Abstract
Development of the Caenorhabditis elegans foregut (pharynx) is regulated by a network of proteins that includes the Retinoblastoma protein (pRb) ortholog LIN-35; the ubiquitin pathway components UBC-18 and ARI-1; and PHA-1, a cytoplasmic protein. Loss of pha-1 activity impairs pharyngeal development and body morphogenesis, leading to embryonic arrest. We have used a genetic suppressor approach to dissect this complex pathway. The lethality of pha-1 mutants is suppressed by loss-of-function mutations in sup-35/ztf-21 and sup-37/ztf-12, which encode Zn-finger proteins, and by mutations in sup-36. Here we show that sup-36 encodes a divergent Skp1 family member that binds to several F-box proteins and the microtubule-associated protein PLT-1/τ. Like SUP-35, SUP-36 levels were negatively regulated by UBC-18–ARI-1. We also found that SUP-35 and SUP-37 physically associated and that SUP-35 could bind microtubules. Thus, SUP-35, SUP-36, and SUP-37 may function within a pathway or complex that includes cytoskeletal components. Additionally, SUP-36 may regulate the subcellular localization of SUP-35 during embryogenesis. We carried out a genome-wide RNAi screen to identify additional regulators of this network and identified 39 genes, most of which are associated with transcriptional regulation. Twenty-three of these genes acted via the LIN-35 pathway. In addition, several S-phase kinase-associated protein (Skp)1–Cullin–F-Box (SCF) components were identified, further implicating SCF complexes as part of the greater network controlling pharyngeal development.
Keywords: sup-36, sup-35, sup-37, pha-1, lin-35, ubc-18, ari-1, ptl-1, F-box, Skp1 pharynx, morphogenesis, Caenorhabditis elegans
THE analysis of suppressor and enhancer mutations is an invaluable tool for elucidating the molecular pathways and networks that control animal development. Using this approach, we have identified a multigene module that controls organogenesis of the Caenorhabditis elegans pharynx (foregut). This network includes the C. elegans Retinoblastoma family ortholog, LIN-35/pRb, and a conserved E2–E3 ubiquitin modification complex, UBC-18/UBCH7–ARI-1/AR1H1 (Fay et al. 2003; Qiu and Fay 2006). LIN-35 and UBC-18–ARI-1 negatively regulate a Zn-finger protein, SUP-35/ZTF-21, at the level of transcription and protein stability, respectively (Mani and Fay 2009). Furthermore, SUP-35, along with a second Zn-finger protein, SUP-37/ZTF-12, functionally opposes PHA-1, a novel cytoplasmic protein that is required for C. elegans embryonic development (Schnabel and Schnabel 1990; Granato et al. 1994; Fay et al. 2004, 2012). Our working model is that in the absence of both lin-35 and ubc-18 activities, SUP-35 levels are abnormally elevated, which in turn interferes with the ability of PHA-1 to carry out essential functions during embryogenesis.
Loss-of-function (LOF) mutations in pha-1 lead to gross defects in pharyngeal development and body morphology (Schnabel and Schnabel 1990; Fay et al. 2004, 2012). Specifically, PHA-1 plays a role in both establishing and maintaining a stable attachment between the anterior epithelial cells of the developing pharynx and the arcade cells that compose the future buccal cavity (mouth) (Fay et al. 2004). This is in part due to the failure of anterior pharyngeal epithelial cells to undergo stereotypical changes in shape and apical–basal polarity, a step that has been termed reorientation (Portereiko and Mango 2001; Fay et al. 2004). When reorientation fails, a connection between the pharynx and arcade cells does not form, which precludes the generation of an intact intestinal tract (Portereiko et al. 2004). The result is a highly penetrant pharynx unattached (Pun) phenotype, whereby animals are unable to feed and therefore arrest as L1 larvae. In addition, strong LOF mutations in pha-1 lead to gross defects in body morphogenesis, which prevent animals from completing embryogenesis and hatching. Similar phenotypes are also observed in lin-35; ubc-18 mutants, as well as in other related compound mutants, and in strains that overexpress SUP-35 (Fay et al. 2003; Mani and Fay 2009).
Notably, LOF mutations in sup-35 and sup-37 completely suppress the lethality of pha-1 LOF mutants, including several molecular nulls (Schnabel et al. 1991; Mani and Fay 2009; Fay et al. 2012). This observation, together with other data, has led to the hypothesis that SUP-35 and SUP-37 act in a pathway or complex that is in opposition to PHA-1, possibly through the regulation of a mutual downstream target or biological processes. In addition, LOF mutations in sup-35 and sup-37 suppress the synthetic lethality of lin-35; ubc-18, lin-35; pha-1, and ari-1; pha-1 double mutants (Fay et al. 2004, 2012; Qiu and Fay 2006; Mani and Fay 2009). Here we describe the molecular characterization of sup-36, a gene in which mutations are capable of suppressing the lethality of pha-1 and lin-35; ubc-18 mutants. sup-36 encodes a Skp1-related protein that binds to several F-box proteins and to the microtubule-associated protein PTL-1/τ. In addition, by carrying out a genome-wide RNAi screen for suppressors of lin-35; ubc-18 larval lethality, we have identified 37 additional genes whose functions intersect with this regulatory network.
Materials and Methods
Strains and maintenance
C. elegans strains were maintained according to standard methods (Stiernagle 2005). Strains used in this study include GE24 [pha-1(e2123) III], WY83 [lin-35(n745) I; ubc-18(ku254) III; kuEx119 (lin-35+; sur-5::GFP)], WY119 [lin-35(n745) I; pha-1(fd1) III; kuEx119], WY163 [pha-1(e2123) III; sup-36(e2217) IV], GE341 [pha-1(e2123) dpy-18(e499) III; sup-36(e2217) IV], GE342 [pha-1(e2123) dpy-18(e499) III; sup-36(e2218) IV], GE391 [pha-1(e2123) dpy-18(e499) III; sup-37(t1012) IV], GE397 [pha-1(e2123) dpy-18(e499) III; sup-36(t1956) IV], GE398 [pha-1(e2123ts) dpy-18(e499) III; sup-36(t1957) IV], WY160 [pha-1(e2123) backcrossed five times to CB4856], WY158 [pha-1(e2123) III; dpy-13(e184) unc-24(e138) IV], WY805 [pha-1(e2123) III; sup-36(e2217) IV; fdEx201], WY512 [sup-36(t1956) IV; fdEx57(rol-6(gf) + SUP-35::GFP)], WY865 [ari-1(tm2549) I; pha-1(e2123) III; fdEx201(pha-1+; sur-5::RFP)], WY729 [pha-1(e2123) III; fdEx121(wild-type sup-36 + surp-5::GFP)], WY730 [pha-1(e2123) dpy-18(e499); fdEx121(sup-36 genomic locus + sur-5::GFP)], WY862 [sup-35(tm1810) III; sup-36(e2217) IV; sup-37(e2215) V; Ex(Ppha-1::GFP)], WY925 [fdEx115(rol-6(gf); SUP-36::GFP)], WY999 [fdEx57], and WY1000 [sup-35(tm1810) III; sup-36(e2217) IV; sup-37(e2215) V; fdEx238(rol-6(gf); SUP-36::GFP)]. Deletion alleles for sup-36 (tm3912), pha-1 (tm3569 and tm3671), and ari-1 (tm2549) were obtained from the National BioResource Project (NBRP) Tokyo, Japan.
Genetic mapping and molecular identification of sup-36
Preliminary mapping placed sup-36 on LGIV (data not shown) (Schnabel et al. 1991). To narrow the sup-36 genomic region, three-point mapping was performed using the balanced strain pha-1(e2123) III; dpy-13(e184) unc-24(e138)/sup-36(e2217) IV. Of the progeny, 6/53 Dpy non-Unc and 82/87 Unc non-Dpy animals acquired the sup-36 mutation. For single-nucleotide polymorphism (SNP) mapping, pha-1(e2123) hermaphrodites were backcrossed five times to CB4856 Hawaiian males, and the resulting pha-1(e2123); CG4856–5× males (WY160) were crossed to pha-1(e2123); dpy-13sup-36(e2217) unc-24 hermaphrodites, followed by standard SNP mapping procedures (Fay 2006). After an analysis of ∼500 Dpy non-Unc and Unc non-Dpy recombinants, the sup-36 mutation was isolated to an ∼80-kb region containing 38 genes between SNPs located on cosmids C01G5 and C01B10. Rescue experiments were performed by amplifying a 1984-bp PCR fragment (chromosome IV, 6,600,477–6,602,461) encompassing the sup-36 genomic region from wild-type animals, using primers 5′-GAAGAGTCTAATTAAGAGTACTGCCGA-3′ and 5′-AGATTTGTGAGCACATTTCGAGA-3′.
Sequence alignments
Alignments were carried out using MAFFT (http://www.ebi.ac.uk/Tools/msa/mafft/) and MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/). After the alignments were reformatted in Phylip4 (http://www.ebi.ac.uk/cgi-bin/readseq.cgi), model testing was carried out (http://darwin.uvigo.es/software/prottest2_server.html), and optimal models were used to generate trees (http://www.atgc-montpellier.fr/phyml/). Both MAFFT and MUSCLE alignment programs produced nearly identical dendrograms (data not shown).
