Abstract
Crossovers play mechanical roles in meiotic chromosome segregation, generate genetic diversity by producing new allelic combinations, and facilitate evolution by decoupling linked alleles. In almost every species studied to date, crossover distributions are dramatically nonuniform, differing among sexes and across genomes, with spatial variation in crossover rates on scales from whole chromosomes to subkilobase hotspots. To understand the regulatory forces dictating these heterogeneous distributions a crucial first step is the fine-scale characterization of crossover distributions. Here we define the wild-type distribution of crossovers along a region of the C. elegans chromosome II at unprecedented resolution, using recombinant chromosomes of 243 hermaphrodites and 226 males. We find that well-characterized large-scale domains, with little fine-scale rate heterogeneity, dominate this region’s crossover landscape. Using the Gini coefficient as a summary statistic, we find that this region of the C. elegans genome has the least heterogeneous fine-scale crossover distribution yet observed among model organisms, and we show by simulation that the data are incompatible with a mammalian-type hotspot-rich landscape. The large-scale structural domains—the low-recombination center and the high-recombination arm—have a discrete boundary that we localize to a small region. This boundary coincides with the arm-center boundary defined both by nuclear-envelope attachment of DNA in somatic cells and GC content, consistent with proposals that these features of chromosome organization may be mechanical causes and evolutionary consequences of crossover recombination.
Keywords: recombination, Caenorhabditis elegans, heterogeneity, hotspots
MEIOTIC recombination creates novel combinations of parental alleles and is a powerful force shaping the genetic variation observed within a population. The frequency of recombination has a profound impact on the efficacy of natural selection in both purging deleterious mutations and in the formation of beneficial allelic combinations (Hill and Robertson 1966; Cutter and Payseur 2013). However, the intensity of recombination in different parts of a genome is rarely constant. Heterogeneity in the meiotic recombination rate has been observed across chromosome lengths and at finer scales of a few kilobase in diverse organisms.
In Caenorhabditis elegans, meiotic recombination rates show a striking and reproducible chromosome-wide pattern of variation. Genetic maps of the five autosomes and X chromosome show that each has a low-recombination central domain flanked by high-recombination arm domains (Barnes et al. 1995; Rockman and Kruglyak 2009). This domain structure has dramatic effects on genetic variation and evolution (Cutter and Payseur 2003; Rockman et al. 2010; Andersen et al. 2012), but the underlying causes generating this pattern are not completely understood.
The domain structure relates to unusual characteristics of the C. elegans chromosomes. The chromosomes lack localized centromeres and exhibit holocentric segregation during mitosis, with kinetochore proteins assembling along the length of each chromosome (Albertson and Thomson 1982). The center and arm regions are defined not by cytology but by recombination rate: gene-dense central clusters emerged from the species’ earliest genetic map (Brenner 1974). With the genome sequence (C. elegans Sequencing Consortium 1998), extensive annotation (Gerstein et al. 2010), and characterization of chromatin modification (Liu et al. 2011), it has become clear that the low-recombination central regions of C. elegans chromosomes are gene dense and enriched for highly expressed genes and markers of open chromatin, while the high-recombination arms are enriched for repetitive sequences, genes with low expression, and markers of closed chromatin.
C. elegans chromosomes exhibit nearly complete crossover interference, with elaborate regulatory mechanisms that ensure that each pair of chromosomes receives exactly one crossover per meiosis (Hillers and Villeneuve 2003; Hammarlund et al. 2005). Variation in crossover frequency along the chromosomes therefore reflects the location preferences of the single crossover event. The preference for chromosome arms may relate to the mechanical requirements for meiotic chromosome segregation in a species lacking discrete centromeres. An off-center crossover defines an acting “short arm” and “long arm” of each bivalent, and different proteins localize to each of these arms to facilitate appropriate regulation of sister-chromatid cohesion (reviewed by Schvarzstein et al. 2010). Because the short and long arm are defined randomly in each meiosis, their designation depends on chomosome-wide communication of the crossover location and a clear distinction between short and long.
The mechanisms that define the arms and centers are unknown, although both structural modifications to chromosomes (e.g., chromosome fusions, Hillers and Villeneuve 2003; translocations, McKim et al. 1988; and insertions, Hammarlund et al. 2005) and perturbations of trans-acting factors [e.g., condensins (Tsai et al. 2008; Mets and Meyer 2009), xnd-1 (Wagner et al. 2010), him-5 (Meneely et al. 2012), slx-1 (Saito et al. 2013), and rec-1 (Zetka and Rose 1995)] can alter crossover location distributions. Many of these factors work in part by affecting the chromatin state of the DNA, which is known to influence crossover distribution in many species (Buard et al. 2009; Pan et al. 2011; Henderson 2012).
The fine-scale distribution of crossovers within chromosomal domains is also poorly characterized. Whether C. elegans chromosomes contain crossover hotspots (1- to 2-kb regions of high recombination in a background of low recombination; Paigen and Petkov 2010), like most well-studied eukaryotes, is unknown, and no recombination-associated cis-acting elements have been identified. If hotspots and cis-acting elements exist, high-resolution crossover location data would facilitate their discovery. In many species, high-resolution crossover distribitions have been inferred from population-genomic data (e.g., McVean et al. 2004), but C. elegans has too little outcrossing—and consequently too much linkage disequilibrium—for those methods to work (Stumpf and McVean 2003). Only direct measurement can reveal the crossover distribution.
Here, we provide an unparalleled view of the crossover distribution across a single region spanning 2.275 Mb of the C. elegans genome. We have used dense molecular genotyping and targeted genetic crosses to generate location data for 243 hermaphrodite- and 226 male-specific crossovers. These data yield a map of crossover distribution across the center-right arm boundary of C. elegans chromosome II. These empirical measures of the meiotic recombination rate allow us to examine the heterogeneity in this regional crossover landscape and to test for the existence of hotspots. We also compare our high-resolution recombination rate map to currently known distributions of genomic features to explore the relationship of these variables with both the position of the recombination rate boundary and local heterogeneity in the C. elegans meiotic crossover rate.
Materials and Methods
Previous work suggests that the chromosome II center-right arm boundary resides at approximately position 12.02 Mb (Rockman and Kruglyak 2009). This boundary is a good candidate for our empirical investigation as it appears to be relatively sharp (Barnes et al. 1995; Rockman and Kruglyak 2009) and is flanked by easily scored phenotypic markers (unc-4 and rol-1).
