Abstract
Patterns of linkage disequilibrium, homoplasy, and incompatibility are difficult to interpret because they depend on several factors, including the recombination process and the population structure. Here we introduce a novel model-based framework to infer recombination properties from such summary statistics in bacterial genomes. The underlying model is sequentially Markovian so that data can be simulated very efficiently, and we use approximate Bayesian computation techniques to infer parameters. As this does not require us to calculate the likelihood function, the model can be easily extended to investigate less probed aspects of recombination. In particular, we extend our model to account for the bias in the recombination process whereby closely related bacteria recombine more often with one another. We show that this model provides a good fit to a data set of Bacillus cereus genomes and estimate several recombination properties, including the rate of bias in recombination. All the methods described in this article are implemented in a software package that is freely available for download at http://code.google.com/p/clonalorigin/.
Keywords: bacteria, homologous recombination, biased recombination, homology-dependent recombination, linkage disequilibrium (LD), homoplasy, four-gamete test (G4)
BACTERIA are organisms that reproduce clonally, but they occasionally exchange fragments of DNA with one another. This process can lead to two outcomes, nonhomologous and homologous recombination (Vos 2009). Nonhomologous recombination occurs when a novel segment of DNA from the donor cell is inserted into the genome of the recipient cell. On the other hand, homologous recombination happens when the DNA from the donor cell replaces its homologous counterpart in the genome of the recipient cell. In this study we are concerned only with the “core” genome of regions present in all sampled genomes (Medini et al. 2008), and therefore only homologous recombination is relevant. Foreign DNA can be taken up by the recipient cell through one of three mechanisms: conjugation (transfer of DNA from one cell to another when they are in physical contact), transduction (bacteriophage-mediated DNA transfer), or transformation (uptake of DNA from the environment by the recipient cell) (Thomas and Nielsen 2005). In homologous recombination, the recipient cell then replaces the homologous section of its DNA with the foreign DNA segment.
A first concept that has helped researchers to appreciate the role of recombination in bacteria is linkage disequilibrium (LD) or the nonrandom association of alleles at different loci (Maynard Smith et al. 1993). LD between a pair of sites is expected to decrease as more and more recombination events affect exclusively one or the other site, so that LD is a function of the distance between pairs of sites. In bacteria on average LD decreases down to a plateau level when pairs of sites are considered that are farther and farther away from each other on the genome, and this is often represented graphically (e.g., Namouchi et al. 2012; Takuno et al. 2012). Another important concept is homoplasy, which is said to occur when given a known tree, a site could not have arisen without either recombination or repeat mutation (Maynard Smith and Smith 1998; Maynard Smith 1999). The probability of a site being homoplasic increases with the number of recombination events affecting the site. For this reason, homoplasy is commonly used as an indicator of the prevalence of recombination (e.g., Nübel et al. 2008; Harris et al. 2010). A related notion is incompatibility between pairs of sites [also known as the four-gamete test (G4)], which occurs when two sites cannot be explained by a shared phylogenetic tree without either recombination or repeat mutation (Hudson and Kaplan 1985; Maynard Smith 1999). Incompatibility between pairs of sites is often used to identify recombination events (e.g., Takuno et al. 2012; Yahara et al. 2012).
Recombination plays a key role in shaping the patterns of all these summary statistics, but they are also crucially affected by other factors, which makes them difficult to interpret. This includes the population structure underlying the relationships between the individuals under study (McVean et al. 2002; Wakeley and Lessard 2003), and this effect is likely to be especially important in bacteria because of their clonal mode of reproduction. Another factor likely to be important in bacterial population genetics is biased recombination, which we define in contrast to free recombination where all individuals in the population are equally likely to recombine. There are many factors contributing to recombination being biased rather than free. Laboratory experiments have shown that the recombination process is homology dependent so that it tends to happen more often between individuals that are less diverged (Roberts and Cohan 1993; Zawadzki et al. 1995; Majewski et al. 2000; Majewski 2001). Furthermore, the geographical and ecological structures observed in many bacterial populations imply a greater opportunity of recombination for pairs of cells that are closely related (Feil and Spratt 2001; Majewski 2001; Cohan 2002; Didelot and Maiden 2010). Purifying selection may also effectively prevent recombination between distantly related bacteria. All these effects would clearly be hard to disentangle, and here we group them all under the single concept of biased recombination. The strength of this bias is an important factor to take into account to understand recombination in bacteria. In particular, this determines how often recombination happens within the diversity of the population under study rather than from other sources. Such recombination events from external sources would strongly affect LD, but have little or no effect on homoplasy and G4 since they introduce what is in effect new polymorphism from the viewpoint of the studied population.
Here we introduce a new statistical framework for inferring the recombination parameters, including the rate of bias in recombination, from a sample of bacterial genomes. Our starting point is an evolutionary model of free recombination that describes the ancestral recombination process given the clonal relationships in the sample. We show how this can easily be extended to allow recombination to be biased. We describe how data can be efficiently simulated under the model, which is crucial to allow the use of approximate Bayesian computation (ABC) techniques (Pritchard et al. 1999; Beaumont et al. 2002) to estimate parameters. We use informative summary statistics about the recombination process such as LD, homoplasy, and G4 to infer parameters. Applications are presented on simulated data sets as well as on a real data set of Bacillus cereus genomes.
Model and Methods
Free recombination model
The process of homologous recombination in bacteria is asymmetric in terms of the genetic contributions made by donor and recipient cells, since typically a small segment of DNA from the donor in the order of a few hundreds or thousands of nucleotides in length is incorporated into the genome of the recipient that is much longer (Didelot and Maiden 2010). This asymmetry contrasts with the well-studied mechanism of crossing over in eukaryotic sexual reproduction where the two parents contribute equally. Consequently, it is possible to consider the (potentially empty) set of genomic sites that have not been affected by recombination since a sample of isolates evolved from a common ancestor, and the ancestral relationships between the isolates at these sites is called the clonal genealogy (Guttman 1997). Alternatively, the clonal genealogy of a set of isolates can be defined as the ancestral tree obtained by tracing the ancestry of the isolates back in time and following the ancestral line of the recipient cell (rather than the donor cell) whenever a recombination event occurred.
The coalescent model with gene conversion describes the ancestry of a bacterial sample subject to homologous recombination (Wiuf 2000; Wiuf and Hein 2000; McVean et al. 2002; Didelot et al. 2009b). A useful approximation of this process is the ClonalOrigin model (Didelot et al. 2010), where given the clonal genealogy the recombinant lines of ancestry are assumed to be independent of each other. This means that given the clonal genealogy the recombinant lines of ancestry are not allowed to recombine and are allowed to coalesce only with the clonal genealogy. Consequently, the clonal and recombination processes can be separated. Here, however, we exploit another property of this model, namely the fact that it has a simple Markovian structure along the genome, similar to that of the sequentially Markov coalescent in approximating the crossing-over ancestral recombination graph (McVean and Cardin 2005; Marjoram and Wall 2006). Given the clonal genealogy this allows for the simulation of pairs of sites at a given physical distance from each other on the genome. As both LD and G4 are defined for pairs of sites, we use this Markovian property of the model to simulate these summary statistics in a computationally efficient manner. A formal description of this model follows, and the mathematical symbols used are summarized in Table 1.