Microscopy
Quantitative fluorescence microscopy was performed using a Nikon Eclipse microscope. Quantification of GFP fluorescence in embryos was carried out using Open Lab Software Version 5.0.2. All images were captured using identical exposure times, and all embryos used in our analysis were of similar developmental stages (∼200–300 cells). Averages of the mean fluorescence were calculated to compare expression levels. P-values were determined using a two-tailed Student’s t-test. Confocal images were acquired using a 100× (1.4 NA) or a 60× (1.49 NA) objective on an Olympus IX-71 inverted microscope. The microscope was equipped with a CSU-X1 spinning disc head (Yokogawa) and a cooled EMCCD camera (ImagEM, Hamamatsu). Image acquisition and microscope automation were controlled using Metamorph software (Molecular Devices).
Yeast two-hybrid analysis
Physical interactions between SUP-36 and FBXC-53, FBXC-32, FBXC-20, and PTL-1 were tested by yeast two-hybrid (Y2H) analysis, using the ProQuest Two-Hybrid System (Invitrogen, Carlsbad, CA). Bait and prey plasmids were generated using the Gateway cloning system. Gateway entry clones for fbxc-53, fbxc-32, and fbxc-20 were a generous gift from Michael Calderwood and Mark Vidal, Harvard Medical School, Boston, MA (Li et al. 2004; Simonis et al. 2009). To generate a Gateway entry clone for ptl-1, a ptl-1a cDNA was PCR amplified using the following primers: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCCATGTCAACCCCTCAATCAG-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTTCATAACGAGCTGATGTC-3′. The resulting amplicon was cloned into the donor vector pDONR221. Genes in the entry clones were transferred to the yeast expression vectors in a Gateway LR recombination reaction with the bait destination vector pDEST32 (sup-36) or the prey destination vector pDEST22 (fbxc-53, fbxc-32, fbxc-20, and ptl-1a). All plasmids were sequenced and tested for self-activation of the reporter HIS3 by growth on histidine-minus (histidine-dropout) medium in the presence of varying levels of 3-amino-1,2,4-triazole (3-AT). Yeast strain MaV203 was cotransformed with the sup-36 bait vector (or the control empty bait vector) and each of the prey vectors (or the control empty prey vector). Transformants were patched onto a SC-Leu-Trp master plate along with the negative activation controls (bait with empty prey vector and preys with empty bait vector) and positive and negative interaction controls supplied by Invitrogen. After 18 hr of incubation at 30°, patches were replica plated to SC-Leu-Trp-Ura and SC-Leu-Trp-His + 25 mM 3-AT plates, and the latter were immediately replica cleaned. Plates were then incubated at 30° for 24 hr, replica cleaned, and incubated for 48 hr at 30°. To test for inhibition of growth on 5-fluoroorotic acid (5-FOA) plates, candidate yeast colonies were streaked onto 0.2% 5-FOA selection plates and the extent of colony growth was determined after 72 hr. The quantitative assay for β-galactosidase (β-gal) activity was performed following the manufacturer’s instructions in liquid cultures, using chlorophenol red-β-d-galactopyranoside (CPRG) as a substrate. β-gal levels were calculated using the following equation: β-gal = (1000 × absorbance at 574 nm)/[(duration of reaction) × (sample volume) × (absorbance at 600 nm)]. Controls supplied by Invitrogen were as follows: strong positive interaction, Krev1/RalGDS-wt; weak positive interaction, Krev1/RalGDS-m1; and negative interaction control, Krev1/RalGDS-m2.
SUP-36 reporter plasmid construction
A SUP-36::GFP translation fusion reporter was generated by amplifying coding sequences along with the 5′-regulatory region of sup-36 from N2 genomic DNA, using primers 5′-CTGCCGAGTTTGAATAAAGAT-3′ and 5′-CATGAGTCTAGATAGTGCAAGTTCTGAAATGAT-3′. The DNA fragment was digested with HindIII and XbaI and was then cloned into corresponding sites in pPD95.75 (Addgene). Sequence analysis was used to confirm the absence of mutations. A Psup-36::GFP transcriptional fusion reporter was generated by amplifying with primers 5′-CTGCCGAGTTTGAATAAAGAT-3′ and 5′-CATGAGTCTAGATAGTGCAAGTTCTGAAATGAT-3′, digesting with HindIII and XbaI, and cloning into the corresponding sites in pPD95.75.
RNA interference
RNA interference (RNAi) was carried out using strains from the Geneservice Library, following standard feeding protocols (Ahringer 2005). Whole-genome RNAi suppressor screening was carried out as described in Polley and Fay (2012). All positive RNAi clones were confirmed by carrying out at least three independent repeats, and clones were sequenced to verify their molecular identities (Supporting Information, Table S1). Double-stranded (ds)RNA for sup-36(RNAi) injection was prepared using RiboMAX Large Scale RNA Production Systems (T7), using primers 5′-TAATACGACTCACTATAGGGGCCATCTTGCGAGATCAAAT-3′ and 5′-TAATACGACTCACTATAGGGCCTTAAAATCAATTTGAAAAAGTTCTG-3′. dsRNA was injected into young adults at ∼1.0 µg/µl, and F1 progeny were scored for viability.
Immunoprecipitation assays
A full-length complementary (c)DNA of SUP-35 was cloned into the BamHI and XhoI sites of pET-26b(+) (Novagen) to create an in-frame fusion with the 6× His tag, using the primer set 5′-ATTAAGGATCCGTGGCTCACACTTTTGCGTGCC-3′ and 5′-ACACGGCTCGAGAATTGAGCACAAGTCAAGGG-3′. His-tagged SUP-35 was expressed in Rosetta (DE3; EMD Biosciences); expression was induced with 0.5 mM isopropyl 1-thio-β-d-galactopyranoside (IPTG) at 37° for 4 hr.
Two primers (5′-GGATCCTAATACGACTCACTATAGGGAACAGCCACCATGCCATCTTGCGAGATCAAATTC-3′ and 5′-CTATTGGACATTCTGGAAAGCGTCTGCT-3′) were used to amplify a full-length sup-36 cDNA containing an upstream bacteriophage T7 promoter and a Kozak consensus sequence, using Pfu DNA polymerase (Thermo Scientific). Likewise, a second set of primers (5′-GGATCCTAATACGACTCACTATAGGGAACAGCCACCATGAGCATCAGCGGAGAGGACAAC-3′ and 5′-TTAATCAGAAGAGACACCATCATCATCTTCTTCCATC-3′) was used to amplify sup-37. Both PCR products were sequenced and used to generate radiolabeled protein in vitro with [35S]methionine, using the TNT T7 Quick for PCR DNA system (Promega, Madison, WI). Expression of correctly sized 35S-labeled proteins was confirmed by SDS–PAGE and autoradiography. Binding assays were performed using the Pierce His Protein Interaction Pull-Down Kit (Thermo Scientific). Eluted proteins were analyzed by SDS–PAGE (7% acrylamide) and detected by autoradiography (SUP-36 and SUP-37) and Coomassie staining (SUP-35).
Microtubule binding assays
pET-26b–SUP-35 was constructed and expressed as described above. His-tagged SUP-35 was purified using the Pierce His Protein Interaction Pull-Down Kit. To increase its solubility, purified protein was dialyzed against Tris buffer (pH 8.0) and then was centrifuged at 100,000 × g for 40 min, using a Beckman (Fullerton, CA) TL-100 ultracentrifuge. The supernatant was saved for the microtubule-binding assay. Microtubule-binding assays were performed using the Microtubule Binding Protein Spin-Down Assay Biochem Kit (Cytoskeleton). SUP-35 binding was analyzed by SDS–PAGE (7% acrylamide) and detected with SYPRO Ruby Protein Gel Stain (Molecular Probes, Eugene, OR). Negative and positive controls for microtubule binding were analyzed by SDS–PAGE (7% acrylamide) and detected by Coomassie staining.
Results
sup-36 encodes a divergent member of the Skp1 family of proteins
LOF mutations in the C. elegans gene pha-1 lead to a fully penetrant embryonic and larval arrest and display gross defects in pharyngeal development and body morphogenesis (Figure 1, A and B) (Schnabel and Schnabel 1990). Recessive mutations in the sup-36 locus were identified in a genetic screen for suppressors of pha-1 LOF lethality (Schnabel et al. 1991). sup-36 mutations suppressed two temperature-sensitive alleles of pha-1, including pha-1(e2123) [pha-1(ts)] (Figure 1C). We mapped sup-36 to a region on chromosome IV that contains 38 genes, using conventional genetic and SNP-mapping methods. Sequence analysis of several candidate genes identified a region that was deleted in strains carrying the canonical allele of sup-36, e2217. We determined that the e2217 deletion encompasses six contiguous genes (nspb-3, F38A5.6, nspb-4, nspb-5, F38A5.8, and F38A5.7). Although the precise endpoints of the e2217 deletion were not determined, F38A5.11/irld-7 and F15B10.3, which bracket the six deleted genes, were not affected by e2217, and thus the total size of the e2217 deletion is likely to be <7 kb.
