Abstract
Self-fertilization is generally seen to be disadvantageous in the long term. It increases genetic drift, which subsequently reduces polymorphism and the efficiency of selection, which also challenges adaptation. However, high selfing rates can increase the fixation probability of recessive beneficial mutations, but existing theory has generally not accounted for the effect of linked sites. Here, we analyze a model for the fixation probability of deleterious mutants that hitchhike with selective sweeps in diploid, partially selfing populations. Approximate analytical solutions show that, conditional on the sweep not being lost by drift, higher inbreeding rates increase the fixation probability of the deleterious allele, due to the resulting reduction in polymorphism and effective recombination. When extending the analysis to consider a distribution of deleterious alleles, as well as the average fitness increase after a sweep, we find that beneficial alleles generally need to be more recessive than the previously assumed dominance threshold (h < 1/2) for selfing to be beneficial from one-locus theory. Our results highlight that recombination aiding the efficiency of selection on multiple loci amplifies the fitness benefits of outcrossing over selfing, compared to results obtained from one-locus theory. This effect additionally increases the parameter range under which obligate outcrossing is beneficial over partial selfing.
Keywords: adaptation, background selection, dominance, mating system evolution, selfing
ALTHOUGH the majority of species reproduce via outcrossing, self-fertilization has been observed to be fairly common in many groups, including angiosperms (Igic and Kohn 2006) and some animals (Jarne and Auld 2006). In particular, different species of flowering plants consistently show a transition from outcrossing to self-reproduction (reviewed in Wright et al. 2013). Selfing is believed to have several short-term evolutionary advantages. These include having a 50% transmission advantage over outcrossers as they can fertilize their own ovules, as well as those of outcrossers (Fisher 1941). Selfing species also have reproductive assurance under pollen limitation, where they are more able to create seeds when mates are rare. This effect leads to an increased ability to colonize new areas (Baker 1955). At the genetic level, selfing can create homozygotes more quickly from recessive beneficial mutations compared to outcrossers, increasing selection acting on them (Pollak 1987; Caballero and Hill 1992; Charlesworth 1992). Selfing can thus rapidly evolve unless it is counteracted by sufficiently high inbreeding depression (Lande and Schemske 1985).
In spite of these short-term advantages, selfing is generally seen as an evolutionary dead-end. For example, Goldberg et al. (2010) (see also Wright and Barrett 2010) demonstrated that self-compatible plants in the nightshade (Solanaceae) family have higher extinction rates than self-incompatible lineages. The general reasoning behind this idea is that natural selection is less efficient overall because the effective population size is reduced by a factor of 1/(1 + F) in a panmictic population (Nordborg 2000), where F is Wright’s inbreeding coefficient [which he denoted FIS (Wright 1951)]. In particular, selfers are thought to be less fit due to drift load, where deleterious mutants build up in selfing populations because of a lack of recombination, analogous to ‘Muller’s ratchet’ in asexual organisms (Muller 1964; Charlesworth et al. 1993). This build-up of deleterious mutations can cause a “mutational meltdown,” where the fitness of selfing lineages are constantly reduced, leading to their extinction (Lynch et al. 1995; Willi 2013).
However, evidence supporting these theoretical disadvantages to selfing is limited (reviewed in Igic and Busch 2013). Relaxed selection against deleterious mutations should lead to an increase in the Dn/Ds ratio along selfing lineages, where Dn and Ds are substitution rates at nonsynonymous and synonymous sites, respectively (Glémin 2007). Yet little evidence for this hypothesis has been found, in contrast to what is observed in purely asexual populations (reviewed in Glémin and Galtier 2012). Some evidence of relaxed purifying selection in selfers has been found by comparing ratio of nonsynonymous over synonymous polymorphism or by calculating the relative frequency of preferred over unpreferred codons. Such studies have been performed on Arabidopsis (Qiu et al. 2011); Capsella (Qiu et al. 2011; Slotte et al. 2013; Brandvain et al. 2013); and Collinsia (Hazzouri et al. 2013). On the other hand, no evidence for relaxed selection was found when comparing divergence rates between species, such as in a separate study of Arabidopsis (Wright et al. 2002), as well as in studies of Triticeae (Haudry et al. 2008; Escobar et al. 2010), with mixed evidence arising in selfing Caenorhabditis species (Cutter et al. 2008). Together, this evidence suggests that selection has been relaxed recently, indicating that selfing is of recent origin and deleterious mutation accumulation is likely to be too weak to be the main cause of their higher extinction rates.
One overlooked idea is that selfing can be disadvantageous as the reduction in polymorphism it causes can lead to a lack of adaptability, especially when colonizing a new environment. Indeed, this was the original hypothesis of Stebbins (1957) as to why self-fertilization should be an “evolutionary blind alley”. Interestingly, selfing is not universally deleterious with regard to selection on adaptive traits. For example, the probability of fixation of recessive beneficial mutations is higher in selfing populations than outcrossing ones (Pollak 1987; Caballero and Hill 1992; Charlesworth 1992); this lies in contrast to the fact that dominant mutations are more likely to fix in outcrossers [the so-called Haldane’s sieve mechanism (Haldane 1927)]. Additionally, while outcrossing populations are more likely to fix mutations from standing variation than in selfers, in line with Stebbins’ hypothesis, selfing populations always fix beneficial alleles more quickly due to the ensuing reduction in heterozygosity (Glémin 2012; Glémin and Ronfort 2013).
In addition, selfing populations suffer from a reduced effective recombination rate due to greater homozygosity (Nordborg 2000); furthermore, it is well known that reduced recombination impedes the efficacy of selection at a specific locus [Hill–Robertson effects (Hill and Robertson 1966; Charlesworth et al. 2009)]. Therefore, the effective population size Ne should be further reduced in selfers, which can broaden the conditions for when adaptation rates should be lower in selfers than in outcrossers. However, the explicit effect of selfing on the dynamics of selection at several loci has been poorly explored up to now (with a few exceptions, such as Pollak and Sabran 1996).
Here, we rectify this situation by analyzing a model of selective sweeps in partially selfing populations and the effect it has on linked deleterious mutations. Hitchhiking of deleterious mutations associated with selective sweeps is not a purely theoretical concept, since it has been documented in outcrossing populations such as humans (Williamson et al. 2007; Chun and Fay 2011). It is therefore likely to have a stronger impact in partially selfing populations as the overall recombination rate is reduced. Recently, Hartfield and Otto (2011) analyzed a model for an outcrossing haploid population and used it to demonstrate how the presence of linked deleterious mutations reduced signals of sweeps in the genome. We extend that model to consider a diploid population that can also be subject to partial self-fertilization, to investigate the possibility that an adaptive substitution of a beneficial allele in selfers results in smaller improvement in fitness than in outcrossers, as linked deleterious mutations hitchhike along with the beneficial allele. We obtain analytical approximations for the probability that a deleterious allele hitchhikes to fixation, instead of being purged following recombination. These solutions are subsequently used to determine what effect selfing has on adaptation in the presence of linked deleterious mutation and how this compares to classic results on adaptation in selfing organisms.