Strains
We acquired CB4856, a wild isolate from Hawaii, and SP419, which carries unc-4 (e120) rol-1 (e91) II in the N2 genetic background, from the Caenorhabditis Genetics Center. To precisely localize the causal mutations in unc-4 and rol-1, we sequenced the SP419 genome by Illumina HiSeq. We generated 100-bp paired-end reads and mapped them to the N2 reference genome using stampy (Lunter and Goodson 2011). We called variants using samtools (Li et al. 2009). We identified a C > T mutation at 9,897,945 (ws200) in a splice-acceptor site in the unc-4 gene as e120, consistent with its original molecular description (Winnier et al. 1999). In rol-1, the causal mutation underlying e91 has not been described. We found a nonsynonymous C > T mutation in rol-1 at position 12,173,584 (ws200). The resulting amino acid replacement (R80C) occurs in an evolutionarily conserved site and is computationally predicted by SIFT (Kumar et al. 2009) and PolyPhen-2 (Adzhubei et al. 2010) to damage ROL-1 function.
Cross design and experimental conditions
Worms were thawed, cleaned by bleaching (Stiernagle 2006), and maintained for several generations at 20° on NGM agar plates seeded with OP50 bacteria before experiments were initiated. All experiments were performed at 20°.
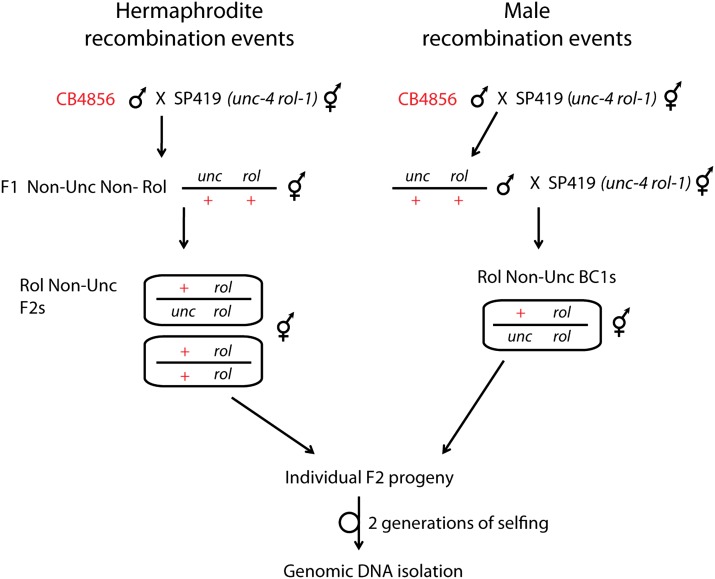
To score for hermaphrodite-specific recombination events, heterozygous F1 hermaphrodite progeny from a cross between SP419 hermaphrodites and CB4856 males were picked as L4 larvae and moved to new NGM plates seeded with OP50 bacteria. Recombinant F2 Rol non-Unc adults were singled and allowed to self for approximately two generations to generate large populations for genomic DNA extraction. Figure 1 shows a schematic of the cross.
Figure 1.
Overview of genetic cross design. Hermaphrodite crossovers are recovered from Rol non-Unc F2s, which may carry one or two recombinant chromosomes. Male crossovers are recovered from Rol non-Unc backcross progeny, which carry a single recombinant chromosome.
To score for male-specific recombination events, heterozygous male F1 progeny were collected and backcrossed to SP419 hermaphrodites. Rol non-Unc backcross progeny were singled and samples prepared as above for genomic DNA extraction.
Genomic DNA extraction and Illumina Golden Gate-based genotype assay
Genomic DNA was extracted using a standard salting-out protocol, as in Rockman and Kruglyak (2009). The DNA was genotyped by Illumina GoldenGate Assay at 183 N2/CB4856 SNPs in the interval between unc-4 and rol-1. The SNPs were selected to focus on the recombination rate boundary region. The majority of SNPs are between positions 11.6 Mb and rol-1 (e91). Genotyping was performed by the DNA Sequencing and Genomics Core Facility of the University of Utah, Salt Lake City.
Genotype data quality control and scoring of recombination breakpoints
Fluorescence intensity scatterplots for all SNP genotypes were inspected manually (Supporting Information, Figure S1). SNPs that passed the rigorous quality control steps were then used to identify crossover positions in each of the samples. Exploiting the fact that C. elegans shows complete crossover interference, all the SNPs sorted by physical position were examined to look for a switch in genotype from heterozygosity to homozygosity. The final data set was trimmed to 122 SNPs that were genotyped reliably in every sample, to allow for identical marker sets for all analyses.
Analyses
All statistical analyses were performed in R (R Core Team 2013). The data set consists of counts of recombination breakpoints observed between each pair of markers, with each breakpoint indexed by the sex of the animal in which the recombination occurred (File S1). To accommodate the structure of the data set—unequal marker-interval sizes and small numbers of observations per interval—we used permutations and simulations to test hypotheses about the underlying process that generated the data.
Sex differences
We first tested for sex-specific recombination rate patterns by two-sample two-tailed Kolmogorov–Smirnov test. The null distribution for the test statistic was determined by 100,000 permutations of each breakpoint’s sex. The resulting permuted data sets have the same numbers of breakpoints per marker interval but their sexes are randomized.
To test for more localized sex-specific rate variation, we performed a likelihood-ratio test of the observed configuration of breakpoints. We compared a shared-rate model, in which the two sexes share a single recombination rate specific to each interval, to a model in which each sex has its own rate. The likelihood of the data under either model is the product, over the marker intervals (indexed by i), of the binomial probability that ki of the ni breakpoints observed in the interval were from a particular sex:
In the one-rate model, the probability p that a breakpoint was generated in that sex is shared across intervals and is simply the fraction of all breakpoints, across all intervals, that were generated in that sex. In the two-rate model, each interval has its own pi, equal to its maximum-likelihood estimate, the fraction of observed breakpoints within that interval that were generated in that sex (i = ki/ni).
The analysis is conditioned on the observed breakpoint distribution, so we exclude marker intervals with zero observed breakpoints from consideration. The resulting analysis includes 100 marker intervals. The null distribution of the LRT statistic was determined by analysis of 100,000 sex-permuted data sets. R functions for these analyses are available as File S2.
Boundary position
We used the R package strucchange 1.4-6 (Zeileis et al. 2002) to localize the center-arm domain boundary. We regressed the per-interval estimate of recombination rate (i.e., the count of breakpoints divided by physical length of the marker interval) on physical position, and we used the Bayesian Information Criterion to identify the optimal number of segments, i.e., nonoverlapping domains defined by different regression coefficients.
Annotation analysis
To identify molecular correlates of recombination rates along the chromosome, we acquired data on several classes of sequence annotation for each marker interval.
GC content was calculated from genome sequence using the GC function in the R package seqinR (Charif and Lobry 2007).