Table 1. List of symbols.
| Symbol | Description |
|---|---|
| Symbols used for the data | |
| A | Aligned sequence data |
| L | Total length of the alignment |
| B | No. blocks in the alignment |
| Wi | ith summary statistic of data |
| Symbols used for the clonal genealogy | |
| Clonal genealogy | |
| T | Sum of branch lengths of the clonal genealogy |
| Symbols used for the recombination events | |
| R1 | No. recombination events affecting the first site |
| No. recombination events affecting the first site that survive to the second site | |
| No. recombination events that start between the two sites and affect the second site | |
| ai | Where the ancestry of the donor meets the clonal genealogy for event i (departure point) |
| bi | Where the transfer of the DNA fragment from donor to recipient occurs for event i (arrival point) |
| L(ai, bi) | Sum of branch lengths on the clonal genealogy between the departure and the arrival of event i |
| D(ai, bi) | Distance in coalescent unit of time between donor and recipient cells of event i |
| Symbols used for the global parameters | |
| θ/2 | Rate of mutation on the branches of the clonal genealogy and the recombinant edges |
| θs/2 | Per-site rate of mutation |
| ρ/2 | Rate of recombination on the branches of the clonal genealogy |
| ρs/2 | Per-site rate of recombination |
| λ | Rate of bias in the recombination process |
| δ | Mean of the geometric distribution modeling the length of recombinant segments |
In the ClonalOrigin model (Didelot et al. 2010), recombination events are independent of one another given the clonal genealogy and the total number of recombination events R given the total branch length of the clonal genealogy T and the population recombination rate ρ is Poisson distributed:
| (1) |
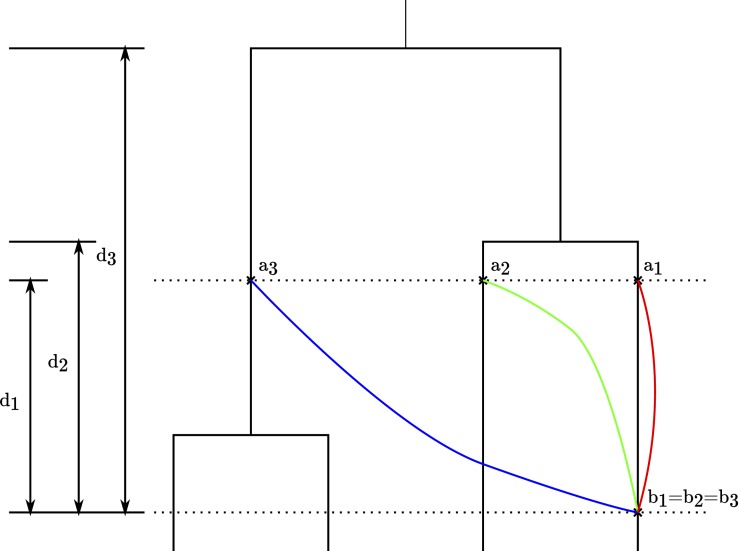
Each recombination event i has four properties: the departure point on the clonal genealogy ai where the ancestry of the donor cell meets the clonal genealogy, the arrival point on the clonal genealogy bi where the recombination occurs, the site on the chromosome where recombination starts xi, and the site on the chromosome where recombination ends yi. Figure 1 shows three recombinations with their arrival and departure points on the clonal genealogy. The three events have the same arrival points, but different departure points on the clonal genealogy.
Figure 1.
Illustration of the recombination model. Consider three recombination events arriving at points b1 = b2 = b3 and departing from points a1, a2, and a3 on the clonal genealogy. In the ClonalOrigin model (free recombination, Equation 2) these three departure points are equally likely because the sums of branch lengths between the times of each bi and ai are equal: L(a1, b1) = L(a2, b2) = L(a3, b3). The amount of evolutionary distance between the donor and recipient cells for the three recombination events is given by D(a1, b1) = 2d1, D(a2, b2) = 2d2, and D(a3, b3) = 2d3. In the biased recombination model (Equation 3), the probability of departing from a1 is higher than that from a2, which is higher than that from a3, because the amount of evolutionary distance between the donor and the recipient cells is increasing: D(a1, b1) < D(a2, b2) < D(a3, b3).
The arrival points bi are uniformly distributed on the clonal genealogy as recombination happens at a constant rate on the branches of the clonal genealogy. A recombinant edge reconnects with the clonal genealogy at a rate equal to the number of ancestors in the clonal genealogy as in the standard coalescent model (Kingman 1982). Thus ai conditional on bi is distributed as
| (2) |
where is the clonal genealogy and L(ai, bi) is the sum of branch lengths on the clonal genealogy between the time of ai and bi. In addition we assume that the recombination events are uniformly distributed along the observed sequences and that their length is geometrically distributed with mean δ.
Extending the model to include biased recombination
We extend the ClonalOrigin model to incorporate the bias in recombination and modify Equation 2 such that a recombinant edge coalesces with the clonal genealogy at a rate that depends on both the number of ancestors in the clonal genealogy and the amount of evolutionary distance between donor and recipient cells. Therefore we propose the following distribution for ai,
| (3) |
where D(ai, bi) is the evolutionary distance in coalescent unit of time between the donor and recipient cells for recombination i and λ is the strength of the recombination bias. Free recombination is nested in this model, as setting λ = 0 results in Equation 2. For values of λ greater than zero, we have that the probability of recombination decreases with the evolutionary distance between donor and recipient. Figure 1 shows the relationship between D(ai, bi) and L(ai, bi) for three recombination events with the same arrival points, but three different departure points on the clonal genealogy. Under a free recombination model, the three recombination events would have the same probability because the sums of branch lengths of clonal genealogy between the arrival and departure points on the clonal genealogy are the same. However, the amount of evolutionary distance between the donor and recipient cells increases from recombination events 1 to 2 to 3. Thus in the model of biased recombination described by Equation 3 with λ > 0, the probability of event 1 is more than that of event 2, which is more than that of event 3.
Simulating pairs of sites
The sequentially Markovian property of our model allows us to simulate pairs of sites at a given physical distance from each other given the clonal genealogy. The simulation is done in three steps. First, we simulate recombination events affecting the first site and their properties. In the second step, we simulate recombination events affecting the second site. This includes some of the recombination events from the first site that are long enough to affect the second site and some new recombinations initiated between the two sites. In the third step the local trees for the two sites are computed and mutations are added.
The sequence data are made of B independent blocks with total length L and subject to mutation and recombination at population rates θ and ρ, respectively. A recombination event may start before a block and be long enough to affect the beginning of a block, so that the probability of observing the recombination start at the beginning of a block is δ times greater than within a block (Didelot and Falush 2007). There are B sites at the beginning of blocks and L − B sites within blocks; thus the recombination rate per site is defined as ρs = ρ/(δB + L − B), and since mutation affects any site with equal probability, the mutation rate per site is θs = θ/L (Didelot et al. 2010).
Given the clonal genealogy , recombination rate per site ρs, mean length of recombination tract δ, the rate of bias in recombination λ, the physical distance between the two sites on the chromosome k, and the mutation rate per site θs, a pair of sites is simulated as follows:
-
Step 1:Simulate recombination events for the first site.
- a. We assume that recombinations start between nucleotides and that they are at least 1 nucleotide long. As the lengths of recombination events are geometrically distributed with mean δ, the rate at which a site i nucleotides before the first site initiates a recombination that survives to the first site is
Summing over all sites before the first site, we get the expected rate of recombination affecting the first site:Therefore the number R1 of recombination events affecting the first site is Poisson distributed:(4) - b. For each recombination event i, the arrival point on the clonal genealogy bi is uniformly distributed and the departure point ai is drawn from Equation 3. To simulate from Equation 3, we use rejection sampling where the proposal distribution is Equation 2 and the simulated ai is accepted with probability
-
Step 2:
Simulate recombination events for the second site. Two types of recombination events can affect the second site. Some events affecting the first site may have survived to the second site and new recombinations could have started between the two sites and have survived to the second site.
