Abstract
The Drosophila melanogaster genome has been extensively characterized, but there remains a pressing need to associate gene products with phenotypes, subcellular localizations, and interaction partners. A multifunctional, Minos transposon-based protein trapping system called Hostile takeover (Hto) was developed to facilitate in vivo analyses of endogenous genes, including live imaging, purification of protein complexes, and mutagenesis. The Hto transposon features a UAS enhancer with a basal promoter, followed by an artificial exon 1 and a standard 5′ splice site. Upon GAL4 induction, exon 1 can splice to the next exon downstream in the flanking genomic DNA, belonging to a random target gene. Exon 1 encodes a dual tag (FLAG epitope and mCherry red fluorescent protein), which becomes fused to the target protein. Hto was mobilized throughout the genome and then activated by eye-specific GAL4; an F1 screen for abnormal eye phenotypes was used to identify inserts that express disruptive fusion proteins. Approximately 1.7% of new inserts cause eye phenotypes. Of the first 23 verified target genes, 21 can be described as regulators of cell biology and development. Most are transcription factor genes, including AP-2, CG17181, cut, klu, mamo, Sox102F, and sv. Other target genes [l(1)G0232, nuf, pum, and Syt4] make cytoplasmic proteins, and these lines produce diverse fluorescence localization patterns. Hto permits the expression of stable carboxy-terminal subfragments of proteins, which are rarely tested in conventional genetic screens. Some of these may disrupt specific cell pathways, as exemplified by truncated forms of Mastermind and Nuf.
Keywords: Minos vector, RNA splicing, GAL4/UAS, ectopic expression, transcription factor
A large portion of Drosophila melanogaster’s 13,967 protein-coding genes have been characterized during the past century of genetic analysis, making it one of the best-understood animals (Ashburner and Bergman 2005; modENCODE Consortium et al. 2010; Bellen et al. 2011; McQuilton et al. 2012). However, most genes have not yet been linked to informative mutant phenotype, protein localization, pathway, or structure–function data. Systematic approaches to alleviate this “phenotype gap” (Dow 2003) include the EP system and its variations, in which a transposon with a GAL4-responsive promoter is mobilized to random loci and then used to ectopically express downstream target genes (Rørth 1996; Rørth et al. 1998). Hundreds of publicly available GAL4 (“driver”) lines offer fine spatial control of target gene expression in the organism, and some temporal control is also possible (Duffy 2002; Elliott and Brand 2008; del Valle Rodríguez et al. 2011). The resulting dominant, inducible, and often visible phenotypes are widely used for performing genetic screens and analyzing genetic interactions. This approach could be expanded in three ways. First, the target protein could be biochemically or fluorescently tagged, as in protein-trapping techniques (below). Second, subfragments of the target protein could be expressed, rather than just the intact wild-type protein; this could provide in vivo structure–function information and yield novel pathway-modulating reagents. Third, most fly misexpression constructs have used the P-element vector, which is not able to provide complete coverage of the genome due to insertional biases (Bellen et al. 2011; Spradling et al. 2011). This can be remedied by using the Minos transposon as a vector, since it has little insertion site specificity beyond a TA dinucleotide and thus provides access to P-element insertional “coldspots” (Metaxakis et al. 2005; Pavlopoulos et al. 2007; Bellen et al. 2011).
We designed the Minos-based Hostile takeover (Hto) transposon system to address these three issues. Hto combines targeted expression of endogenous genes with protein tagging and permits expression of stable C-terminal protein fragments, which have rarely been subjected to genetic screens in the past. Importantly, since Minos can transpose in a broad range of organisms, the Hto system could potentially be applied to other species (Pavlopoulos et al. 2007; de Wit et al. 2010; Hozumi et al. 2010; Sasakura et al. 2010).