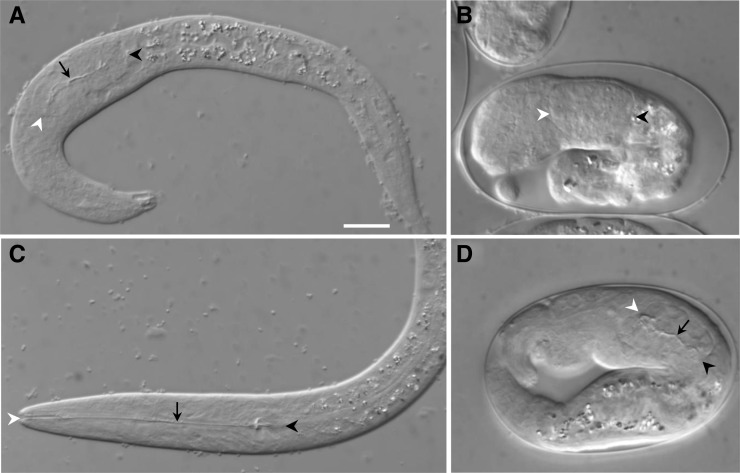
Figure 1.
Pharyngeal phenotypes of pha-1 mutants and suppressed strains. (A–D) DIC micrographs of L1-stage larvae (A and C) and embryos (B and D) are shown. A and B show pha-1(ts) mutants grown at the nonpermissive temperatures of 22° and 25°, respectively. Note abnormal, compressed pharyngeal structures at both temperatures, whereas body morphogenesis defects are more severe at the higher temperature. C shows a suppressed pha-1(ts); sup-36(e2217) double mutant grown at 25° with a normal pharynx and body morphology. D shows a pha-1 embryo grown at the permissive temperature of 16° with overexpression of sup-36 by an extrachromosomal array (fdEx121). Open and solid arrowheads indicate anterior and posterior boundaries of the pharynx, respectively. Solid arrows delineate the pharyngeal lumen. Bar in A, 10 µm for A–D.
Further analysis of the six genes within the deleted region specifically implicated F38A5.7 as sup-36. First, expression of wild-type copies of F38A5.7 from an extrachromosomal array completely reverted the viability of pha-1(ts); sup-36(e2217) double mutants at 25° in three of three independent lines tested. Moreover, pha-1(ts); sup-36(e2217) animals that contained the F38A5.7 array (marked by sur-5::GFP) arrested as embryos with a Pun phenotype that was identical to that of pha-1(ts) single mutants at 25° (data not shown). Second, dsRNA injection of F38A5.7 [sup-36(RNAi)] suppressed pha-1(ts) lethality at 25° (Table 1). Third, sequencing of additional alleles of sup-36 identified multiple independent mutations within F38A5.7 (see below), whereas mutations in other genes within the e2217-deleted region were not detected (data not shown). We therefore designated F38A5.7 as sup-36.
Table 1. Suppression of pharyngeal defects by sup-36.
| Genotype (temperature) | Fertile adults (%) |
|---|---|
| pha-1(e2123ts) (16°) | 91 (n = 285) |
| pha-1(e2123ts) (25°) | 0 (n = 310) |
| pha-1(e2123); sup-36(e2217) (25°) | 95 (n = 320) |
| pha-1(e2123); sup-36(RNAi) (25°) | 97 (n = 438) |
| pha-1(tm3671) (20°) | 0 (n = 252) |
| pha-1(tm3671); sup-36(e2217) (20°) | 76 (n = 695) |
| pha-1(tm3569) (20°) | 0 (n = 279) |
| pha-1(tm3569); sup-36(e2217) (20°) | 81 (n = 349) |
| lin-35(n745); pha-1(fd1) (20°) | 0 (n = 248) |
| lin-35(n745); pha-1(fd1); sup-36(RNAi) (20°) | 75 (n = 276) |
| lin-35(n745); ubc-18(ku254) (20°) | 0 (n = 137) |
| lin-35(n745); ubc-18(ku254); sup-36(e2217) (20°) | 70 (n = 118) |
| ari-1(tm2549); pha-1(e2123) (16°) | 0 (n = 255) |
| ari-1(tm2549); pha-1(e2123); sup-36(RNAi) (16°) | 90 (n = 811) |
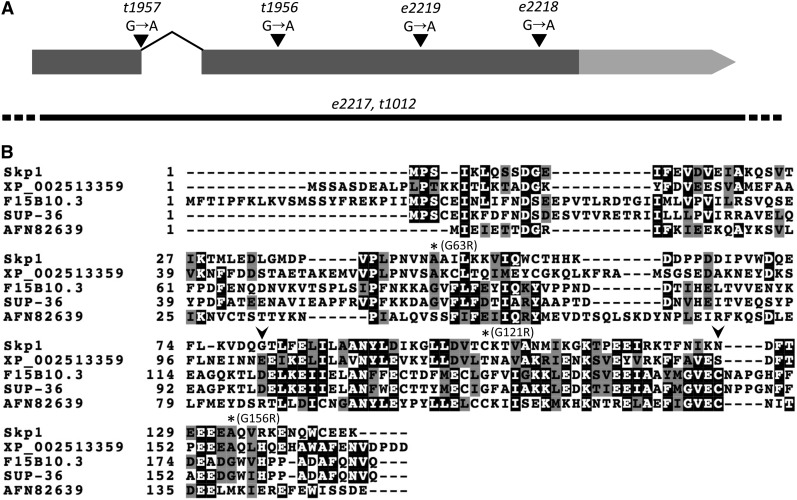
sup-36 contains two exons and encodes a predicted protein of 169 amino acids (Figure 2). Sequence analysis identified a single paralog of sup-36 in C. elegans, F15B10.3, which resides immediately downstream of sup-36 on LGIV and is the result of an apparent gene duplication event within the C. elegans lineage. SUP-36 is 65% identical and 83% similar to F15B10.3, but, unlike SUP-36, F15B10.3 is encoded by three exons (Figure 2B and data not shown). Sequence homology searches also identified a number of SUP-36–like proteins in related nematode species as well as several proteins with homology to SUP-36 in nonnematode organisms (Figure 2B and data not shown). Interestingly, two of the genes from nonnematode species are annotated members of the conserved family of S-phase kinase-associated proteins (Skp). Skp1 proteins form multisubunit E3 (ubiquitin ligase) complexes with F-box proteins and cullins [Skp1–Cullin–F-Box (SCF) complexes] and target a wide range of proteins for degradation by the proteasome (Deshaies 1999; Willems et al. 2004; Kipreos 2005). SUP-36 is 21% identical and 40% similar to the microsporidia Skp1-like protein AFN82639 and 18% identical and 44% similar to XP_002513359 from castor bean (Figure 2B). In addition, SUP-36 is 18% identical and 39% similar to human Skp1, whereas AFN82639 and XP_002513359 are both 33% identical to human Skp1 (Figure 2B). Consistent with these observations, Pfam analysis identified Skp1 dimerization domains spanning amino acid residues 100–145 of SUP-36 (P = 0.00088) and 121–167 of F15B10.3 (P = 0.0066; Figure 2B). Phylogenetic analysis further suggested that SUP-36 and F15B10.3 are indeed divergent members of the Skp1 family of proteins (Figure S1).
Figure 2.
sup-36 genomic locus and SUP-36 protein sequence. (A) Schematic representation of the sup-36 genomic locus. Coding regions are indicated by regions with dark shading, and the 3′-UTR is shown as a region with light shading. Six sequenced sup-36 alleles are shown. e2217 and t1012 are deletion alleles (solid line) with endpoints that are not precisely defined (dashed lines). (B) Clustal alignment of SUP-36 and F15B10.3 from C. elegans, Skp1 from human, and two Skp1-like proteins from Encephalitozoon romaleae (microsporidia; AFN82639) and Ricinus communis (castor bean; XP_002513359). Solid blocks with open letters indicate identical amino acid residues; shaded blocks with solid letters indicate similar residues. Three missense mutations are indicated by asterisks. Arrowheads indicate boundaries of the conserved Skp1 dimerization domain.