General Approach
We commence by describing a general branching process method that forms the basis of our analysis. The model is an extension of that used in Hartfield and Otto (2011), which calculated the fixation probability of hitchhiking deleterious mutations in haploids subject to recombination. This method considers that a beneficial mutation emerges while linked to a deleterious allele, but the haplotype remains beneficial overall (with selective advantage snet). This haplotype spreads through the population according to a deterministic model, and at each timepoint, recombination could occur to create a fitter hapolotype without the deleterious mutant. This recombinant can either fix or go extinct in the long term; we then integrate over the entire time of the sweep to calculate the probability that successful recombination never occurs, so that the deleterious mutant fixes with the sweep. Hartfield and Otto (2011) showed that the branching process approximation is accurate if Nsnet ≳ 10, where snet is the overall selective advantage of the haplotype carrying both the advantageous and deleterious mutations. Therefore its application is quite general except when selection is very weak, in which case drift strongly affects the fixation probability of beneficial mutations (mathematically, this occurs when Nsnet approaches 1).
In our study, we account for deleterious hitchhiking in a diploid genomes, with arbitrary dominance within each loci and with individuals subject to a fixed inbreeding rate F. The full description of the model would normally involve 10 genotypes, which also takes into account the gametic phase of double heterozygotes mutants and two-locus inbreeding coefficients alongside the inbreeding rate F (see, for example, Golding and Strobeck 1980). To simplify the problem, the method of Hartfield and Otto (2011) allows the reduction of the equations to single-locus cases. However, the full solution for arbitrary selfing rate and dominance at each locus turns out to be intractable analytically, but can be solved numerically. Therefore, we subsequently examine special cases that are solvable and shed light on the process of deleterious hitchhiking with inbreeding.
Table 1 outlines the notation used in our analysis. Consider a population of N diploid individuals, so there are 2N haploid chromosomes in the population. A beneficial allele arises at a locus A; we designate this allele as A1 and the wild type as A0. This allele has a fitness advantage of hasa when present as a heterozygote and sa as a homozygote (relative to a wild-type fitness of 1). We focus on the most closely linked deleterious allele at locus B and assume that all others are likely to be shed over the course of the sweep and have negligible chance of fixation. We denote the deleterious allele as B1 and the wild type as B0. This assumption also implies that no other deleterious mutants exist between A0B0 loci on other wild-type chromosomes. This is a reasonable assumption if the recombination rate r is low, but other deleterious mutants could exist on other wild-type chromosomes that will increase the probability of deleterious hitchhiking; in these cases, our calculation can be viewed as a conservative estimate. The focal deleterious allele is selected against with strength hdsd as a heterozygote and sd as a homozygote. We assume that selection is additive over both loci, so that the fitness of the advantageous-deleterious haplotype when it appears is 1 + hasa − hdsd. We must also assume that hasa > hdsd to ensure that the haplotype is beneficial when rare and that (1 − ha)sa > (1 − hd)sd to ensure that the haplotype remains beneficial and will fix when frequent (so to avoid overdominant behavior).
Table 1. Glossary of notation.
| Symbol | Usage |
|---|---|
| 2N | Overall (diploid) population size |
| A0, A1 | Wild-type allele, beneficial mutation at locus A |
| B0, B1 | Wild-type allele, deleterious mutation at locus B |
| sa, sd | Fitness coefficients of advantageous, deleterious allele |
| ha, hd | Dominance coefficients of advantageous, deleterious allele |
| p | Frequency of advantageous-deleterious (A1B1) haplotype |
| σ | Proportion of matings that are self-fertilizing |
| F | Wright’s (1951) inbreeding coefficient, σ/(2 − σ) |
| , Δw | Population mean fitness, change in fitness following sweep |
| r | Recombination rate between two loci |
| r0 | Baseline recombination rate if distance between alleles is exponentially distributed |
| θ | Relative fitness of recombinant (advantageous-only) haplotype |
| Π | Fixation probability of recombinant haplotype |
At a given time, the advantage-deleterious haplotype (denoted A1B1) is at frequency p, with the wild-type haplotype present at frequency 1 − p. The A0B1 haplotype that carries only the deleterious allele is assumed to be present at a negligible frequency once the sweep commences and is not considered in the model. During reproduction of each generation, a proportion σ of matings are caused by self-fertilization, while 1 − σ are outcrossing. In this case, the steady-state level of inbreeding is equal to F = σ/(2 − σ) (Pollak 1987; Caballero and Hill 1992; Charlesworth 1992), where F is Wright’s inbreeding coefficient (Wright 1951). Therefore the population consists of homozygote A1B1 genotypes at frequency p2 + p(1 − p)F, A0B0/A1B1 genotypes at frequency 2p(1 − p)(1 − F), and wild-type A0B0 homozygotes at frequency (1 − p)2 + p(1 − p)F.
Let the A1B1 haplotype appear at initial frequency p0; usually this is 1/2N. We are interested in the cases where this haplotype is not lost stochastically when it is rare and proceeds to sweep through the population. It is also assumed that the population size is large enough so that drift effects do not completely determine the trajectory of the A1B1 haplotype. Mathematically, this condition arises when Nestot > 1, where stot = hasa − hdsd is the selective advantage of the haplotype when it appears. This condition further implies that stot > (1 + F)/N. Accordingly, prior to the formation of the advantageous-only haplotype, the change in frequency of the A1B1 haplotype can be given deterministically by the term:
| (1) |
which also assumes that selection is weak (sa, sd ≪ 1), where is the mean fitness of the population:
| (2) |
In Equation 1, Δp (on the left-hand side) should also be multiplied by ; we can neglect this term in our model since if sa, sd are small. Note also that the expressions in Equation 1 and 2 are equivalent to a one-locus diploid model with two alleles, where sa − sd is the selective advantage s of the fitter allele, and (hasa − hdsd)/(sa − sd) is the degree of dominance. Further details of the derivation are outlined in supporting information, File S1.
During the time course of this sweep, the A1B1 haplotype could recombine with the wild-type A0B0 haplotype to produce the advantageous mutation on its own (A1B0). Additionally a deleterious haplotype A0B1 is produced, which we assumed to be quickly lost after its formation. This process occurs with probability (1/2) ⋅ r ⋅ 2p(1 − p)(1 − F); it is clear from this term that higher levels of inbreeding reduce the effective recombination rate, and so it is likelier for the deleterious allele to hitchhike to fixation.
When rare, the advantageous-only haplotype (A1B0) segregates with A0B0 haplotypes to form diploid genotypes with frequency (1 − F)(1 − p); with the advantageous-deleterious A1B1 haplotype with frequency p(1 − F); and with itself with frequency F. By comparing the mean fitness of these genotypes with the population’s mean fitness prior to the introduction of the recombinant, we obtain the relative fitness of the recombinant haplotype, which we denote by θ(p):
| (3) |
Using these terms, we can calculate the general fixation probability of a deleterious mutation hitchhiking with a sweeping beneficial mutation.
Fixation Probability of a Recombinant Haplotype, Π
When the recombinant (A1B0) haplotype forms, its relative fitness advantage over time changes due to the continuing spread of the preexisting A1B1 haplotype. That is, when the A1B1 haplotype is rare, the recombinant haplotype has a fitness advantage of around hasa compared to the population mean fitness. However, when A1B1 is common, since the advantageous allele is widespread, the fitness advantage of the A1B0 recombinant is hdsd. Therefore, when calculating the fixation probability of A1B0, these changes need to be taken into consideration. Hartfield and Otto (2011) found that for outcrossing haploids, if the selective advantage is weak (s(t) ≪ 1 for all time t), Π is the solution to the following differential equation, which was derived using time-inhomogeneous branching-process methods (similar to those used in Kimura and Ohta 1970),
| (4) |
where p is the frequency of A1B1. To extend this solution to partially selfing diploids, we use Equation 4 but with s(t) = θ(p) (Equation 3), dp/dt = Δp (Equation 1), and multiply the Π2 term by (1 + F), to indicate how inbreeding magnifies the effect of drift and reduces the effective population size by this factor. The full differential equation to be solved is therefore
| (5) |
Overall Fixation Probability of a Hitchhiking Deleterious Allele
A deleterious allele fixes with an advantageous allele, if a recombinant does not form over the entire course of the sweep that subsequently emerges in the population. Equation 4 of Hartfield and Otto (2011) showed that this probability PHH can be given by
| (6) |
Here we use 2N in Equation 6 since we now have a diploid population, and κ(p) is the probability of the advantageous-only recombinant forming then emerging; κ(p) = r(1 − F)p(1 − p)Π(p), where r(1 − F) is the effective recombination rate in partially selfing organisms, caused through the reduction in heterozygosity (Golding and Strobeck 1980). As with the equation for dΠ/dp, we additionally replace dp/dt with Δp (Equation 1).