- a. As the length of recombination events is geometrically distributed, the probability of a recombination that is affecting the first site to have survived to the second site is
Thus the number of recombination events from the first site that survive to the second site is binomially distributed:(5) - b. The number of recombination events that start between the two sites and that affect the second site is distributed as
(6)
This is because there are only k positions between the two sites where recombination could have started.
c. For each of the recombination events affecting the second but not the first site, the departure and arrival points on the clonal genealogy are simulated as detailed in step 1.
-
Step 3:
For both sites, extract the local trees backward in time (from tips to root), given the clonal genealogy and the recombination events. Mutations are then simulated on these local trees as follows.
-
The number Mj of mutations affecting the local tree at site j is distributed as
(7) where Tj is the total branch length of the local tree at site j.
- We are interested in simulating polymorphic sites and Equation 7 only for plausible values of θs and Tj leads to many nonpolymorphic sites. To remedy this problem, we use an importance sampling strategy (Fearnhead 2007). A local tree is in the target distribution if Equation 7 leads to at least one mutation on that local tree. The proposal distribution is made of all local trees simulated by steps 1 and 2. Therefore the importance sampling weight is
(8)
The simulated local trees are importance sampled using the weights from Equation 8, and the number of mutations on the local tree is simulated from the truncated Poisson with one or more mutations.
c. Mutations are uniformly distributed on the local trees. For simplicity we use the Jukes–Cantor model where all mutations are equally likely, but any mutation model could be used (Whelan et al. 2001).
-
Inference using whole genomes
Whole genomes can be compared using Mauve that detects and aligns the conserved genomic regions in the presence of rearrangements (Darling et al. 2004, 2010). Given a core alignment A of whole bacterial genomes and the clonal genealogy [estimated, for example, using ClonalFrame (Didelot and Falush 2007)], we want to infer the posterior density of the model parameters P(ρs, δ, λ, θs|A, ). However, due to the complexity of the model, the likelihood function is intractable and therefore we cannot use standard approaches such as a Markov chain Monte Carlo (MCMC). One solution would be to use data augmentation techniques as in Didelot et al. (2010). Instead here we use ABC (Pritchard et al. 1999; Beaumont et al. 2002), where the likelihood does not have to be computed, but simulation from the model has to be efficient. In effect, the likelihood is approximated through a distance metric on a set of informative summary statistics between the simulated and observed data. There are several implementations of the ABC algorithm (reviewed in Beaumont 2010; Csilléry et al. 2010) and we have implemented and tested both ABC-MCMC (Marjoram et al. 2003) and ABC-SMC (Sequential Monte Carlo; Beaumont et al. 2009) approaches. The results presented used a parallel ABC-MCMC implementation where given the current chain state θj, n states are proposed independently and for each one data are simulated in parallel (where n is the number of cores available). The proposed states and their simulated data are examined sequentially in the ABC-MCMC algorithm. For each rejected proposed state, the MCMC stays at θj. If a proposed state is accepted, then the remaining proposed states are discarded. If all proposed states are rejected, then the MCMC has stayed at θj for n states and the process is repeated with a proposal of n new states. This parallelization scheme is similar to that of prefetching, which was developed for MCMC with known likelihood (Brockwell 2006).
Summary statistics and distance metric
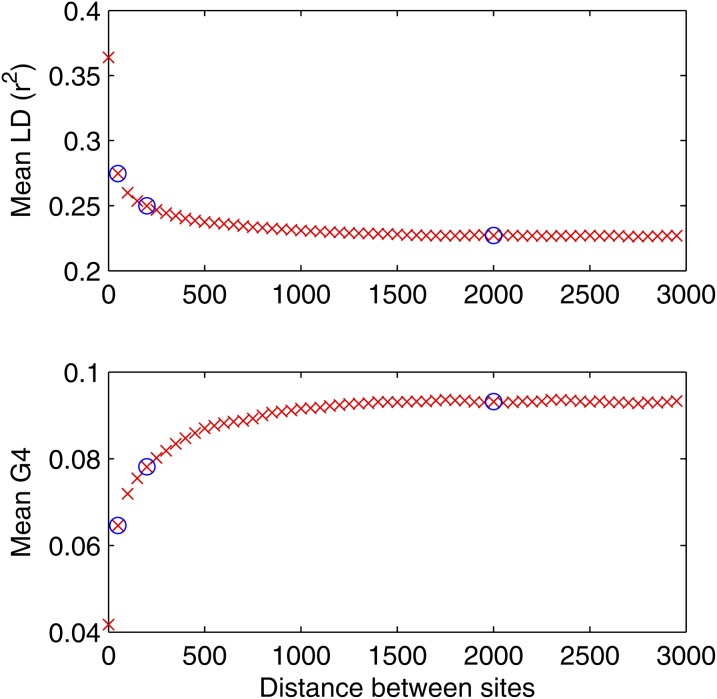
Since one of the parameters we need to infer is the mutation rate θs, we included in the summary statistics the proportion of segregating sites S that is highly informative about this parameter (Watterson 1975). To calculate S from the simulated data, Equation 8 was used, which gives the probability that a simulated site is polymorphic. The most widely used summary statistics that are informative about the recombination process are LD, homoplasy, and incompatibility between pairs of sites (meaning for biallelic sites, all four possible haplotypes are present, G4). To measure LD, r2 was used, which quantifies the amount of association between a pair of biallelic sites (Hill and Robertson 1968). As r2 and G4 are distance dependent, for empirical data sets, we plot the mean of r2 and G4 against distance between the sites. Figure 2 shows an example for a sample of 13 B. cereus whole genomes (Didelot et al. 2010). As summary statistics we choose three points on the LD and G4 plots that capture the decay and the constant part of the plots. These points are shown with blue circles in Figure 2 and here correspond to pairs of sites at distances of 50, 200, and 2000 nucleotides from each other. These distances need to be chosen according to the r2 and G4 plots of the given empirical data set. Background LD can be affected by other factors than recombination such as genetic drift (Falush et al. 2003), although these would not affect the variation in LD at different distances. To account for this, and since we are here interested in recombination, we used the differences in LD as summary statistics, i.e., LD100 − LD2000 and LD100 − LD200. We also included as a summary statistic the proportion of homoplasic sites relative to the clonal genealogy and a new variable that we called clade homoplasy, which is calculated as follows: Given a clonal genealogy, it is divided into its two largest clades and for biallelic sites if both alleles are present in both clades, we say that the site is clade homoplasic. We introduce this new summary statistic as an indicator of the amount of recombination between the clades, which will be informative about the rate of bias in the recombination process.
Figure 2.
LD and G4 plots for 13 Bacillus cereus whole genomes, as a function of the distance between pairs of sites. LD decreases and G4 increases until they both plateau at ∼1000 bp. The blue circles indicate the three values of LD and G4 that were used as summary statistics in the inference procedure.
In total, we therefore use eight summary statistics: the proportion of segregating sites, two distance-based differences in LD, three distance-based values of G4, one value for homoplasy, and one value for clade homoplasy. These summary statistics are compared between the observed and simulated data sets, using a metric equal to the sum of the squared normalized distances,
| (9) |
where x′ is the simulated data, x is the observed data, and Wi is the ith summary statistic of the data.
