Abstract
We employed deep genome sequencing of two parents and 12 of their offspring to estimate the mutation rate per site per generation in a full-sib family of Drosophila melanogaster recently sampled from a natural population. Sites that were homozygous for the same allele in the parents and heterozygous in one or more offspring were categorized as candidate mutations and subjected to detailed analysis. In 1.23 × 109 callable sites from 12 individuals, we confirmed six single nucleotide mutations. We estimated the false negative rate in the experiment by generating synthetic mutations using the empirical distributions of numbers of nonreference bases at heterozygous sites in the offspring. The proportion of synthetic mutations at callable sites that we failed to detect was <1%, implying that the false negative rate was extremely low. Our estimate of the point mutation rate is 2.8 × 10−9 (95% confidence interval = 1.0 × 10−9 − 6.1 × 10−9) per site per generation, which is at the low end of the range of previous estimates, and suggests an effective population size for the species of ∼1.4 × 106. At one site, point mutations were present in two individuals, indicating that there had been a premeiotic mutation cluster, although surprisingly one individual had a G→A transition and the other a G→T transversion, possibly associated with error-prone mismatch repair. We also detected three short deletion mutations and no insertions, giving a deletion mutation rate of 1.2 × 10−9 (95% confidence interval = 0.7 × 10−9 − 11 × 10−9).
Keywords: mutation, Drosophilla, Genome sequencing
ACCURATE knowledge of the spontaneous mutation rate is fundamental for advancing the understanding of many key questions in evolutionary biology. The rate of spontaneous mutation provides the base line for inferring the rate of molecular evolutionary change in the absence of natural selection or biased gene conversion. If an estimate of the neutral nucleotide diversity for a population is available, then it is also possible to estimate its recent effective population size. The spontaneous mutation rate per site appears in many aspects of evolutionary theory, such as the prediction of nucleotide diversity as a function of genetic distance in models of background selection (Hudson and Kaplan 1995; Nordborg et al. 1996) and the prediction of the equilibrium genomic base composition (Charlesworth and Charlesworth 2010).
Indirect approaches to estimating the mutation rate have relied on the equilibrium frequencies of dominant phenotypes caused by mutations at single loci or on the molecular divergence between species at putatively neutrally evolving sites (reviewed by Keightley 2012). Both approaches rely on parameter estimates that may be inaccurate, such as the number of sites in a gene that can produce a mutant phenotype or the generation time and divergence date of a species pair. Estimates are therefore subject to considerable uncertainty. In the past decade, it has become feasible to estimate the spontaneous mutation rate by applying mutation detection technology or direct sequencing of amplicons or complete genomes of mutation accumulation (MA) lines that have built up spontaneous mutations for many generations. This approach has been applied to the microbes Saccharomyces cerevisiae (Lynch et al. 2008), Dictyostelium discoideum (Saxer et al. 2012), and Chlamydomonas reinhardtii (Ness et al. 2012; Sung et al. 2012); the invertebrates Caenorhabditis elegans (Denver et al. 2004, 2009) and Drosophila melanogaster (Haag-Liautard et al. 2007; Keightley et al. 2009; Schrider et al. 2013); and the flowering plant Arabidopsis thaliana (Ossowski et al. 2010). These studies have generated valuable information of relevance to evolutionary theory, but there are a number of drawbacks to using MA lines for the characterization of the spontaneous mutation process. First, in outbreeding diploid species, MA lines are normally founded from an inbred progenitor, the production of which could fix deleterious recessive mutations that modify the mutation rate. It would be expected that such mutations would tend to increase the mutation rate, assuming that natural selection drives the mutation rate toward a physiochemical minimum in sexual species (Sniegowski et al. 2000). Second, deleterious recessive mutations, such as recessive lethals in diploids, will not become fixed in MA lines maintained by selfing or full-sib mating. If large insertion or deletion events (indels) tend to produce recessive lethality, for example, then these will be underrepresented in the spectrum of mutations detected. Third, premeiotic clusters of mutations (Woodruff and Thompson 1992) are not amenable to analysis using MA lines, because lines are expected either to have or not to have a single fixed mutation at a given site. Finally, MA lines are time consuming to produce and it is not feasible to produce them in most species.
Studying the rate and properties of new spontaneous mutations in a more natural setting is therefore desirable, and has recently been attempted by whole-genome sequencing of parent–offspring trios (Roach et al. 2010; Conrad et al. 2011; Kong et al. 2012; Michaelson et al. 2012). By these means, the genomes of tens of human trios have been sequenced and several thousands of new mutations detected, giving a detailed picture of the rate, age, and sex dependency and spectrum of new mutations in our own species. Difficulties with this approach are its relatively high cost if applied on a large scale (though costs continue to fall), the large number of false positives called that need to be checked, the possibility of missing genuine mutations if filtering is applied to reduce false positive calls, and the problem of obtaining an unbiased estimate of the number of callable sites, which is necessary to calculate the rate of mutation per site.