The general solution to find Π(p), as given by Equation 5, is long and unwieldy, and therefore it is intractable to use with Equation 6 to find a solution with arbitrary dominance and inbreeding values. However, numerical integration of Equations 5 and 6 can be performed using the NDSolve and NIntegrate functions of the Mathematica software (Wolfram Research, Inc. 2010) (File S2.1). Alternatively, by investigating special cases, analytical solutions can be obtained for the hitchhiking probability, which we show to be accurate over a wide parameter space.
Outline of Simulation Methods
Throughout this article, we compare our analytical and numerical solutions to stochastic simulations to verify their accuracy. Simulations were written in R and are available online (File S3; results are available in File S4). Initially, the advantageous-deleterious haplotype was introduced at an initial frequency of 1/2N (paired with a wild-type haplotype), with all other genotypes set to the wild type. The frequency of genotype gi was then changed deterministically by a factor due to selection, where wi is the fitness of the genotype and is the population mean fitness. Reproduction due to selfing and outcrossing was then determined using recursion equations outlined in Hedrick (1980, Equation 3). N genotypes were then resampled from a multinomial distribution, to calculate frequency changes due to fluctuations caused by random drift. This action completed one life cycle of the simulation. This cycle was repeated until a single haplotype was fixed and the simulation repeated until the advantageous allele A1 fixed 10,000 times. After this number of fixations was reached, it was noted how many times each haplotype fixed overall, so one could calculate the fixation probability of the advantageous-deleterious haplotype, given that the advantageous allele ultimately reached fixation. If we assume an exponential distribution of recombination lengths, then for each reintroduction of the advantageous-deleterious haplotype the recombination length is chosen from an exponential distribution, then scaled using Haldane’s mapping function, (1 − exp(− 2r))/2, to ensure that r does not exceed 1/2. The fixation probabilities of each haplotype are also used to determine the average fitness gain to a population following the sweep.
Specific Examples
ha = hd = 1/2, arbitrary F
The easiest case to analyze is that in which selection at both loci is additive, so heterozygote loci have half the selective effect of homozygote loci. As selfing has no effect on the probability of fixation at a single locus for this case (Caballero and Hill 1992; Charlesworth 1992), it allows one to see how inbreeding acting on recombination affects the hitchhiking probability. By solving Equation 5 with both dominance values equal to 1/2, and further assuming that selection is weak (that is, sa and sd ≪ 1), we obtain
| (7) |
This is half the value of Π obtained in Hartfield and Otto (2011, Equation 2) for a haploid, fully outcrossing population; the reduction by half is due to the dominance coefficient reducing selection on heterozygotes by this factor. Therefore, with additive selection, inbreeding has no effect on the fixation probability of recombinant haplotypes as they appear. The reason for this behavior is that inbreeding affects both the rate of change of p and the selective advantage of A1B0 by a factor (1 + F) due to increased drift effects. Since this extra drift affects all terms equally, it cancels out in Equation 5.
Substituting Equation 7 into Equation 6 and solving, we obtain the following hitchhiking probability
| (8) |
with
| (9) |
That is, the hitchhiking probability is equal to that for a haploid outcrossing population, raised to the power 2(1 − F)/(1 + F). We can use Equation 8 to ascertain how inbreeding affects the fixation probability. If F = 0, then the probability is squared, indicating how the likelihood of deleterious hitchhiking is reduced due to the higher population size (2N in diploid populations, compared to N in haploid populations), increasing the net recombination rate. As F increases, the power term is reduced, increasing the probability of deleterious hitchhiking. This is due to two consequences of inbreeding; first, it increases the speed at which the A1B1 haplotype sweeps through the population (see also Glémin 2012), and it also reduces the number of A1B1/A0B0 heterozygote genotypes, reducing the effective recombination rate by a factor (1 − F). We can demonstrate this process formally by noting that
| (10) |
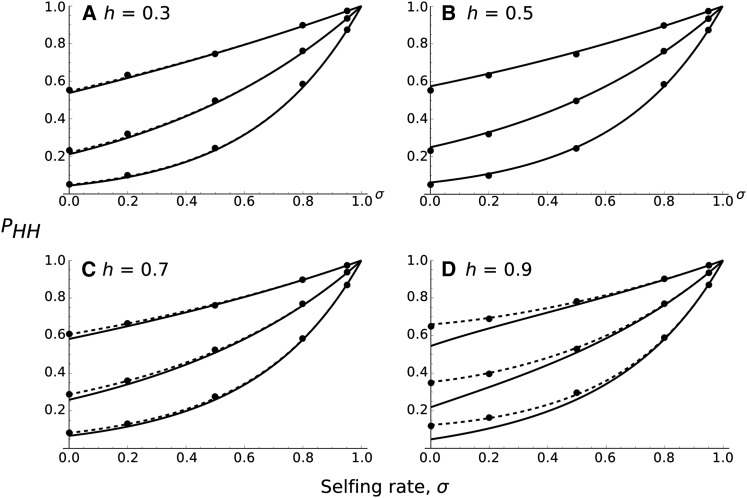
so the power term always decreases with higher inbreeding, until it reaches a value of 0 when F = 1. Finally, by substituting F = σ/(2 − σ) above, for σ the selfing rate, we see that (1 − F)/(1 + F) = 1 − σ, so the power to which the haploid probability is raised decreases linearly with the selfing rate. The only approximation used in Equation 8 is to assume weak selection, as in Hartfield and Otto (2011) (see File S2.1) so that it almost perfectly matches the numerical solution. Equation 8 also very accurately matches stochastic simulations (Figure 1).
Figure 1.
Probability of hitchhiking of the deleterious allele (PHH) as a function of the selfing rate, σ, assuming a fixed recombination distance between both loci. N = 10,000, sa = 0.02, sd = 0.01, and from bottom to top in each plot: 4Nr = 0.4, 1, 2. The dominance levels of each allele are equal to 0.3 (A), 0.5 (B), 0.7 (C), or 0.9 (D). Curves correspond to analytical approximations (Equation 13 for F near one, except for h = 0.5, where Equation 8 is used instead). Points correspond to 10,000 stochastic simulations for which the beneficial allele has fixed. Confidence intervals are small enough to lie within each point. The dashed curve corresponds to numerical integration of Equation 6.
ha = hd near 1/2, arbitrary F
We can come up with a more complete formula for the deleterious hitchhiking probability by taking a series expansion of Equation 5 around ha = hd = 1/2. By doing so, we obtain a new differential equation for Π:
| (11) |
By solving this equation (while assuming weak selection), we obtain
| (12) |
Note that inbreeding affects the fixation probability in this case, and also that if h = 1/2 then we retrieve the previous solution for Π (Equation 7).