The number of bases covered by repeats was calculated from the RepeatMasker track of c_elegans.WS200.annotations.gff2, downloaded from WormBase (Yook et al. 2012), which annotates 2390 repeat elements in the studied region. Repeats are classed into 594 categories, which include several types of simple sequence repeats and low-complexity tracts (e.g., TTTAn, polypyrimadine, A rich, etc.), dispersed repeats identified de novo computationally (e.g., Ce000163, etc.), and identifiable copies of known transposons (e.g., MARINER5_CE, LINE2B_CE, HELICOP2, etc.). For our calculation of base coverage, we included only the 1323 instances of dispersed repeats, i.e., excluding simple sequence repeats and low complexity tracts, which are predominantly composed of the “AT-rich” class (928 of 1067). We also analyzed the 32 individual repeat types that occur more than 10 times each in the unc-4–rol-1 region. For each, we tested whether the number of occurrences in the arm was as expected from a binomial distribution with an arm probability of 9.2%, corresponding to the proportion of the physical region located in the arm, and used a Bonferroni correction to control for the 32 tests with family-wise error rate of 0.05.The number of gene start sites per interval was calculated from data retrieved from WormBase ws220 using WormMart and remapped to our reference ws200 coordinates. Processed ChIP-chip data on marks associated with open chromatin or meiosis were downloaded the from http://modencode.org in the form of GBrowse.wig tracks. We used the data sets in Table 1.
Table 1. Data from modENCODE.
| Mark | Track | DCCid |
|---|---|---|
| LEM2 | SDQ3891_LEM2_N2_MXEMB | modENCODE_2729 |
| H3K4ME2 | H3K4ME2_N2_L3 | modENCODE_2402 |
| H3K4ME3 | WA30534819_H3K4ME3_N2_YA_s7 | modENCODE_3552 |
| H3K9ME2 | H3K9ME26D11_N2_L3 | modENCODE_2406 |
| H4K8ac | AB15823_H4K8AC487128_N2_L3 | modENCODE_2774 |
| H4K16ac | MP07329_H4K16ACDAM1612187_N2_EEMB | modENCODE_3182 |
To test for arm-center differences, we used multivariate logistic regression. The binary response variable was arm or center, defined by the crossover domain boundary found by strucchange analysis, and the predictors were the signal intensities from the wig tracks, averaged across the SNP marker intervals. To plot the .wig track data, we first recentered the values on the chromosome II average values. To magnify the signal, we then cubed the values. This transformation shrinks small values toward zero and amplifies extremes, making them easier to visualize.
Modeling constant-rate domains
To test whether our observations could be generated by discrete domains with constant rates, we used a parametric bootstrapping approach. We simulated data according to a two-domain model with a rate change at the strucchange-estimated boundary. The two rates were fixed by linear interpolation on the Marey map, which plots genetic position (i.e., the proportion of all observed recombination breakpoints that are to the left of a given physical position) as a function of physical position, yielding a graph in which the slope represents the local recombination rate (Chakravarti 1991). Our simulated data derive from a Marey map consisting of a straight line between the left edge of our studied region and the boundary marker and a straight line from that marker to the right edge of the region.
To test whether larger numbers of constant-rate domains could explain our data, we applied first-order (i.e., piecewise linear) regression splines to the Marey map of the observed data. Our analysis is designed to identify the simplest smoothing function for the Marey map that is consistent with our observations. We modeled increasing numbers of constant-rate domains by increasing the number of knots (i.e., rate changes) in a constrained median b-spline regression implemented in the R package cobs (Ng and Maechler 2007). The regression is constrained to pass through the boundaries of our data [i.e., genetic position 0, physical position unc-4 (e120), and genetic position 1, physical position rol-1 (e91)] and to be monotonically nondecreasing (i.e., negative recombination rates are prohibited). For each number n of knots, we calculated the likelihood ratio of the spline-estimated Marey map compared to a model in which each marker interval has its own rate. We generated a null distribution by simulation, generating data sets of breakpoints distributed according to the spline-estimated Marey map and observed in the same marker intervals as our real data. We calculated the LRT statistic for each simulation. We then asked whether our real data reject a model with n knots at P < 0.05. The analysis is complicated by the challenge of identifying optimal knot locations. In running cobs on our data, we specify the maximum number of knots to allow in the regression. The software then reduces this number by deleting knots whose elimination is justified by the Akaike information criterion (Ng and Maechler 2007). As a result of the knot selection algorithm, the final model for a maximum of n knots may have m knots (m < n) yet have a better fit to the data than a run in which m knots was specified as the maximum. We considered n up to 40. R functions for these analyses are available as File S3.
Modeling hotspots by simulation
A typical description of recombination rate heterogeneity takes the form “y% of recombination occurs in x% of the physical genome.” A simple summary statistic that integrates across the whole range of x’s and y’s is the Gini coefficient. Widely used in studies of income or wealth inequality in the social sciences, the Gini coefficient ranges from zero (perfect equality of rates along the chromosome) to one (all recombination events occur at a single location).
To calculate Gini coefficients, we sorted marker intervals by their recombination rates to generate curves that show the proportion (y) of genetic distance covered by the least recombinant proportion (x) of physical distance for each x (i.e., Lorenz curves; Gastwirth 1972; cf. Figure 2B in Paigen et al. 2008). If recombination rates are constant, the curve will approach y = x, and as rates become less equal along the interval the curve falls below y = x to an extent measured by the Gini coefficient. We used a trapezoidal approximation to calculate the area under the curve (AUC). The Gini coefficient is 1 – (2 × AUC).
Figure 2.
(A) Crossover distributions in the unc-4—rol-1 interval do not differ between hermaphrodites and males. In these Marey map representations, each marker’s relative genetic position (the cumulative fraction of crossover breakpoints to the left of the marker) is plotted as a function of its physical position on the chromosome. Triangles indicate the positions of the SNP markers. (B) Structural change analysis localizes the center-arm boundary to 11.96 Mb. The point estimate of the boundary is shown as a vertical black line on the Marey map of combined hermaphrodite and male data. The red dashed lines mark the confidence interval (the right limit is concealed behind the point estimate). Inset: blue, the improvement of fit induced by adding additional rate domains in the regression of recombination rate on physical position. Black: the tradeoff between explanation and overfitting measured by the Bayesian information criterion. The optimum is at one rate boundary.
