Abstract
The Bloom syndrome helicase, BLM, has numerous functions that prevent mitotic crossovers. We used unique features of Drosophila melanogaster to investigate origins and properties of mitotic crossovers that occur when BLM is absent. Induction of lesions that block replication forks increased crossover frequencies, consistent with functions for BLM in responding to fork blockage. In contrast, treatment with hydroxyurea, which stalls forks, did not elevate crossovers, even though mutants lacking BLM are sensitive to killing by this agent. To learn about sources of spontaneous recombination, we mapped mitotic crossovers in mutants lacking BLM. In the male germline, irradiation-induced crossovers were distributed randomly across the euchromatin, but spontaneous crossovers were nonrandom. We suggest that regions of the genome with a high frequency of mitotic crossovers may be analogous to common fragile sites in the human genome. Interestingly, in the male germline there is a paucity of crossovers in the interval that spans the pericentric heterochromatin, but in the female germline this interval is more prone to crossing over. Finally, our system allowed us to recover pairs of reciprocal crossover chromosomes. Sequencing of these revealed the existence of gene conversion tracts and did not provide any evidence for mutations associated with crossovers. These findings provide important new insights into sources and structures of mitotic crossovers and functions of BLM helicase.
Keywords: mitotic recombination, BLM helicase, crossovers
MEIOTIC recombination was discovered 100 years ago by T. H. Morgan and his students in classic studies of Drosophila genetics (Morgan 1911). Since that time, a great deal has been learned about the functions, molecular mechanisms, and regulation of meiotic recombination. This process is initiated through the introduction of programmed DNA double-strand breaks (DSBs), which are then repaired through highly regulated homologous recombination (HR) pathways such that a substantial fraction of repair events produce reciprocal crossovers (reviewed in Kohl and Sekelsky 2013). The chiasmata that form at sites of crossovers help to ensure accurate segregation of homologous chromosomes. In addition, crossovers generate chromosomes with novel combinations of alleles at linked loci, leading to increased genetic diversity.
A quarter century after the discovery of meiotic recombination, Curt Stern, also working with Drosophila, found that crossovers can occur in somatic cells (Stern 1936). This phenomenon is usually called “mitotic recombination,” although most such events are thought to occur during interphase rather than in mitosis per se. Compared to meiotic recombination, little is known about mitotic recombination. Except in some specialized cases, like antibody gene rearrangement, mitotic recombination occurs in response to DNA damage (spontaneous or exogenously induced). Mitotic recombination, like meiotic recombination, can be initiated by DSBs, but it is unclear whether DSBs constitute a substantial fraction of the events that initiate spontaneous mitotic recombination.
There are crucial differences in how DSB repair proceeds in mitotically proliferating cells compared to meiotic cells (reviewed in Andersen and Sekelsky 2010). First, meiotic DSB repair uses HR exclusively, whereas mitotically proliferating cells use both HR and homology-independent mechanisms. Second, mitotic HR typically involves use of the sister chromatid as a repair template rather than the homologous chromosome, as in meiosis. Third, a substantial fraction of meiotic DSBs are repaired as crossovers, but HR in mitotic cells tends to occur through pathways that do not produce crossovers.
Structure-selective DNA helicases are major contributors to the prevention of crossovers in mitotic cells. Foremost among these is BLM helicase, so-named because mutations in BLM cause the hereditary disorder Bloom syndrome. The predominant clinical features of Bloom syndrome are small size and a high risk for early onset of a broad range of cancers (German and Ellis 1998). Genetic and biochemical studies have shown that BLM and its orthologs can disassemble recombination intermediates that might otherwise be processed through pathways that produce crossovers (van Brabant et al. 2000; Adams et al. 2003; Ira et al. 2003; Wu and Hickson 2003; Oh et al. 2007; De Muyt et al. 2012). The strong anticrossover functions of BLm are evident in cellular phenotypes associated with loss of BLM, including an elevation in crossovers between sister chromatids (sister chromatid exchange, SCE), homologous chromosomes, and heterologous chromosomes (German 1964; Chaganti et al. 1974).
Drosophila melanogaster has advantages as a metazoan model for studying mitotic crossovers. The absence of meiotic crossovers in the males (Morgan 1912) means that mitotic crossovers that occur in the male germline can be easily detected among progeny. Also, in Dipteran insects, pairing of homologous chromosomes is not restricted to meiotic cells, but occurs in somatic and premeiotic germline cells (Stevens 1908). Consequently, the homologous chromosome is frequently used as a template during DSB repair (Rong and Golic 2003). Thus, a substantial fraction of recombination events that give rise to SCEs in other species may instead result in crossovers between homologous chromosomes in Drosophila; crossovers between homologous chromosomes are much more amenable to genetic and molecular analyses than SCEs.
We took advantage of these features of Drosophila to investigate mitotic crossovers that occur in Drosophila BLM (formerly mus309) mutants. Spontaneous mitotic crossovers are highly elevated in Blm mutants (Johnson-Schlitz and Engels 2006; McVey et al. 2007), suggesting that these mutants may be a good model for discovering the origins of spontaneous mitotic crossovers in Bloom syndrome cells. To investigate these origins, we treated Blm mutants with a variety of DNA damaging agents to determine which types of damage induce mitotic crossovers. As a complementary approach, we knocked out specific repair pathways in Blm mutants to determine which of these remove spontaneous damage that can lead to crossovers if left unrepaired. We also mapped spontaneous mitotic crossovers that occur in Blm mutants and found that the distribution is nonrandom, suggesting that some sites or regions of the genome are more prone to damage than others. Finally, we sequenced the exchange sites of pairs of reciprocal crossovers. Our findings reveal important new information about sources and structures of mitotic crossovers and functions of BLM helicase.
Materials and Methods
Mitotic crossover assays
Unless otherwise noted, mutants were heteroallelic or hemizygous for amorphic alleles (supporting information, Table S1). Premeiotic mitotic crossovers in the male germline were measured as in McVey et al. (2007). Crosses were done to generate males of the desired genotype that were heterozygous for st and e markers on chromosome 3. DNA damaging agents were added to food containing larvae from these crosses, as in Yıldız et al. (2002). Doses used (expressed as concentration of stock solution added to food) were 0.01% methyl methanesulfonate (MMS), 0.004% nitrogen mustard mechlorethamine (HN2), 0.01% camptothecin (CPT) (in DMSO), and 120 mM hydroxyurea (HU). Ultraviolet (UV) dose was 100 J/m2. For HU, we also measured sensitivity to killing, since this had not previously been reported. Sensitivity was measured as in Yıldız et al. (2002). In untreated vials, there were 357 control adults and 228 Blm mutants. In vials treated with 100 mM HU, there were 113 control adults and 17 mutants (two-tailed P < 0.0001 by Fisher’s exact test).
To score mitotic crossovers, single adult males of the desired genotype that emerged from these cultures were crossed to ste virgin females and the progeny were scored as being parental or recombinant (Figure 1A). An average of 50–100 progeny were obtained from each male; vials with progeny counts at least two standard deviations below the mean (generally <10–15 progeny) were discounted. Because crossovers are predominantly or exclusively premeiotic, single crossover events can give rise to clusters of progeny. We therefore treated each single male as a separate experiment. One-way ANOVA tests were done using Prism 6.03 (GraphPad), with Bonferroni correction for multiple comparisons. MMS, UV, and HU treatments were done at the same time as the untreated control shown in Figure 1B. HN2 was done several years later with a simultaneous untreated control. This was not significantly different from the original control, but statistical significance was determined by an unpaired t-test to the contemporaneous control set. CPT treatment had its own untreated control in which DMSO (the solvent used to dissolve CPT) was added to the food. An unpaired t-test was done to compare treated to control.