In this article, we apply deep, whole-genome sequencing of parents and multiple offspring from a single family of D. melanogaster originating from recently caught wild flies, thereby minimizing the impact of inbreeding in the laboratory. We assign genotypes at each site in the genome for each individual using the Genome Analysis Toolkit (GATK) (Depristo et al. 2011). Like other currently available genotype callers, mapping error leads GATK to falsely assign many sites in some or all individuals as heterozygous. Using the genotype calls, together with read depth and mapping quality at each site, we develop a software pipeline to call candidate mutations, incorporating a minimal amount of filtering of variants present in the offspring, allowing mutations to be called in a nearly unbiased manner, and the number of callable sites to be accurately estimated. The set of candidate mutations that passes automated filtering is then manually curated by viewing their sequencing reads in the Integrated Genomics Viewer (IGV) (Thorvaldsdóttir et al. 2012). The IGV facilitates the identification of false positives caused by mapping errors manifest as single nucleotide polymorphisms (SNPs) and/or indels in perfect association, almost invariably affecting multiple offspring. We check that the automated filtering and manual curation do not remove genuine mutations by determining whether synthetic mutations incorporated into the real data pass our filters. We then check each plausible candidate mutation using Sanger sequencing to identify genuine mutations and obtain a direct estimate of the mutation rate.
Materials and Methods
Flies
A D. melanogaster full-sib family was produced by crossing individuals from isofemale lines derived from a population collected in Ghana in January 2010 (Verspoor and Haddrill 2011). A single male and female parent taken from different lines were allowed to mate for 3 days and then separated. Both the male and female parent were virgins and eclosed within 8 hr of each other. Offspring were collected from the mating vial over the course of 1 wk. Parents and offspring were individually snap frozen in liquid nitrogen and stored at −80° until DNA extraction. Genomic DNA for whole-genome sequencing was obtained from the parents and 12 offspring (10 females and two males) by phenol-chloroform extraction. DNA from a second panel of 8 offspring, used to help confirm mutations, was obtained using a Qiagen Gentra Puregene Cell kit.
Whole-genome sequencing
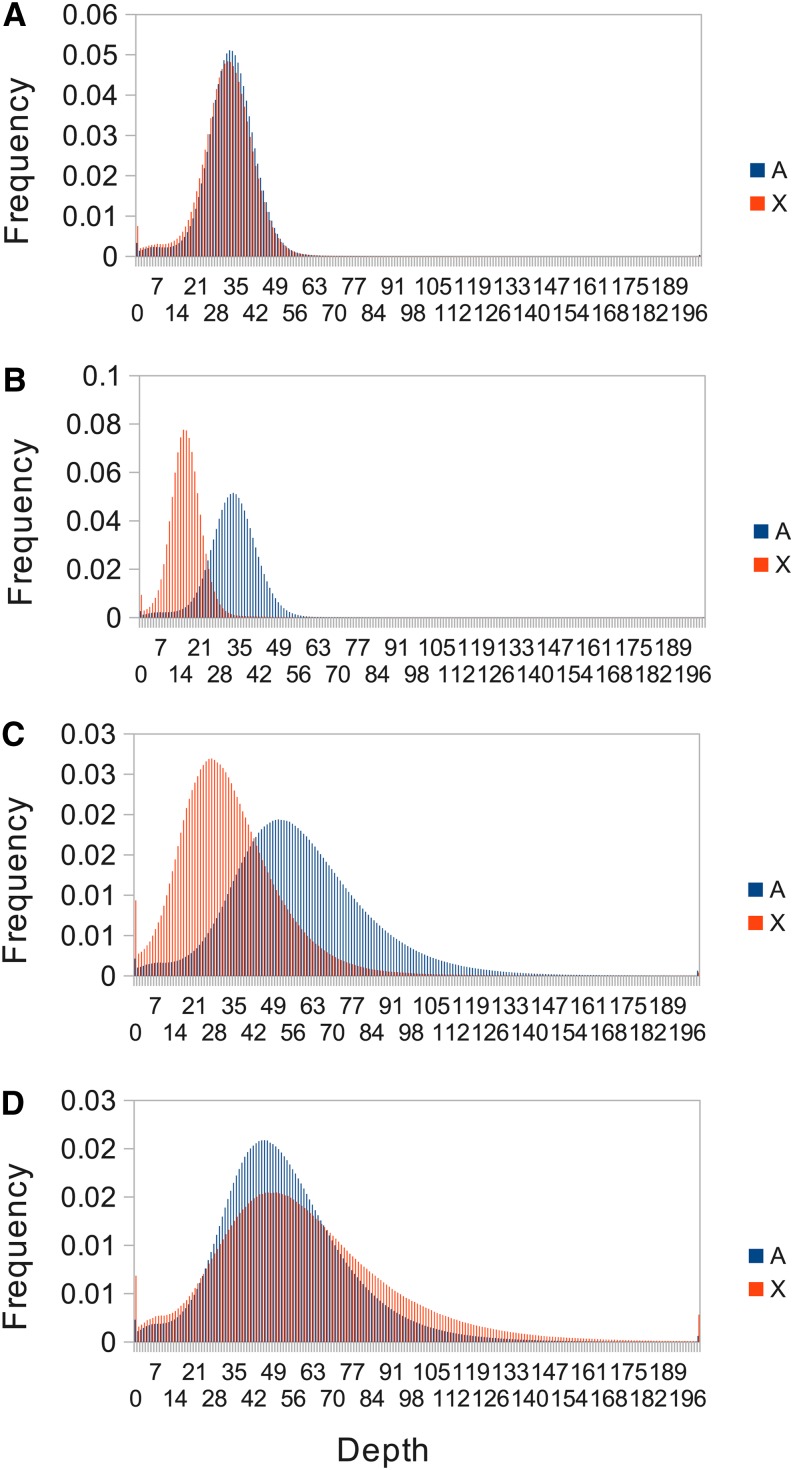
Genome sequencing, including associated library production, was carried out at the Beijing Genomics Institute, Hong Kong. A single Illumina library with a mean insert size of ∼470 bp was prepared for each individual fly. Library production was successful from unamplified DNA for the two parents, but was unsuccessful for the offspring. We therefore had libraries prepared from whole-genome amplified DNA from the offspring using a Qiagen REPLI-g Mini kit prior to library preparation. This method, using a high-fidelity polymerase (Phi 29) to generate fragments of up to 100 kb, is believed to generate a relatively even representation of sites across the genome and previously has been successfully used for Illumina sequencing of D. melanogaster (Langley et al. 2011). Paired-end sequencing of 100-bp reads to a mean read depth of ∼50 times (Figure 1) was carried out on the Illumina HiSequation 2000 instrument.
Figure 1.
Distributions of read depth of the two parents, (A) female and (B) male and two representative offspring, (C) male and (D) female for the autosomes (blue) and X chromosome (red). Duplicate reads were removed.
Sequence alignment