To study models of heterogeneous recombination rates, we simulated data according to a recombination map with gamma-distributed rates on a 1-kb scale. That is, each kilobase of simulated chromosome draws its recombination rate from a gamma distribution. We then assign the observed number of crossovers to each simulated kilobase according to their relative rates, positioning the crossovers uniformly within each kilobase. Rates are thus constant within each kilobase but vary among kilobase intervals. To consider different degrees of rate heterogeneity, we used gamma distributions with different shape parameters, ranging from highly heterogeneous rates (shape parameter, 0.01) to relatively constant rates (shape parameter, 100). For each gamma distribution, we simulated 5000 data sets of crossovers. For each data set, we then modeled our observation process by counting how many crossovers occur in each of the bins defined by our actual genetic markers. From these simulated observations we then calculated Gini coefficients. R functions for estimating Gini coefficients and simulating crossover observations are available as File S4.
Results
High-resolution recombination breakpoint data
We used visible markers to select for chromosomes with crossovers in a 2.275-Mb region previously shown to contain the boundary between the center and right arm of C. elegans chromosome II. From crosses between the N2 reference strain, carrying the visible marker mutations, and the Hawaiian wild isolate CB4856, we recovered chromosomes with breakpoints between the markers from 243 hermaphrodite and 226 male meioses. We localized breakpoints by genotyping 122 SNP markers (File S1). The median marker interval is 5.7 kb, the median interval size for observed breakpoints is 15 kb, and more than a third of the breakpoints are mapped to intervals of less than 10 kb.
No sex differences in recombination rate
The patterns of recombination rates for males and hermaphrodites are globally indistinguishable in our studied region (Figure 2A). The cumulative breakpoint distributions along the region are not significantly different (two-sample two-tailed Kolmogorov–Smirnov test, D = 0.13, P = 0.669 by permutation).
The globally similar crossover distributions could mask subtle, fine-scale differences between the sexes. To test for such localized sex differences, we performed a likelihood-ratio test comparing the probability of the data under a model with a sex-specific recombination rate for each SNP marker interval to a nested model with a single rate per interval, shared by the sexes. The likelihood-ratio test failed to reject the shared-rate model (P = 0.448 by permutation). Tested individually, none of the intervals exhibits significant sex differences after correcting for multiple tests, and the distribution of P-values over the intervals is not skewed to low values. In the absence of sex differences, we pooled the recombination breakpoint observations for subsequent analyses.
Location of the recombination rate domain boundary
The boundary between the center and right arm of chromosome II is conspicuous in our data. We employed structural change analysis to localize its position. This regression analysis, typically applied to time-series data, seeks the number and location of boundaries between domains with different regression coefficients (here, mean recombination rate as a function of physical position). As expected, a one-boundary model is favored by Bayesian information criterion (Figure 2B). The interval coincident with the estimated boundary is between the SNP markers at 11,959,442 and 11,965,092, and the 95% confidence interval is flanked by the markers at 11,923,477 and 11,967,491. The average recombination rate per marker interval is more than five times greater to the right of the boundary than it is to the left. The estimated boundary position is ∼60 kb to the left of the position estimated previously from sparser data (Rockman and Kruglyak 2009). The region harbors genes and is superficially ordinary.
Correlates of crossover rate boundary position
Many features of the C. elegans genome differ between chromosomal centers and arms (Barnes et al. 1995; C. elegans Sequencing Consortium 1998; Liu et al. 2011). We asked whether the center-arm boundary that we have localized for crossover rate coincides with boundaries defined by DNA sequence and chromatin features.
Because promoter regions are known to harbor crossover hotspots in many species (Pan et al. 2011), we characterized the number of 5′ gene ends in each interval. As is well established, the arm region has lower gene density than the center. On average, the crossover-defined central domain of our study region contains a gene start site every 3045 bp vs. 1 per 4964 bp in the arm. The density does not exhibit a discrete break, however, and structural change analysis fails to identify gene-density domain structure (i.e., the Bayesian information criterion favors a one-domain model; note, however, that we have little power given the small number of genes per interval).
GC content exhibits a small but significant shift, coincident with the shift in crossover rate to within a few kilobases. Structural change analysis identifies two domains of GC content in our studied region, with the boundary falling between the SNP markers at 11,971,266 and 11,982,291 and a confidence interval that spans 11,949,774 to 11,986,926 and includes the point estimate of the crossover boundary. The studied region averages 36.8% GC in the center domain and 34.6% in the arm. Although GC content and 5′ gene end count are correlated (r2 = 0.065, F1,121 = 8.5, P = 0.004), the residual GC content after regressing out the effect of 5′ gene ends exhibits the same pattern and inferred boundary.
Dispersed repeat sequences occupy more of the arm (19.4%) than they do the center (9.8%), but as with promoters there is not a statistically detectable boundary. Most individual repeat classes are present too few times in the region to permit statistical analysis, but 32 classes are present at least 10 times each. Of these, 5 exhibit a statistically significant arm-center bias, with all 5 enriched in the arm. One of the annotated repeats is simply defined as AT-rich, which is the complement to the GC bias already described. Of the other four repeats, two, Ce0000051 and CELE14B, are enriched on both arms of chromosome II, and indeed on the arms of every chromosome (Figure S2). The others, CELE1 and Ce000256, do not show enrichments on both arms. Ce000256 has a very striking genomic distribution, with 14 of its 48 total instances in the region between 11.98 and 12.30 Mb on chromosome II and none at all on chromosome IV or X. The individual repeat types are not sufficiently numerous within our study region to allow structural change analysis.