Protein trapping is a powerful method that was developed in flies by several groups, including large-scale screens and stock collections from the FlyTrap group (http://flytrap.med.yale.edu) (Morin et al. 2001; Clyne et al. 2003; Kelso et al. 2004; Buszczak et al. 2007; Quiñones-Coello et al. 2007; Aleksic et al. 2009; Neumuller et al. 2012). A protein trap transposon carries the GFP-coding region flanked by a 3′ splice site (ss) and a 5′ ss. This GFP exon can splice just upstream of, or within, a target gene’s coding region, producing a chimera of GFP and the target protein. These protein traps ideally report the wild-type expression and localization patterns of the target protein, rather than create mutant phenotypes.
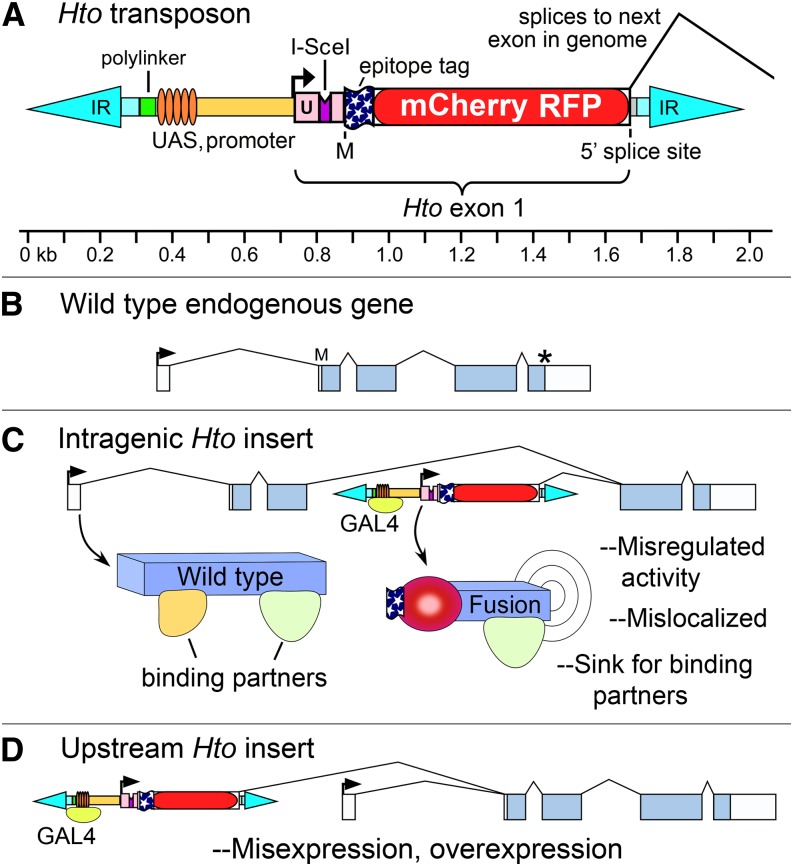
To add an inducible promoter to a protein trap, one must essentially build a new “front end” for the target gene in the following form: UAS-promoter-(5′ UTR-start codon-tag ORF)-5′ ss, where the “artificial exon 1” in parentheses splices into the target gene (Figure 1). A potential challenge to this strategy is that random insertions of the vector will generate novel introns spanning sequence that is not normally intronic, i.e., either intergenic regions or parts of exons. Since these regions have not evolved to be treated as introns, they may include sequences that confuse the splicing machinery. Intergenic regions may also contain transcriptional barriers, e.g., poly(A) signals from remnants of transposons, which would limit splicing to the target gene. Thus, we chose to test the “artificial exon 1” strategy in conjunction with a phenotypic screen, so inserts that make defective transcripts would not be recovered.
Figure 1.