Monte Carlo estimation of r/m
An important quantity in bacterial population genetics is the ratio r/m of rates at which nucleotides are substituted due to recombination and mutation (Guttman and Dykhuizen 1994; Vos and Didelot 2009). In our model this is equal to
| (10) |
Given a recombination on the clonal genealogy, the probability of a substitution being introduced due to the recombination event at the site is given by
| (11) |
Where (D) is the expected distance between the donor and recipient cells in a coalescent unit of time given a recombination event. Therefore for a given set of parameters, the probability of substitution given a recombination event is estimated using Equation 11 by simulating many recombination events on the clonal genealogy and computing the average distance between donors and recipients. Equation 10 is then used to estimate r/m. This Monte Carlo procedure is applied for each value of the parameters in the posterior sample to obtain a sample from the posterior distribution of r/m.
Results
Relationship between parameters and summary statistics
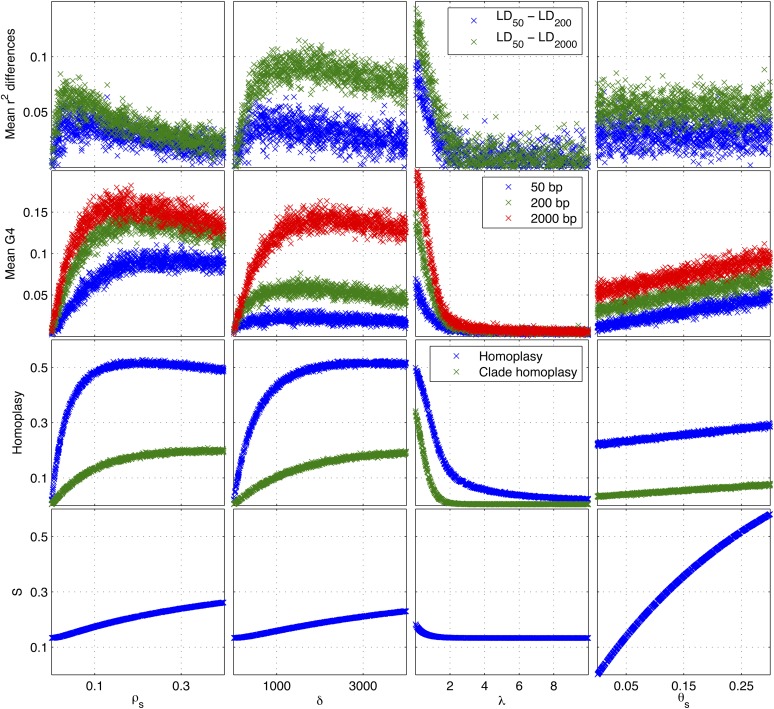
We used simulated data to investigate the relationship between the model parameters and the summary statistics. A clonal genealogy with 15 taxa was simulated under the coalescent model (Supporting Information, Figure S1) and the parameters ρs = 0.02, δ = 300, λ = 1.2, and θs = 0.05 were used, which represent reasonable values for a real bacterial population (Fraser et al. 2007; Didelot et al. 2010). We then changed one parameter at a time in the intervals ρs ∈ [0, 0.4], δ ∈ [0, 4000], λ ∈ [0, 10], θs ∈ [0, 0.3] and simulated the summary statistics to see how they varied with the parameters. For each parameter value, we simulated 2000 pairs of sites distant from each other by 50, 200, and 2000 bp.
Figure 3 shows how the summary statistics change with the model parameters. ρs, δ, and λ have a large influence on r2, G4, and homoplasies and a relatively small effect on the proportion of segregating sites S. On the other hand θs has little impact on r2, G4, and homoplasies, but it has a large influence on S. In the absence of recombination (ρs = 0) the differences in mean r2 are zero, which indicates r2 is independent of distance between pairs of sites. As ρs increases, the differences in mean r2 increase to a maximum, beyond which, as ρs increases, the differences in mean r2 decrease and for very high values of ρs, the differences approach zero, which indicates r2 again becomes independent of distances between pairs of sites. Increasing ρs increases homoplasies and G4 up to a maximum beyond which the mean G4 and homoplasy slightly decrease. δ has a similar but nonidentical effect on r2, G4, and homoplasies. However, λ has the opposite effect on r2, G4, and homoplasies. This is because as λ increases, the effect of the recombination decreases as the donor cells tend to have a smaller evolutionary distance relative to the recipient cells and therefore local trees become more and more similar to the clonal genealogy. For extremely high values of λ this results in no differences in mean r2, no homoplasies, and zero incompatible pairs of sites (G4), which is similar to those observed in the absence of recombination. θs has the largest influence on the proportion of segregating sites S, but ρs, δ, and λ also slightly affect it. This is because as the number of recombination events increases, the probability that a recombination edge reattaches itself higher up the clonal genealogy increases and that would increase the total branch length of local trees relative to the clonal genealogy.
Figure 3.
Relationship between model parameters and the summary statistics. For a given clonal genealogy (shown in Figure S1), the four model parameters were changed one at a time and the summary statistics were simulated. When unchanged, the parameters were ρs = 0.02, δ = 300, λ = 1.2, and θs = 0.05.
It is important to note that the clonal genealogy has a large impact on the observed patterns of LD and homoplasy. To illustrate this, we performed the same sensitivity analysis as above but using a different clonal genealogy (Figure S2). The resulting relationships between model parameters and summary statistics are shown in Figure S3. These relations are quantitatively the same as described above based on Figure 3, but the exact values differ significantly. It is therefore essential to account for the clonal genealogy as we do here to correctly interpret the values of the summary statistics. Having done this, there are strong relationships between model parameters and the summary statistics (Figure 3), which means that inference via ABC on the basis of these statistics should provide good statistical power to infer parameter values.
Application to simulated data sets
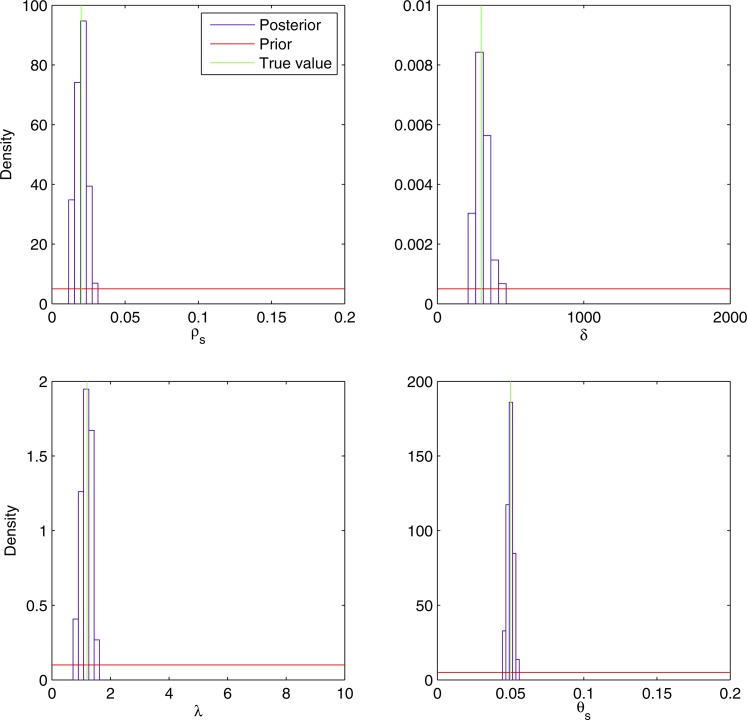
We first applied our inference methodology to a data set simulated under our model. Fifteen genomes of length 1,000,000 bp were simulated, based on the clonal genealogy shown in Figure S4, and using the following parameters: ρs = 0.02, δ = 300, λ = 1.2, and θs = 0.05. Figure S5 shows the LD and G4 plots for this data set. The LD r2 for these simulated data were (0.1970, 0.1605, 0.1339) for pairs of sites distant by (50, 200, 2000) bp, respectively. The proportions of G4 were (0.0242, 0.0613, 0.1002) for pairs of sites distant by the same respective amounts. The proportions of homoplasic and clade homoplasic sites were, respectively, (0.3067, 0.0931). Finally, the proportion of segregating sites was S = 0.1235.