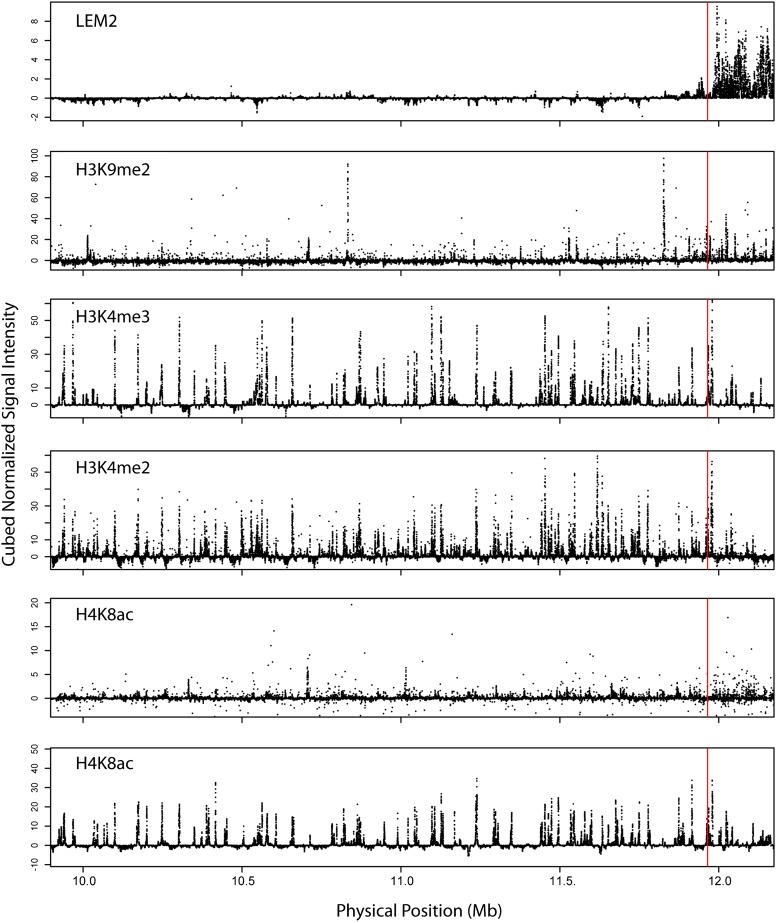
Several chromatin marks are known to correlate with recombination in many species, but the distribution of these marks in meiotic cells in C. elegans has not been reported. Histone modifications, measured primarily in embryonic and larval worms, exhibit strong center-arm dichotomous distributions (Liu et al. 2011). In an analysis of modENCODE data for our SNP marker intervals, we found that ChIP-chip signals for H3K9me2 and H4K16Ac are significantly higher in the arm intervals than in the center, while H4K8Ac shows the opposite pattern. H3K4me2 and H3K4me3 show no significant difference. None of these marks shows any abrupt changes in distribution near the crossover domain boundary, as assessed by visual inspection (Figure 3). We also considered an aspect of higher-order chromosome structure, attachment of chromatin to the nuclear lamina, measured in embryos by LEM2 chromatin immunoprecipitation (Ikegami et al. 2010; Liu et al. 2011). LEM2, uniquely among the studied marks, exhibits a bimodal distribution of ChIP signal intensities. The signal is low across the center and then shifts to a consistently high level in the arm, starting at a position roughly coincident with the crossover domain boundary (Figure 3). By multivariate logistic regression, incorporating the six modENCODE data sets as predictors, LEM2 is the best predictor of arm vs. center status for our SNP intervals, with H3K9me2, H4K16, and H3K4me3 also significant. Within the arm region, however, the substantial fluctuations in LEM2 ChIP-chip signal exhibit absolutely no correlation with our per-interval estimates of recombination rate, nor do any of the other marks.
Figure 3.
Nuclear lamina attachment domains colocalize with crossover domains. Cubed signal intensities from six modENCODE ChIP-chip data sets are plotted along the studied region on chromosome II. The red line indicates the estimated position of the crossover domain boundary. Only LEM2 shows a coincident change in signal intensity.
Fine-scale recombination rate heterogeneity
The dramatic rate difference between the chromosomal center and arm domains, detected by our analysis of rate variation among marker intervals, does not exclude the possibility of fine-scale rate heterogeneity within these domains. To explicitly test for additional rate variation, we simulated breakpoint data according to a simple model of constant recombination probabilities within domains but different probabilities between domains. Our data decisively reject the two-rate model (P < 0.00001).
The rejection of a two-rate model could be due to the presence of many small subdomains, within the center and arm domains, whose rates differ only slightly from the domain averages and hence are not detected by structural change analysis. To test such models of constant-rate subdomains, we fitted our data with piecewise linear regression of genetic position on physical position. The simplest model of constant-rate subdomains that our data fail to reject requires eight subdomains (Figure S3), with five in the center and three in the arm (P = 0.065; all simpler models rejected at P < 0.01 based on 100,000 simulated data sets per model).
Absence of strong recombination hotspots
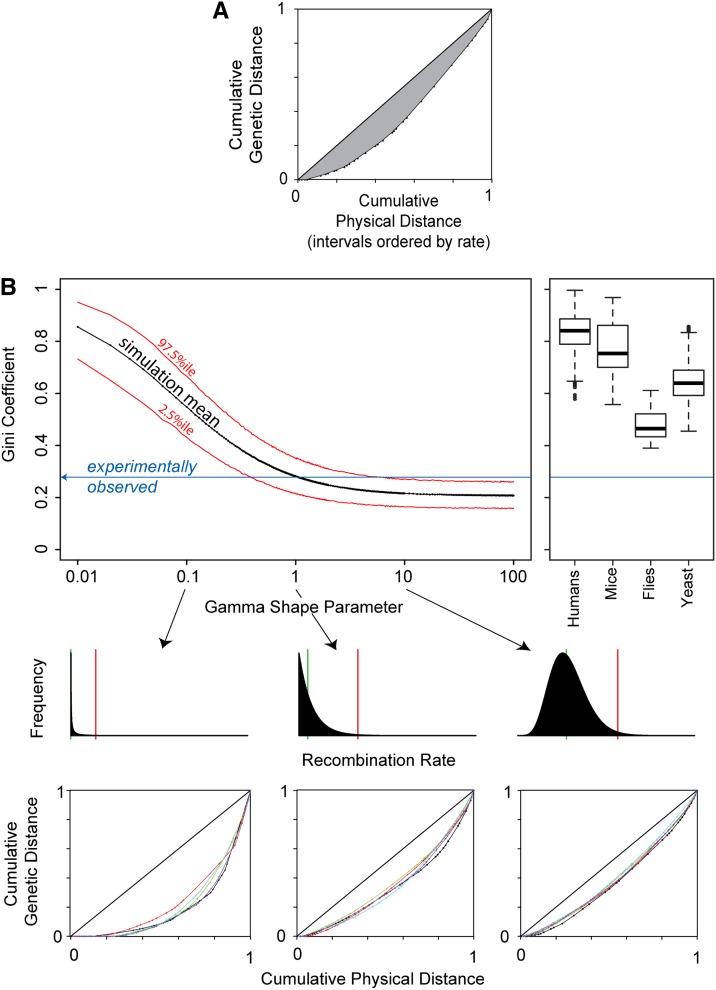
The regression model in the previous section assumes that adjacent marker intervals tend to have similar recombination rates. An alternative is that recombination rate is highly heterogeneous on a finer scale, such that adjacent intervals often have very different recombination rates. A recombination map punctuated with hotspots could produce such heterogeneity. To examine such a scenario, we focused on the arm domain of our data set, which includes observations of 218 breakpoints within a 214 kb region, genotyped with 4.4-kb median marker spacing. As an evaluation statistic, we use the Gini coefficient, which ranges from zero to one and measures the unevenness of the distribution of the crossover breakpoints (see Materials and Methods). For the observed data, the estimated Gini coefficient is 0.278, and Figure 4A provides a representation of the underlying data. It shows that, for example, 80% of breakpoints fall in the most recombinant 60% of the physical interval. Comparable data for humans (Kong et al. 2010) and mice (Paigen et al. 2008) have Gini coefficients ∼0.8, consistent with the prominent role of hotspots in these mammals (see File S5, Figure 4B, and Figure S4). The model species Saccharomyces cerevisiae (Mancera et al. 2008) and Drosophila melanogaster (Singh et al. 2013) have intermediate Gini coefficients, lower than the mammals but higher than we observe in the C. elegans chromosome II arm (see File S5, Figure 4B, and Figure S5). The central domain section of our studied region, although its breakpoint density is lower (251 breakpoints in 2061 kb), also exhibits a distinctly low Gini coefficient, 0.400.