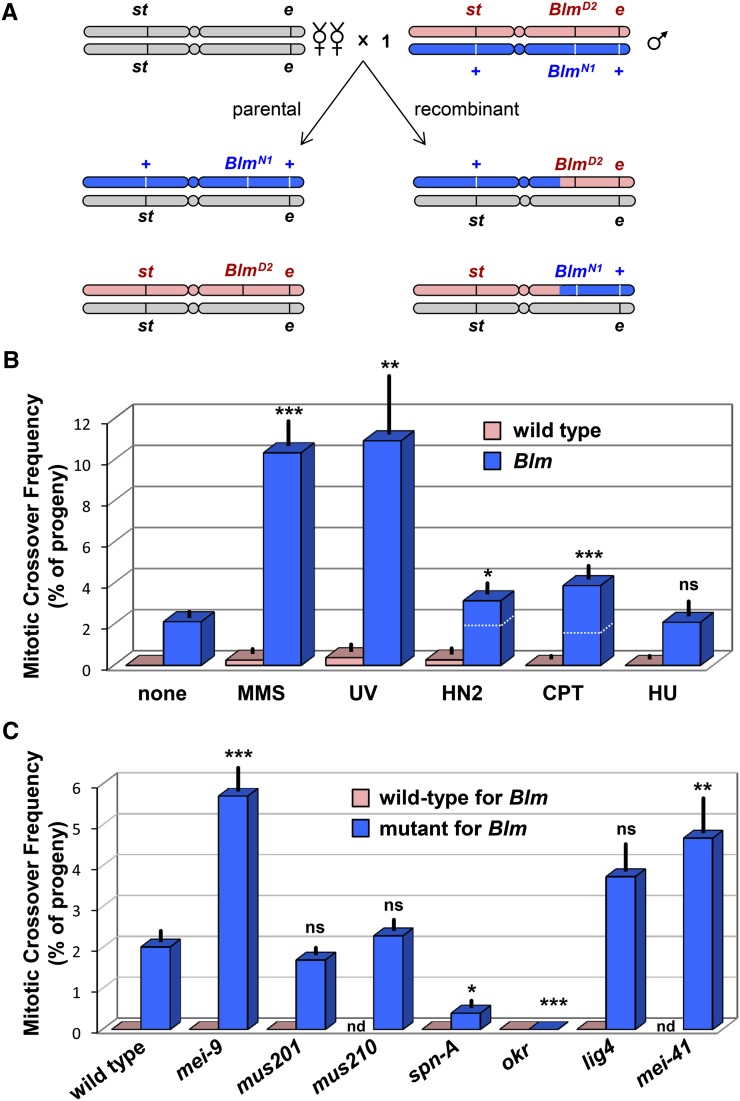
Figure 1.
Effects of DNA damaging agents and DNA repair defects on mitotic crossover rate in Blm mutants. (A) Schematic of method to measure mitotic crossovers. Single males heteroallelic for amorphic Blm mutations and heterozygous for st and e are crossed to tester females. Progeny are scored as being parental (left) or recombinant (right) for st and e. The two recombinant classes are drawn with a crossover in the same position, because we can sometimes recover the two reciprocal products of a single crossover. (B) Frequency of crossovers between st and e in wild-type (pink) and Blm (blue) male germlines after treatment of larvae with the indicated DNA damaging agents. See Materials and Methods for doses. (C) Frequency of male germline crossovers between st and e in various single mutants (pink) and in double mutants with Blm (blue). nd, not done. Error bars are standard error of the mean (n = 16, 35, 9, 14, 25, and 9 males for treatments of Blm in B, left to right; n = 16, 22, 41, 33, 21, 22, 20, and 17 males for Blm mutant genotypes in C). One-way ANOVA test were done to compare each treatment to untreated Blm (B, P < 0.0001) and each double mutant to the Blm single mutant (C, P < 0.0001), with Bonferroni correction for multiple comparisons. Dotted white lines on HN2 and CPT bars indicate values of matched controls. In these cases, unpaired t-tests were done to compare to the matched control (see Materials and Methods). NS, P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001.
Crossover distribution assays
Crosses between balanced stocks generated males homozygous or heteroallelic for Blm and heterozygous for markers on 2L. The experiment depicted in Figure 2A used males of genotype netdppd-hodpbprcn; BlmN1/TM6B and females of genotype P{SUPor-P}GlcATSKG01446; BlmN1/TM6B. Male progeny that were homozygous for BlmN1 and heterozygous for chromosome 2 were crossed to netdppd-hodpbprcn females, and the progeny of this cross were scored for mitotic crossovers. Crossovers that occurred between dp and b were further characterized via PCR to determine whether they occurred proximal or distal to the P-element. In such cases, DNA was obtained via single-fly preps and amplified with the primers GTCTAGTGCCAGGCTACTCG and GCGGACCACCTTATGTTATTTC.
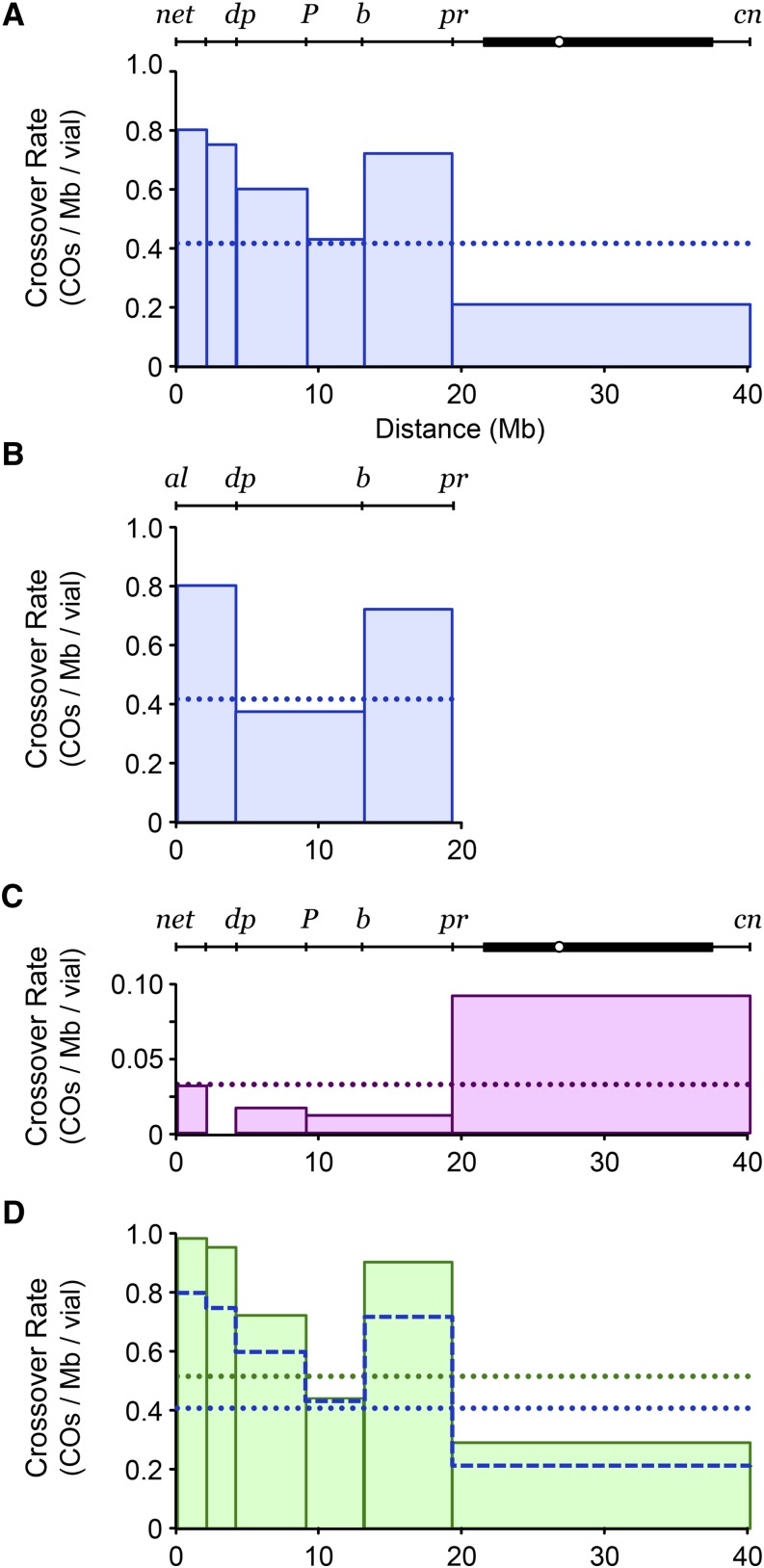
Figure 2.