Hto vector map and protein-trapping strategy. (A) Schematic diagram of the Hto transposon. IR, Minos inverted repeats; black arrow, transcription start; U, 5′ UTR; M, start codon. 3xFLAG epitope tag and mCherry RFP coding regions are indicated. Hto exon 1 can splice to the next downstream genomic exon as indicated. See Figure S1 for sequence and complete annotation. (B) Hypothetical wild-type gene structure. Boxes, exons; arrow, transcription start; asterisk, stop codon. (C) Intragenic Hto insertion. When Hto inserts within the coding range, GAL4 induction of Hto can lead to expression of a tagged, C-terminal fragment of the endogenous target protein (“Fusion”), which may cause phenotypes by various mechanisms as indicated (see text). (D) Upstream insertions of Hto may express a tagged version of the full-length target protein, if there is no stop codon between the 5′ end of the target gene’s exon 2 and its start codon. Note that, in C and D, the wild-type gene products may still be produced using the target gene’s endogenous promoter, since Hto is minimally disruptive.
We report the implementation of the Hto system and describe 23 target genes recovered using a simple F1 screen for eye defects. We show proof-of-principle that Hto lines are effective for generating scorable phenotypes, analyzing subcellular protein distribution, localization on polytene chromosome spreads, immunoblotting, and physical and genetic interaction analyses. In addition, the system can identify new kinds of dominant negative proteins that may be broadly useful additions to the genetic toolkit.
Materials and Methods
Drosophila lines
The transposase line w1118; snaSco/SM6a, P{hsILMiT}2.4 (here called Hsp70-MiT) was obtained from the H. Bellen/Gene Disruption Project; it carries the Minos transposase construct PhsILMiT (w+) inserted on a Cy-marked balancer (Metaxakis et al. 2005). The following lines were obtained from the Bloomington Drosophila Stock Center (Indiana University): UAS-GFP-Rab11 (Bloomington #8506); “FLP-out” stocks P[hsFLP]12 and P[GAL4-Act5C(FRT.CD2).P]; GAL4 drivers GMR-GAL4; eyeless (ey)-GAL4; pannier (pnr)-GAL4; and heat-shock driver P[GAL4-Hsp70.PB]89-2-1 (Bloomington #1799, here called Hsp70-GAL4).
Production of Hto vector
The annotated sequence of the Hto-WP transposon (1983 bp) is available at GenBank (accession no. JN049642) and Supporting Information, Figure S1; all base numbers within Hto refer to this sequence record. The polylinker (bases 309–354) and right side of Hto (bases 736–1983) were produced using total gene synthesis and cloned into pUC57 (GenScript, Piscataway, NJ). The left side of Minos (bases 1–308) was amplified from genomic DNA of fly stock carrying a Mi[ET1] insert (H. Bellen/Gene Disruption Project) and cloned into the Hto polylinker as an EcoRI-NheI fragment. The remaining sequence (bases 355–735), consisting of the UAS and basal promoter, was amplified from pUAST (Brand and Perrimon 1993) and cloned into the polylinker as a BamHI-XbaI fragment, yielding the complete Hto transposon in pUC57. The Hto transposon was next cloned into Drosophila P-element vector pCaSpeR4 as an EcoRI-XhoI fragment to generate the final clone, pCaSpeR4-P{w+, Mi[Hto-WP]}, which was then used for germline transformation. An insert on chromosome 2R: 8,824,114, called Starter2, was used as the starter element for the GMR-GAL4-based screens.