Figure 4.
Recombination rate in the chromosomal arm region is not highly heterogeneous. (A) With the intervals ordered by their recombination rates, the cumulative proportion of genetic distance covered is plotted as a function of the cumulative proportion of physical distance covered. The Gini coefficient, equal to the proportion of the plot above the curve and below the diagonal (i.e., the shaded region), provides a summary statistic for rate heterogeneity among intervals. (B) The observed Gini coefficient (blue) is consistent with an exponential distribution of recombination rates at the kilobase scale (i.e., shape = 1). The black curve shows the mean Gini coefficient for data sets simulated with rates for each kilobase drawn from a gamma distribution with the indicated shape parameter (plotted on a log scale), from highly heterogeneous rates (0.01) to nearly constant (100). The red curves encompass 95% of simulations at each parameter value. At right, we show distributions of estimated Gini coefficients for comparable data from humans and the model species Mus musculus, Drosophila melanogaster, and Saccharomyces cerevisiae (see File S5). Plotted below are examples to illustrate the simulations. In the first row are gamma distributions of recombination rates for three values of the shape parameter (0.1, 1, and 10). The green bar indicates the median observation of random draws from the distribution, and the red bar indicates the 99th percentile. At low values of the shape parameter, the distribution has much of its density near zero and a long tail of rare high rates (hotspots). At a value of one, the distribution is exponential, and the median rate is well above zero. At higher values, the distribution becomes bell shaped. Below these distributions are curves of cumulative genetic distance as a function of cumulative physical distance, as in A, here shown for simulated data. Five simulations are shown for each distribution (five colors per plot).
To better understand the distribution of recombination rates that could generate our observed data, we simulated observations under a range of rate heterogeneities. We drew recombination rates from the gamma distribution, whose shape parameter allows for a simple continuum from high heterogeneity to low heterogeneity (Figure 4B). Our arm data reject highly heterogeneous distributions of recombination rate (shape parameter <0.39), such as would be found in a species with pronounced hotspots. Similarly, our data reject narrow distributions of rates (shape parameter >6.0), consistent with the requirement for multiple-rate subdomains in the piecewise-linear model detailed in the preceding section. The distribution that produces Gini coefficients most similar to the observed coefficient is approximately exponential (shape parameter, 1.06). Under this model, the most recombinant kilobase (of the 214 simulated) is expected to have a recombination rate ∼5.5 times higher than the average rate. The observed data fall outside the 95% confidence interval of models whose most recombinant kilobase is expected to be >10 times the average or <2.5 times the average. Although our analysis implies a landscape of few relatively high- and many relatively low-recombination rate segments, the variability within these segments is much more modest than that expected for an intensely hotspot-rich landscape.
Discussion
We have generated a high-resolution map of crossover frequency across a 2.275-Mb region spanning the boundary between the center and right arm of C. elegans chromosome II. A major finding is that heterogeneity in crossover frequency in this region is dominated by the effects of large chromosomal domains, with little or none of the fine-scale hotspot-type heterogeneity that characterizes most other well-studied species. Using the Gini coefficient as a summary statistic, we found that this region of the C. elegans genome has the least heterogeneous fine-scale crossover landscape of any species for which comparable data are available.
In humans and mice, most recombination happens in hotspots, many specified by cis-acting sequences that interact with the chromatin modifying protein PRDM9 (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). Species that lack PRDM9, including dogs (Axelsson et al. 2012), flies (Cirulli et al. 2007; Comeron et al. 2012; McGaugh et al. 2012; Singh et al. 2013), Arabidopsis (Yelina et al. 2012), and budding yeast (Mancera et al. 2008), also exhibit patterns of fine-scale rate heterogeneity. C. elegans, which lacks functional PRDM9 (Oliver et al. 2009), now joins the fission yeast Schizosaccharomyces pombe as a prominent exception. In S. pombe, crossover density is roughly constant, at least across a few well-studied intervals on chromosome I (Young et al. 2002). These species show that hotspots are not an inherent feature of all crossover distributions.
By simulation, we found that the C. elegans chromosome II right-arm region that we examined in this study has recombination rates that are neither highly heterogeneous nor perfectly constant. The data are consistent with modest rate variation along the sequence, due either to small subdomains with constant rates or fine-scale rate variation of modest magnitude. Our results underscore the notion that recombination rate variation is not simply a matter of hotspots and coldspots but is a quantitative continuum of crossover probability (Pan et al. 2011).
The molecular causes for crossover hotspots vary among species, determined by both the distribution of double-strand breaks (DSB) and the relative probabilities of crossover and noncrossover repair pathways. In some species, DSB distributions closely match crossover distributions, despite a large excess of DSBs in each meiosis (Pan et al. 2011; Smagulova et al. 2011). In others, including S. pombe, DSB hotspots do not translate into crossover hotspots because of potent regulation at the DSB repair step (Hyppa and Smith 2010). In C. elegans, the distribution of DSBs is not fully characterized. Strong crossover interference allows only one DSB to resolve as a crossover, and current data suggest that each pair of homologs receives multiple DSBs (Rosu et al. 2011; Saito et al. 2013), leaving unresolved the molecular basis for the species’ nonuniform crossover distribution.
Despite the modest fine-scale crossover rate heterogeneity we have detected, C. elegans exhibits dramatic broad-scale rate variation. We localized a boundary between a low-rate chromosomal center and a high-rate chromosomal arm to an interval of just a few kilobases. In C. elegans, a single off-center crossover plays a critical role in specifying chromatid cohesion and orientation during prophase (Schvarzstein et al. 2010), and the crossover rate domain structure may be a signature of underlying structural features that quantitatively exclude crossovers from the chromosome centers. The discrete boundary we have identified holds promise for identifying these structural features.