Mitotic crossover distribution on chromosome 2L. (A) Distribution in Blm mutant males (532 crossovers from 313 males). The drawing at the top depicts the region assayed. Circle, centromere; thick line, pericentric heterochromatin. Bars indicate the crossover frequency in each interval. The dotted line shows the mean frequency across the entire region. Scale is in millions of base pairs (Mbp) from the left end of 2L. (B) Distribution in Blm mutant males (634 crossovers from 391 males) using a different set of chromosome 2 markers. (C) Distribution of mitotic crossovers in the female germline (12 crossovers from 13 vials). Note the different scale than in other panels. (D) Distribution in Blm mutant males (334 crossovers from 157 males) that are heterozygous for a DNApolα-180 mutation. The superimposed dashed blue line is the distribution from A.
For the experiment depicted in Figure 3B, the marker chromosome stock was changed to netdppd-hodpbprcn; ru BlmN1 DNApol-α180 ca/TM6B. Subsequent crosses remained the same.
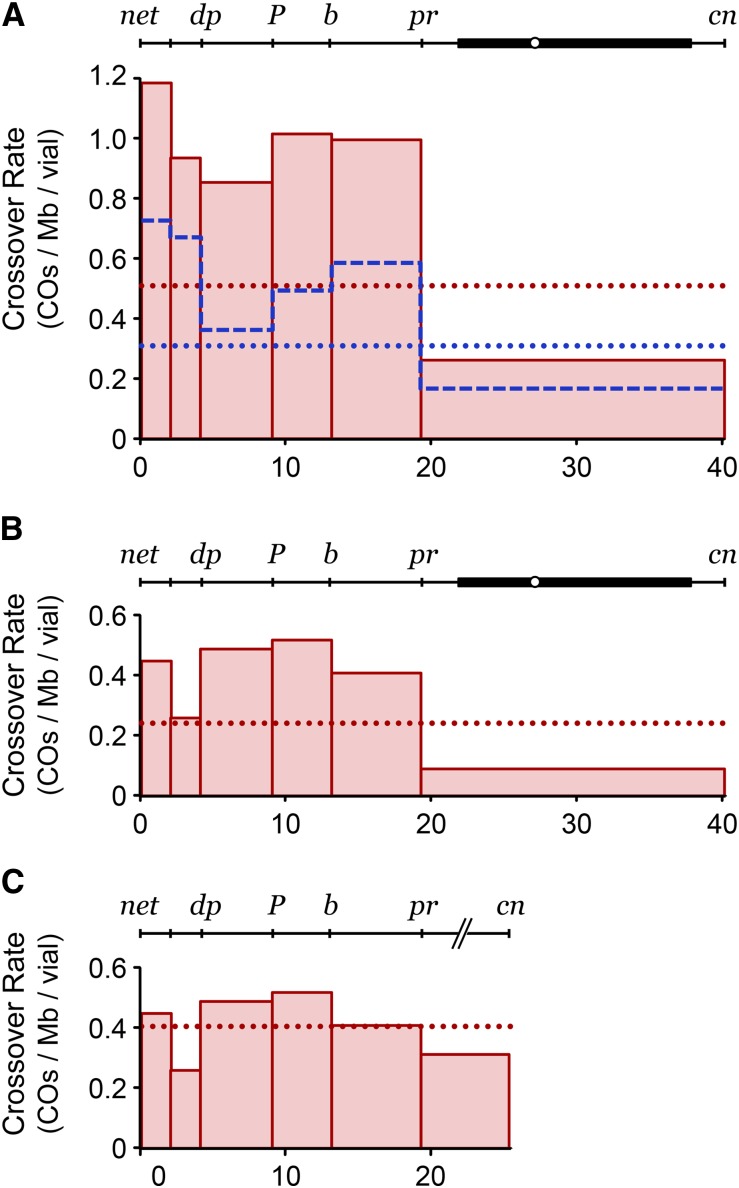
Figure 3.
Distribution of irradiation-induced crossovers. (A) Distribution of crossovers in Blm mutant males exposed to 250 rads of gamma irradiation during larval development; thick, dotted red line shows the mean frequency across the interval assayed (232 crossovers from 101 males). The thin, dashed blue line shows the distribution from unirradiated control males done at the same time; the thick, dotted blue line is the mean frequency in controls (202 crossovers from 140 males). (B) Distribution of crossovers resulting from irradiation. The unirradiated frequency was subtracted from each interval in A, removing spontaneous crossovers and leaving only irradiation-induced crossovers. (C) Data in B were regraphed to exclude the pericentric heterochromatin between pr and cn.
The experiment depicted in Figure 2D began with parental males of genotype al dp b pr cn/SM6a; BlmD2/TM6B and parental females of genotype w; cn bw sp; BlmN1/TM6B. The 2nd chromosome 2 of the females is the reference sequence chromosome, derived from stock no. 2057 from the Bloomington Stock Center. Male progeny that were heteroallelic for Blm and heterozygous for chromosome 2 were crossed to al dp b pr cn females, and progeny of that cross were scored for mitotic crossovers.
To measure mitotic crossovers in the female germline (Figure 2C), females, mutant for mei-P22, which is required to make meiotic DSBs (Liu et al. 2002) and Blm, were used. To overcome the requirement for maternal BLM protein in embryonic development, we expressed BLM from a UASp::Blm transgene using a Matα::GAL4 driver that turns on expression after meiotic recombination is complete, as in Kohl et al. (2012). Due to the low fecundity of mutants that do not do meiotic recombination, we placed 18–22 females into each vial, but still counted each vial as a separate experiment.