Illumina sequences of each individual fly were aligned to the D. melanogaster reference genome (release 5.44, March 2012) using Burrows-Wheeler Aligner (BWA) version 0.6.2-r126 (Li and Durbin 2009), and duplicate reads were removed using Picard tools (http://picard.sourceforge.net). We then used the IndelRealigner in GATK (Depristo et al. 2011) to carry out local realignment around insertion/deletion events (indels), with all individuals included in the realignment. We have previously found that this is an effective procedure for removing the majority of false SNP and indel calls caused by misaligned indels (Ness et al. 2012).
Genotype calling
A high proportion of false SNP and indel calls are known to be caused by mismapping (Li 2011). To reduce potential miscalls, we removed sites for which mapping quality was <20 using SamTools mpileup (Li et al. 2009). We also insisted that both parents were well covered and of high purity (i.e., having consistent base calls at a given site). To achieve this, we disregarded sites at read depth <10 in either parent and at which either parent was impure (i.e., we excluded sites that did not have the same base calls in each read in each parent). Finally, we also disregarded sites at which both parents were homozygous for the nonreference allele. Filtering on the parents is not expected to affect genotype calls in the offspring.
We used the UnifiedGenotyper in GATK (Depristo et al. 2011) to assign genotypes at each site in the parents and the offspring, assuming a heterozygosity parameter of 0.01. With high read depth, as in the present case, changing this parameter has a negligible impact on genotype calls (Depristo et al. 2011; Ness et al. 2012). We called variants across all individuals within the same GATK run. As recommended in the documentation, we ran GATK while treating X-linked sites in males (i.e., the male parent and the two male offspring) as if they were autosomal and then used a gender-aware algorithm to call mutations in the offspring. Sites marked as low quality by GATK, i.e., containing the LowQual flag in the variant call format (VCF) file output by GATK were disregarded.
Calling candidate mutations
We wrote a custom script to process the VCF file to call candidate mutations in the offspring. Sites at a read depth >10 were marked as potentially having a mutation if at least one offspring was called by GATK as a heterozygote (or a male offspring was called as a homozygote for the nonreference allele in the case of X-linked loci). For such a site to pass filtering, no more than three impure offspring were allowed (where impurity is defined as the presence of reads containing nonreference alleles in any offspring). In addition, we disregarded candidates where the largest number of nonreference reads within any individual was three or less.
Manual curation of candidate mutations
The aligned reads of each individual surrounding each candidate mutation were examined using the Integrated Genomics Viewer (IGV) (Thorvaldsdóttir et al. 2012). Candidates showing either of the following properties were assumed to be false positives:
-
(1)
Candidates present in more than one offspring that contained at least one single nucleotide polymorphism (SNP) or indel in complete association and in the same reads as the candidate mutation. That is, none of the reads containing the wild-type allele at the candidate mutation site carried any of the associated SNP or indel alleles. This is a hallmark of an alignment artifact due to mismapping of a duplicated region (Li 2011).
-
(2)
Cases where, by carrying out a local realignment, we could resolve a candidate SNP mutation by showing that the reads carrying the SNP could be aligned perfectly to an indel already present in multiple individuals, including one or both parents.