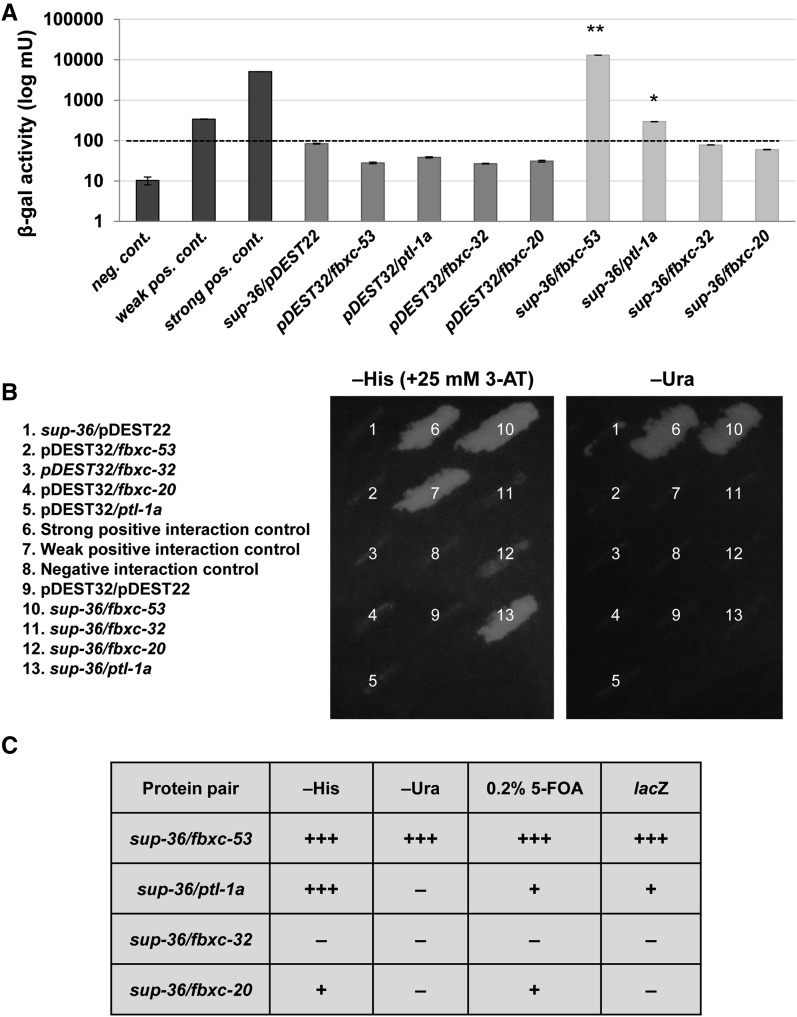
A hallmark of Skp1 family members is their association with F-box proteins (Deshaies 1999; Kipreos and Pagano 2000; Willems et al. 2004). Notably, SUP-36 exhibits high-confidence (termed “core”) physical interactions with three C. elegans F-box proteins (FBXC-20, FBXC-32, and FBXC-53), based on Y2H analyses (Li et al. 2004; Simonis et al. 2009). Furthermore, SUP-36 binds to FBXC-20 in in vitro immunoprecipitation assays (Simonis et al. 2009). Using independently generated Y2H clones and strains, we found that SUP-36 indeed showed physical interactions with FBXC-20 and FBXC-53 (Figure 3 and Figure S2). Among the three F-box proteins, interactions between SUP-36 and FBXC-53 were the strongest, leading to positive outcomes in His−, Ura−, 5-FOA, and β-gal assays (Figure 3 and Figure S2). Interactions between SUP-36 and FBXC-20 were weaker but nevertheless were scored as positive in two of the four standard Y2H assays. These findings further implicate SUP-36 as a divergent member of the Skp1 family of proteins and suggest a possible role for SUP-36 in ubiquitin-mediated proteolysis.
Figure 3.
SUP-36 Y2H interactions. (A) Results from a quantitative β-gal assay. Assays were carried out in triplicate, and error bars indicate 95% confidence intervals. Dashed line indicates the value of the highest negative control (sup-36/pDEST22). P-values were obtained using a Student’s t-test by comparing the indicated yeast strain to the control sup-36/pDEST22 (**P < 0.01, *P < 0.05). (B) Growth of the indicated yeast strains on His− (+25 mM 3-AT) and Ura− plates. C summarizes results for the four Y2H reporters. +++, strong reporter activation; +, weak interaction; and –, no interaction.
Sequence analysis of three additional alleles of sup-36 (e2218, e2219, and t1956) identified mutations that lead to nonconservative substitutions at positions G156(R), G121(R), and G63(R), respectively (Figure 2, A and B). All three glycine residues are conserved between SUP-36 and F15B10.3, whereas human Skp1 and XP_002513359 contain alanine residues at the equivalent positions of G63 and G156 of SUP-36 (Figure 2B). The substitution G121R lies within the predicted Skp1 dimerization domain, where equivalent residues from other species are noncharged (Figure 2B). A fourth allele, t1957, contains a mutation (G to A) in the first nucleotide of the single sup-36 intron and is predicted to abolish splicing (Figure 2A) (Blumenthal and Steward 1997). Because the 60-nucleotide intron contains no stop codons and the frame is preserved, this mutation is predicted to result in an insertion of 20 novel amino acids between positions 38 and 39 of the SUP-36 peptide. A fifth allele of sup-36, t1012, contains a deletion of the entire sup-36 coding region as well as neighboring sequences, similar to e2217 (Figure 2A). The identification of mutations affecting the open reading frame of F38A5.7 in all six sup-36 alleles strongly supports the conclusion that sup-36 is encoded by F38A5.7. We also note that a strain containing a consortium-generated sup-36 deletion allele, tm3912, is strongly impaired for growth and that this phenotype was not rescued by expression of wild-type sup-36 from an extrachromosomal array (data not shown). This finding, along with our observation that the sup-36 deletion mutants e2217 and t1012 are not obviously growth impaired, indicates that a closely linked mutation is likely to be responsible for the tm3912 slow-growth phenotype.
Genetic analysis of sup-36 suppression
LOF mutations in sup-36 can suppress the embryonic and larval-lethal phenotype of pha-1(ts) mutants at 25° (Schnabel et al. 1991). We have confirmed these findings along with our previous observations that sup-36 LOF can suppress the synthetic lethal phenotypes of lin-35(n745); ubc-18(ku354) and lin-35(n745); pha-1(fd1) double mutants (Table 1) (Fay et al. 2004). In addition, the synthetic lethality of ari-1(tm2549); pha-1(ts) mutants at 16° was also suppressed by sup-36(RNAi) (Table 1). These findings are consistent with genetic suppression data previously obtained for sup-35 and sup-37 and further link these three suppressors to a common cellular function (Qiu and Fay 2006; Mani and Fay 2009; Fay et al. 2012). Furthermore, we find that similar to LOF mutations in sup-35 and sup-37 (Fay et al. 2012), loss of sup-36 suppressed two null deletion alleles of pha-1 (tm3671 and tm3569) (Table 1). These latter findings support a model whereby SUP-35/-36/-37 act in parallel to PHA-1 but in a manner that is functionally antagonistic.
During the course of conducting transgenic rescue experiments with wild-type sup-36, we noted that overexpression of SUP-36 via extrachromosomal arrays caused lethality in pha-1(ts) mutants at 16° but showed no deleterious effect on wild type (Figure 1D and data not shown). pha-1(ts) mutants that overexpressed sup-36 arrested as either embryos or L1 larvae and displayed pharyngeal morphogenesis defects typical of pha-1(ts) mutants at nonpermissive temperatures. Because the gene product encoded by pha-1(ts) has a partially impaired function at the permissive temperature of 16° (Fay et al. 2004), it seems likely that reduction of PHA-1 activity rendered the animals more sensitive to toxic effects caused by the overexpression of a protein that normally inhibits or otherwise counteracts PHA-1 downstream functions. Moreover, the toxic effects of SUP-36 overexpression were very similar to those previously observed for overexpression of SUP-35 (Mani and Fay 2009). Notably, LOF of either sup-35 or sup-37 suppressed the lethality of sup-36 overexpression from an extrachromosomal array in the pha-1(ts) background (Figure S3), suggesting that sup-35 and sup-37 function genetically downstream of sup-36. We previously observed, however, that both sup-36 and sup-37 can suppress the toxic effects of SUP-35 overexpression (Mani and Fay 2009). A plausible explanation for these conflicting genetic findings is that SUP-35/-36/-37 act at a single common step.
SUP-36 is expressed in a dynamic pattern during embryogenesis
To determine the pattern of sup-36 expression during development, we generated a construct that encodes the full-length SUP-36 protein fused to GFP at its C terminus. The construct also contains ∼1200 bp of sequences upstream of the sup-36 initiator codon. Importantly, five of five independent extrachromosomal arrays expressing SUP-36::GFP led to lethality of pha-1(ts); sup-36(e2217) embryos at 25°, indicating that the SUP-36::GFP fusion protein is functional (Figure S4). In addition, we note that a transcriptional reporter construct (Psup-36::GFP), containing sup-36 upstream sequences only, showed a similar pattern of expression (data not shown).
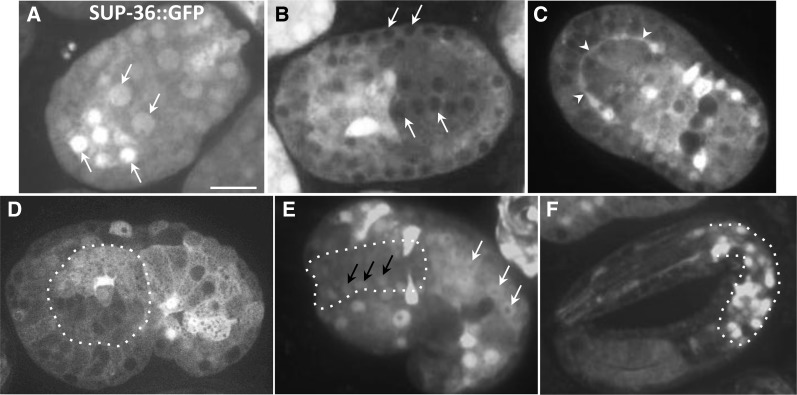
SUP-36::GFP was first detected at approximately the 50-cell stage (∼100 min postfertilization), at which point it was broadly expressed and localized primarily within nuclei (Figure 4A). Nuclear localization persisted until cells reached the ∼200-cell stage (∼200 min), at which time SUP-36::GFP was observed predominantly in the cytoplasm (Figure 4B). Beginning in early morphogenesis (∼350 min), a ring of SUP-36::GFP expression was often detected in the region abutting the basement membrane that circumscribes the anterior pharyngeal primordium (Figure 4C). As embryonic morphogenesis initiated, localization of SUP-36::GFP became more nuclear and by later stages was enriched in pharyngeal nuclei, particularly within the posterior bulb (Figure 4, D–F). Although the functional significance of the dynamic expression pattern of SUP-36 is currently unclear, the presence of SUP-36 in pharyngeal cells throughout embryogenesis is consistent with an autonomous function in this tissue.