Chromatin state is well established as a determinant of DSB and crossover position (e.g., Reddy and Villeneuve 2004; Wagner et al. 2010; Pan et al. 2011; Smagulova et al. 2011), and chromatin state in somatic cells is often well correlated with DSB and crossover distributions (de Castro et al. 2012; Lichten and de Massy 2011; Wu and Lichten 1994). In C. elegans, recombination and somatic chromatin are broadly correlated via enrichments or depletions on arms vs. centers (Liu et al. 2011; Lui and Colaiácovo 2013). Our data show that H3K9me2, a histone modification previously associated with DSB formation in C. elegans (Reddy and Villeneuve 2004), and signatures of open chromatin (H3K4me2, H3K4me3, H4K8Ac, and H4K16Ac) exhibit no conspicuous changes in distribution across the crossover rate boundary. A marker of nuclear-envelope-associated DNA (LEM2) (Ikegami et al. 2010), conversely, shows an abrupt shift from uniformly low levels to uniformly high levels at the crossover boundary. The spatial coincidence of the boundary is precise to the resolution of our data, consistent with a possible functional role for the nuclear lamina in defining domains of recombination regulation (Ikegami et al. 2010; Saito et al. 2013). It is tempting to speculate that the boundaries set up in interphase nuclei via LEM2 sequestering chromosome arms to the nuclear membrane might modulate the meiotic recombination rate domain structure. However, we see no correlation between LEM2 and fine-scale recombination rate variation in the arm region we examined. The absence of strong concordance between crossover rate and histone modifications may be due to differences between the meiotic cells and the somatic nuclei in which these modifications were measured. We also note that the chromosomal distribution of H2AK5Ac, a leading candidate for chromatin-state determinant for crossover regulation (Wagner et al. 2010; Couteau and Zetka 2011; Lui and Colaiácovo 2013), is unknown.
GC content exhibits a slight but precise shift at the crossover boundary, with lower GC content in the arm than in the center. In many species, GC content is positively correlated with recombination rate, possibly as a result of biased gene conversion (BGC), whereby gene conversion favors the transmission of GC alleles over AT alleles (Eyre-Walker 1993; Galtier et al. 2001; Birdsell 2002). We observe the opposite pattern. If noncrossover pathways are the favored resolution of DSBs in the chromosome centers (Saito et al. 2013), and if noncrossover gene conversions disproportionately exhibit GC-biased transmission, BGC could be a mechanism that generates the observed GC domain structure. In yeast, BGC is actually restricted to crossover-associated gene conversion tracts (Lesecque et al. 2013), but the relationship between BGC and recombination pathway clearly evolves and varies among species (Capra and Pollard 2011; Poh et al. 2012). Recent dissection of the partially redundant Holiday junction resolvases in C. elegans (Saito et al. 2013; O’Neil et al. 2013; Agostinho et al. 2013) leads us to the hypothesis that slx-1, which is disproportionately responsible for noncrossover gene conversion in chromosome centers (Saito et al. 2013), may be more prone to BGC than the other resolvases. BGC is expected to have very weak effects on equilibrium GC content in selfing organisms (Marais et al. 2004), due to the scarcity of heterozygotes, but it could persist in C. elegans from evolution in its outcrossing ancestor (Cutter et al. 2008).
Our findings provide a high-resolution basis for future studies of crossover control in C. elegans. By characterizing crossover distributions and chromosome structure under perturbation, we hope to identify the molecular causes for the distinctive pattern of recombination in this species. At the same time, our detailed account of recombination in wild-type worms sets the stage for investigations of the population-genetic and molecular evolutionary consequences of linkage.
Supplementary Material
Acknowledgments
This work is supported by the National Institutes of Health, NIH (R01GM089972 to M.V.R.), and the National Science Foundation (Doctoral Dissertation Improvement Grant 1210762 to T.K.). We thank David Riccardi, Luke Noble, and the NYU Center for Genomics & Systems Biology Genome Core facility for assistance in identifying the mutations in SP419 and David Riccardi, Jasmine Nicodemus, and Jia Shen for assistance in the lab. We thank Andreas Hochwagen and members of the Rockman lab for helpful comments on manuscript drafts. We also thank three anonymous reviewers for their comments and suggestions to improve the manuscript. We thank the Caenorhabditis Genetics Center, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440), for strains.
Footnotes
Communicating editor: M. P. Colaiácovo
Literature Cited
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., et al. , 2010. A method and server for predicting damaging missense mutations. Nat. Methods 7: 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinho , A., B. Meier, R. Sonneville, M. Jagut, A. Woglar et al, 2013. Combinatorial regulation of meiotic Holliday junction resolution in C. elegans by HIM-6 (BLM) helicase, SLX-4, and the SLX-1, MUS-81 and XPF-1 nucleases. PLoS Genet. 9: e1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson D. G., Thomson J. N., 1982. The kinetochores of Caenorhabditis elegans. Chromosoma 86: 409–428. [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Gerke J. P., Shapiro J. A., Crissman J. R., Ghosh R., et al. , 2012. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat. Genet. 44: 285–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E., Webster M. T., Ratnakumar A., Ponting C. P., Lindblad-Toh K., 2012. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res. 22: 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes T. M., Kohara Y., Coulson A., Hekimi S., 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F., Buard J., Grey C., Fledel-Alon A., Ober C., et al. , 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsell , J. A., 2002. Integrating genomics, bioinformatics, and classical genetics to study the effects of recombination on genome evolution. Mol. Biol. Evol. 19: 1181–1197. [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J., Barthes P., Grey C., de Massy B., 2009. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 28: 2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, 1998. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282: 2012–2018. [DOI] [PubMed] [Google Scholar]
- Capra , J. A., and K. S. Pollard, 2011. Substitution patterns are GC-biased in divergent sequences across the metazoans. Genome Biol. Evol. 3: 516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti A., 1991. A graphical representation of genetic and physical maps: the Marey map. Genomics 11: 219–222. [DOI] [PubMed] [Google Scholar]
- Charif, D., and J. R. Lobry, 2007 SeqinR 1.0–2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis, pp. 207–232 in Structural Approaches to Sequence Evolution: Molecules, Networks, Populations, edited by H. E. Roman, U. Bastolla, M. Port,o and M. Vendruscolo. Springer-Verlag, New York. [Google Scholar]
- Cirulli E. T., Kliman R. M., Noor M. A., 2007. Fine-scale crossover rate heterogeneity in Drosophila pseudoobscura. J. Mol. Evol. 64: 129–135. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couteau F., Zetka M., 2011. DNA damage during meiosis induces chromatin remodeling and synaptonemal complex disassembly. Dev. Cell 20: 353–363. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Payseur B. A., 2003. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol. Biol. Evol. 20: 665–673. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Payseur B. A., 2013. Genomic signatures of selection at linked sites: unifying the disparity among species. Nat. Rev. Genet. 14: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutter , A. D., J. D. Wasmuth and N. L. Washington, 2008. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics 178: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castro E., Soriano I., Marin L., Serrano R., Quintales L., et al. , 2012. Nucleosomal organization of replication origins and meiotic recombination hotspots in fission yeast. EMBO J. 31: 124–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker , A., 1993. Recombination and mammalian genome evolution. Proc. Biol. Sci. 252: 237–243. [DOI] [PubMed] [Google Scholar]