We chose uniform priors for all model parameters in the following ranges: ρs ∈ [0, 0.2], δ ∈ [0, 2000], λ ∈ [0, 10], and θs ∈ [0, 0.2]. We ran a parallel ABC-MCMC chain of 300,000 iterations with the ABC threshold ε = 0.015 and the proposal density tuned so that the acceptance rate was 0.4%. The histograms in Figure 4 show the marginal distribution of posterior samples for each of the four parameters. The posterior distribution of the recombination rate per site ρs had a mean of 0.020 with a 95% credibility interval (CI) = 0.012–0.028. The posterior of the mean recombination tract length δ had a mean of 309 with CI = 226–449. The posterior of the rate of bias of recombination λ had a mean of 1.18 with CI = 0.81–1.47. The posterior of the mutation rate θs had a mean of 0.050 with CI = 0.046–0.054. For each of the four parameters, the true value that was used for simulation was well within the 95% credibility interval and in each case close to the mean of the posterior distribution. Furthermore, Figure 4 shows that the posterior distributions are much tighter than the prior distributions for each of the four parameters. This means that the summary statistics upon which inference is based carry significant information about the underlying values of the parameters, as had previously been suggested by the correlations between parameters and summary statistics in simulated data sets (Figure 3).
Figure 4.
Estimated marginal posterior densities of the parameters for the simulated data set. The values used in simulation are shown in green and are ρs = 0.02, δ = 300, λ = 1.2, and θs = 0.05. The red lines show the uniform prior densities used for the model parameters and the blue histograms show the marginal posterior densities estimated using ABC-MCMC.
Running this inference procedure on a cluster of 12 Intel 3.33-GHz cores took ∼70 hr. The computing time of the inference procedure depends on the range of parameters being inferred as higher values of ρs and δ lead to slower simulations. As the inference procedure is time consuming, testing our model on hundreds of simulated data sets is not possible and we tested our algorithm on 11 additional simulated data sets with a range of parameters. We limited our parameter ranges to biologically meaningful values. The parameter ranges used are ρs = [0, 0.07], δ = [0, 1000], λ = [0, 2], and θs = [0.02, 0.08]. We used the clonal genealogy of Figure S4 and used different parameter values to simulate 11 data sets, each made of 15 whole genomes of 1,000,000 bp. We then used our method to infer the parameter values for each of the 11 data sets. Figure S6 and Figure S7 show the marginal posterior density for each of the 11 data sets. For values of ρs or δ equal to zero, there are no recombination events. In such cases either ρs and δ can be close to zero while the other parameter and λ can change freely. In addition, extremely high values of λ lead to patterns similar to those of the scenario without recombination. Such instances are easily recognized as LD and G4 plots are straight lines and therefore could be excluded from further analysis. For all other reasonable values of ρs, δ, λ, and θs, the posterior ranges are much tighter than the prior ranges. This shows that our inference method works as expected and that inference is possible for a wide range of parameter values.
To assess the effect of inferring the clonal genealogy incorrectly, we performed two additional simulations. Given the clonal genealogy of Figure S4, the distance matrix li,j was computed between all pairs of leaves. A modified distance matrix was then computed by replacing each li,j with a uniform draw from the interval [0.75li,j, 1.25li,j], and a modified tree was computed using Unweighted Pair Group Method with Arithmetic Mean (UPGMA) on the modified distance matrix. The two resulting genealogies are shown in Figure S8, and these differ from the true clonal genealogy of Figure S4 in both tree topology and branch lengths. These two incorrect genealogies were then used to infer the model parameters. The posterior marginal densities are shown in Figure S9, indicating that that our model and inference procedure are robust to slight misspecification of the clonal genealogy. In both cases the true parameters used for simulation of data are well within the 95% credible interval of the posterior densities.
Application to B. cereus
We applied our method to estimate recombination properties based on a core alignment of 13 whole genomes of B. cereus, including the first genome from this species to be fully sequenced (Ivanova et al. 2003). These are the same data as previously analyzed by Didelot et al. (2010), thus allowing comparisons between the two analyses to be drawn. This previous analysis was performed using the ClonalOrigin model, which does not account for the bias in recombination. Nevertheless, the posterior distribution of recombination events contained a clear excess of recombination between close relatives (cf. Figure 5 of Didelot et al. 2010).
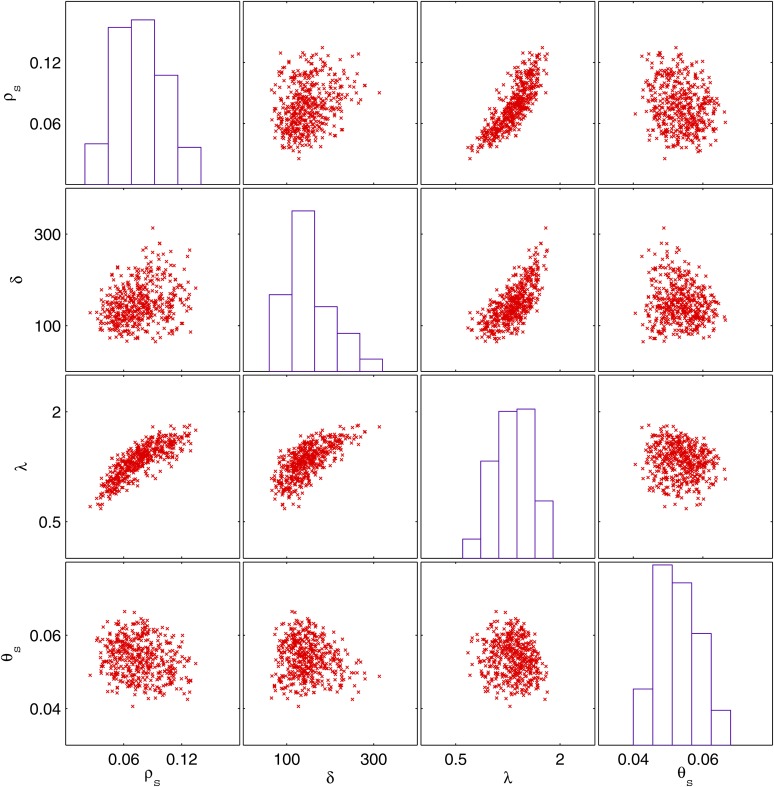
Figure 5.
Posterior distributions of model parameters for the B. cereus data set. The histograms show the marginal posterior distributions of each parameter whereas the scatter plots show their joint posterior distributions.
Figure S10 shows the clonal genealogy of the data that was estimated by Didelot et al. (2010), using ClonalFrame (Didelot and Falush 2007). A unique tree topology with little uncertainty in the branch lengths was reconstructed. Figure 2 shows the LD and G4 plots for this data set. Three points on the plots were selected to be used as summary statistics, with distance between the pairs of sites at 50, 200, and 2000 bp. The mean r2 and G4 for pairs of sites at these distances were, respectively, (0.2738, 0.2493, 0.2270) and (0.0679, 0.0808, 0.0932); the proportion of segregating sites in this data set was S = 0.174; and the proportions of homoplasic and clade homoplasic sites were, respectively, 0.29 and 0.15.