Phenotypic screens
Hopping of the Hto element from Starter2 to new chromosomal sites was mediated by Minos transposase, expressed from the heat-inducible transgene, Hsp70-MiT, located on the SM6a balancer. To drive transposition, Starter2 was crossed to SM6a, Hsp70-MiT, and the progeny were heat-shocked 1 hr at 37° at day 2–4 after hatching (schematic in Figure S2). The resulting mosaic males, SM6a, Hsp70-MiT*/Starter2*; +*/+*, were then crossed to a retinal GAL4 driver, GMR-GAL4, also on chromosome 2 (an asterisk indicates a potentially mutagenized chromosome). The F1 progeny of the mosaic flies were either Starter2*/GMR-GAL4; +/+* or SM6a, Hsp70-MiT*/GMR-GAL4; +/+*. All F1 flies were screened en masse by dissecting microscope for defects. Individual F1 progeny with abnormal phenotypes were recovered and crossed to either the balancer line TM3,Sb/TM6,Tb or the wing driver ms1096w-GAL4 (Park and Edwards 2004) to recover the causative Hto insert. Each Hto line was crossed to balancer chromosomes to remove the other original chromosomes from the F1 fly, along with any extraneous Hto inserts that they may carry. Hto lacks a white+ marker; thus an Hto element was scored by the presence of its GAL4-dependent phenotype or by GAL4-driven expression of mCherry red fluorescent protein (RFP) until it was balanced. Thereafter an Hto insert can be treated like any other unmarked mutation; the elements tested so far have all remained stable (>40 generations for the oldest lines). The REM insert was on the SM6a, Hsp70-MiT chromosome; the presence of the MiT transgene led to mosaicism, so MiT was removed using the Δ2-3 P-transposase construct, resulting in a stable Hto insert.
Estimation of hop rate
Random Curly females from the F1 screen were scored for GMR-GAL4-induced RFP expression in the retina using an epifluorescence dissecting scope. Since the Curly flies inherited SM6a, Hsp70-MiT instead of the Starter2* chromosome, any RFP signal indicates the presence of one or more new hops in trans (to a different chromosome than Starter2). The resulting hop rate therefore excludes any “local” hops or other cis hops. In practice, we did recover cis hops in the screen, but they were less common than hops to chromosome 3, so local hopping does not appear to be a major concern. In the scored females, hops to X, 2, and 3 could be observed. Their male sibs were not scored, but presumably only ∼81% of these hops could be recovered in the Curly males, since they do not inherit the paternal X, which is 19% of the genome. The overall trans hop rate (among an equal mix of males and females) was adjusted accordingly to 91% of the female-only rate. For screen 2, with an RFP-positive rate of 5.6%, double inserts were ignored. For screen 3, with an RFP-positive rate of 20.0%, we estimated that, if all hops are independent, 18% of females would carry one insert, 2% would carry two inserts, and 0.15% would carry three inserts, leading to ∼11% more inserts than RFP-positive flies. To convert the number of trans hops to the total number of hops, we assume no bias between cis and trans hops and count the Starter2 chromosome as 20% of the total euchromatin present in the mosaic male fly (44 Mb/218 Mb); thus the total number of hops equals 1.25 times the number of trans hops.
Adult phenotype documentation (Figure 2 and Figure 3)
Figure 2.
Driving expression of each Hto line in the developing eye and wing gives a unique combination of adult phenotypes. (Left column of each set) Wings with ms1096-GAL4 (wing expression). (Middle column of each set) Eyes/heads with ey-GAL4 (early eye expression). (Right column of each set) Eyes with GMR-GAL4 (retinal expression). (Top left row) Wild type: flies with each GAL4 driver but no Hto insert appear normal. Left bar, for all wings, 500 µm; right bar, for all heads, 200 µm. Each subsequent row shows the typical set of ms1096-, ey-, and GMR-GAL4 phenotypes for the line indicated (three-letter code); the line’s target gene is given at the bottom of each row. Chromosome X and 3 lines are shown here in genomic order; chromosome 2 and 4 lines are shown in Figure 3. All GMR-GAL4 eyes carry a single copy of w+ (the marker present in GMR-GAL4), and so the variations in eye color are due to Hto. See also Materials and Methods.
Figure 3.
Adult phenotypes of Hto lines from chromosomes 2 and 4: DRE, POV, REM, WRS, HJF, and OMD; presented as in Figure 2.
Each designated line was crossed to the indicated driver at 25°, except ms1096>GND (21°) and ey>GND (18°). Female progeny are shown in all cases. Wings were mounted as in Edwards et al. (2007). To display extended depth of field, eye/head images were composited manually in Photoshop from ∼5 focal planes.