Figure 4.
Embryonic expression pattern of SUP-36::GFP. (A–F) Confocal fluorescence images of full-length functional SUP-36::GFP in embryos. A and B show embryos prior to morphogenesis (∼100–350 min postfertilization), C and D show embryos at early stages of morphogenesis (∼350–400 min), E shows an embryo at the 1.5-fold stage (∼430 min), and F shows an ∼3-fold-stage embryo (>600 min). Arrows in A, B, and E indicate the positions of nuclei. Arrowheads in C indicate expression of SUP-36::GFP along the basal surface of primordial pharyngeal cells in the region abutting the basement membrane. Dotted outlined regions indicate the developing pharynx. Bar in A, 10 µm for A–F.
Regulation of SUP-36 by upstream factors
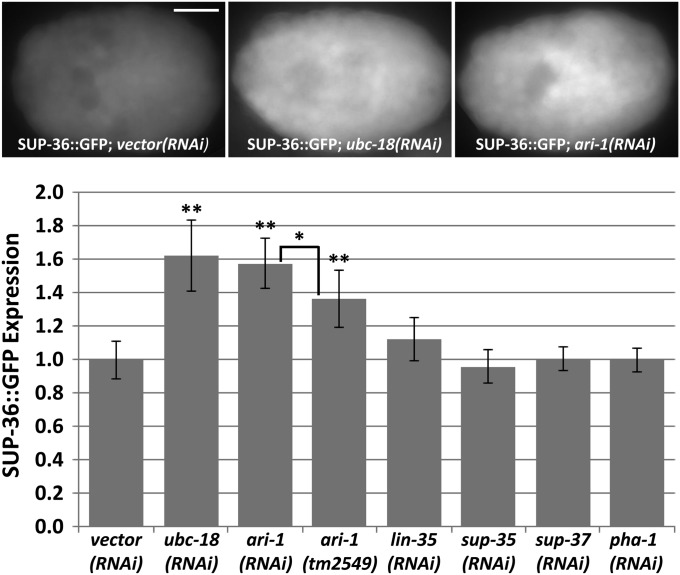
sup-35 is regulated at the level of transcription by LIN-35 and HCF-1 (Mani and Fay 2009; Fay et al. 2012). Specifically, LIN-35 functions as a negative regulator of sup-35 expression, whereas HCF-1 promotes sup-35 transcription. In addition, SUP-35 is negatively regulated by UBC-18 and ARI-1, which form a conserved E2–E3 complex (Qiu and Fay 2006; Mani and Fay 2009). In contrast, sup-37 expression is not regulated by LIN-35, HCF-1, or UBC-18–ARI-1 (Fay et al. 2012). We found that whereas sup-36 expression was not regulated by LIN-35 or HCF-1, SUP-36 levels were negatively regulated by UBC-18–ARI-1 (Figure 5). SUP-36::GFP was upregulated ∼1.6-fold following RNAi of ubc-18 or ari-1, which is slightly less than the ∼2-fold increase previously observed for SUP-35::GFP under these conditions (Fay et al. 2012). Consistent with this, we detected an ∼1.4-fold increase in SUP-36::GFP in ari-1(tm2549) null deletion mutants (Figure 5). The somewhat lower fold change in expression observed in ari-1(tm2549) vs. ari-1(RNAi) mutants likely reflects the residual activities of the ari-1 paralogs C27A12.6 and C27A12.7, whose expression is inhibited by ari-1(RNAi) through “off-target” effects (Qiu and Fay 2006). Thus, the three ari-1-like genes in C. elegans are likely to be partially redundant. Finally, SUP-36::GFP expression levels were not altered following reduction of sup-35 or sup-37 by RNAi (Figure 5).
Figure 5.
SUP-36::GFP expression regulation by upstream regulators. Expression levels of a full-length SUP-36::GFP reporter were assayed in embryos at the ∼300-cell stage under the indicated conditions. Top panels show representative fluorescence images of wild-type, ubc-18(RNAi), and ari-1(RNAi) embryos. Bottom panel shows quantification of embryos (n > 50 for each). Error bars indicate 95% confidence intervals (C.I.’s). Statistical analysis was carried out using Students t-test; **P < 0.001 relative to vector control, *P < 0.05. Bar, 10 µm.
Regulation of pha-1 expression by SUP-36
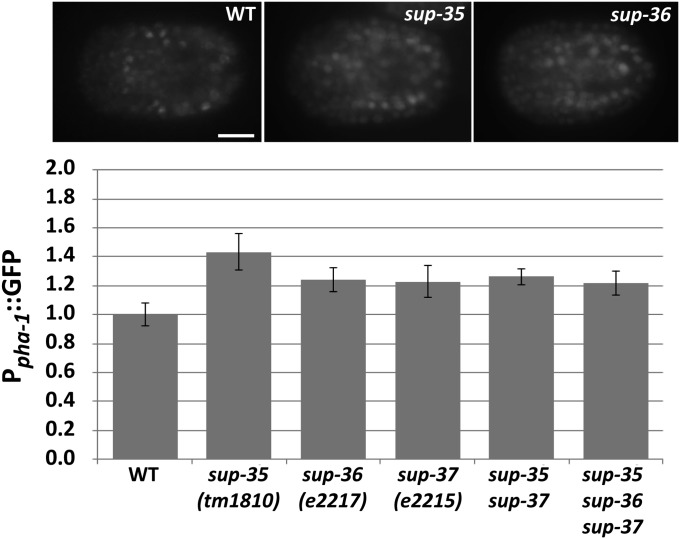
Both SUP-35 and SUP-37 negatively regulate pha-1 expression at the level of transcription (Mani and Fay 2009; Fay et al. 2012). However, this regulation is relatively weak and cannot account for the ability of mutations in sup-35 and sup-37 to suppress null alleles of pha-1 (Fay et al. 2012). Nevertheless, we examined pha-1 expression in sup-36 mutants to see whether sup-36 may also negatively regulate pha-1 transcription. Consistent with sup-35 and sup-37 mutants, sup-36 mutants showed a small but statistically significant increase in the expression of a Ppha-1::GFP reporter (Figure 6), indicating that SUP-36 also plays some role in the regulation of pha-1 expression. To test for additive effects, we examined compound mutants with sup-36, including a sup-35; sup-36; sup-37 triple mutant. We failed, however, to observe enhanced effects on pha-1 expression in compound mutants (Figure 6). The absence of any additive effects is consistent with the model that SUP-35/-36/-37 may act together as a functional unit.
Figure 6.
pha-1 regulation by SUP-35, SUP-36, and SUP-37. Expression levels of a Ppha-1::GFP reporter were assayed in embryos at the ∼300-cell stage under the indicated conditions. Error bars indicate 95% C.I.’s. P < 0.05 based on Student’s t-test for all backgrounds relative to wild type.
Physical interactions between SUP-35, SUP-37, and SUP-36
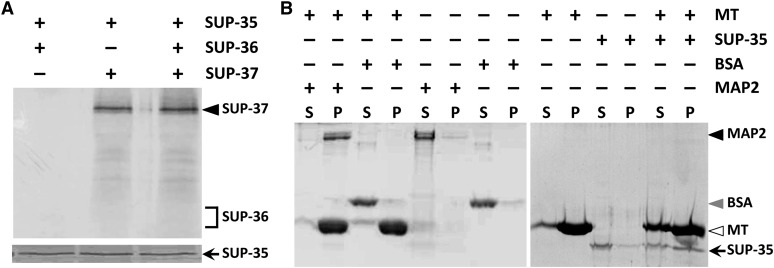
Our genetic data and expression analyses suggested that SUP-35, SUP-36, and SUP-37 may physically interact, possibly within the context of a macromolecular complex or pathway. To further test this model, we assayed purified His-tagged SUP-35 for binding to in vitro-translated SUP-36 and SUP-37. SUP-37 coprecipitated with SUP-35 (Figure 7A), suggesting that these two Zn-finger proteins associate in vivo. In contrast, incubation of SUP-35 and SUP-36 did not lead to the coprecipitation of SUP-36 with SUP-35 (Figure 7A); this was also true for additional tests using a bacterially expressed GST–SUP-36 fusion protein (data not shown). Furthermore, incubation of both SUP-36 and SUP-37 with purified SUP-35 resulted in the coprecipitation of SUP-37 only (Figure 7A). These findings suggest that SUP-36 does not bind directly to either SUP-35 or SUP-37.