We chose uniform priors for all model parameters in the ranges ρs ∈ [0, 0.2], δ ∈ [0, 2000], λ ∈ [0, 4], and θs ∈ [0, 0.2]. Several independent ABC-MCMC chains were run with similar results. The histograms on Figure 5 show the marginal posterior densities for the estimated parameters. The posterior mean for the recombination rate ρs was 0.077 with CI = 0.036–0.127. The posterior mean of the recombination tract length δ was 152 bp with CI = 74–279. The posterior mean of the rate of bias in recombination λ was estimated to be 1.32 with CI = 0.812–1.788. The posterior mean of the mutation rate θs was 0.0528 with CI = 0.0437–0.0640. The estimates of θs and δ were in agreement with previous estimates [median of θs = 0.0438 and δ = 236 (Didelot et al. 2010)]. However, this previous analysis estimated that recombination was significantly less frequent [ρs = 0.017 (Didelot et al. 2010)]. In this previous study, the recombination rate was probably underestimated as a result of not accounting for the bias in recombination. In a model with biased recombination, a larger fraction of recombination events are between close relatives and therefore have little effect and would tend to go undetected by methods that do not account for it. The relative impact of recombination to mutation r/m (Guttman and Dykhuizen 1994; Vos and Didelot 2009) was estimated using Equations 10 and 11. r/m had a mean of 3.4 with CI = 1.6–6.7 (Figure S11). This is slightly higher than the previous estimate from ClonalFrame [mean of r/m = 2.41 (Didelot et al. 2010)].
The posterior distributions of the four model parameters were significantly correlated as shown by the scatter plots in Figure 5. θs had moderate levels of negative correlation with ρs (Pearson’s linear correlation coefficient, r = −0.26, P = 1.3 × 10−16), δ (r = −0.12, P = 2.5 × 10−4), and λ (r = −0.16, P = 3.5 × 10−7). The strongest associations, however, were the positive correlation of ρs with λ (r = 0.83, P = 2.0 × 10−253) and δ with λ (r = 0.75, P = 1.3 × 10−178). ρs and δ were also slightly correlated (r = 0.34, P = 3.2 × 10−29). Since higher values of λ translate into a higher bias in recombination (where recombination occurs between more similar isolates) and therefore a smaller effect of recombination, it is logical that there is to some extent a trade-off between smaller ρs and λ on one hand (meaning less recombination with more effect per recombination) and higher ρs and λ on the other hand (meaning more recombination with less effect per recombination). Likewise, higher values of δ indicate larger recombination events and therefore a higher effect per event, which explains the trade-off between λ and δ.
To test the fit of our model with biased recombination to the observed data, we considered the posterior predictive distribution of three additional summary statistics, i.e., their distribution when parameters are drawn from the posterior sample (Gelman et al. 1996). These summary statistics had not been used in inference, but were similar in principle to the clade homoplasy statistic previously defined. The B. cereus clonal genealogy was divided into four distinct clades. One of these clades had a single member, which was ignored. We measured the amount of clade homoplasy between the other three clades and used them as posterior predictive summary statistics. This Bayesian model criticism approach has been used in several previous ABC studies (Thornton and Andolfatto 2006; Morelli et al. 2010). We found that the observed values of the three summary statistics were contained within the boundaries of the posterior predictive distributions (Figure S12). Our model with biased recombination is therefore able to reproduce the observed summary statistics and represents a good fit to the data.
Comparison with experimental studies
Several experimental studies have demonstrated a log-linear relationship between sequence divergence and frequency of recombination (Roberts and Cohan 1993; Zawadzki et al. 1995; Vulić et al. 1997; Majewski et al. 2000). These results are summarized in Figure 1A of Fraser et al. (2007), which shows that different bacterial species have a similar log-linear relationship, with a coefficient of ∼20. To compare our results on biased recombination to these previous experimental studies, we need to compute the relative rate of recombination between two isolates as a function of their homology. Since this equates to considering recombination between two cells living at the same time, the first part of Equation 3 is equal to one and the probability of recombination is proportional to exp(−λD), where D is the distance between the donor and the recipient cells in a coalescent unit of time. The expected amount of sequence divergence per site π between two genomes separated by a branch of length D is π = θsD/2, which implies that D = 2π/θs, and therefore we obtain that the rate of recombination between two cells is proportional to exp(−2λπ/θs). The frequency of recombination has therefore a log-linear relationship with sequence divergence in our biased recombination model, with a coefficient (measured on a log of base 10 as in previous studies) of 2λ/(θsln(10)). In the case of the B. cereus application above, this coefficient had a mean of 22.1 with CI = 12.5–32.0. Our estimate for the rate of bias in recombination is therefore slightly higher than the rate of homology dependency of recombination that was found in previous experimental studies.
Discussion
Linkage disequilibrium, G4, and homoplasy are often interpreted informally as evidence of recombination. We have introduced a flexible statistical framework to interpret the values of these statistics calculated from whole bacterial genomes. Our underlying model is based on an approximation to the coalescent with gene conversion (Didelot et al. 2010), which has the advantage of being sequentially Markovian along the genomes. This allows us to simulate patterns of LD and G4 through sampling of many pairs of sites at given distances, which takes only a small fraction of the computational power that would be needed to simulate large segments of DNA. Approximate Bayesian computation (Pritchard et al. 1999; Beaumont et al. 2002) was used to perform inference under this bacterial population genomic model. This approach offers great flexibility to implement extensions of the model like the one we presented in Equation 3 to account for the biased recombination, simply by modifying the way simulation is performed without the need to compute a new likelihood function. We applied our method to simulated data sets and a real data set consisting of 13 whole genomes of B. cereus. We showed that these data contain evidence that the recombination process depends on the evolutionary distance between donors and recipients and measured the strength of this relationship. Our model is robust to slight misspecification of the clonal genealogy, but gross inaccuracies would lead to misleading results.
Evidence for a higher rate of recombination within than between the three major clades of B. cereus was first presented using multilocus sequence typing data, by searching for the most likely origin of ClonalFrame recombination segments (Didelot et al. 2009a). This approach was also applied to genomic data from Salmonella enterica, and more recombination was found within five lineages than between them (Didelot et al. 2011). However, this method is not very powerful, because ClonalFrame does not look for potential donors of the recombination events and therefore is better able to detect recombination coming from farther away (Didelot and Falush 2007). A better approach is the one implemented in ClonalOrigin (Didelot et al. 2010), where the source of recombined fragments is inferred jointly with the recombination events rather than relying on a postprocessing step. By comparison of the number of recombination events observed between pairs of branches and expected under the prior model, recombination was found to happen more often between members of the same B. cereus clades (Didelot et al. 2010). Similar results have been obtained using the same technique in other organisms, such as Sulfolobus islandicus (Cadillo-Quiroz et al. 2012) and Escherichia coli (Didelot et al. 2012). However, this is still not fully satisfactory from a statistical point of view, since the analysis is done using a prior model where recombination does not depend on evolutionary distance, which is proved to be incorrect by the posterior distribution of events. For this reason, this approach does not allow us to estimate the strength of bias in recombination, since the posterior depends on both the prior (where this parameter is zero) and the observed data (which contain evidence that this parameter is nonzero). The best statistical approach is therefore the one we presented here, where the model explicitly incorporates this important parameter, so that we can use Bayesian statistics to formally estimate its value.