We used the DEVIAT program (Cirulli et al. 2007) to perform bootstrapping to test whether crossover distributions were significantly nonuniform. P-values reported were obtained by running 100,000 bootstrapping trials. Correcting for multiple tests did not affect the significance of any results.
Molecular analysis of reciprocal crossover products
Crossover structure analysis was carried out on reciprocal crossovers derived from the experiment depicted in Figure 2B. In vials where male siblings with reciprocal marker configurations were present, each was crossed to y; Pin/SM6a, al dp sp females. The al, dp, and sp markers on SM6a were used to identify presence of the crossover chromosome in progeny. Siblings that carried both the crossover chromosome and SM6a were crossed to each other to make a balanced stock, which were later used to generate multiple individuals with an identical chromosome 2 genotype of al dp b pr cn/CO.
In the case that al and dp were both present on the initial crossover chromosome, it was possible that sp had been crossed off in an unrelated mitotic crossover; as such, the male was first crossed to net dppd-ho dp wgSp-1 b pr cn/SM6a. Male progeny of this cross that were not balanced for chromosome 2 were crossed to y/y+Y; Pin/SM6a, al dp sp females, and the appropriate progeny were crossed to make a stock, as above.
Males that were to be used for single nucleotide polymorphism (SNP) mapping via high-throughput sequencing were taken from these balanced stocks and crossed to al dp b pr cn. Progeny of the genotype al dp b pr cn/CO were collected and frozen at −80°. Genomic DNA was isolated and libraries were prepared for sequencing on the Illumina HiSeq 2000. Four sequencing libraries corresponded to each half of two reciprocal crossovers. SNPs were detected by comparison to the Drosophila reference sequence (release 5). The SNP information gleaned from the sequencing was used to narrow down the location of these crossovers, first by testing restriction fragment-length polymorphisms, and then by sequencing over regions with multiple SNPs. This SNP information was later used to characterize eight additional reciprocal crossovers.
Because each individual chromosome from a reciprocal recombination event was crossed to a reference stock, it was possible to narrow down the region where the crossover event occurred by finding the region where known heterologies switch from being heterozygous to homozygous or vice versa. For each reciprocal recombination pair, primer sets were designed for SNPs located within the region determined to contain the exchange based on phenotypic mapping. PCR and sequencing of these SNP-containing regions was performed until the site of exchange was narrowed to less than the distance between two available heterologies. Then, the region between the two nearest heterologies was amplified and sequenced to search for any insertions, deletions, inversions, or other heterologies that could be used for further mapping.
Results
Agents that block replication fork progression increase mitotic crossovers in Blm mutants
Spontaneous mitotic crossovers in the male germline are elevated by orders of magnitude in Blm mutants (Johnson-Schlitz and Engels 2006; McVey et al. 2007). Treatment of larvae with ionizing radiation (IR), which generates DSBs, causes a further increase, suggesting that DSBs can be a source of these crossovers (McVey et al. 2007). To determine whether BLM prevents crossovers induced by damage other than DSBs, we treated Blm mutant larvae with a variety of agents: CPT, an inhibitor of topoisomerase I, which generates replication-associated DSBs (Liu et al. 2000); MMS, which alkylates bases (Beranek 1990); UV light, which induces primarily pyrimidine dimers and 6,4-photoproducts; HN2, which generates base adducts and interstrand crosslinks (Wijen et al. 2000); and HU. HU inhibits ribonucleotide reductase, leading to depleted deoxyribonucleotide triphosphate (dNTP) pools and consequent slowing and/or stalling of replication (Alvino et al. 2007). Blm mutants are hypersensitive to killing by each of these agents (Boyd et al. 1981; McVey et al. 2007).
Treatment with CPT resulted in increased crossovers (Figure 1B), consistent with a previous study that found elevated crossovers after IR (McVey et al. 2007), and suggesting that DSBs that occur in the context of replication can lead to interhomolog crossovers when BLM is absent. We also detected elevated mitotic crossovers after treatment with MMS, UV, and HN2 (Figure 1B). There was no increase in mitotic crossovers after treatment with HU (Figure 1B), even though Blm mutants are hypersensitive to killing by HU at the dose used (24% survival relative to control; P < 0.0001; see Materials and Methods). Together, our results suggest that BLM is important in responding to broken (CPT), blocked (MMS, UV, and HN2), and slowed or stalled (HU) forks, and that broken or blocked forks may be processed through pathways that can lead to crossovers when BLM is absent.
Effects of eliminating DNA repair pathways on mitotic crossover frequencies in Blm mutants
We next asked whether removing specific DNA repair pathways would affect mitotic crossover frequencies in Blm mutants. We hypothesized that knocking out nucleotide excision repair (NER), a process responsible for removing damage caused by UV and some MMS and HN2 damage, would lead to increased crossover frequency. We used null mutations in mei-9 and mus201, which encode the orthologs of XPF/Rad1 and XPG/Rad2, endonucleases that make nicks 5′ and 3′ of the damaged base, respectively (Sekelsky et al. 2000). Crossovers were significantly elevated in mei-9; Blm mutants relative to Blm single mutants, but were not elevated in mus201; Blm mutants (Figure 1C). Rad1 has NER-independent DNA repair functions (Klein 1988; Fishman-Lobell and Haber 1992; Ivanov and Haber 1995); the crossover elevation caused by removing MEI-9 might be a consequence of disrupting pathways other than NER. We also tested the effect of removing the NER damage recognition protein XPC, which is encoded by the mus210 gene (Sekelsky et al. 2000). Removal of XPC, like removal of XPG, had no effect on crossover rate (Figure 1C).
DSBs can be repaired by HR or by nonhomologous end joining (NHEJ). We knocked out both HR and NHEJ to determine the relative contributions of these pathways in responding to the spontaneous lesions that lead to crossovers in Blm mutants. To knock out HR, we used mutations in spn-A, which encodes Rad51 (Staeva-Vieira et al. 2003), and okr, which encodes Rad54 (Ghabrial et al. 1998). Crossovers were eliminated in okr; Blm and significantly reduced in Blm spn-A mutants (Figure 1C). The residual crossovers in Blm spn-A double mutants (only 4 of 21 males had recombinant progeny) probably result from maternally loaded Rad51 protein and/or transcript (McVey et al. 2004a). These data indicate that, as expected, most or all mitotic crossovers are generated through HR pathways.
We knocked out the canonical NHEJ pathway with a mutation in the DNA ligase 4 gene lig4. If some spontaneous DSBs are repaired through NHEJ, then when NHEJ is compromised, these DSBs might be channeled into BLM-dependent HR pathways, leading to an elevation in crossovers in double mutants with Blm. There was no significant elevation in crossovers in these double mutants, suggesting that NHEJ does not normally play a major role in repairing damage that leads to crossing over when BLM is absent.
Finally, we eliminated the G2-M DNA damage checkpoint with a mutation in mei-41, which encodes the ortholog of ATR (Hari et al. 1995); this led to a significant increase in crossovers (Figure 1C).