After substituting Equation 12 into Equation 6, followed by some algebraic simplifications (File S2.1), we obtain a more general formula for P around h = 1/2, which can be written in a similar form as Equation 8,
| (13) |
with
| (14) |
and
| (15) |
| (16) |
where is the “effective” dominance level experienced by an allele at a low frequency, and is the effective dominance level experienced by an allele at a high frequency (see Glémin 2012). Note that Equation 8 can be written as Equation 13 with Ca = Cd = 1 + F.
ha = hd, F close to one
A final approximation can be obtained if we take a series expansion of Equation 5 around F = 1, to obtain an accurate approximation for highly selfing populations. In this case the differential equation for Π is
| (17) |
which can be solved to produce the following solution:
| (18) |
As before, this yields Equation 7 for h = 1/2. By solving the ensuing Equation 6 after making simplifications, we obtained a formula for PF≈1 of the same form as Equation 13 with
| (19) |
and
| (20) |
The two approximations (ha and hd near 1/2, and F close to one) give similar results and are accurate except if h is too far from 1/2 and F close to 0 (see Figure 1 and File S2.1).
Different Levels of Dominance for the Two Mutations
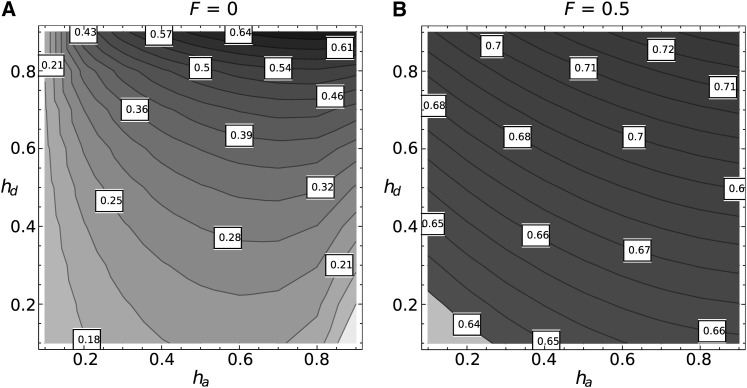
So far, we have considered that the two mutations shared the same dominance levels, which is quite restrictive. Using numerical integration of Equation 6, we can explore more general cases. In fully outcrossing populations, Figure 2A shows that dominant deleterious alleles are more likely than recessive ones to hitchhike. This is somewhat counterintuitive because we have expected dominant deleterious alleles to be more easily purged and therefore contribute less to the fixation probability.
Figure 2.
Probability of hitchhiking of the deleterious allele for the combination of dominance levels for the beneficial (ha) and the deleterious (hd) alleles. Values were obtained through numerical integration of Equation 6. N = 10,000, sa = 0.01, sd = 0.001, Nr = 1, with F = 0 (A) or F = 0.5 (B).
However, what matters is the relative fitness of the recombinant when it occurs. At low frequency, the relative fitness of the recombinant weakly depends on the level of dominance of the deleterious allele; it is equal to hasa when p = 0. On the contrary, at high frequency, the relative fitness of the recombinant is dominated by the level of dominance of the deleterious allele. The more dominant the allele, the lower the relative fitness of the recombinant, which converges toward (1 − hd)sd for p = 1. Moreover, the relative fitness of the recombinant haplotype decreases over time, and recombinants are formed earlier when the deleterious allele is recessive. Because of this asymmetry, the difference between recessive and dominant deleterious alleles becomes apparent during the second part of the sweep (this is illustrated in File S2.1). Therefore, the net probability of successful recombinant formation is lowered if hd is higher.
The effect of the level of dominance of the beneficial allele is somewhat different. The probability of hitchhiking is higher for intermediate levels of dominance, for which the length of the sweeps are the shortest (van Herwaarden and van der Wal 2002; Glémin 2012). If we increase the inbreeding level F to 0.5 (Figure 2B), this intermediate behavior disappears, so hitchhiking probability increases with ha. This behavior arises because homozygotes are formed more quickly with inbreeding, so the sweeps act more like a haploid system, which goes to fixation in a shorter period of time as selection acting on genotypes increases from hasa to sa (Glémin 2012, and the next section). This effect reduces the opportunity for recombination to arise, leading to an increased probability of deleterious hitchhiking.
Surveys of deleterious mutations across several studies of different species predominantly show them to be partly recessive (see Manna et al. 2011 for a recent review). Given this observation, we also specifically explored varying ha for a fixed hd < 1/2. We see (File S2.1) that the effect of dominance is rather weak compared to the effect of selfing on recombination, so that our previous approximations for ha = hd (and even ha = hd = 1/2) give a reasonable picture of what could happen under realistic biological conditions, especially if recombination rates are sufficiently high.
Accounting for the Distribution of Deleterious Alleles Throughout the Genome
For the general formulation, we assumed that there was a fixed difference between the advantageous allele and the nearest segregating deleterious mutant. We can alter the model to account for a distribution of deleterious alleles, and therefore also the distance to the nearest allele, throughout the genome.
The frequency of a strongly selected allele in the genome, at mutation–selection balance in an infinite population, is where u is the per-locus mutation rate (Caballero and Hill 1992). Hence the expected number of deleterious mutants per haploid genome is for U the genomic deleterious mutation rate. By comparing this result to mutation–selection balance in an outcrossing diploid population , we see that the mean distance to a deleterious allele in a selfing species is times that for an outcrossing species. Assuming that the net genetic distance between the beneficial mutation and the first deleterious allele across the population is exponentially distributed with mean and R = 4Nr (also R0 = 4Nr0), we can integrate the probability of hitchhiking over this distribution as
| (21) |
where Pfix is the fixation probability of the hitchhiking deleterious mutant, given a fixed recombination rate r. It is clear that due to the exp(−R) term in the function that this integral equals zero if R → ∞, since this functions remains following integration. Therefore, the total hitchhiking probability equals
| (22) |
The solution takes the general form (see File S2.2 for full details of the derivation)
| (23) |
Note that r0 is also proportional to sd, since the deleterious allele strength determines the average distance between mutant alleles. However, because sd is not affected by the selfing rate, it is directly included in r0.
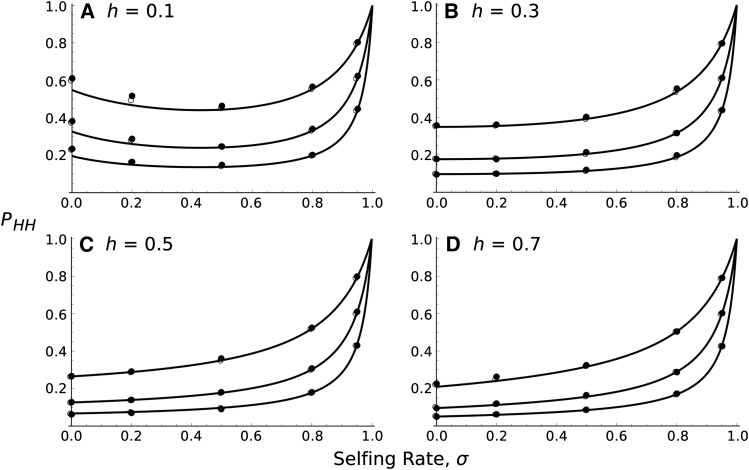
Figure 3 plots profile curves for Equation 23, with Pfix set to PF≈1 (Equation 13 combined with Equations 19 and 20) since it proved to be most accurate compared to simulations under a general parameter range. From the results we see that if h is high enough, the previous result holds that higher inbreeding levels aid fixation of the hitchhiking deleterious mutation. However, we also see that for low h, there appears to be a nonzero optimum inbreeding rate that minimizes the hitchhiking probability. This plot suggests that if deleterious alleles are sufficiently recessive, a nonzero rate of inbreeding efficiently purges them more quickly, as reduced drift increases the probability of formation of deleterious homozygotes following creation of the recombinant haplotype (Charlesworth and Charlesworth 1987). Furthermore, low rates of inbreeding do not reduce recombination to a disadvantageous low level.