- Galtier , N., G. Piganeau, D. Mouchiroud and L. Duret, 2001. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics 159: 907–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastwirth J. L., 1972. The estimation of the Lorenz curve and Gini index. Rev. Econ. Stat. 54: 306–316. [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M., Davis M. W., Nguyen H., Dayton D., Jorgensen E. M., 2005. Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans. Genetics 171: 1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I. R., 2012. Control of meiotic recombination frequency in plant genomes. Curr. Opin. Plant Biol. 15: 556–561. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Hillers K. J., Villeneuve A. M., 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13: 1641–1647. [DOI] [PubMed] [Google Scholar]
- Hyppa R. W., Smith G. R., 2010. Crossover invariance determined by partner choice for meiotic DNA break repair. Cell 142: 243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami K., Egelhofer T. A., Strome S., Lieb J. D., 2010. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol. 11: R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Gudbjartsson D. F., Masson G., Sigurdsson A., et al. , 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103. [DOI] [PubMed] [Google Scholar]
- Kumar P., Henikoff S., Ng P. C., 2009. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4: 1073–1081. [DOI] [PubMed] [Google Scholar]
- Lesecque , Y., D. Mouchiroud and L. Duret, 2013. GC-biased gene conversion in yeast is specifically associated with crossovers: molecular mechanisms and evolutionary significance. Mol. Biol. Evol. 30: 1409–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., de Massy B., 2011. The impressionistic landscape of meiotic recombination. Cell 147: 267–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Rechtsteiner A., Egelhofer T. A., Vielle A., Latorre I., et al. , 2011. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 21: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui D. Y., Colaiacovo M. P., 2013. Meiotic development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 133–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G., Goodson M., 2011. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 21: 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancera E., Bourgon R., Brozzi A., Huber W., Steinmetz L. M., 2008. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marais , G., B. Charlesworth and S. I. Wright, 2004. Recombination and base composition: the case of the highly self-fertilizing plant Arabidopsis thaliana. Genome Biol. 5: R45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh S. E., Heil C. S., Manzano-Winkler B., Loewe L., Goldstein S., et al. , 2012. Recombination modulates how selection affects linked sites in Drosophila. PLoS Biol. 10: e1001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S., Howell A. M., Rose A. M., 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A., Myers S. R., Hunt S., Deloukas P., Bentley D. R., et al. , 2004. The fine-scale structure of recombination rate variation in the human genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Meneely P. M., McGovern O. L., Heinis F. I., Yanowitz J. L., 2012. Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets D. G., Meyer B. J., 2009. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S., Bowden R., Tumian A., Bontrop R. E., Freeman C., et al. , 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P., Maechler M., 2007. A fast and efficient implementation of qualitatively constrained quantile smoothing splines. Stat. Model. 7: 315–328. [Google Scholar]
- Oliver P. L., Goodstadt L., Bayes J. J., Birtle Z., Roach K. C., et al. , 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 5: e1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil , N. J., J. S. Martin, J. L. Youds, J. D. Ward, M. I. Petalcorin et al, 2013. Joint molecule resolution requires the redundant activities of MUS-81 and XPF-1 during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K., Petkov P., 2010. Mammalian recombination hot spots: properties, control and evolution. Nat. Rev. Genet. 11: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K., Szatkiewicz J. P., Sawyer K., Leahy N., Parvanov E. D., et al. , 2008. The recombinational anatomy of a mouse chromosome. PLoS Genet. 4: e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Sasaki M., Kniewel R., Murakami H., Blitzblau H. G., et al. , 2011. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144: 719–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov E. D., Petkov P. M., Paigen K., 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh , Y. P., C. T. Ting, H. W. Fu, C. H. Langley and D. J. Begun, 2012. Population genomic analysis of base composition evolution in Drosophila melanogaster. Genome Biol. Evol. 4: 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team , 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Reddy K. C., Villeneuve A. M., 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452. [DOI] [PubMed] [Google Scholar]
- Rockman M. V., Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., Skrovanek S. S., Kruglyak L., 2010. Selection at linked sites shapes heritable phenotypic variation in C. elegans. Science 330: 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S., Libuda D. E., Villeneuve A. M., 2011. Robust crossover assurance and regulated interhomolog access maintain meiotic crossover number Science 334: 1286–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito , T. T., D. Y. Lui, H.-M. Kim, K. Meyer and M. P. Colaiacovo, 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M., Wignall S. M., Villeneuve A. M., 2010. Coordinating cohesion, co-orientation, and congression during meiosis: lessons from holocentric chromosomes. Genes Dev. 24: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N. D., Stone E. A., Aquadro C. F., Clark A. G., 2013. Fine-scale heterogeneity in crossover rate in the garnet-scalloped region of the Drosophila melanogaster X chromosome. Genetics 194: 375–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F., Gregoretti I. V., Brick K., Khil P., Camerini-Otero R. D., et al. , 2011. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472: 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006). WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org.
- Stumpf M. P., McVean G. A., 2003. Estimating recombination rates from population-genetic data. Nat. Rev. Genet. 4: 959–968. [DOI] [PubMed] [Google Scholar]
- Tsai C. J., Mets D. G., Albrecht M. R., Nix P., Chan A., et al. , 2008. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 22: 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C. R., Kuervers L., Baillie D. L., Yanowitz J. L., 2010. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467: 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnier A. R., Meir J. Y., Ross J. M., Tavernarakis N., Driscoll M., et al. , 1999. UNC-4/UNC-37-dependent repression of motor neuron-specific genes controls synaptic choice in Caenorhabditis elegans. Genes Dev. 13: 2774–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. C., Lichten M., 1994. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263: 515–518. [DOI] [PubMed] [Google Scholar]
- Yelina N. E., Choi K., Chelysheva L., Macaulay M., de Snoo B., et al. , 2012. Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 8: e1002844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K., Harris T. W., Bieri T., Cabunoc A., Chan J., et al. , 2012. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 40: D735–D741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. A., Schreckhise R. W., Steiner W. W., Smith G. R., 2002. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol. Cell 9: 253–263. [DOI] [PubMed] [Google Scholar]
- Zeileis A., Leisch F., Hornik K., Kleiber C., 2002. strucchange: an R package for testing for structural change in linear regression models. J. Stat. Softw. 7: 1–38. [Google Scholar]
- Zetka M. C., Rose A. M., 1995. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics 141: 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.