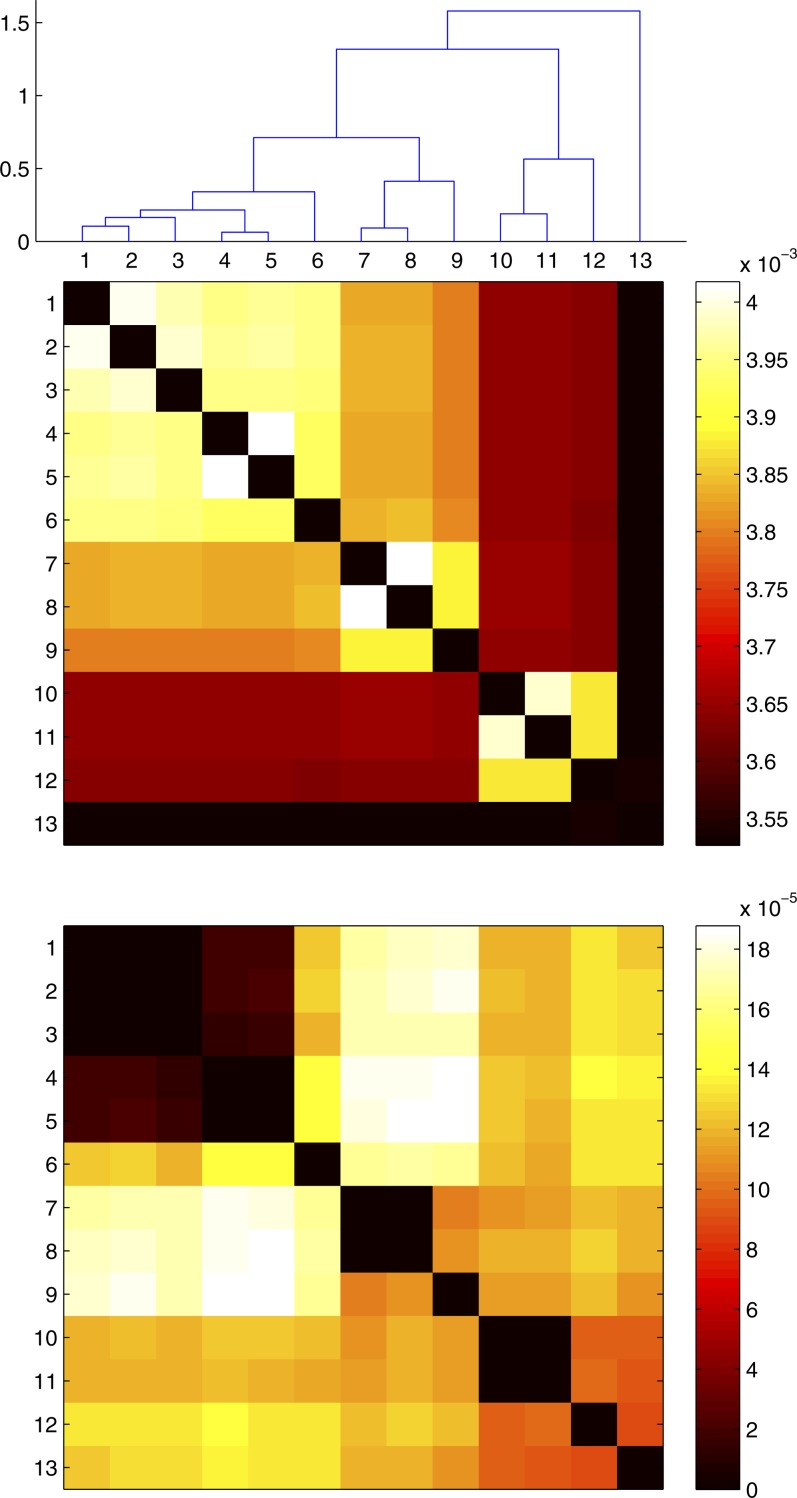
We estimated the coefficient for the log-linear relationship between recombination rate and the effective sequence divergence to be ∼22 in B. cereus. This is slightly higher than previous estimates based on laboratory experiments, which were ∼20 (Fraser et al. 2007). This higher coefficient could be due to the fact that laboratory experiments measure the rate of recombination between two bacteria only when they are brought into contact, whereas there are factors in nature, such as geographical or ecological structuring of the population, that would increase the sexual isolation between distantly related bacteria (Majewski 2001; Didelot and Maiden 2010). Yet this coefficient is far lower than the value of 300 predicted by population genetics models to be required for recombination to be on one hand a strong cohesive force between highly homologous bacteria and on the other hand very rare between diverged bacteria, thus resulting in clusters of diversity that could be considered to represent separate species (Falush et al. 2006; Hanage et al. 2006; Fraser et al. 2007, 2009; Achtman and Wagner 2008). To test this hypothesis further, we used a Monte Carlo simulation to see the effect of the next evolutionary events likely to happen to any one of the B. cereus genomes in our data set. We found that for all except the most closely related pairs of genomes, future recombination events would result in convergence, i.e., a reduction of the genetic distance (Figure 6, bottom). However, we also found that mutation would increase the genetic distance between any pair of genomes at a much higher rate than recombination would reduce it (Figure 6, top). We therefore conclude that all pairs of genomes are likely to diverge in the near future, since the convergence effect of recombination will not be sufficient to compensate for the divergence effect of mutation. Convergence via recombination is likely to be restricted to rare situations where strong selective or ecological factors are involved, such as found in the convergence of S. enterica serovars Typhi and Paratyphi A (Didelot et al. 2007) or the convergence of Campylobacter jejuni and coli (Sheppard et al. 2008).
Figure 6.
Prediction of the future effect of mutation and recombination on the genetic distance between pairs of B. cereus genomes. The heat map at the top indicates the rate at which mutation will increase the distance between all pairs of genomes (i.e., pairwise divergence). The heat map at the bottom indicates the rate at which recombination will decrease these same distances (i.e., pairwise convergence). For closely related isolates, recombination leads to divergence, which is shown as zero convergence. The rate at which mutation causes divergence is an order of magnitude higher than the rate at which recombination leads to convergence. Thus in these isolates, the overall short-term impact of recombination and mutation is divergence of the isolates.
Supplementary Material
Acknowledgments
We thank Rory Bowden, Alison Etheridge, Richard Everitt, Daniel Falush, Simon Myers, and Daniel Wilson for useful ideas and helpful discussions. We also thank two anonymous reviewers whose comments improved an earlier version of this manuscript. M. Azim Ansari received a scholarship from the Life Sciences Interface Doctoral Training Centre, which is funded by the Engineering and Physical Sciences Research Council. This study was partly funded by the UK Clinical Research Collaboration (UKCRC) Modernising Medical Microbiology Consortium, which is supported by the Wellcome Trust (grant 087646/Z/08/Z), and by the Medical Research Council, the Biotechnology and Biological Sciences Research Council, and the National Institute for Health Research on behalf of the Department of Health (grant G0800778).