The distribution of mitotic crossovers in the absence of BLM is nonrandom
The elevation in mitotic crossovers due to loss of BLM occurs even in the absence of exogenous damage, presumably in response to spontaneous problems (Johnson-Schlitz and Engels 2006; McVey et al. 2007). There are many potential sources of spontaneous problems, including random DNA damage, failure to complete replication before entry into mitosis, and collisions between replication forks and transcription complexes. Some of these events may be more prone to occur in some regions of the genome than others, and thus crossovers might occur more frequently in these regions. To test this idea, we mapped the distribution of crossovers within a 43-Mbp region (∼20% of the Drosophila genome). We used visible markers to divide the region from net, at the left end of 2L, to cn, toward the left end of 2R, into six intervals, and determined rates of crossing over in each interval (Figure 2A). We recovered 532 independent crossovers from males that were homozygous for the deletion allele BlmN1. These crossovers were distributed nonrandomly (P < 0.0001 by bootstrapping), with the net-dp and b-pr intervals having the highest frequencies and the pr-cn region, which includes the centromere and ∼16 Mb of pericentric heterochromatin, having a substantially lower frequency than other intervals. Using a different set of markers, we mapped an additional 634 crossovers from males heteroallelic for BlmN1 and the nonsense allele BlmD2 (Figure 2B). The distribution was also significantly nonrandom (P = 0.0002) in this background. Notably, the regions with the highest frequencies of crossing over were similar in the two experiments.
We also mapped mitotic crossovers in the female germline. Previous mapping of crossovers in Blm mutants revealed an apparently random distribution across the euchromatin, but it is thought that most of these are meiotic crossovers (McVey et al. 2007; Kohl et al. 2012). To determine the contribution of mitotic recombination to this set, we measured crossovers in double mutants with mei-P22, a gene whose product is required to generate meiotic DSBs (Liu et al. 2002). Crossovers are not detected in mei-P22 single mutants (Liu et al. 2002), but do occur in mei-P22 Blm double mutants (Figure 2C). These occur at a much lower frequency than in the male germline (compare the scales in Figure 2, A and C). The distribution of mitotic crossovers in the male germline is strikingly different than the distribution in the female germline. The difference is most prominent in the pr–cn interval, which consists of ∼6.6 Mb of euchromatin and 16 Mb of pericentric heterochromatin. In the male germline, crossovers are least frequent in this interval, whereas in the female germline they are most frequent in this region. Although we mapped only 12 independent crossovers in the female germline, compared to 532 in the male germline, the fraction occurring in the centromere-spanning interval is significantly different between these samples (8 of 12 in the female germline, 134 of 532 in the male germline; P = 0.0034 by two-tailed Fisher’s exact test).
Nonrandom distribution of mitotic crossovers might arise if some regions of the genome are more likely to experience spontaneous problems. In mammalian cells, common fragile sites (CFSs) are regions with an elevated incidence of chromosome breaks when DNA replication is partially impeded, which is usually achieved by growing cells in the presence of a low dose of the DNA polymerase inhibitor aphidicholin (APH) (Debatisse et al. 2012). To test the idea that regions of higher mitotic crossovers in Blm mutants might correspond to or contain CFSs, we genetically mimicked APH treatment by reducing the dosage of the catalytic subunit of DNA polymerase α (Polα), a condition that affects genome stability (LaRocque et al. 2007). Heterozygosity for a null mutation in DNApol-α180 caused an increase in the male germline crossover frequency of flies lacking BLM, but the distribution of crossovers remained strikingly similar (Figure 2D). This result supports the hypothesis that many of the mitotic crossovers recovered in the absence of BLM result from problems encountered during replication.
An alternative explanation for the nonrandom distribution of mitotic crossovers is that BLM-dependent pathways are used to different degrees in different regions of the genome. For example, DSBs in highly repetitive sequences might be repaired through single-strand annealing or end joining pathways that would not be compromised by the absence of BLM. To test this possibility, we treated Blm mutant larvae with ionizing radiation to induce DSBs and then measured germline mitotic crossovers in the resulting adult males. The distribution of IR-induced crossovers was substantially different from the distribution of spontaneous crossovers (Figure 3A). The distribution is still significantly nonrandom (P < 0.0001), but this appears to be driven by the low number of crossovers recovered in the pr-to-cn interval. Since the majority of this interval is made up of the centromere and the pericentric heterochromatin, we hypothesized that crossovers are either absent from or rare within these regions. In support of this hypothesis, when we consider only the euchromatic distance between pr and cn (6.6 Mb instead of 23 Mb; Figure 3C), the distribution is not significantly different from random (P = 0.2133). This is not true for the spontaneous events (without IR treatment), which are significantly nonrandomly distributed even if we omit the heterochromatic length (P = 0.0011 for data in Figure 2A; P = 0.0020 for Polα reduction in Figure 2D) or consider only the five intervals wholly within the euchromatic part of 2L (P = 0.0005; P = 0.0019 for DNA Polα reduction). These findings suggest that when a DSB is induced by IR and repaired in the absence of BLM, the probability that a crossover will be produced is the same across the euchromatin, at least at low resolution. In the heterochromatin, however, either DSB repair is independent of BLM or a noncrossover pathway is used (see Discussion). We conclude that the nonrandom distribution of spontaneous crossovers that occurs in the absence of BLM is most likely due to a nonrandom distribution of initiating lesions.
Molecular structures of mitotic crossovers
Additional insights into sources of mitotic crossovers can be obtained from molecular analysis of crossover chromosomes. In our male germline assays, crossovers arise during premeiotic mitotic proliferation. This can result in an individual crossover being recovered multiple times in a cluster of progeny. In some cases, the presumptive reciprocal product is present in siblings. This permits molecular analysis of reciprocal mitotic recombination products, something that has not been possible in previous studies of metazoan mitotic recombination.
We isolated 10 independent pairs of siblings with reciprocal crossover marker configurations. Two pairs were subjected to Illumina sequencing. This allowed us to determine crossover positions, which were within 10 kilobase pairs (kbp) of one another in both cases (Figure 4), and to identify SNPs between the two parental chromosomes. We used these SNPs to determine crossover positions in the remaining 8 pairs. In two cases, the crossover sites were separated by several megabases, suggesting that these chromosomes were derived from different recombination events in the same germline. These were not analyzed further. In another example, both crossover sites fell within an 80-kb region within which no additional SNPs were identified. This pair was also not analyzed further. In the remaining five cases, the two crossover sites were near one another, consistent with them being bona fide reciprocal crossover products.
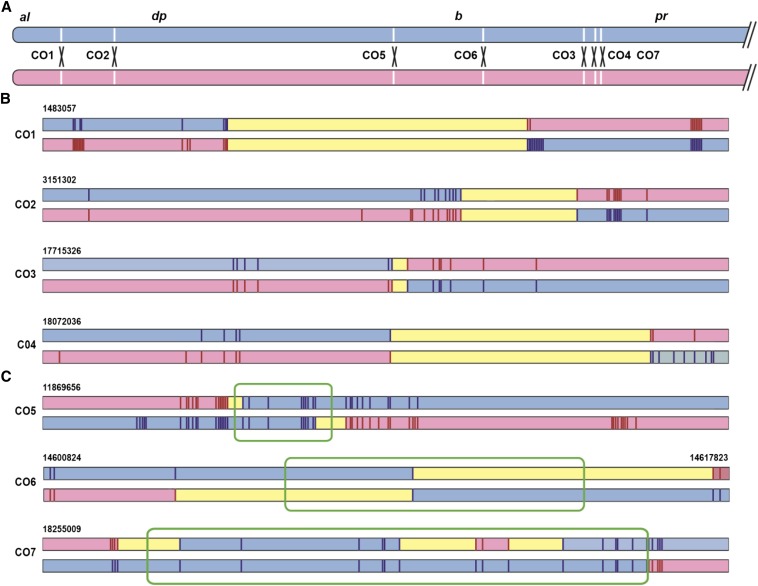
Figure 4.