Figure 7.
SUP-35 interacts with both SUP-37 and microtubules. (A) Coprecipitation experiments using purified full-length His-tagged SUP-35 and full-length 35S-labeled SUP-36 and SUP-37 in vitro-translated proteins. The presence of correctly sized 35S-labeled SUP-36 and SUP-37 was confirmed in separate experiments (data not shown). SUP-36 and SUP-37 were detected by autoradiography, whereas SUP-35 was detected by Coomassie staining. (B) Microtubule (MT) pull-down binding assays that were carried out using full-length His-tagged SUP-35 along with positive (MAP2) and negative (BSA) controls as indicated. Note that the large majority of SUP-35 is contained within the supernatant (S) in the absence of microtubules but is enriched in the pellet fraction (P) in the presence of microtubules.
SUP-35 contains a conserved regulator of microtubule dynamics (RMD) domain (Mani and Fay 2009), which confers binding to microtubules (Oishi et al. 2007). To test whether SUP-35 can bind to microtubules, we used purified His-tagged SUP-35 in a microtubule pull-down assay. SUP-35 was consistently enriched within the microtubule-concentrated pellet fractions (Figure 7B). We note that our incubation conditions, which were necessary to maintain SUP-35 in a soluble form, slightly inhibited the enrichment of microtubules within the pelleted fraction relative to conditions used for our microtubule-binding (MAP2) and nonbinding (BSA) controls (Figure 7B). Nevertheless, the enrichment of SUP-35 within the pelleted fraction in the presence of microtubules indicates that SUP-35 binds to microtubules and is consistent with the presence of an RMD domain in SUP-35. Interestingly, SUP-36 and PTL-1, the C. elegans ortholog of the mammalian microtubule-associated protein τ/MAP2/MAP4, display high-confidence interactions based on two-hybrid studies (Li et al. 2004; Simonis et al. 2009). Using independently generated SUP-36 bait and PTL-1 prey constructs, we have confirmed binding between SUP-36 and PTL-1 in the yeast system (Figure 3). It is therefore possible that PTL-1 and microtubules could bridge an association between SUP-35–SUP-37 and SUP-36.
SUP-35 is mislocalized in sup-36 mutants
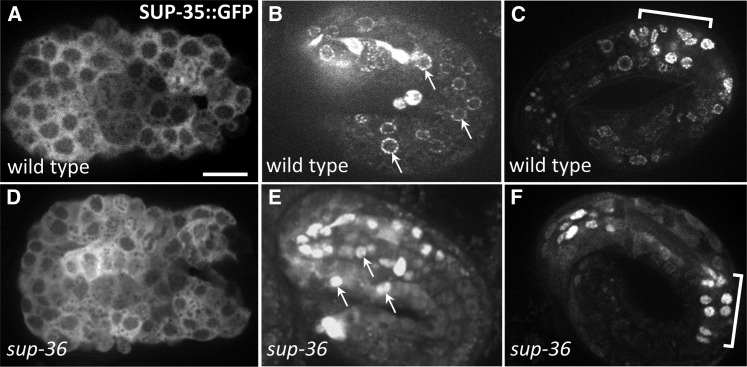
Overexpression of SUP-35 or SUP-35::GFP can be toxic in a wild-type background (Mani and Fay 2009). For this reason, our previous analysis of SUP-35::GFP expression was carried out in a sup-36 mutant background, in which SUP-35 overexpression was not toxic. More recently, we have been able to generate strains that express SUP-35::GFP in a wild-type background, which may be due to lower levels of SUP-35 expression from this array. SUP-35 expression in wild type and sup-36 mutants was identical at early stages of development, in which the large majority of SUP-35::GFP is cytoplasmic (Figure 8, A and D). Beginning at the time of morphogenesis, however, significant differences in SUP-35::GFP localization were observed between wild type and sup-36 mutants. Whereas SUP-35::GFP expression in wild type was punctate and largely confined to the region coinciding with the nuclear membrane or periphery, expression in sup-36 mutants, as previously reported (Mani and Fay 2009), was predominantly nuclear. This suggests that SUP-36 may directly or indirectly regulate SUP-35 subcellular localization. We did not detect any obvious differences in the cell types that expressed SUP-35::GFP in wild type vs. sup-36 mutants, suggesting that SUP-36 specifically regulates SUP-35 localization within the cell. To see whether SUP-36 expression may be regulated by SUP-35 or SUP-37, we examined SUP-36::GFP expression in wild type (Figure 4) and in the sup-35(tm1810); sup-36(e2217); sup-37(e2215) triple-mutant background (Figure S5), but we detected no obvious differences.
Figure 8.
SUP-35::GFP localization is altered in sup-36 mutants. (A–F) Confocal fluorescence images of full-length functional SUP-35::GFP in embryos. A and D show embryos prior to morphogenesis (∼300–350 min postfertilization), B and E show twofold-stage embryos (∼450 min), and C and F show approximately threefold-stage embryos (>600 min). Both wild-type and sup-36(e2217) strains contained the same SUP-35::GFP extrachromosomal array (fdEx57). Arrows in B and E indicate the positions of nuclei. Brackets in C and F indicate the region containing posterior pharyngeal cells. Bar in A, 10 µm for A–F.
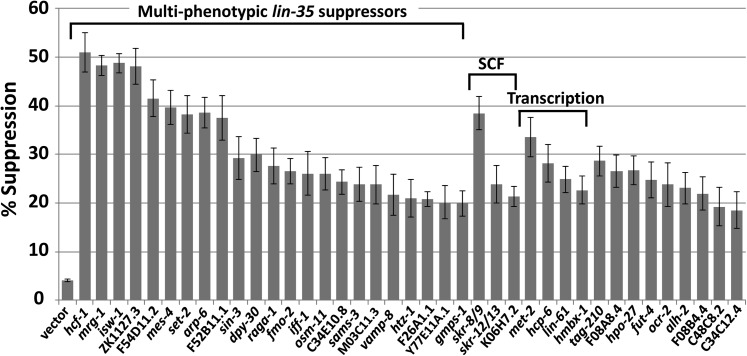
A whole-genome RNAi suppressor screen identifies transcriptional regulators and SCF components as mediators of pharyngeal development
In a saturating genetic screen carried out by Schnabel and colleagues, only three loci (sup-35, sup-36, and sup-37) were identified that could suppress pha-1 LOF lethality (Schnabel et al. 1991). All three pha-1 suppressors also suppress the synthetic-lethal phenotypes of lin-35; ubc-18, lin-35; pha-1, and ari-1; pha-1 double mutants (Table 1) (Fay et al. 2004, 2012; Qiu and Fay 2006; Mani and Fay 2009). We reasoned, however, that additional regulators of this pharyngeal network are likely to exist and may be identified by their ability to suppress lin-35; ubc-18 mutants, which display a less severe phenotype than do strong LOF pha-1 mutants. We therefore carried out a whole-genome RNAi feeding screen for suppressors of lin-35; ubc-18 synthetic lethality. In addition to finding several expected suppressors, such as sup-35 and sup-37, we identified 39 additional clones that consistently suppressed lin-35; ubc-18 larval arrest (Figure 9; Table S1). Whereas <5% of lin-35; ubc-18 mutants normally bypassed larval arrest, the identified suppressors typically led to 20–50% of double mutants reaching the adult stage (Figure 9). Molecular descriptions of the suppressors are shown in Table S1. The single largest class of suppressors contains 23 genes and corresponds to many previously identified multiphenotypic lin-35 suppressors (MPLS) (Polley and Fay 2012). These genes are thought to act antagonistically to LIN-35 and to regulate the expression of transcriptional targets at the level of chromatin structure (Cui et al. 2006b; Polley and Fay 2012). Our screen identified six new MPLS genes, including F26A1.1, which suppressed four distinct lin-35-synthetic phenotypes but not lin-35; slr-2 and lin-15a/b (Figure S6), thus accounting for the absence of this gene from previous suppressor screens. Somewhat surprisingly, most lin-35; ubc-18 suppressors failed to significantly suppress the phenotypically related synthetic lethality of lin-35; pha-1(fd1) mutants (Figure S6 and data not shown). This may be attributable to the somewhat stronger and more highly penetrant phenotype of lin-35; pha-1 double mutants (Fay et al. 2004), a conclusion that is consistent with results obtained from the analysis of MPLS genes identified through a lin-35; slr-2 suppressor screen (Polley and Fay 2012). Furthermore, a small-scale lin-35; pha-1 RNAi suppressor screen failed to identify suppressors that were distinct from those identified by the lin-35; ubc-18 screen (data not shown).
Figure 9.
lin-35; ubc-18 suppressors identified by a whole-genome RNAi screen. Shown is percentage of suppression of larval arrest by 39 sequence-confirmed RNAi clones. Vector denotes a control RNAi. Suppressors have been organized into functional classes. The six new multiphenotypic lin-35 suppressor genes that were identified are arp-6, fmo-2, vamp-8, F26A1.1, Y77E11A.1, and gmps-1. Error bars indicate 95% C.I.’s. For all suppressor RNAi clones, P < 0.001 (relative to vector control) based on the exact test.