Choice of samples: the GMR-GAL4 eye phenotypes were extremely uniform, with little variation from fly to fly. The ms1096-GAL4 wing phenotypes were also fairly uniform, although the mutant wings were often prone to secondary defects, especially blistering or crumpling; relatively flat wings from each cross were chosen to avoid obscuring the primary defect. The ey-GAL4 phenotypes were the most variable; in many cases, the left and right eyes of one fly differed in size and shape, and for some lines (e.g., BRW) a shape defect had a reduced penetrance (but always >10%). An eye phenotype that is typical among penetrant flies is shown in each case. For lines GND, XTC, HJF, and OMD, 95–100% of the ey-GAL4>Hto flies had severe reductions in head tissue, making them unable to eclose from the pupal case; instead, live, mature pharate adults were dissected from the pupal case and photographed. For ey-GAL4>BRO and ey-GAL4>SMH, most were pupal lethal, and escapers are shown.
Identification of inserts and transcript analysis
To determine the chromosomal insertion site for each line, genomic DNA was isolated and subjected to thermal asymmetric interlaced-PCR (TAIL-PCR) (Liu and Chen 2007) to amplify the region that flanks the Hto element. TAIL-PCR employs nested Hto-specific forward primers, used in succession, and an arbitrary degenerate (AD) primer that binds to various locations in the genome. The TAIL-PCR procedure was followed essentially as in Quinones-Coello et al. (2007), using their AD primers AD2–AD6. The nested Hto primers were TAIL-1 (5′-CCATCGTGGAACAGTACGAAC), TAIL-2 (GCATGGACGAGCTGTACAAG), and TAIL-3 (GTAAATCACATTACGCCGCGTTC). The PCR products were gel-purified and sequenced. The flanking genomic DNA was identified on the Drosophila genome using BLASTN and FlyBase genome maps, and the putative target gene was predicted by inspection of the downstream gene models. To test for the predicted splicing event, RNA was recovered from Hto-expressing flies and RT-PCR was used to amplify the joint between Hto exon 1 and the downstream target exon. An Hto line was crossed to Hsp70-GAL4, and 15 adult progeny were heat-shocked for 45 min at 37° and left to express at room temperature for 4 hr. RNA was then isolated and reverse transcribed using the primer T18VAdaptor-r (GAAGACAGACACCGGACTTTTTTTTTTTTTTTTTTV) or, in some cases, primers specific to downstream exons of the putative target gene. PCR was then performed on this complementary DNA (cDNA) product using a forward primer in Hto and either Adaptor-r (GAAGACAGACACCGGAC) or a target gene-specific reverse primer. The resulting amplicon was then sequenced to identify the splice; in all cases, Hto exon 1 spliced cleanly to a downstream exon in the form ....mCherry coding-cggccgcaggcg/target exon..., where “cggccgcaggcg” is the linker sequence at the end of Hto exon 1 and “/” is the splice junction.
Nomenclature of tagged genes
An Hto insert can sometimes lie in one gene but express the neighboring gene in response to GAL4; thus an insert may be phenotypically associated with two genes. We also note gene name changes for CG11071 and CG17181. Analysis of the GND insert near CG11071 led us to conclude that CG11071 is not an independent gene, but an alterative 3′ end for the mamo locus; this was validated by FlyBase scientists and the mamo annotation was corrected. The QSR insert tags CG17181, which we rename as follows. CG17181 is one of six Drosophila members of the Snail superfamily of zinc (Zn)-finger transcription factors, sharing 54–62% identity to the others in the ∼140-aa Zn-finger region. The Snail superfamily has been divided into the snail proper and scratchA and scratchB subfamilies (Kerner et al. 2009). CG17181 is the sole scratchA gene in Drosophila and was referred to as both scratch 3 and scratch-like 1 in Kerner et al. (2009). However, the scratchA genes lack the so-called Scratch domain typical of the scratchB subfamily, so we prefer to avoid that terminology for this gene. To instead reflect its membership in the Snail superfamily, we propose the name kahuli (kah), referring to the Hawaiian tree snail.
