Tyr-Trp crosslink, as KatG[Tyr229Phe] lacks this modification required for catalase activity 26, 28, 29). In fact, in our biomimetic assay, a low level (3 μM/min) of H2O2 is provided; with INH as a reducing substrate in excess (and low micromolar enzyme) catalase turnover will be suppressed because of a low rate of reaction expected for the second turnover by H2O2 required for formation of oxyferrous heme, the steady-state species in the KatG catalase reaction.30 Replacement of Trp321 proposed to be the site of an initial radical in a pathway from heme to surface amino acid radicals including one formed on Trp135,10 had no impact on INH activation.
Table 1.
Rates (Vmax) and apparent KM values for formation of IN-NAD in a biomimetic assay. a
| KatG/mutants | Vmax | K M | kcat/KM |
|---|---|---|---|
| (μM/min) | (μM) | (mM−1min−1) | |
| WT | 1.52 ± 0.02 | 192 ± 8 | 7.9 |
| Asp137Ser | 1.12 ± 0.02 | 17.5 ± 1 | 64 |
| Ser315Thr | 0.16 ± 0.01 | 8400 ± 700 | 0.02 |
| Arg418Leu | 0.76 ± 0.02 | 165 ± 16 | 4.6 |
| Tyr229Phe | 1.03 ± 0.03 | 101 ± 19 | 10.2 |
| Trp321Phe | 1.30 ± 0.02 | 227 ± 13 | 5.7 |
KatG (1 μM), NAD ( 200 μM), H2O2 (3.0 μM/min), and varying INH.
The Vmax values do not vary widely except for KatG[Ser315Thr]. We speculate that the low value results from a slow rate of INH entry into the restricted active site such that hypervalent heme intermediates are not efficiently utilized, and are instead reduced in reactions producing amino acid radicals.17, 31 Other possibilities such as slow product release may also play a role (as suggested by a reviewer).
In summary, this study demonstrates that altering the dimensions of the bottleneck in the substrate access channel in KatG can impede or enhance INH peroxidation rates relative to the WT enzyme. The biomimetic assay kinetics in parallel with the structural findings are best interpreted as evidence for INH reacting near the heme edge in KatG rather than at amino acid radicals at surface sites.
Supplementary Material
Footnotes
Electronic Supplementary Information (ESI) available: [Experiental details, kinetics fitting and crystallization data are included]. See DOI: 10.1039/b000000x/
The coordinates and structure factors of KatG[D137S] and KatG[R418L] have been deposited in the Protein Data Bank through EMBL-EBI (http://www.pdbe.org/) with acess codes 4C50 and 4C51.
This work was supported by NIH grant 2R56AI060014-06A1 (NIAID) and NSF grant CHE-1058116 to R.S.M, and by The Research Council of Norway (RCN) grants 214239/F20 and 218412/F50 and Cost Action CM1003 to K.K.A with additional support by RCN grant 216625/F50 and European Community’s Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant 283570) on project 1760. We gratefully acknowledge the ESRF (MX-1201, MX-1377) and SLS (20110683, 20111245) for providing beamtime, and the team at ID29 at the ESRF, the team at X10SA (SLS) and Dr. Guillaume Pompidor for valuable help.
references
- 1.Ng VH, Cox JS, Sousa AO, MacMicking JD, McKinney JD. Mol. Microbiol. 2004;52:1291–1302. doi: 10.1111/j.1365-2958.2004.04078.x. [DOI] [PubMed] [Google Scholar]
- 2.Wakamoto Y, Dhar N, Chait R, Schneider K, Signorino-Gelo F, Leibler S, McKinney JD. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 3.Wilming M, Johnsson K. Angew. Chem. Int. Ed. Engl. 1999;38:2588–2590. doi: 10.1002/(sici)1521-3773(19990903)38:17<2588::aid-anie2588>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y. Res. Microbiol. 1993;144:143–149. doi: 10.1016/0923-2508(93)90029-2. [DOI] [PubMed] [Google Scholar]
- 5.Rozwarski DA, Grant GA, Barton DHR, Jacobs WR, Jr., Sacchettini JC. Science. 1998;279:98–102. doi: 10.1126/science.279.5347.98. [DOI] [PubMed] [Google Scholar]
- 6.Slayden RA, Barry CE., 3rd Microbes Infect. 2000;2:659–669. doi: 10.1016/s1286-4579(00)00359-2. [DOI] [PubMed] [Google Scholar]