The lin-35; ubc-18 suppressor screen also identified four putative regulators of transcription that are not MPLS genes (Figure 9). Two of these genes, met-2/SETDB1 and lin-61/L(3)MBTL2, were previously identified as class B SynMuv genes (Poulin et al. 2005; Harrison et al. 2007). The synthetic multivulval (SynMuv) phenotype is produced by mutations in two genes (one class A gene and one class B gene) that act redundantly to inhibit the formation of multiple vulval structures (Fay and Yochem 2007). (Wild-type hermaphrodite worms have but one vulva.) Class B SynMuv genes function redundantly with class A SynMuv genes to repress the expression of LIN-3/EGF in the hypodermis (Cui et al. 2006a; Fay and Yochem 2007). Class B SynMuv genes include LIN-35/pRb and its binding partner EFL-1/E2F (Lu and Horvitz 1998). Notably, many class B SynMuv genes are conserved in mammals and function within several structurally related transcriptional repressor complexes (Fay and Yochem 2007; van den Heuvel and Dyson 2008). LIN-61 contains four malignant brain tumor (MBT) repeats, which bind to modified histones and are found in transcriptional repressors including the Polycomb-group proteins (Harrison et al. 2007; Bonasio et al. 2010). LIN-61 binds to histone H3 when this protein is di- or trimethylated on lysine 9 (Koester-Eiserfunke and Fischle 2011). Correspondingly, MET-2 is a histone-methyltransferase that promotes H3K9 dimethylation (Andersen and Horvitz 2007; Bessler et al. 2010; Towbin et al. 2012). Thus, LIN-61 and MET-2 are closely linked functionally and together promote transcriptional repression. This differs from the proposed mode of action for MPLS genes, which are thought to act primarily as transcriptional activators (Cui et al. 2006a; Petrella et al. 2011; Xiao et al. 2011; Polley and Fay 2012). The identification of class B SynMuv genes as suppressors of a lin-35-synthetic phenotype was somewhat surprising given that in the context of LIN-3 regulation, class B SynMuv genes act in concert with each other and in some cases can even show enhancer-type interactions (Andersen et al. 2008).
Another class of lin-35; ubc-18 suppressors includes genes that encode putative components of SCF complexes. This includes one RNAi clone that targets the closely related Skp1 family members skr-8 and skr-9 as well as a separate RNAi clone that targets both skr-12 and skr-13 (Figure 9). A third clone targets K06H7.2, which encodes an F-box protein. Given that sup-36 encodes a protein that is related to Skp1 family members, suppression by SCF-type clones may occur through a mechanism that is related to suppression by sup-36. This is consistent with these clones not suppressing unrelated lin-35-synthetic phenotypes. The identification of SCF components as suppressors of lin-35; ubc-18 further supports the hypothesis that the molecular function of SUP-36 is linked to that of other Skp1 family members.
Discussion
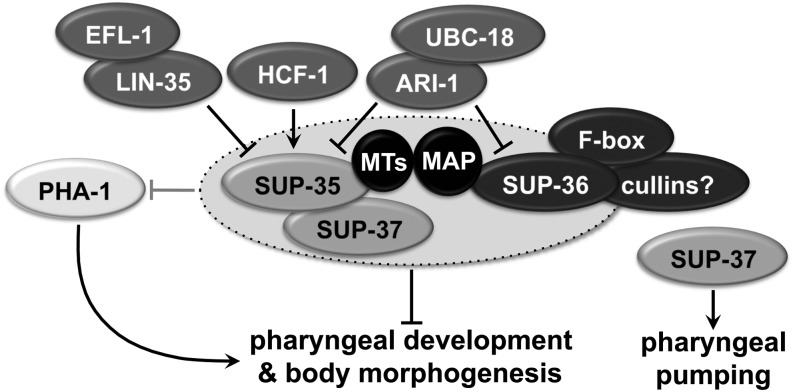
Our collective findings support the model for pharyngeal development depicted in Figure 10. In this model, LIN-35–EFL-1, which forms a transcriptional repressor complex, and HCF-1, which acts as a positive transcriptional regulator, mutually control the expression levels of sup-35 mRNA (Mani and Fay 2009; Fay et al. 2012). Direct control of SUP-35 expression by these upstream factors is further supported by the recent finding that LIN-35 associates with the promoter of SUP-35 in vivo (Kudron et al. 2013). In addition, ARI-1–UBC-18 negatively regulates both SUP-35 and SUP-36 post-translationally, presumably through ubiquitin-mediated proteolysis (Figure 5) (Mani and Fay 2009). Thus, in lin-35; ubc-18 double mutants, levels of both SUP-35 and SUP-36 are elevated, which then interferes with PHA-1 activity. Consistent with this, overexpression of either SUP-35 or SUP-36 leads to pha-1 LOF phenotypes in a genetic background in which pha-1 activity is partially compromised, i.e., pha-1(ts) at 16° (Figure 1D) (Mani and Fay 2009). In addition, SUP-35/-36/-37 negatively regulate pha-1 at the level of transcription, although this effect is weak (Figure 6). Finally, separate from its role with SUP-35 and SUP-36 in opposing PHA-1 functions, SUP-37 functions independently to regulate pharyngeal pumping and ovulation (Fay et al. 2012). We note that the hypothesized common target of SUP-35/-36/-37 and PHA-1 is not currently known, nor is the basic molecular function of PHA-1 understood.
Figure 10.
Model of pharyngeal regulation. Although binding between various binary components such as SUP-35–SUP-37 and SUP-35–microtubules was demonstrated by experimentation, it is unknown whether simultaneous interactions between multiple components also occur. For additional details, see text.
Our observation that purified SUP-35 and SUP-37 directly associated (Figure 7A) supports a model whereby these proteins function as a dimer, possibly within the context of a larger protein complex. Although SUP-36 did not bind directly to either SUP-35 or SUP-37 in our assays, SUP-36 bound to the conserved microtubule-associated protein PTL-1/τ in Y2H assays (Figure 3) (Li et al. 2004; Simonis et al. 2009). Based on our finding that SUP-35 bound directly to microtubules (Figure 7B; likely via the RMD domain), we speculate that SUP-35/-36/-37 may be capable of forming a macromolecular complex, with cytoskeletal components bridging the connection between SUP-35–SUP-37 and SUP-36 (Figure 10). Complex formation by SUP-35/-36/-37 is also largely consistent with genetic and phenotypic data, as well as with their coexpression during certain stages of embryogenesis (Mani and Fay 2009; Fay et al. 2012). Nevertheless it is important to point out that although two-way binding between various components (e.g., SUP-35–SUP-37 and SUP-35–microtubules) was demonstrated (Figure 3 and Figure 7), it is unknown whether simultaneous interactions between multiple components occur. In addition, other possibilities for functional relationships exist, as suggested by the observation that SUP-36 may regulate the subcellular localization of SUP-35 (Figure 8). Furthermore, the SUPs are likely to have functions that are independent of each other. For example, SUP-37 functions independently of SUP-35 and SUP-36 to promote pharyngeal pumping and somatic gonad functions (Fay et al. 2012). Thus, although complex formation between the SUPs may occur, it is unlikely to account for all of their activities.
We have confirmed that SUP-36 binds to at least two C. elegans F-box proteins (FBXC-20 and FBXC-53) and was reported to bind to a third (FBXC-32) based on Y2H assays (Figure 3) (Li et al. 2004; Simonis et al. 2009). This finding is consistent with SUP-36 functioning as a Skp1 family member and supports the idea that SUP-36 is a component of SCF complexes and may function in protein ubiquitylation. Although all three F-box proteins are expressed during embryogenesis, their specific functions are currently unknown (Levin et al. 2012). Although RNAi of these genes failed to suppress either pha-1 or lin-35; ubc-18 mutants (data not shown), there is likely to be a high degree of genetic redundancy among F-box proteins in C. elegans, thereby making their functional analysis difficult. For example, whereas the human and Drosophila melanogaster genomes are predicted to encode 38 and 22 F-box proteins, respectively, C. elegans is predicted to encode 322 (Kipreos and Pagano 2000). Our finding that SUP-36 can regulate SUP-35::GFP subcellular localization suggests that SUP-35 could be a target of SUP-36-mediated ubiquitylation, which is also consistent with our observation that SUP-35 is ubiquitylated in vivo (Mani and Fay 2009). Notably, ubiquitin modification is known to regulate the nuclear export of multiple mammalian proteins including p53, DCN1, RUNX3, and SHMT1 (Lohrum et al. 2001; Chi et al. 2009; Wu et al. 2011; Anderson et al. 2012).