Figure 3.
Probability of hitchhiking of the deleterious allele as a function of the selfing rate, σ, by taking into account an exponential distribution of deleterious alleles across the genome. N = 10,000, sa = 0.02, sd = 0.01, and from bottom to top in each plot: 4Nr0 = 0.4, 1, 2. The dominance levels of each allele are equal to 0.1 (A), 0.3 (B), 0.5 (C), or 0.7 (D). Curves correspond to Equation 23. Solid circles correspond to 10,000 stochastic simulations for which the beneficial allele has fixed and open circles to solutions based on numerical integration of the hitchhiking probability (Equation 6 with r determined by an exponential distribution).
However, the other result from this analysis implies that inbreeding is not always better in fixing recessive beneficial mutations. For a single locus under free recombination, the probability of fixation is higher in a selfing population if ha < 1/2 and lower if ha > 1/2. Our analysis shows that with our model, selfing offers an advantage only if h is low enough, and even then only a partially selfing population offers the greatest benefit to reducing the deleterious allele’s fixation probability.
Total Fitness Gains After the Appearance of the Beneficial Mutation
We can also use the previous analysis to determine to what extent inbreeding affects the mean fitness of the population following the appearance of a beneficial mutation. There is a tradeoff between how the dominance level of the beneficial mutant affects the outcome of the sweep. Recessive beneficial alleles (h ≈ 0) take a longer period of time to fix than those with intermediate dominance (0 < h < 1), due to weaker selection acting on them when rare. This is reflected in the fact that recessive mutants usually leave weaker signatures of selection in the genome in outcrossing species (Teshima and Przeworski 2006; Ewing et al. 2011). Furthermore, this delay increases the probability that a recombinant haplotype arises and the deleterious allele does not fix (see also Hartfield and Otto 2011). However, dominant alleles are more likely to fix in outcrossing populations (Haldane’s sieve; Haldane 1927; Pannell et al. 2005), yet selfing can create recessive beneficial homozygotes rapidly, giving them the same fixation probability as dominant mutants (Haldane 1927; Charlesworth 1992). Therefore, it should be determined to what extent inbreeding can increase the population’s fitness after a sweep, given a certain dominance level of mutations.
The mean fitness increase following a sweep can be calculated by Pemerge × [PHH(sa − sd) + (1 − PHH)sa]. Pemerge is the probability that the advantageous-deleterious haplotype is not lost when rare; if the overall strength of selection sa − sd is strong, then a good approximation is given by (Caballero and Hill 1992; Charlesworth 1992; Glémin and Ronfort 2013)
| (24) |
and we use PHH from Equation 23. Note that, here, we assume that the beneficial allele necessarily appears closely linked to a deleterious allele, such that the initial fate of the beneficial allele is fully determine by the fate of the advantageous-deleterious haplotype. This is a reasonable assumption to make, given the large number of deleterious alleles that are likely to segregate in a genome (see quantitative arguments in Hartfield and Otto 2011).
We can also propose an approximation for the general case (ha ≠ hd) by using Equation 24 after replacing h by (hasa − hdsd)/(sa − sd). In the preceding section, we showed that the effect of dominance on the probability of hitchhiking is weak as compared to the effect of reducing effective recombination rate by selfing and that the additive case captures the main pattern quite well (File S2.1). We can thus simply use PHH(h = 1/2) to obtain (see also File S2.3)
| (25) |
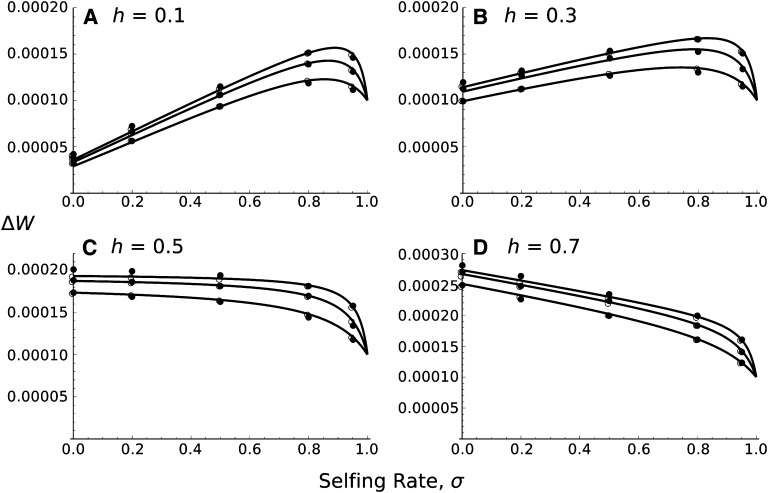
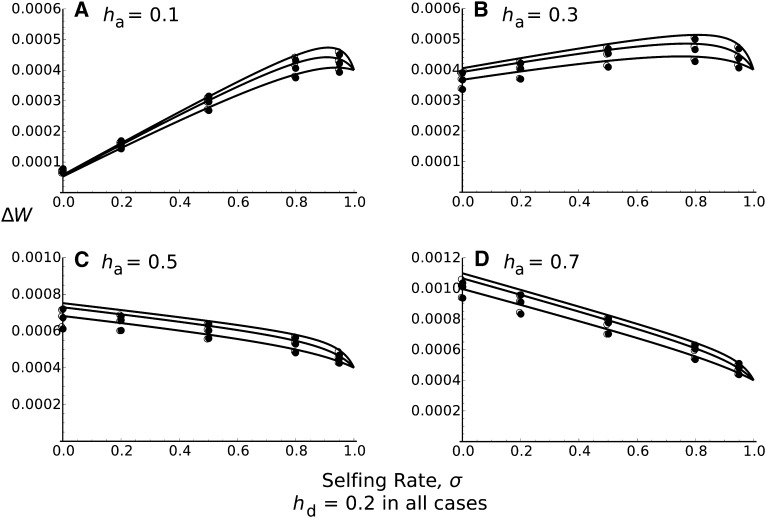
Figure 4 plots the mean fitness increase as a function of σ for different levels of dominance. Although rather crude, this approximation gives accurate results. Figure 4 shows that the fitness gain can be higher for obligate outcrossers than for complete selfers, even for partially recessive beneficial alleles, contrary to what is expected under single locus theory, where the dominance threshold is 1/2 (Caballero and Hill 1992; Charlesworth 1992). This result highlights how outcrossing can be beneficial through recombination removing the linked deleterious mutation, which would also be fixed in fully selfing populations. Hence in Figure 4, A and B, the optimal selfing rate is not 1, as predicted with one-locus theory, but slightly less than that. Equation 25 is also accurate for ha ≠ hd as compared to stochastic simulations (Figure 5 and File S2.3).
Figure 4.
Average fitness increase in the population following a successful sweep, as a function of the selfing rate σ. N = 10,000, sa = 0.02, sd = 0.01, and from bottom to top in each plot: 4Nr0 = 0.4, 1, 2. The dominance levels of each allele are equal to 0.1 (A), 0.3 (B), 0.5 (C), or 0.7 (D). Curves correspond to Equation 25. Solid circles correspond to 10,000 stochastic simulations for which the beneficial allele has fixed and open circles to solutions based on numerical integration of the hitchhiking probability (Equation 6 with r determined by an exponential distribution).