Estimation of the rate of failure to detect mutations
To check whether the automated filtering or manual curation of candidate mutations might lead to the removal of genuine mutations, we developed an approach based on generating synthetic mutations in the sequencing reads to estimate the false negative rate for base pair changes. We generated 1,000 synthetic mutations at random positions in the genome in randomly picked offspring with no conditioning on the read or genotype state of any individual. If the offspring at the selected site had read depth x, we randomly sampled a number, y, of nonreference bases from the empirical distribution for read depth x (which we generated as described below). We then changed y reads from the major base to a randomly selected, different base. To maintain sequencing errors that might be present in the data, we did not change any nonmajor bases. The complete data set of reads, including the synthetic mutations, was then remapped to the reference genome and mutations called using the identical pipeline as described above, including the manual curation step for a random sample of mutations. We calculated the rate of failure to detect mutations from 1 minus the number of synthetic mutations called, divided by the number of mutated sites at which a mutation could have been called. This number of callable sites is less than the total number of mutated sites because some sites do not pass the quality controls of our pipeline (i.e., because they are of low quality or of insufficient read depth in a parent or offspring).
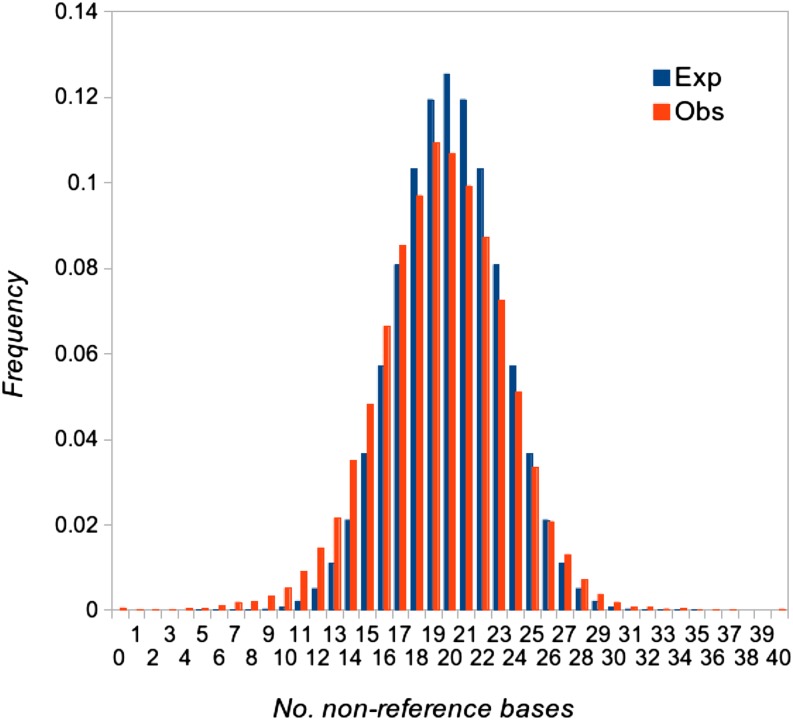
To sample numbers of nonreference reads from realistic distributions, we used empirical distributions of the number of nonreference alleles in sites of offspring that have a high probability of being heterozygous for natural variants present in the parents. We considered autosomal sites where the parents were called as homozygous for alternate alleles, at which both parents were sequenced at a read depth ≥30, and at which the reads for both parents were pure. In compiling such sites, the offspring read states were ignored. We generated distributions for read depth of 1–100. Numbers of sites used to generate these distributions is listed in Supporting Information, Table S1. An example of the empirical distribution for read depth of 40, based on 38,000 putatively heterozygous sites in the offspring, and its expectation for a binomial distribution with equal frequencies of reference and nonreference bases, is shown in Figure 2.
Figure 2.
Empirical (red) and expected (blue) distributions of the number of nonreference bases at heterozygous sites for offspring having read depths of 40. The empirical distribution shows the relative frequencies of sites having numbers of nonreference bases from 0 to 40. The expected distribution is binomial for equal frequencies of reference and nonreference bases.
Number of callable sites
Our estimate of the mutation rate is the number of confirmed mutations divided by the number of callable sites in each of the individual offspring (there are therefore potentially up to 12 callable sites at a given position in the genome). Callable sites exclude sites of low mapping quality, where the parents have a nonreference allele or have insufficient read depth (<10) and where a given offspring has insufficient read depth (≥10).
Large-scale events
We used Pindel (Ye et al. 2009) to search for deletions, short insertions, inversions, tandem duplications, and long insertions that were supported by at least five reads in one offspring and not supported in either parent or any other offspring. There were very large numbers of candidate mutations called if we allowed a candidate mutation to affect more than a single individual. We therefore needed to employ a more stringent filter, and it was not feasible to identify premeiotic cluster mutations using Pindel.