Structures of reciprocal mitotic crossover products. (A) The euchromatic left arm of chromosome 2 is depicted with the locations of crossovers analyzed at the sequence level. The marker chromosome is blue and the reference chromosome is pink. (B) Molecular structures of reciprocal crossover products in which gene conversion tracts were not detected. Each line represents a 10-kb region surrounding the crossover site. Regions inferred to be derived from the marker chromosome are shaded in blue and those from the reference chromosome in pink. Yellow segments represent regions within which the chromosomal origin cannot be determined; exchanges occurred with these regions. Vertical lines indicate polymorphisms that were definitively genotyped. In some cases, DNA samples were exhausted before all polymorphisms could be genotyped on both products. (C) Molecular structures of reciprocal crossover products with evidence for associated gene conversion tracts. Colors are as in B. Green boxes indicate regions of gene conversion. Note that for CO6, the region included is 17 kb instead of 10 kb as in all other cases in panels B and C.
We used Sanger sequencing to sequence crossover regions for the seven pairs of reciprocal crossover chromosomes, including the two analyzed previously by Illumina sequencing. In four of these pairs the exchange sites on both chromosomes fell between the same pair of adjacent SNPs, supporting the inference that these are reciprocal products of single crossover events. These crossovers did not have detectable gene conversion tracts. The distances between SNPs in these cases, which represents the maximum possible size of undetectable conversion tracts, ranged from 573 bp to 4420 bp (mean = 2536 bp). In the other three pairs, crossover sites were in different SNP intervals, revealing the existence of gene conversion tracts associated with these crossovers. In CO5, the conversion tract, which includes nine SNPs, is between 1057 and 1748 bp. In CO6, the tract includes only a single SNP, but the nearest identified polymorphisms are 5876 bp to the left and 7539 bp to the right; therefore, the length of this tract is between 1 and 13,415 bp. CO7 has a complex tract. The conversion tract is between 6847 and 7831 bp long, but on one chromosome the converted region is interrupted by an unconverted segment of 697–2385 bp, spanning three SNPs. Potential origins of this structure are outlined in Discussion.
In this analysis, we sequenced >28,000 bp of DNA in regions encompassing crossover points (i.e., between the nearest flanking SNPs), and >70,000 bp in regions within 10 kb of a crossover site. We did not detect any de novo sequence changes, such as new SNPs, insertions, or deletions. Based on these data, the rate of mutation associated with these crossovers is <10−4 per base pair.
Discussion
Functions of BLM in preventing mitotic crossovers
Our data indicate that DSBs and damage that is predicted to block replication forks induce mitotic crossing over in mutants lacking BLM. Similarly, treatment of Bloom syndrome patient-derived cells with the alkylating agent ethyl methanesulfonate (EMS) leads to elevated SCEs (Krepinsky et al. 1979). These findings support models in which BLM is important in managing forks when DNA synthesis is blocked. Damage that occurs outside of S phase can certainly also lead to mitotic crossovers. For example, DSBs generated enzymatically and gaps resulting from P-element excision are associated with mitotic crossing over when BLM is absent (Johnson-Schlitz and Engels 2006; S. L. Andersen and J. Sekelsky, unpublished data). This is likely to reflect roles of BLM in directing noncrossover outcomes of DSB repair. Since this topic that has been discussed at length elsewhere (e.g., Andersen and Sekelsky 2010), we restrict the discussion below to the less well understood roles of BLM in replication fork repair.
It has been proposed that BLM catalyzes regression of blocked forks, a process that is thought to both stabilize the fork against breakage and allow repair complexes to access the damage (Ralf et al. 2006; Wu and Hickson 2006). An alternative suggested by genetic experiments in Drosophila is that another enzyme catalyzes regression and that BLM reverses the regression to allow fork restart after repair (Andersen et al. 2011). Both models propose that forks that cannot be regressed or reversed may either break spontaneously or be cleaved by structure-selective endonucleases. In the absence of BLM, DSB repair often leads to crossing over, resulting in elevated SCEs (for repair using the sister) or mitotic crossing over (for repair using the homologous chromosome).
Interestingly, treatment with hydroxyurea, which is thought to slow or stall fork progression, was not associated with increased crossover frequency in our studies. Blm mutants are hypersensitive to killing by the doses used, so BLM does participate in the response to slowed or stalled fork progression. There are a number of possible explanations. One is that BLM-independent mechanisms of dealing with stalled forks do not involve DSB induction and therefore are unlikely to result in crossovers. There may be one or more other helicases that can partially compensate for the absence of BLM at paused forks instead of nuclease-mediated DSB formation. Candidates include FANCM and MARCAL1, as orthologs of these proteins have been implicated in fork reversal in vertebrates (Gari et al. 2008; Bétous et al. 2012). Another possibility is that HU-induced recombination occurs only between sister chromatids and would therefore not be detected in our assay. It is also possible that the reduction in dNTP pools precludes recombinational processes that require DNA synthesis. Given that about half the Blm larvae survive to adulthood at the HU doses used, extensive DNA replication must be possible, although recombination may still be inhibited by local or transient reductions in dNTP pools. Finally, cells that lack BLM may have no other pathway for managing HU-stalled forks, triggering apoptosis. Our assay requires that cells go through meiosis and make mature, functional sperm. We did not observe any decrease in the number of progeny produced by Blm males when they were treated with HU (data not shown), suggesting that cell death was not pervasive, but modest elevations in cell death frequency might still go undetected due to rapid proliferation in the germline.
Knocking out NHEJ had no effect on crossover frequency (Figure 1C), despite previous studies in Drosophila that have revealed roles for both NHEJ and HR in repairing DSBs in the male germline (Preston et al. 2006; Bozas et al. 2009; Beumer et al. 2013). These experiments involved enzymatic induction of DSBs, probably throughout the cell cycle. Numerous studies in yeast and mammalian cells indicate that NHEJ predominates during G1 and HR predominates during S and G2 (reviewed in Chapman et al. 2012), so it is perhaps not surprising that roles for both NHEJ and HR are observed. NHEJ is rarely used to repair breaks produced by P-element excision, except in the absence of Rad51 (McVey et al. 2004a). It was suggested that excision occurs primarily or exclusively during S and G2, when HR predominates. Similarly, if our crossover assay is responding to DSBs or other lesions that occur during S phase, they would normally be repaired by HR.
Based on this discussion and previously proposed models, we hypothesize that the extreme elevation in crossovers observed when BLM is absent is explained by a combination of altered processing of replication fork lesions (e.g., production of DSBs by cleavage of regressed forks that cannot be reversed, as in Andersen et al. 2011) and loss of a major anticrossover activity during DSB repair by HR (reviewed in Andersen and Sekelsky 2010).