Figure 5.
Average fitness increase in the population following a successful sweep, as a function of the selfing rate σ, when ha ≠ hd. N = 10,000, sa = 0.02, sd = 0.01, and from bottom to top in each plot: 4Nr0 = 0.4, 1, 2. The dominance levels of the advantageous allele ha = 0.1 (A), 0.3 (B), 0.5 (C), or 0.7 (D), with hd = 0.2 in all cases. Curves correspond to Equation 25. Solid circles correspond to 10,000 stochastic simulations for which the beneficial allele has fixed and open circles to solutions based on numerical integration of the hitchhiking probability (Equation 6 with r determined by an exponential distribution).
Solving Δw|F = 0 = Δw|F = 1 for ha, we can obtain the threshold dominance level for which the fitness gain is higher in outcrossers than in selfers
| (26) |
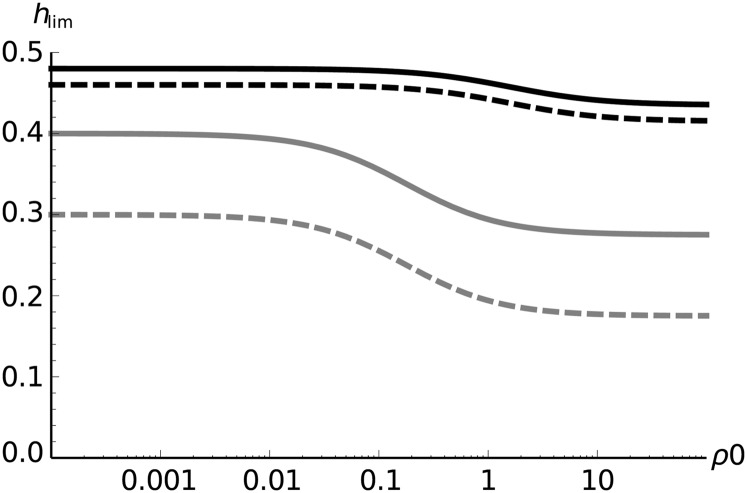
where S = sd/sa and ρ0 = Nr0. This threshold can be much lower than 1/2, especially for high ρ0 for which Equation 26 converges toward
| (27) |
Even in the absence of recombination (ρ0 = 0), linkage to deleterious alleles is disadvantageous to selfing, provided that deleterious alleles are recessive (hd < 1/2). With full linkage, only the fixation of the advantageous-deleterious haplotype matters. In the limit ρ0 → 0, Equation 26 reduces to
| (28) |
which is lower than 1/2 when hd < 1/2. This is because while selfing increases the apparent dominance of the beneficial allele, which helps its fixation and increases the fitness gains following a sweep, it also raises the apparent dominance of the deleterious allele, which decreases population fitness.
This behavior is shown in Figure 6. So in highly recombining species, outcrossing can offer the highest fitness increases, even if beneficial mutants are partly recessive, as it will also enable more efficient purging of the deleterious mutant. This result widens the parameter space under which strict outcrossing is beneficial over complete selfing.
Figure 6.
Limit levels of dominance for which the fitness gain is higher in outcrossers as a function of ρ0 = Nr0, as given by Equation 26. S = sa/sd = 0.1 (black) or 0.5 (gray); hd = 0.3 (solid lines) or 0.1 (dashed lines).
Discussion
Selfing is thought to be an evolutionary dead-end due to its inability to purge deleterious mutations and to adapt as quickly as outcrossing populations, especially in new environments (Stebbins 1957). However, it is currently unclear how the two processes of deleterious mutation removal and beneficial allele fixation interact and how outcrossers and selfers fix adaptive mutations in the presence of deleterious mutations, which can be more efficiently purged in selfing organisms if they are recessive (Charlesworth and Charlesworth 1987). To this end, we extended the model of Hartfield and Otto (2011), which determined the probability of fixation of deleterious alleles that could hitchhike with selective sweeps in haploid genomes. This new model considered a diploid population, so both the advantageous and deleterious allele have possible dominance effects on fitness, and also includes the possibility that organisms could partially self-fertilize and subsequently inbreed.
One-locus theory predicts that selfing should favor the fixation of recessive beneficial mutations (ha < 1/2) (Pollak 1987; Caballero and Hill 1992; Charlesworth 1992). Here, we show that linkage to deleterious allele favors outcrossing, even for partly recessive alleles. This is due to selfing exposing both the beneficial allele and the linked deleterious allele to increased selection, thus favoring the initial stochastic loss of the haplotype if the deleterious allele is recessive and the beneficial one not too recessive. Overall, this reduces the dominance level above which outcrossing is favored (Equation 26).
We found that, conditional on the initial haplotype not being lost stochastically, inbreeding increases the fixation probability of hitchhiking deleterious alleles, due to a decreased effective population size speeding up the rate of fixation of the sweep and reducing the effective recombination rate. These mechanisms are most clearly demonstrated using Equation 8 (which assumed ha = hd = 1/2). This finding is especially true if the underlying genomic recombination rate is high (Figure 6). This is because the fitness advantage of outcrossers, due to recombination disentangling beneficial alleles from poor backgrounds, is much stronger than the advantage of selfing of exposing beneficial alleles to selection. These results verify our intuition that the effect of linkage to multiple sites favors outcrossing over a larger parameter range than considered in just one-locus models, due to the further reductions in effective population size caused by selection interference at linked sites (Comeron et al. 2008). However, once we condition on the possible distribution of deleterious mutations present, we found that if both the advantageous and deleterious mutations are recessive enough, then there exists a nonzero rate of inbreeding that minimizes fixation probability or maximizes mean fitness accordingly (Figure 3, Figure 4, and Figure 5). This result suggests that intermediate selfing rates could be an optimal strategy in breeding programs for maximizing crop yield. More generally, our results suggest that the interaction between deleterious and advantageous mutations should be taken into account in models of breeding system evolution, especially the evolution of mixed-mating systems.
Our results also bear implications for the signatures that selection can leave on neutral variation in a genome. In selfers, selective sweeps are expected to be less frequent than in outcrossers, as adaptation is thought to be less efficient. However, if they do arise then they should leave a stronger signature of a sweep because they fix in a shorter timeframe, and the reduction in linked neutral variation should extend over longer genomic regions due to reduced recombination (Glémin 2012). Linked deleterious alleles increase the time taken for a sweep and also increase the opportunity for recombination, compared to when linked neutral variation only is present, hence reducing the signature of selective sweeps (Hartfield and Otto 2011). Given that most deleterious alleles are recessive, our model unexpectedly reinforces the idea that linked deleterious mutations blur signals of selective sweeps. Figure 2A shows that recessive deleterious alleles are less easily hitchhiked. This is because once the advantageous-deleterious haplotype has reached a sufficiently high frequency, homozygotes are formed, exposing deleterious alleles to selection and slowing down the course of the sweep. This effect allows more opportunity for recombination to arise, increasing neutral variation around the site of a sweep (Hartfield and Otto 2011). Although linked deleterious alleles should also increase sweep duration in highly selfing species, leading to a weakened signal of selection, signatures of selective sweep should remain stronger compared to outcrossers. This is due to the strong effect of selfing on reducing effective recombination rates.