Checking candidate mutations by Sanger sequencing
Nongenome-amplified genomic DNA from the offspring was used as template for PCR amplification and Sanger sequencing of amplicons containing candidate mutations. We took advantage of our knowledge of the Illumina sequences to design PCR primers that exactly matched the relevant region of the individual in question and thereby avoided heterozygosity in the primer sites and the possibility of allele-specific amplification. We also verified that allele-specific amplification had not occurred by checking for the presence of heterozygous SNPs (manifest as double peaks in the Sanger chromatograms) that were called at nearby sites in the Illumina reads (we were able to perform this check in all cases). Sequencing was carried out on both strands.
Results
We employed Illumina technology to sequence the genomes of a pair of D. melanogaster parents and 12 of their offspring. The parents were sequenced from unamplified genomic DNA to a mean read depth of ∼50 times (at nonhemizygous sites), and the vast majority of sites were sequenced at read depth of >20 times (Figure 1). Sequencing libraries made from unamplified genomic DNA from the offspring did not pass quality control, so we used whole-genome amplification prior to Illumina library preparation and sequencing for the offspring. We found that whole-genome amplification led to somewhat higher variation in read depth in the offspring than the parents (Figure 1), but this disadvantage was partly offset by the presence of substantially fewer duplicate reads in the offspring (averaging 2%) than the parents (42% in the male parent and 22% in the female parent). We removed duplicate reads from the data, so their presence leads to a lower effective read depth.
Read mapping and identification of candidate mutations
We used BWA (Li and Durbin 2009) to align the reads of each individual to the D. melanogaster reference genome sequence and we then used GATK (Depristo et al. 2011) to realign the reads around indels and to call the genotype of each individual. We assigned candidate mutations by using a pipeline incorporating strong filtering on the parents with the objective of excluding sites within mismapped reads and minimal filtering on the offspring. There was a total of 88 sites containing candidate single nucleotide or small indel mutations that were taken forward for detailed examination.
Manual curation of candidate mutations
The 88 candidate mutations were taken forward for analysis using the IGV (Thorvaldsdóttir et al. 2012). The IGV allows visual inspection of the base calls within each read mapped in a region of the chromosome in every individual. Two general patterns indicated the presence of a mapping error:
-
(1)
The presence of SNPs or indels in perfect association with the candidate mutation in two or more offspring. There were 69 sites showing this pattern. Examples of screenshots from the IGV showing representative candidates of this type are shown in Figure S1 and Figure S2. We believe that such cases are a consequence of mapping duplicated regions present in the samples but not the reference. The alternative explanation of premeiotic clusters involving several tightly linked sites is unlikely for two reasons. First, we never observed the pattern of tightly linked variants exemplified in Figure S1 and Figure S2 around a candidate mutation in a single individual, so mutations occurring at multiple linked sites would always have to produce premeiotic clusters. Second, to our knowledge there is no mechanism that could produce mutation clusters conforming to the pattern illustrated in Figure S1 and Figure S2.
-
(2)
In five cases, we were able to find an alternative local alignment by changing the position of an indel variant that removed a candidate single nucleotide mutation. One of these cases (Figure S3) involved the alignment around a deletion mutation (3L: 10,514,561) that we subsequently confirmed by Sanger sequencing. In the remaining cases (example, Figure S4), multiple individuals including the parents had an indel variant, and we postulate that the individual(s) carrying the candidate single nucleotide mutation has the indel rather than the mutation.
All candidates falling into these two patterns were assumed to be false positives and were not taken forward for further detailed analysis.
Confirmation of candidate mutations
There were 10 sites called by GATK that had a read pattern strongly consistent with the presence of a genuine mutation (Table 1) and 4 sites where the evidence for a genuine mutation was judged to be weaker. We used Sanger sequencing of nongenome-amplified genomic DNA from the offspring to check these 14 candidates. None of the four weak candidate mutations that were amplified were confirmed by Sanger sequencing. Of the 10 candidate mutations called with high confidence, 8 were confirmed by Sanger sequencing. The remaining two candidates gave easily interpretable Sanger sequencing chromatograms in at least one direction, but the focal sites proved to be clearly wild type homozygous. Mutations were not undetected due to allele-specific amplification, because nearby heterozygous sites called by GATK in the Illumina sequences were apparent in the corresponding Sanger sequencing chromatograms in all cases. We then tested whether detection failure could be explained by a sample mix-up by sequencing amplicons containing the two undetected candidate mutations in all 12 of the offspring. All of these individuals proved to be homozygous wild type at the positions of the two candidate mutations and, moreover, nearby SNPs showed the same genotype pattern across the individuals in the Illumina and Sanger sequences in the cases of both candidates. This confirms that these two candidate mutations were false positives, a surprising result considering the large numbers of nonreference reads present in the Illumina sequences (Table 1).