Common fragile sites and the distribution of spontaneous mitotic crossovers
We mapped spontaneous mitotic crossovers in the germlines of males that lack BLM (Figure 2 and Figure 3). Within the region analyzed (∼20% of the genome) crossover distribution was highly nonrandom. Crossovers likely occur near the location of the initiating event, suggesting that some regions of the genome are more prone to experiencing these initiating events. We hypothesize that these regions may constitute CFSs in Drosophila. In mammalian cells, CFSs are defined as regions that frequently experience chromosome breakage when cells experience inhibition of DNA polymerases, typically accomplished by growing cells in a low dose of APH (reviewed in Durkin and Glover 2007). In support of our hypothesis, genetically reducing DNA polymerase alpha resulted in a higher rate of mitotic crossovers while retaining the same nonrandom distribution. Breakage at CFSs is also increased in ATR mutants (Casper et al. 2002); similarly, mitotic crossovers were highly elevated by removal of Drosophila ATR (Figure 1C), although we did not measure distribution in this background.
The relationship between BLM, CFSs, and crossovers is complex. Sister chromatid exchange is elevated at CFSs (Glover and Stein 1987; Hirsch 1991; Gaddini et al. 1995). Elevated SCEs is a hallmark of Bloom syndrome cells (Chaganti et al. 1974), but whether the elevation occurs preferentially at CFSs has not been reported. Nonetheless, there is a clear connection between BLM and CFSs. Mammalian cells in culture frequently have ultrafine DNA bridges (UFBs) that are decorated with BLM protein (Chan et al. 2007). One class of UFB is associated with CFSs and is induced by APH (Chan et al. 2009). BLM is present at these sites in the absence of DSBs and the number of UFBs increases in cells lacking BLM. Because of this, Chan et al. (2009) hypothesized that BLM helps to resolve connections between sister chromatids that arise after replication stress, particularly at regions with intrinsic replication difficulties, like CFSs. In the absence of BLM, linkages at CFSs are more likely to persist and break. In this scenario, the elevation in crossovers is due to a combination of increased DSBs and differences in the outcome of DSB repair. This is similar to the models for fork blockage described above, where BLM may have a role first in preventing DSBs and second in promoting noncrossover repair of any DSBs that do arise.
The existence of CFSs in Drosophila offers a parsimonious explanation for the nonrandom distribution of mitotic crossovers in Blm mutants. Given the resolution of our mapping we cannot say whether each of the elevated regions has a single CFS or merely a higher density of CFSs than other regions. High-resolution mapping of a large number of mitotic crossovers will answer the question of CFS density and perhaps provide unique insights into causes of fragility.
In the male germline, crossovers were lowest in the region that spans that centromere and pericentric heterochromatin (Figure 2 and Figure 3). Chan et al. (2007) noted that BLM does decorate a class of UFB associated with centromere regions. They hypothesized that these occur at regions that have not completed replication due to the late timing of replication of heterochromatic sequences, and that BLM helps to decatenate such unreplicated regions to allow mitosis to proceed. The absence of BLM would be expected to lead to more DSBs in heterochromatin, and therefore more crossovers. The paucity of crossovers in heterochromatic regions may result from the use of BLM-independent DSB repair pathways in these regions. Given the repetitive nature of heterochromatic sequences, one might expect that most HR repair of DSBs in heterochromatin will occur through the single-strand annealing (SSA) pathway, which does not require BLM (Johnson-Schlitz and Engels 2006). However, Chiolo et al. (2011) found that repair of heterochromatic DSBs in Drosophila Kc167 cells is dependent on Rad51 and Rad54, suggesting that repair occurs through HR. Interestingly, breaks were moved out of the heterochromatin compartment of the nucleus before loading of Rad51, possibly to prevent recombination with other chromosome regions with the same repetitive sequences. The authors suggest that HR using sister chromatids or perhaps homologous chromosomes, if they are relocated with the broken chromosome, will ensure genome stability. Our finding that crossovers between homologous chromosomes are rare in heterochromatic regions suggests that the homolog is not a frequent template for repair, at least in the male mitotic germline, perhaps because it does not relocate with the broken chromosome.
In contrast to the situation in the male germline, crossovers in the female germline appear to be elevated in the interval that spans the centromere. The markers we used did not allow us to determine whether these crossovers are occurring within the heterochromatin vs. the centromere-proximal euchromatin. Likewise, we cannot say what fraction of the male germline crossovers in this interval are in euchromatin vs. heterochromatin. Nonetheless, we speculate that differences in chromatin structure, perhaps related to the fact that chromosomes undergo synapsis and recombination only in female meiosis, are a major contributor to differences in mitotic crossover maps.
Molecular structures of mitotic crossovers
Our system for studying spontaneous mitotic crossovers allowed us to sequence both reciprocal products of individual crossover events. Most of the crossovers we analyzed had structures compatible with current models of crossover formation via an intermediate with Holliday junctions—either no detectable gene conversion tract or a single tract of conversion. The exception is CO7, which had a complex conversion tract. This type of tract could be the result of multiple cycles of strand invasion, synthesis, and dissociation. In this case, there would have been at least one round of DNA repair synthesis using the homologous chromosome as a template, followed by at least one round using the sister chromatid, and then again using the homologous chromosome. Previous studies demonstrated that repair of large double-stranded gaps in Drosophila involves multiple such cycles (McVey et al. 2004a). Although BLM is required for the dissociation step, there is residual dissociation in Blm mutants, due either to maternally loaded BLM that has persisted in the germline or to other helicases that can weakly compensate for the absence of BLM (Adams et al. 2003).
As discussed above, BLM is thought to help to decatenate replication forks that experience problems when converging in regions susceptible to replication difficulties, such as CFSs. In the absence of BLM, such regions may spontaneously break during anaphase or they may be cut by structure-selective endonucleases. If cuts are introduced at both forks, this may lead to a double-stranded DNA gap. Repair of gaps in the absence of BLM often results in deletions extended into adjacent sequences (Adams et al. 2003; McVey et al. 2004b). We did not detect any deletions among the crossovers we analyzed, but our sample size was small. Analysis of additional crossovers, particularly those associated with CFSs or produced in backgrounds that lack BLM and additional DNA repair proteins, is therefore likely to yield important insights into both sources of spontaneous lesions and mechanisms of repair.
Acknowledgments
We thank Mohamed Noor for providing the DEVIAT program, Susan Cheek for technical assistance, and Xiaojun Guan for analysis of Illumina sequence for identification of SNPs. This work was supported by grants from the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health to J.S., under awards R01 GM099890 and R01 GM061252. M.C.L., S.L.A., J.K.H., and K.P.K. were supported in part by NIGMS award T32 GM007092. E.P.S. was supported by a grant from the NIGMS division of Training, Workforce Development, and Diversity under the Institutional Research and Academic Career Development award K12 GM000678.