Care must be taken though when evaluating our model under very general circumstances, since little is known about the rate and strength of beneficial mutations in partial selfers, especially compared to data obtained from outcrossing species (Eyre-Walker 2006; Eyre-Walker and Keightley 2007). In Arabidopsis thaliana, a study of the underlying distribution of fitness effects estimated that half of all mutations are beneficial (Shaw et al. 2002), although this result has proved controversial (Keightley and Lynch 2003; Bataillon 2003). Barrier et al. (2003) estimated that ∼5% of genes were under adaptive selection, with further work verifying that adaptive evolution is rare overall, but is present in genes coding for stress and immune responses (Slotte et al. 2011; see also Clark et al. 2007). Similarly, ∼1% of sampled genes show evidence of positive selection in the legume Medicago truncatula (Paape et al. 2013). In general, mutations are deleterious, so these adaptive alleles are likely to arise in close linkage with deleterious sites. Estimates of the deleterious mutation rate range from 0.07 to 0.1 mutations per generation per genome in Arabidopsis (Ossowski et al. 2010; Rutter et al. 2012), 0.4 in Caenorhabditis elegans (Denver et al. 2004), and upper limits of 0.6–0.8 in Daphnia pulex (Lynch et al. 1998; Deng et al. 2006). Further empirical estimates of the distribution of fitness effects in facultative sexuals, and partially selfing organisms, will be important to determine the effect of advantageous-deleterious interactions on the evolution of mating systems.
In addition, there is also a lack of empirical dominance data in selfing and outcrossing populations. In general populations, deleterious mutations have been found to be recessive (Simmons and Crow 1977; Halligan and Keightley 2009; Agrawal and Whitlock 2011), and it has recently been shown theoretically that deleterious mutations should have an average dominance value of 1/4 (Manna et al. 2011). However, Vassilieva et al. (2000) found that for a laboratory population of Caenorhabdits elegans set up to be mostly outcrossing, most traits exhibited a value of h ≈ 1/2. Data are even more lacking for dominance levels of advantageous mutations. Existing evidence suggests that these are generally additive or recessive (Orr 2010), but studies have only generally been performed on outcrossing species, such as Drosophila. Recently, a metaanalysis of quantitative trait loci studies in domesticated plant species showed that fixed adaptive traits in selfing plants tended to be recessive, while those in outcrossers tended to be dominant (Ronfort and Glémin 2013). These data match up with theory on adaptation in partially selfing plants, including our results, that selfing species can more ably fix recessive beneficial mutations. However, studies of quantitative traits do not inform on the fine-scale interactions between the underlying loci.
Finally, the results of our model pose the question as to how other linkage effects, such as between two beneficial mutations or between epistatically interacting pairs, affect fitness gains in partially selfing organisms. We therefore plan to extend this model to other adaptive cases of interest in a future research article. We also hope that the results of this work, as well as others, will motivate genomics research into the fitness and dominance effects of mutations in selfing and outcrossing species, to determine how selective effects, linkage, and genomic segregation might all interact to affect mating system evolution.
Supplementary Material
Acknowledgments
We thank Marcy Uyenoyama and two anonymous reviewers for constructive comments on the manuscript. M.H. is funded by an ATIP-Avenir grant from Centre National de la Recherche Scientifique (CNRS) and Institut National de la Santé et de la Recherche Médicale (INSERM) to Samuel Alizon, and acknowledges additional support from the CNRS and the IRD. This work was also supported by a grant from the Agence Nationale de la Recherche to S.G. (ANR-11-BSV7-013-03). This is publication ISEM 2013-160.
Footnotes
Communicating editor: M. K. Uyenoyama
Literature Cited
- Agrawal A. F., Whitlock M. C., 2011. Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics 187: 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker H. G., 1955. Self-compatibility and establishment after ‘long-distance’ dispersal. Evolution 9: 347–349. [Google Scholar]
- Barrier M., Bustamante C. D., Yu J., Purugganan M. D., 2003. Selection on rapidly evolving proteins in the Arabidopsis genome. Genetics 163: 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bataillon T., 2003. Shaking the ‘deleterious mutations’ dogma? Trends Ecol. Evol. 18: 315–317. [Google Scholar]
- Brandvain Y., Slotte T., Hazzouri K. M., Wright S. I., Coop G., 2013. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 9: e1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A., Hill W. G., 1992. Effects of partial inbreeding on fixation rates and variation of mutant genes. Genetics 131: 493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1992. Evolutionary rates in partially self-fertilizing species. Am. Nat. 140: 126–148. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Betancourt A. J., Kaiser V. B., Gordo I., 2009. Genetic recombination and molecular evolution. Cold Spring Harb. Symp. Quant. Biol. 74: 177–186. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., 1987. Inbreeding depression and its evolutionary consequences. Annu. Rev. Ecol. Evol. Syst. 18: 237–268. [Google Scholar]
- Charlesworth D., Morgan M., Charlesworth B., 1993. Mutation accumulation in finite outbreeding and inbreeding populations. Genet. Res. 61: 39–56. [Google Scholar]
- Chun S., Fay J. C., 2011. Evidence for hitchhiking of deleterious mutations within the human genome. PLoS Genet. 7: e1002240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. M., Schweikert G., Toomajian C., Ossowski S., Zeller G., et al. , 2007. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317: 338–342. [DOI] [PubMed] [Google Scholar]
- Comeron J. M., Williford A., Kliman R. M., 2008. The Hill–Robertson effect: evolutionary consequences of weak selection and linkage in finite populations. Heredity 100: 19–31. [DOI] [PubMed] [Google Scholar]
- Cutter A. D., Wasmuth J. D., Washington N. L., 2008. Patterns of molecular evolution in Caenorhabditis preclude ancient origins of selfing. Genetics 178: 2093–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H.-W., Li J., Pfrender M. E., Li J.-L., Deng H., 2006. Upper limit of the rate and per generation effects of deleterious genomic mutations. Genet. Res. 88: 57–65. [DOI] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Lynch M., Thomas W. K., 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682. [DOI] [PubMed] [Google Scholar]
- Escobar J. S., Cenci A., Bolognini J., Haudry A., Laurent S., et al. , 2010. An integrative test of the dead-end hypothesis of selfing evolution in Triticeae (Poaceae). Evolution 64: 2855–2872. [DOI] [PubMed] [Google Scholar]
- Ewing G., Hermisson J., Pfaffelhuber P., Rudolf J., 2011. Selective sweeps for recessive alleles and for other modes of dominance. J. Math. Biol. 63: 399–431. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., 2006. The genomic rate of adaptive evolution. Trends Ecol. Evol. 21: 569–575. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A., Keightley P. D., 2007. The distribution of fitness effects of new mutations. Nat. Rev. Genet. 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Fisher R. A., 1941. Average excess and average effect of a gene substitution. Ann. Eugen. 11: 53–63. [Google Scholar]
- Glémin S., 2007. Mating systems and the efficacy of selection at the molecular level. Genetics 177: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glémin S., 2012. Extinction and fixation times with dominance and inbreeding. Theor. Popul. Biol. 81: 310–316. [DOI] [PubMed] [Google Scholar]