Confocal analysis
Subcellular localization of Hto fusion proteins was documented by confocal microscopic observation of the mCherry RFP tag in epithelia.
Ovaries:
Flies of the genotype P[hsFLP]12, P[GAL4-Act5C(FRT.CD2).P]; Hto were heat-shocked for 1 hr at 37° to induce FLP-out clones (Pignoni and Zipursky 1997). These flies carry an Act5C>CD2>GAL4 construct; heat-shock induction of FLP recombinase triggers excision of the CD2 stuffer fragment, allowing high-level GAL4 production from the Act5C promoter in random clones of cells. After 2.5 days at 25°, to allow induction of Hto and growth of the clones, the ovaries were dissected, fixed in 2% formaldeyde/PBS, washed in PBS and then in PBS with 0.2% Triton X-100, and stained with SYBR Green to label DNA. In some cases, Alexa 633-phalloidin was added to label f-actin, or Alexa 633-wheat germ agglutinin (WGA) was added to stain the nuclear envelope and other structures. For the colocalization study, UAS-GFP-Rab11 on chromosome 2 was crossed to BRW/+ flies bearing the FLP-out chromosome; the progeny thus yielded ovary FLP-out clones of GFP-Rab11 either with or without BRW, and these were processed together so they could be directly compared; colocalization was analyzed using Colocalization Finder in ImageJ (National Institutes of Health).
Salivary glands:
Larvae of the genotype Hsp70-GAL4; Hto were fixed and stained as above. The Hsp70-GAL4 construct from Bloomington stock #1799 expresses strongly and specifically in salivary glands without heat induction. For chromosome spreads, Hsp70-GAL4; Hto salivary glands were fixed 8 min in 2% formaldeyde/PBS, washed in PBS and then in PBS with 0.2% Triton X-100, and stained with SYBR Green. Glands were then placed in a drop of Vectashield (Vector Labs, Burlingame, CA) on a slide and topped with a coverslip, and the polytene chromosomes were released and spread by tapping the coverslip; the slides were then directly observed (this procedure modified from DiMario et al. 2006). Images were collected with a Leica SP2 confocal microscope using sequential scanning and processed in Photoshop; in all cases, the mCherry RFP channel is presented with the original contrast (no sigma curve).
Western blot analysis
To express an Hto fusion protein, the Hto line was crossed to Hsp70-GAL4; adult progeny were heat-shocked 1 hr at 37° and allowed to express for 8 hr. Total protein extracts were prepared by homogenizing 20 male flies in 100 μl of grinding buffer (125 mM Tris, 1% Triton X-100, pH 6.8). Insoluble material was pelleted and supernatants were incubated with SDS/PAGE sample buffer prior to loading onto 10% polyacrylamide gels. Protein samples were resolved at 30 mA and transferred to nitrocellulose membranes. Membranes were incubated 1 hr in a 2% BSA blocking solution followed by an overnight incubation with anti-FLAG monoclonal M2 (Sigma-Aldrich). Blots were probed with an alkaline phosphatase (AP)-linked secondary antibody and visualized by using 5-bromo-4-chloro-3’-indolyphosphate (BCIP)/nitro-blue tetrazolium liquid substrate system (Sigma-Aldrich). Sizes were estimated using EZ-Run prestained protein ladder (Fisher); each band was recalibrated by direct comparison to an unstained protein ladder under our SDS-PAGE conditions; the recalibrated sizes are given.