Table 1. Ten candidate mutations called by GATK (i.e., sites having a read pattern consistent with a genuine single nucleotide or indel mutation) and one confirmed deletion mutation (X 2,693,102) called only by Pindel.
| Read depth | Read depth in mutant individual | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Location | Detected by GATK (G) or Pindel (P) | Mutation event | Offspring code | Female parent | Male parent | Offspring mean | Wild-type base | Mutant base | Confirmed |
| 2L 2,301,848 | G | A→C | 90 | 28 | 39 | 50.4 | 30 | 11 | Yes |
| 2L 5,955,655 | G | C→T | 88 | 42 | 47 | 33.0 | 15 | 16 | Yes |
| 2L 19,399,106 | G | G→T, A (two individuals) | 74, 88 | 23 | 36 | 13.8 | 10, 4 | 11, 8 | Yes |
| 2R 6,136,602 | G | G→A | 89 | 33 | 35 | 61.3 | 21 | 24 | Yes |
| 2R 14,887,552 | G | C→A | 84 | 39 | 32 | 64.5 | 45 | 9 | No |
| 2R 16,372,704 | G | C→T | 89 | 26 | 27 | 49.4 | 32 | 17 | Yes |
| 3L 10,514,561 | G, P | Deletion TAAAAATGCTCT | 94 | 32 | 17 | 52.2 | 16 | 14 | Yes |
| 3R 1,431,265 | G | C→T | 89 | 32 | 34 | 44.8 | 33 | 6 | Yes |
| 3R 7,755,276 | G | G→A | 79 | 26 | 56 | 55.3 | 50 | 20 | No |
| 3R 1,2126,610 | G, P | Deletion TCTCCGAAATAGG | 84 | 28 | 34 | 48.7 | 22 | 15 | Yes |
| X 2,693,102 | P | Deletion TGTT | 94 | 34 | 21 | 59.3 | 39 | 12 | Yes |
GATK, Genome Analysis Toolkit.
Does our pipeline remove genuine mutations?
When trying to identify very rare mutant sites, current genotype calling algorithms, including GATK, tend to throw up false positives. It is necessary, therefore, to incorporate automated filtering and/or manual curation as part of a mutation identification pipeline. It is possible that filtering/curation might remove genuine mutations from the data, leading to downwardly biased estimates of the mutation rate. To our knowledge, with the exception of Ness et al. (2012), the issue of the rate of failure to detect mutations in the context of the analysis of short read genome sequence data has not previously been addressed.
To estimate the rate of failure to detect genuine mutations (the false negative rate), we generated synthetic point mutations (i.e., new heterozygous sites present in one individual offspring) by altering the sequencing read data. To model the distribution of read number in the novel mutations, we used the empirical distributions of the number of nonreference bases observed at heterozygous SNPs in the offspring where the parents were called with high confidence as homozygous for different alleles. At such sites, 99.3% of offspring are called as heterozygous. Figure 2 shows an example of the empirical distribution for a read depth of 40. The mean of the observed distribution is 19.4, i.e., slightly lower than expected mean (20), presumably because reads containing nonreference bases are less likely to be mapped. The variance of the observed distribution is also somewhat higher than the variance of a binomial distribution (an appropriate null distribution), possibly because of nonuniform amplification, either at the whole-genome amplification or Illumina library preparation stage. Using the empirical distributions for read depth of 1–100, we generated 1,000 synthetic point mutations at random positions in the genome in randomly picked offspring as described in Materials and Methods. Of these, 859 synthetic mutations were callable (i.e., they occurred at a site of sufficient depth where the parents were pure, so a mutation could have been called, according to the rules of our pipeline), and 99.4% of the callable synthetic mutations were positively identified by our pipeline. We then used the IGV to check whether or not a random sample of 50 of the callable synthetic mutations would have been taken forward for verification by Sanger sequencing. We found that mutations would have been unequivocally taken forward in all cases, and none would have been rejected because they occurred in a region subject to mismapping. The rate of false negatives for single nucleotide mutations therefore appears to be very low in this experiment.
Single nucleotide mutation events
Of the mutations confirmed by Sanger sequencing, there were five single nucleotide mutations (SNMs) affecting a single individual, one of which was a transversion and four of which were transitions. This ratio is consistent with an overall transition mutational bias in Drosophila (Moriyama and Powell 1996; Haag-Liautard et al. 2007; Keightley et al. 2009; Schrider et al. 2013). We also detected SNMs at one site on chromosome 2L (position 19,399,106) affecting two individuals, i.e., there was a G/C→T/A transversion in one individual and a G/C→A/T transition in a second. Mutation clusters resulting from events occurring early in the germline lineage are an expected feature of the mutation process in multicellular eukaryotes (Woodruff and Thompson 1992), but a cluster involving two kinds of event is somewhat unexpected. One possible explanation is that the transition mutation resulted from error-prone repair of a premeiotic mutation (Goodman 2002). We investigated this cluster further by Sanger sequencing of eight additional sibs from the same family, and these were wild type at the site. This suggests that the parents were truly homozygous at the site (note the parents were both sequenced at high depth (Table 1), making this highly likely a priori), and points to the mutation event having occurred late in the development of the germ line such that it only affects a small proportion of offspring.