Footnotes
Communicating editor: Y. S. Song
Literature Cited
- Achtman M., Wagner M., 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6: 431–440. [DOI] [PubMed] [Google Scholar]
- Beaumont M. A., 2010. Approximate Bayesian computation in evolution and ecology. Annu. Rev. Ecol. Evol. Syst. 41: 379–406. [Google Scholar]
- Beaumont M. A., Zhang W., Balding D. J., 2002. Approximate Bayesian computation in population genetics. Genetics 162: 2025–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont M. A., Cornuet J. M., Marin J. M., Robert C. P., 2009. Adaptive approximate Bayesian computation. Biometrika 96: 983–990. [Google Scholar]
- Brockwell A. E., 2006. Parallel Markov chain Monte Carlo simulation by pre-fetching. J. Comput. Graph. Stat. 15: 246–261. [Google Scholar]
- Cadillo-Quiroz H., Didelot X., Held N. L., Herrera A., Darling A., et al. , 2012. Patterns of gene flow define species of thermophilic Archaea. PLoS Biol. 10: e1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan F. M., 2002. Sexual isolation and speciation in bacteria. Genetica 116: 359–370. [PubMed] [Google Scholar]
- Csilléry K., Blum M. G. B., Gaggiotti O. E., François O., 2010. Approximate Bayesian Computation (ABC) in practice. Trends Ecol. Evol. 25: 410–418. [DOI] [PubMed] [Google Scholar]
- Darling A. C. E., Mau B., Blattner F. R., Perna N. T., 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14: 1394–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T., 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5: e11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Falush D., 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175: 1251–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Maiden M. C. J., 2010. Impact of recombination on bacterial evolution. Trends Microbiol. 18: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Achtman M., Parkhill J., Thomson N. R., Falush D., 2007. A bimodal pattern of relatedness between the Salmonella Paratyphi A and Typhi genomes: Convergence or divergence by homologous recombination? Genome Res. 17: 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Barker M., Falush D., Priest F. G., 2009a Evolution of pathogenicity in the Bacillus cereus group. Syst. Appl. Microbiol. 32: 81–90. [DOI] [PubMed] [Google Scholar]
- Didelot X., Lawson D., Falush D., 2009b SimMLST: simulation of multi-locus sequence typing data under a neutral model. Bioinformatics 25: 1442–1444. [DOI] [PubMed] [Google Scholar]
- Didelot X., Lawson D., Darling A., Falush D., 2010. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics 186: 1435–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Bowden R., Street T., Golubchik T., Spencer C., et al. , 2011. Recombination and population structure in Salmonella enterica. PLoS Genet. 7: e1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Méric G., Falush D., Darling A. E., 2012. Impact of homologous and non-homologous recombination in the genomic evolution of Escherichia coli. BMC Genomics 13: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Stephens M., Pritchard J. K., 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D., Torpdahl M., Didelot X., Conrad D. F., Wilson D. J., et al. , 2006. Mismatch induced speciation in Salmonella: model and data. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361: 2045–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead P., 2007. Computational methods for complex stochastic systems: a review of some alternatives to MCMC. Stat. Comput. 18: 151–171. [Google Scholar]
- Feil E. J., Spratt B. G., 2001. Recombination and the population structures of bacterial pathogens. Annu. Rev. Microbiol. 55: 561–590. [DOI] [PubMed] [Google Scholar]
- Fraser C., Hanage W. P., Spratt B. G., 2007. Recombination and the nature of bacterial speciation. Science 315: 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser C., Alm E. J., Polz M. F., Spratt B. G., Hanage W. P., 2009. The bacterial species challenge: making sense of genetic and ecological diversity. Science 323: 741–746. [DOI] [PubMed] [Google Scholar]
- Gelman A., Meng X.-L., Stern H., 1996. Posterior predictive assessment of model fitness via realized discrepancies. Stat. Sin. 6: 733–759. [Google Scholar]
- Guttman D., Dykhuizen D., 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266: 1380–1383. [DOI] [PubMed] [Google Scholar]
- Guttman D. S., 1997. Recombination and clonality in natural populations of Escherichia coli. Trends Ecol. Evol. 12: 16–22. [DOI] [PubMed] [Google Scholar]
- Hanage W. P., Spratt B. G., Turner K. M. E., Fraser C., 2006. Modelling bacterial speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361: 2039–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. R., Feil E. J., Holden M. T. G., Quail M. A., Nickerson E. K., et al. , 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327: 469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 226–231. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1985. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111: 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N., Sorokin A., Anderson I., Galleron N., Candelon B., et al. , 2003. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature 423: 87–91. [DOI] [PubMed] [Google Scholar]
- Kingman J. F. C., 1982. The coalescent. Stoch. Proc. Appl. 13: 235–248. [Google Scholar]
- Majewski J., 2001. Sexual isolation in bacteria. FEMS Microbiol. Lett. 199: 161–169. [DOI] [PubMed] [Google Scholar]
- Majewski J., Zawadzki P., Pickerill P., Cohan F. M., Dowson C. G., 2000. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 182: 1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram P., Wall J. D., 2006. Fast “coalescent” simulation. BMC Genet. 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjoram P., Molitor J., Plagnol V., Tavare S., 2003. Markov chain Monte Carlo without likelihoods. Proc. Natl. Acad. Sci. USA 100: 15324–15328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., 1999. The detection and measurement of recombination from sequence data. Genetics 153: 1021–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Smith N. H., 1998. Detecting recombination from gene trees. Mol. Biol. Evol. 15: 590–599. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J., Smith N. H., O’Rourke M., Spratt B. G., 1993. How clonal are bacteria? Proc. Natl. Acad. Sci. USA 90: 4384–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G., Awadalla P., Fearnhead P., 2002. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160: 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A. T., Cardin N. J., 2005. Approximating the coalescent with recombination. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D., Serruto D., Parkhill J., Relman D. A., Donati C., et al. , 2008. Microbiology in the post-genomic era. Nat. Rev. Microbiol. 6: 419–430. [DOI] [PubMed] [Google Scholar]
- Morelli G., Didelot X., Kusecek B., Schwarz S., Bahlawane C., et al. , 2010. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 6: e1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namouchi A., Didelot X., Schöck U., Gicquel B., Rocha E. P. C., 2012. After the bottleneck: genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res. 22: 721–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nübel U., Roumagnac P., Feldkamp M., Song J.-H., Ko K. S., et al. , 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 105: 14130–14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J. K., Seielstad M. T., Perez-Lezaun A., Feldman M. W., 1999. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol. Biol. Evol. 16: 1791–1798. [DOI] [PubMed] [Google Scholar]
- Roberts M. S., Cohan F. M., 1993. The effect of DNA sequence divergence on sexual isolation in Bacillus. Genetics 134: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard S. K., McCarthy N. D., Falush D., Maiden M. C. J., 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320: 237–239. [DOI] [PubMed] [Google Scholar]
- Takuno S., Kado T., Sugino R. P., Nakhleh L., Innan H., 2012. Population genomics in bacteria: a case study of Staphylococcus aureus. Mol. Biol. Evol. 29: 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C. M., Nielsen K. M., 2005. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3: 711–721. [DOI] [PubMed] [Google Scholar]
- Thornton K., Andolfatto P., 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172: 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos M., 2009. Why do bacteria engage in homologous recombination? Trends Microbiol. 17: 226–232. [DOI] [PubMed] [Google Scholar]
- Vos M., Didelot X., 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3: 199–208. [DOI] [PubMed] [Google Scholar]
- Vulić M., Dionisio F., Taddei F., Radman M., 1997. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl. Acad. Sci. USA 94: 9763–9767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeley J., Lessard S., 2003. Theory of the effects of population structure and sampling on patterns of linkage disequilibrium applied to genomic data from humans. Genetics 164: 1043–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G. A., 1975. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276. [DOI] [PubMed] [Google Scholar]
- Whelan S., Liò P., Goldman N., 2001. Molecular phylogenetics: state-of-the-art methods for looking into the past. Trends Genet. 17: 262–272. [DOI] [PubMed] [Google Scholar]
- Wiuf C., 2000. A coalescence approach to gene conversion. Theor. Popul. Biol. 57: 357–367. [DOI] [PubMed] [Google Scholar]
- Wiuf C., Hein J., 2000. The coalescent with gene conversion. Genetics 155: 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahara K., Kawai M., Furuta Y., Takahashi N., Handa N., et al. , 2012. Genome-wide survey of mutual homologous recombination in a highly sexual bacterial species. Genome Biol. Evol. 4: 628–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawadzki P., Roberts M. S., Cohan F. M., 1995. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics 140: 917–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.