- Glémin, S., and N. Galtier, 2012 Genome evolution in outcrossing versus selfing versus asexual species, pp. 311–335 in Methods Mol. Biol., Vol. 855, Chap. 11, edited by M. Anisimova. Humana Press, Clifton, NJ. [DOI] [PubMed] [Google Scholar]
- Glémin S., Ronfort J., 2013. Adaptation and maladaptation in selfing and outcrossing species: new mutations versus standing variation. Evolution 67: 225–240. [DOI] [PubMed] [Google Scholar]
- Goldberg E. E., Kohn J. R., Lande R., Robertson K. A., Smith S. A., et al. , 2010. Species selection maintains self-incompatibility. Science 330: 493–495. [DOI] [PubMed] [Google Scholar]
- Golding G. B., Strobeck C., 1980. Linkage disequilibrium in a finite population that is partially selfing. Genetics 94: 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. B. S., 1927. A mathematical theory of natural and artificial selection, part V. Selection and mutation. Math. Proc. Camb. Philos. Soc. 23: 838–844. [Google Scholar]
- Halligan D. L., Keightley P. D., 2009. Spontaneous mutation accumulation studies in evolutionary genetics. Annu. Rev. Ecol. Syst. 40: 151–172. [Google Scholar]
- Hartfield M., Otto S. P., 2011. Recombination and hitchhiking of deleterious alleles. Evolution 65: 2421–2434. [DOI] [PubMed] [Google Scholar]
- Haudry A., Cenci A., Guilhaumon C., Paux E., Poirier S., et al. , 2008. Mating system and recombination affect molecular evolution in four Triticeae species. Genet. Res. 90: 97–109. [DOI] [PubMed] [Google Scholar]
- Hazzouri K. M., Escobar J. S., Ness R. W., Killian Newman L., Randle A. M., et al. , 2013. Comparative population genomics in Collinsia sister species reveals evidence for reduced effective population size, relaxed selection, and evolution of biased gene conversion with and ongoing mating system shift. Evolution 67: 1263–1278. [DOI] [PubMed] [Google Scholar]
- Hedrick P. W., 1980. Hitchhiking: a comparison of linkage and partial selection. Genetics 94: 791–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1966. The effect of linkage on limits to artificial selection. Genet. Res. 8: 269–294. [PubMed] [Google Scholar]
- Igic B., Busch J. W., 2013. Is self-fertilization an evolutionary dead end? New Phytol. 198: 386–397. [DOI] [PubMed] [Google Scholar]
- Igic B., Kohn J. R., 2006. The distribution of plant mating systems: study bias against obligately outcrossing species. Evolution 60: 1098–1103. [PubMed] [Google Scholar]
- Jarne P., Auld J. R., 2006. Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution 60: 1816–1824. [DOI] [PubMed] [Google Scholar]
- Keightley P. D., Lynch M., 2003. Towards a realistic model of mutations affecting fitness. Evolution 57: 683–685. [DOI] [PubMed] [Google Scholar]
- Kimura M., Ohta T., 1970. Probability of fixation of a mutant gene in a finite population when selective advantage decreases with time. Genetics 65: 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R., Schemske D. W., 1985. The evolution of self-fertilization and inbreeding depression in plants. I. Genetic models. Evolution 39: 24–40. [DOI] [PubMed] [Google Scholar]
- Lynch M., Conery J., Burger R., 1995. Mutation accumulation and the extinction of small populations. Am. Nat. 146: 489–518. [Google Scholar]
- Lynch M., Latta L., Hicks J., Giorgianni M., 1998. Mutation, selection, and the maintenance of life-history variation in a natural population. Evolution 52: 727–733. [DOI] [PubMed] [Google Scholar]
- Manna F., Martin G., Lenormand T., 2011. Fitness landscapes: an alternative theory for the dominance of mutation. Genetics 189: 923–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H. J., 1964. The relation of recombination to mutational advance. Mutat. Res. 1: 2–9. [DOI] [PubMed] [Google Scholar]
- Nordborg M., 2000. Linkage disequilibrium, gene trees and selfing: an ancestral recombination graph with partial self-fertilization. Genetics 154: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr H. A., 2010. The population genetics of beneficial mutations. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledo J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paape T., Bataillon T., Zhou P., Kono T. J. Y., Briskine R., et al. , 2013. Selection, genome-wide fitness effects and evolutionary rates in the model legume Medicago truncatula. Mol. Ecol. 22: 3525–3538. [DOI] [PubMed] [Google Scholar]
- Pannell J. R., Dorken M. E., Eppley S. M., 2005. ‘Haldane’s Sieve’ in a metapopulation: sifting through plant reproductive polymorphisms. Trends Ecol. Evol. 20: 374–379. [DOI] [PubMed] [Google Scholar]
- Pollak E., 1987. On the theory of partially inbreeding finite populations. I. Partial selfing. Genetics 117: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak E., Sabran M., 1996. On the theory of partially inbreeding finite populations. IV. The effective population size for polyploids reproducing by partial selfing. Math. Biosci. 135: 69–84. [DOI] [PubMed] [Google Scholar]
- Qiu S., Zeng K., Slotte T., Wright S., Charlesworth D., 2011. Reduced efficacy of natural selection on codon usage bias in selfing Arabidopsis and Capsella species. Genome Biol. Evol. 3: 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronfort J., Glémin S., 2013. Mating system, Haldane’s sieve, and the domestication process. Evolution 67: 1518–1526. [DOI] [PubMed] [Google Scholar]
- Rutter M. T., Roles A., Conner J. K., Shaw R. G., Shaw F. H., et al. , 2012. Fitness of Arabidopsis thaliana mutation accumulation lines whose spontaneous mutations are known. Evolution 66: 2335–2339. [DOI] [PubMed] [Google Scholar]
- Shaw F. H., Geyer C. J., Shaw R. G., 2002. A comprehensive model of mutations affecting fitness and interferences for Arabidopsis thaliana. Evolution 56: 453–463. [DOI] [PubMed] [Google Scholar]
- Simmons M. J., Crow J. F., 1977. Mutations affecting fitness in Drosophila populations. Annu. Rev. Genet. 11: 49–78. [DOI] [PubMed] [Google Scholar]
- Slotte T., Bataillon T., Hansen T. T., St. Onge K., Wright S. I., et al. , 2011. Genomic determinants of protein evolution and polymorphism in Arabidopsis. Genome Biol. Evol. 3: 1210–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K. M., Ågren J. A., Koenig D., Maumus F., et al. , 2013. The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835. [DOI] [PubMed] [Google Scholar]
- Stebbins G. L., 1957. Self fertilization and population variability in the higher plants. Am. Nat. 91: 337–354. [Google Scholar]
- Teshima K. M., Przeworski M., 2006. Directional positive selection on an allele of arbitrary dominance. Genetics 172: 713–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Herwaarden O. A., van der Wal N. J., 2002. Extinction time and age of an allele in a large finite population. Theor. Popul. Biol. 61: 311–318. [DOI] [PubMed] [Google Scholar]
- Vassilieva L. L., Hook A. M., Lynch M., 2000. The fitness effects of spontaneous mutations in Caenorhabdits elegans. Evolution 54: 1234–1246. [DOI] [PubMed] [Google Scholar]
- Willi Y., 2013. Mutational meltdown in selfing Arabidopsis lyrata. Evolution 67: 806–815. [DOI] [PubMed] [Google Scholar]
- Williamson S. H., Hubisz M. J., Clark A. G., Payseur B. A., Bustamante C. D., et al. , 2007. Localizing recent adaptive evolution in the human genome. PLoS Genet. 3: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram Research, Inc. , 2010. Mathematica Edition, Version 8.0. Wolfram Research, Champaign, IL. [Google Scholar]
- Wright S., 1951. The genetical structure of populations. Ann. Eugen. 15: 323–354. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Barrett S. C. H., 2010. The long-term benefits of self-rejection. Science 330: 459–460. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Lauga B., Charlesworth D., 2002. Rates and patterns of molecular evolution in inbred and outbred Arabidopsis. Mol. Biol. Evol. 19: 1407–1420. [DOI] [PubMed] [Google Scholar]
- Wright S. I., Kalisz S., Slotte T., 2013. Evolutionary consequences of self-fertilization in plants. Proc. R. Soc. B 280: 20130133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.