- 7.Wei C-J, Lei B, Musser JM, Tu S-C. Antimicrob. Agents Chemother. 2003;47:670–675. doi: 10.1128/AAC.47.2.670-675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao X, Yu H, Yu S, Wang F, Sacchettini JC, Magliozzo RS. Biochemistry. 2006;45:4131–4140. doi: 10.1021/bi051967o. [DOI] [PubMed] [Google Scholar]
- 9.Metcalfe C, Macdonald IK, Murphy EJ, Brown KA, Raven EL, Moody PC. J. Biol. Chem. 2008;283:6193–6200. doi: 10.1074/jbc.M707412200. [DOI] [PubMed] [Google Scholar]
- 10.Wiseman B, Carpena X, Feliz M, Donald LJ, Pons M, Fita I, Loewen PC. J. Biol. Chem. 2010;285:26662–26673. doi: 10.1074/jbc.M110.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrodinger, LLC . The PyMOL Molecular Graphics System. 2010. Version 1.3r1. [Google Scholar]
- 12.Ho B, Gruswitz F. BMC Struct. Biol. 2008;8:49. doi: 10.1186/1472-6807-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu S, Chouchane S, Magliozzo RS. Protein Sci. 2002;11:58–64. doi: 10.1110/ps.09902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh R, Switala J, Loewen PC, Ivancich A. J. Am. Chem. Soc. 2007;129:15954–15963. doi: 10.1021/ja075108u. [DOI] [PubMed] [Google Scholar]
- 15.Wengenack NL, Rusnak F. Biochemistry. 2001;40:8990–8996. doi: 10.1021/bi002614m. [DOI] [PubMed] [Google Scholar]
- 16.Ranguelova K, Suarez J, Magliozzo RS, Mason RP. Biochemistry. 2008;47:11377–11385. doi: 10.1021/bi800952b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chouchane S, Girotto S, Yu S, Magliozzo RS. J. Biol. Chem. 2002;277:42633–42638. doi: 10.1074/jbc.M207916200. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen M, Quemard A, Broussy S, Bernadou J, Meunier B. Antimicrob. Agents Chemother. 2002;46:2137–2144. doi: 10.1128/AAC.46.7.2137-2144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hersleth HP, Ryde U, Rydberg P, Görbitz CH, Andersson KK. J. Inorg. Biochem. 2006;100:460–476. doi: 10.1016/j.jinorgbio.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 20.Johnsson K, Schultz PG. J. Am. Chem. Soc. 1994;116:7425–7426. [Google Scholar]
- 21.Amos RI, Gourlay BS, Schiesser CH, Smith JA, Yates BF. Chem. Commun. 2008:1695–1697. doi: 10.1039/b719570b. [DOI] [PubMed] [Google Scholar]
- 22.Wengenack NL, Todorovic S, Yu L, Rusnak F. Biochemistry. 1998;37:15825–15834. doi: 10.1021/bi982023k. [DOI] [PubMed] [Google Scholar]
- 23.Yu S, Girotto S, Lee C, Magliozzo RS. J. Biol. Chem. 2003;278:14769–14775. doi: 10.1074/jbc.M300326200. [DOI] [PubMed] [Google Scholar]
- 24.Jakopitsch C, Auer M, Regelsberger G, Jantschko W, Furtmuller PG, Ruker F, Obinger C. Biochemistry. 2003;42:5292–5300. doi: 10.1021/bi026944d. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Khajo A, Jarrett S, Suarez J, Levitsky Y, Burger RM, Jarzecki AA, Magliozzo RS. J. Biol. Chem. 2012;287:37057–37065. doi: 10.1074/jbc.M112.401208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, Girotto S, Zhao X, Magliozzo RS. J. Biol. Chem. 2003;278:44121–44127. doi: 10.1074/jbc.M304757200. [DOI] [PubMed] [Google Scholar]
- 27.Carpena X, Wiseman B, Deemagarn T, Herguedas B, Ivancich A, Singh R, Loewen PC, Fita I. Biochemistry. 2006;45:5171–5179. doi: 10.1021/bi060017f. [DOI] [PubMed] [Google Scholar]
- 28.Jakopitsch C, Auer M, Ivancich A, Ruker F, Furtmuller PG, Obinger C. J. Biol. Chem. 2003;278:20185–20191. doi: 10.1074/jbc.M211625200. [DOI] [PubMed] [Google Scholar]
- 29.Cade CE, Dlouhy AC, Medzihradszky KF, Salas-Castillo SP, Ghiladi RA. Protein Sci. 2010;19:458–474. doi: 10.1002/pro.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakopitsch C, Wanasinghe A, Jantschko W, Furtmuller PG, Obinger C. J. Biol. Chem. 2005;280:9037–9042. doi: 10.1074/jbc.M413317200. [DOI] [PubMed] [Google Scholar]
- 31.Ivancich A, Jakopitsch C, Auer M, Un S, Obinger C. J. Am. Chem. Soc. 2003;125:14093–14102. doi: 10.1021/ja035582+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.