Footnotes
Communicating editor: S. Bickel
Literature Cited
- Adams M. D., McVey M., Sekelsky J., 2003. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299: 265–267. [DOI] [PubMed] [Google Scholar]
- Alvino G. M., Collingwood D., Murphy J. M., Delrow J., Brewer B. J., et al. , 2007. Replication in hydroxyurea: it’s a matter of time. Mol. Cell. Biol. 27: 6396–6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Sekelsky J., 2010. Meiotic vs. mitotic recombination: two different routes for double-strand break repair: the different functions of meiotic vs. mitotic DSB repair are reflected in different pathway usage and different outcomes. Bioessays 32: 1058–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. L., Kuo H. K., Savukoski D., Brodsky M. H., Sekelsky J., 2011. Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet. 7: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek D. T., 1990. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 231: 11–30. [DOI] [PubMed] [Google Scholar]
- Bétous R., Mason A. C., Rambo R. P., Bansbach C. E., Badu-Nkansah A., et al. , 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer, K. J., J. K. Trautman, K. Mukherjee, and D. Carroll, 2013 Donor DNA utilization during gene targeting with zinc-finger nucleases. G3 3: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd J. B., Golino M. D., Shaw K. E. S., Osgood C. J., Green M. M., 1981. Third-chromosome mutagen-sensitive mutants of Drosophila melanogaster. Genetics 97: 607–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozas A., Beumer K. J., Trautman J. K., Carroll D., 2009. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 182: 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper A. M., Nghiem P., Arlt M. F., Glover T. W., 2002. ATR regulates fragile site stability. Cell 111: 779–789. [DOI] [PubMed] [Google Scholar]
- Chaganti R. S., Schonberg S., German J., 1974. A manyfold increase in sister chromatid exchanges in Bloom’s syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71: 4508–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. L., North P. S., Hickson I. D., 2007. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 26: 3397–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K. L., Palmai-Pallag T., Ying S., Hickson I. D., 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol. 11: 753–760. [DOI] [PubMed] [Google Scholar]
- Chapman J. R., Taylor M. R., Boulton S. J., 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47: 497–510. [DOI] [PubMed] [Google Scholar]
- Chiolo I., Minoda A., Colmenares S. U., Polyzos A., Costes S. V., et al. , 2011. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell 144: 732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli E. T., Kliman R. M., Noor M. A., 2007. Fine-scale crossover rate heterogeneity in Drosophila pseudoobscura. J. Mol. Evol. 64: 129–135. [DOI] [PubMed] [Google Scholar]
- De Muyt A., Jessop L., Kolar E., Sourirajan A., Chen J., et al. , 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol. Cell 46: 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M., Le Tallec B., Letessier A., Dutrillaux B., Brison O., 2012. Common fragile sites: mechanisms of instability revisited. Trends Genet. 28: 22–32. [DOI] [PubMed] [Google Scholar]
- Durkin S. G., Glover T. W., 2007. Chromosome fragile sites. Annu. Rev. Genet. 41: 169–192. [DOI] [PubMed] [Google Scholar]
- Fishman-Lobell J., Haber J. E., 1992. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science 258: 480–484. [DOI] [PubMed] [Google Scholar]
- Gaddini L., Pelliccia F., Limongi M. Z., Rocchi A., 1995. Study of the relationships between common fragile sites, chromosome breakages and sister chromatid exchanges. Mutagenesis 10: 257–260. [DOI] [PubMed] [Google Scholar]
- Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A., 2008. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29: 141–148. [DOI] [PubMed] [Google Scholar]
- German J., 1964. Cytological evidence for crossing-over in vitro in human lymphoid cells. Science 144: 298–301. [DOI] [PubMed] [Google Scholar]
- German J., Ellis N., 1998. Bloom syndrome, pp. 301–315 in The Genetic Basis of Human Cancer, edited by Vogelstein B., Kinzler K. McGraw-Hill, New York. [Google Scholar]
- Ghabrial A., Ray R. P., Schüpbach T., 1998. okra and spindle-B encode components of the RAD52 DNA repair pathway and affect meiosis and patterning in Drosophila oogenesis. Genes Dev. 12: 2711–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover T. W., Stein C. K., 1987. Induction of sister chromatid exchanges at common fragile sites. Am. J. Hum. Genet. 41: 882–890. [PMC free article] [PubMed] [Google Scholar]
- Hari K. L., Santerre A., Sekelsky J., McKim K. S., Boyd J. B., et al. , 1995. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell 82: 815–821. [DOI] [PubMed] [Google Scholar]
- Hirsch B., 1991. Sister chromatid exchanges are preferentially induced at expressed and nonexpressed common fragile sites. Hum. Genet. 87: 302–306. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J. E., 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov E. L., Haber J. E., 1995. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 2245–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Schlitz D., Engels W. R., 2006. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc. Natl. Acad. Sci. USA 103: 16840–16845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein H., 1988. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics 120: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Sekelsky J., 2013. Meiotic and mitotic recombination in meiosis. Genetics 194: 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl K. P., Jones C. D., Sekelsky J., 2012. Evolution of an MCM complex in flies that promotes meiotic crossovers by blocking BLM helicase. Science 338: 1363–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepinsky A. B., Heddle J. A., German J., 1979. Sensitivity of Bloom’s syndrome lymphocytes to ethyl methanesulfonate. Hum. Genet. 50: 151–156. [DOI] [PubMed] [Google Scholar]
- LaRocque J. R., Dougherty D. L., Hussain S. K., Sekelsky J., 2007. Reducing DNA polymerase alpha in the absence of Drosophila ATR leads to P53-dependent apoptosis and developmental defects. Genetics 176: 1441–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Jang J. K., Kato N., McKim K. S., 2002. mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics 162: 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Desai S. D., Li T. K., Mao Y., Sun M., et al. , 2000. Mechanism of action of camptothecin. Ann. N. Y. Acad. Sci. 922: 1–10. [DOI] [PubMed] [Google Scholar]
- McVey M., Adams M., Staeva-Vieira E., Sekelsky J. J., 2004a Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167: 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Larocque J. R., Adams M. D., Sekelsky J. J., 2004b Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion. Proc. Natl. Acad. Sci. USA 101: 15694–15699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Andersen S. L., Broze Y., Sekelsky J., 2007. Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H., 1911. Random segregation vs. coupling in mendelian inheritance. Science 34: 384. [DOI] [PubMed] [Google Scholar]
- Morgan T. H., 1912. Complete linkage in the second chromosome of the male of Drosophila. Science 36: 719–720. [Google Scholar]
- Oh S. D., Lao J. P., Hwang P. Y., Taylor A. F., Smith G. R., et al. , 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. R., Flores C. C., Engels W. R., 2006. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics 172: 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralf C., Hickson I. D., Wu L., 2006. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281: 22839–22846. [DOI] [PubMed] [Google Scholar]
- Rong Y. S., Golic K. G., 2003. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165: 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelsky J., Hollis K. J., Eimerl A. I., Burtis K. C., Hawley R. S., 2000. Nucleotide excision repair endonuclease genes in Drosophila melanogaster. Mutat. Res. 459: 219–228. [DOI] [PubMed] [Google Scholar]
- Staeva-Vieira E., Yoo S., Lehmann R., 2003. An essential role of DmRad51/SpnA in DNA repair and meiotic checkpoint control. EMBO J. 22: 5863–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern C., 1936. Somatic crossing over and segregation in Drosophila melanogaster. Genetics 21: 625–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens N. M., 1908. A study of the germ cells of certain diptera, with reference to the heterochromosomes and the phenomena of synapsis. J. Exp. Biol. 5: 359–383. [Google Scholar]
- van Brabant A. J., Ye T., Sanz M., German I. J., Ellis N. A., et al. , 2000. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39: 14617–14625. [DOI] [PubMed] [Google Scholar]
- Wijen J. P., Nivard M. J., Vogel E. W., 2000. The in vivo genetic activity profile of the monofunctional nitrogen mustard 2-chloroethylamine differs drastically from its bifunctional counterpart mechlorethamine. Carcinogenesis 21: 1859–1867. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I. D., 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I. D., 2006. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40: 279–306. [DOI] [PubMed] [Google Scholar]
- Yıldız Ö., Majumder S., Kramer B. C., Sekelsky J., 2002. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol. Cell 10: 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]