Across all 12 offspring sequenced we were able to call mutations at a total of 1.23 × 109 sites. Counting the mutation cluster as two events for the purpose of estimating the mutation rate and as one event for estimating its confidence interval (CI), our estimate of the single nucleotide mutation rate is μ = 2.8 × 10−9 (95% CI = 1.0 × 10−9 − 6.1 × 10−9) per site per generation. Assuming a neutral model, and equating autosomal synonymous diversity for African D. melanogaster (0.016; e.g., Campos et al. 2013) to 4Neμ, the effective population size (Ne) of the species is estimated to be ∼1.4 × 106.
Indels
We detected two deletion mutations of 12 and 13 bases in length, which were confirmed by Sanger sequencing (Table 1). We used Pindel to search for candidate deletions, short insertions, inversions, tandem duplications, and long insertions present in one offspring. Pindel successfully identified the candidate deletions called by GATK, and a further candidate deletion, which was confirmed by Sanger sequencing (Table 1). There were no supported short insertions or inversions. Two candidate tandem duplications and three large insertions were taken forward for investigation by PCR, but these were not confirmed, based on the absence of extra bands of the expected size. PCR products were sequenced to confirm that the correct regions had been amplified. Our failure to detect large deletions or tandem duplications is not inconsistent with results of Schrider et al. (2013), who sequenced MA lines that had undergone a total of 1,160 generations of spontaneous mutation accumulation. Their data predict that we would expect to see only 0.6 such events. Overall, our results are consistent with an overall deletion bias, as observed in a previous study (Haag-Liautard et al. 2007), but the number of indel events is too small for meaningful inference. The rate of deletion mutations is 1.2 × 10−9 (95% confidence interval = 0.7 × 10−9 − 11 × 10−9).
Discussion
We were able to detect mutations at 90% of sites in the euchromatic genome and exhaustively checked each plausible candidate mutation by Sanger sequencing or PCR to eliminate false positives. By introducing synthetic mutations into the data, we showed that the rate of failure to detect mutations (the false negative rate) is extremely low. Although we detected a modest number of mutations in the 12 individual D. melanogaster offspring sequenced, our results are suggestive of differences from previous work that employed mutation detection or whole-genome sequencing in MA lines of D. melanogaster. Our estimate of the single nucleotide mutation rate is 2.8 × 10−9, similar to an estimate of 3.5 × 10−9, based on 174 single nucleotide events detected by genome sequencing of MA lines (Keightley et al. 2009). In other studies involving mutation detection by denaturing high performance liquid chromatography (Haag-Liautard et al. 2007) or whole-genome sequencing (Schrider et al. 2013), one line (Florida-33) had a substantially higher single nucleotide mutation rate (7.7 × 10−9), and its 95% confidence limits do not overlap with those of the present experiment or with those of Keightley et al. (2009). It is possible that a mutator allele may have become fixed in the inbred ancestor of these lines. We also detected small deletion events only (i.e., no small insertions), consistent with the deletion bias that has been observed among small indel events in Drosophila (Petrov et al. 1996; Haag-Liautard et al. 2007).
To obtain a precise estimate of the mutation rate, parent–offspring genome sequencing needs to be applied to substantially larger numbers of individuals than genome sequencing of MA lines. There are, however, several advantages to the approach. First, mutations are accumulated in a single generation and remain in the heterozygous state (unless X-linked in males), so only dominant lethal or near-lethal mutations are expected to be underrepresented. Second, premeiotic clusters of mutations can be detected. In the present experiment, we attempted to detect clusters of single nucleotide mutations present in up to three individuals. Our analysis revealed one cluster affecting two individuals, unexpectedly involving two different kinds of base substitution (G→A and G→T). It is highly unlikely that this represents two independent mutation events, but more likely represents an event in the germline lineage that resolved into two different mutations, perhaps involving error-prone repair of one of two premeiotic mutations (Goodman 2002). Third, genome sequencing of offspring and parents can be applied to any species where the parents and offspring can be identified and where a reference genome sequence is available.
Our bioinformatic pipeline identified 88 candidate mutations that needed to be individually checked for plausibility, and we used the IGV (Thorvaldsdóttir et al. 2012), a powerful software tool for this purpose. It is feasible to use the IGV for a few hundred mutations at most, so increasing the scale of an experiment to include several hundreds of individuals could not rely solely on the IGV and would require a different strategy. One possibility would be to write software that can distinguish genuine mutations from mismapped paralogs that contain multiple SNPs in perfect association with the candidate mutation and affecting multiple individuals. Another, more straightforward strategy would be to filter sites that have a low average read depth, since we observed that badly mapped reads tend to have mapped at a low depth of coverage, with many fewer mapped at high quality. Having large numbers of individuals per family is therefore advantageous for either strategy, since mismapping is easier to detect when it affects multiple individuals, and mean read depth is more accurately measured in large families. With such strategies in place, in the near future we expect to see mutation rates estimated by the genome sequencing of offspring and parents in diverse species and in large cohorts of offspring.
Supplementary Material
Acknowledgments
We thank Brian Charlesworth for the suggestion to carry out this experiment, comments on the manuscript, and helpful discussions; and Donald Smith for advice on mutation confirmation. We are grateful to the Biotechnology and Biological Sciences Research Council, the Wellcome Trust, and a Natural Environment Research Council Fellowship (reference NE/G013195/1) to P.R.H. for funding.