In addition to SUP-35/-36/-37, our genome-wide lin-35; ubc-18 RNAi suppressor screen identified 39 genes whose activities integrate with the described network to regulate pharyngeal development (Figure 9). Based on their ability to suppress multiple independent lin-35-synthetic phenotypes, the majority of suppressors are likely to act in a manner that directly opposes LIN-35 functions. Namely, MPLS genes may be positive regulators of targets that are globally repressed by LIN-35. Somewhat to our surprise, we also identified two class B SynMuv genes, LIN-61 and MET-2, which act through histone modification and binding to repress transcription. Like LIN-35, LIN-61 and MET-2 repress LIN-3/EGF expression in conjunction with class A SynMuv genes (Cui et al. 2006a; Andersen and Horvitz 2007). Unlike LIN-35 and EFL-1, however, LIN-61 and MET-2 are not core components of the C. elegans DP–Rb–class B SynMuv (DRM) complex in C. elegans (Harrison et al. 2007). We therefore hypothesize that LIN-61 and MET-2 may regulate some other node within the network (e.g., ubc-18 or ari-1), thereby accounting for their ability to repress the synthetic phenotype when inhibited. Finally, our identification of several SCF components is consistent with SUP-36 functioning as a Skp1-like protein (Figure 9).
Although the regulatory network and genes that we have identified are likely to be fairly comprehensive, a number of key questions remain, including the basic cellular function of PHA-1 and the means by which SUP-35/-36/-37 and PHA-1 oppose each other’s activities. In addition, the meaning behind the dynamic expression patterns of SUP-35 and SUP-36 is unknown, as is the role that the microtubule cytoskeleton might play in either SUP-35/-36/-37 localization or function. Therefore future studies will be required to clarify the mechanistic details of this gene network.
Supplementary Material
Acknowledgements
We thank Amy Fluet from BiomEditor for editing and Ralf Schnabel and the Caenorhabditis Genetics Center for providing strains. Aleksandra Kuzmanov was supported by Wyoming IDeA Networks for Biomedical Excellence (INBRE) grant P20 GM103432. This work was supported by grant GM066868 from the National Institutes of Health.
Footnotes
Communicating editor: P. Sengupta
Literature Cited
- Ahringer, J., 2005 Reverse genetics (April 6, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.47.1, http://www.wormbook.org. [Google Scholar]
- Andersen E. C., Horvitz H. R., 2007. Two C. elegans histone methyltransferases repress lin-3 EGF transcription to inhibit vulval development. Development 134: 2991–2999. [DOI] [PubMed] [Google Scholar]
- Andersen E. C., Saffer A. M., Horvitz H. R., 2008. Multiple levels of redundant processes inhibit Caenorhabditis elegans vulval cell fates. Genetics 179: 2001–2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. D., Eom J. Y., Stover P. J., 2012. Competition between sumoylation and ubiquitination of serine hydroxymethyltransferase 1 determines its nuclear localization and its accumulation in the nucleus. J. Biol. Chem. 287: 4790–4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler J. B., Andersen E. C., Villeneuve A. M., 2010. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 6: e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal, T., and K. Steward, 1997 RNA processing and gene structure, pp. 117–145 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer, and J. R. Priess. Cold Spring Harbor Laboratory Press, Plainview, NY. [PubMed]
- Bonasio R., Lecona E., Reinberg D., 2010. MBT domain proteins in development and disease. Semin. Cell Dev. Biol. 21: 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X. Z., Kim J., Lee Y. H., Lee J. W., Lee K. S., et al. , 2009. Runt-related transcription factor RUNX3 is a target of MDM2-mediated ubiquitination. Cancer Res. 69: 8111–8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M., Chen J., Myers T. R., Hwang B. J., Sternberg P. W., et al. , 2006a SynMuv genes redundantly inhibit lin-3/EGF expression to prevent inappropriate vulval induction in C. elegans. Dev. Cell 10: 667–672. [DOI] [PubMed] [Google Scholar]
- Cui M., Kim E. B., Han M., 2006b Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R. J., 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15: 435–467. [DOI] [PubMed] [Google Scholar]
- Fay D., 2006. Genetic mapping and manipulation: chapter 5–SNPs: three-point mapping (February 17, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.94.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S., Yochem J., 2007. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev. Biol. 306: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay D. S., Large E., Han M., Darland M., 2003. lin-35/Rb and ubc-18, an E2 ubiquitin-conjugating enzyme, function redundantly to control pharyngeal morphogenesis in C. elegans. Development 130: 3319–3330. [DOI] [PubMed] [Google Scholar]
- Fay D. S., Qiu X., Large E., Smith C. P., Mango S., et al. , 2004. The coordinate regulation of pharyngeal development in C. elegans by lin-35/Rb, pha-1, and ubc-18. Dev. Biol. 271: 11–25. [DOI] [PubMed] [Google Scholar]
- Fay D. S., Polley S. R., Kuang J., Kuzmanov A., Hazel J. W., et al. , 2012. A regulatory module controlling pharyngeal development and function in Caenorhabditis elegans. Genetics 191: 827–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M., Schnabel H., Schnabel R., 1994. Genesis of an organ: molecular analysis of the pha-1 gene. Development 120: 3005–3017. [DOI] [PubMed] [Google Scholar]
- Harrison M. M., Lu X., Horvitz H. R., 2007. LIN-61, one of two Caenorhabditis elegans malignant-brain-tumor-repeat-containing proteins, acts with the DRM and NuRD-like protein complexes in vulval development but not in certain other biological processes. Genetics 176: 255–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E. T., 2005. Ubiquitin-mediated pathways in C. elegans (December 01, 2005), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.36.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kipreos E. T., Pagano M., 2000. The F-box protein family. Genome Biol. 1(5): REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koester-Eiserfunke N., Fischle W., 2011. H3K9me2/3 binding of the MBT domain protein LIN-61 is essential for Caenorhabditis elegans vulva development. PLoS Genet. 7: e1002017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudron M., Niu W., Lu Z., Wang G., Gerstein M., et al. , 2013. Tissue-specific direct targets of Caenorhabditis elegans Rb/E2F dictate distinct somatic and germline programs. Genome Biol. 14: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M., Hashimshony T., Wagner F., Yanai I., 2012. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev. Cell 22: 1101–1108. [DOI] [PubMed] [Google Scholar]
- Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., et al. , 2004. A map of the interactome network of the metazoan C. elegans. Science 303: 540–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrum M. A., Woods D. B., Ludwig R. L., Balint E., Vousden K. H., 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21: 8521–8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Horvitz H. R., 1998. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell 95: 981–991. [DOI] [PubMed] [Google Scholar]
- Mani K., Fay D. S., 2009. A mechanistic basis for the coordinated regulation of pharyngeal morphogenesis in Caenorhabditis elegans by LIN-35/Rb and UBC-18-ARI-1. PLoS Genet. 5: e1000510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K., Okano H., Sawa H., 2007. RMD-1, a novel microtubule-associated protein, functions in chromosome segregation in Caenorhabditis elegans. J. Cell Biol. 179: 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella L. N., Wang W., Spike C. A., Rechtsteiner A., Reinke V., et al. , 2011. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138: 1069–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polley S. R., Fay D. S., 2012. A network of genes antagonistic to the LIN-35 retinoblastoma protein of Caenorhabditis elegans. Genetics 191: 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portereiko M. F., Mango S. E., 2001. Early morphogenesis of the Caenorhabditis elegans pharynx. Dev. Biol. 233: 482–494. [DOI] [PubMed] [Google Scholar]
- Portereiko M. F., Saam J., Mango S. E., 2004. ZEN-4/MKLP1 is required to polarize the foregut epithelium. Curr. Biol. 14: 932–941. [DOI] [PubMed] [Google Scholar]
- Poulin G., Dong Y., Fraser A. G., Hopper N. A., Ahringer J., 2005. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 24: 2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Fay D. S., 2006. ARI-1, an RBR family ubiquitin-ligase, functions with UBC-18 to regulate pharyngeal development in C. elegans. Dev. Biol. 291: 239–252. [DOI] [PubMed] [Google Scholar]
- Schnabel H., Schnabel R., 1990. An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science 250: 686–688. [DOI] [PubMed] [Google Scholar]
- Schnabel H., Bauer G., Schnabel R., 1991. Suppressors of the organ-specific differentiation gene pha-1 of Caenorhabditis elegans. Genetics 129: 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis N., Rual J. F., Carvunis A. R., Tasan M., Lemmens I., et al. , 2009. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat. Methods 6: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2005 Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community, WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin B. D., Gonzalez-Aguilera C., Sack R., Gaidatzis D., Kalck V., et al. , 2012. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell 150: 934–947. [DOI] [PubMed] [Google Scholar]
- van den Heuvel S., Dyson N. J., 2008. Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9: 713–724. [DOI] [PubMed] [Google Scholar]
- Willems A. R., Schwab M., Tyers M., 2004. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta 1695: 133–170. [DOI] [PubMed] [Google Scholar]
- Wu K., Yan H., Fang L., Wang X., Pfleger C., et al. , 2011. Mono-ubiquitination drives nuclear export of the human DCN1-like protein hDCNL1. J. Biol. Chem. 286: 34060–34070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Bedet C., Robert V. J., Simonet T., Dunkelbarger S., et al. , 2011. Caenorhabditis elegans chromatin-associated proteins SET-2 and ASH-2 are differentially required for histone H3 Lys 4 methylation in embryos and adult germ cells. Proc. Natl. Acad. Sci. USA 108: 8305–8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.