Footnotes
Communicating editor: D. Begun
Literature Cited
- Campos J. L., Zeng K., Parker D. J., Charlesworth B., Haddrill P. R., 2013. Codon usage bias and effective population sizes on the X chromosome vs. the autosomes in Drosophila melanogaster. Mol. Biol. Evol. 30: 811–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., Charlesworth D., 2010. Elements of Evolutionary Genetics. Roberts & Co., Greenwood Village, Colorado. [Google Scholar]
- Conrad D. F., Keebler J. E. M., DePristo M. A., Lindsay S. J., Zhang Y., et al. , 2011. Variation in genome-wide mutation rates within and between human families. Nat. Genet. 43: 712–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver D. R., Morris K., Lynch M., Thomas W. K., 2004. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature 430: 679–682. [DOI] [PubMed] [Google Scholar]
- Denver D. D., Dolan P. C., Wilhelm L. J., Sung W., Lucas-Lledó J. I., et al. , 2009. A genome-wide view of Caenorhabditis elegans base-substitution mutation processes. Proc. Natl. Acad. Sci. USA 106: 16310–16314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., J. R. Maguire et al, 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M. F., 2002. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71: 17–50. [DOI] [PubMed] [Google Scholar]
- Haag-Liautard C., Dorris M., Maside X., Macaskill S., D. L. Halligan et al, 2007. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature 445: 82–85. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Kaplan N. L., 1995. Deleterious background selection with recombination. Genetics 141: 1605–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., 2012. Rates and fitness consequences of new mutations in humans. Genetics 190: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley P. D., Trivedi U., Thomson M., Oliver F., Kumar S., et al. , 2009. Analysis of the genome sequences of three Drosophila melanogaster spontaneous mutation accumulation lines. Genome Res. 19: 1195–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Frigge M. L., Masson G., Besenbacher S., Sulem P., et al. , 2012. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488: 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley C. H., Crepeau M., Cardeno C., Corbett-Detig R., Stevens K., 2011. Circumventing heterozygosity: sequencing the amplified genome of a single haploid Drosophila melanogaster embryo. Genetics 188: 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2009. Fast and accurate short read alignment with Burrows-Wheeler Transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., et al. , 2009. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Sung W., Morris K., Coffey N., Landry C. R., et al. , 2008. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc. Natl. Acad. Sci. USA 105: 9272–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson J. J., Shi Y., Gujral M., Zheng H., Malhotra D., et al. , 2012. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151: 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama E. N., Powell J. R., 1996. Intraspecific nuclear DNA variation in Drosophila. Mol. Biol. Evol. 13: 261–277. [DOI] [PubMed] [Google Scholar]
- Ness R. W., Morgan A. D., Colegrave N., Keightley P. D., 2012. Estimate of the spontaneous mutation rate in Chlamydomonas reinhardtii. Genetics 192: 1447–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M., Charlesworth B., Charlesworth D., 1996. The effect of recombination on background selection. Genet. Res. 6: 159–174. [DOI] [PubMed] [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledo J. I., Warthmann N., Clark R. M., et al. , 2010. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov D. A., Lozovskaya E. R., Hartl D. L., 1996. High intrinsic rate of DNA loss in Drosophila. Nature 384: 346–349. [DOI] [PubMed] [Google Scholar]
- Roach J. C., Glusman G., Smit A. F. A., Huff C. D., Hubley R., et al. , 2010. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science 328: 636–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxer G., Havlak P., Fox S. A., Quance M. A., Gupta S., et al. , 2012. Whole genome sequencing of mutation accumulation lines reveals a low mutation rate in the social amoeba Dictyostelium discoideum. PLoS ONE 7: e46759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider D. R., Houle D., Lynch M., Hahn M. W., 2013. Rates and genomic consequences of spontaneous mutational events in Drosophila melanogaster. Genetics 194: 937–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski P. D., Gerrish P. J., Johnson T., Shaver A., 2000. The evolution of mutation rates: separating causes from consequences. Bioessays 22: 1057–1066. [DOI] [PubMed] [Google Scholar]
- Sung W., Ackerman M. S., Miller S. F., Doak T. G., Lynch M., 2012. Drift-barrier hypothesis and mutation rate evolution. Proc. Natl. Acad. Sci. USA 109: 18488–18492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., 2012. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verspoor R. L., Haddrill P. R., 2011. Genetic diversity, population structure and wolbachia infection status in a worldwide sample of Drosophila melanogaster and D. simulans populations. PLoS ONE 6: e26318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff R. C., Thompson J. N., Jr, 1992. Have premeiotic clusters of mutation been overlooked in evolutionary theory? J. Evol. Biol. 5: 457–464. [Google Scholar]
- Ye K., Schulz M. H., Long Q., Apweiler R., Ning Z., 2009. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 25: 2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.