Results
A vector for simultaneous tagging and controlled expression of protein fragments: rationale and design
The Minos[Hto-WP] transposon (henceforth referred to as “Hto”) is shown schematically in Figure 1A and with annotated sequence in Figure S1. Between the 254-bp terminal inverted repeats (IR), the internal portion of Hto carries the UAS (GAL4-binding sites) and basal promoter from the EP element, an artificial exon 1, and a canonical 5′ ss. The exon 1 sequence includes a 5′ UTR derived from a strongly expressed gene (sqh), with an added I-SceI endonuclease site (Bellaiche et al. 1999), followed by a start codon and the coding region for the dual tag, 3xFLAG-mCherry RFP (FLAG-RFP). When an Hto-bearing fly line is crossed to a GAL4-expressing line, GAL4 binds the UAS and induces transcription through the Hto exon 1 and the right IR into the flanking genomic DNA. Hto exon 1 can then splice to the next available genomic exon in the forward direction (“target exon”).
Two main classes of inserts have the potential to cause dominant phenotypes: intragenic and upstream. In an intragenic insertion, Hto lies within the target gene’s transcription unit and splices to an internal target exon (Figure 1, B and C). If the target exon has a coding region in reading frame 0, translation will yield a productive fusion, in which the C-terminal portion of the target protein is tagged at its N terminus with FLAG-RFP. If the C-terminal portion includes a full, properly folded protein domain, it may cause a phenotype. With an upstream insertion, Hto can potentially lie tens of kilobases 5′ of the target gene and still produce a fusion protein. The endogenous first exon of the target gene will be bypassed (since it lacks a 3′ ss) and substituted by Hto exon 1 (Figure 1D). If exon 2 begins with a frame 0 ORF, a productive fusion will result as above. If exon 2 begins with a 5′ UTR, a fusion protein can still be made if the UTR is fortuitously in frame with the endogenous start codon. In the latter case, the full-length target protein is made and is connected to FLAG-RFP by a linker derived from translating the UTR sequence.
Hto fusion proteins may cause dominant, GAL4-dependent phenotypes by several mechanisms (see Prelich 2012). A full- or near-full-length protein may be detrimental due to simple overexpression or when expressed in the wrong stage or cell type. For C-terminal fragments, additional mechanisms are possible. The fusion may act as a dominant negative: it could bind and sequester proteins that are required for the wild-type protein to function or make nonproductive complexes with wild type. In other cases, the N-terminal region of the wild-type protein might normally serve to regulate or localize the protein; removal of the N terminus by Hto could leave the protein active, but misregulated or mislocalized (Figure 1C).
Phenotypic screen for disruptive protein traps
The Hto transposon was inserted into the fly genome using a P-element vector, yielding “starter elements” from which new hops of Hto can be generated by exposure to Minos transposase. An insertion on chromosome 2 (Starter2) was used to conduct a series of phenotypic screens (see Materials and Methods and Figure S2 for details). Starter2 flies were crossed to the Hsp70-MiT transposase line, and the progeny were heat-shocked to generate mosaic male flies with new Hto hops in the germ line. These mosaic flies were crossed to females bearing the driver GMR-GAL4. Their F1 progeny expressed any new Hto hops specifically in the developing retina, where GMR-GAL4 is primarily active. All F1 with eye defects were retained, given a unique three-letter code to denote the specific Hto insert, and used to establish balanced stocks.
Three such screens were performed (Table 1). Following screen 1, we determined that new Hto inserts can be readily detected by scoring for red fluorescence in the eyes of GMR>Hto flies, regardless of whether they have an eye defect (Figure S3A). Thus in screens 2 and 3, a large portion of F1 were frozen and scored for new hops to estimate the hop rate (Materials and Methods). In screens 1 and 2, a single heat shock was used to initiate transposase expression in the mosaic parental flies. This led to 5.6% (39/695) of female F1 progeny bearing new trans hops (cis hops were not counted). In screen 3, a double heat shock was administered, and this boosted transposition to 20% of F1 females (but with a concomitant increase in the probability of multiple inserts). Screen 3 was also performed more efficiently than screens 1 and 2; flies were scored rapidly and only striking phenotypes were retained, resulting in a lower rate of phenotypic positives per hop. Despite these variations, the cumulative results for all three screens provide a reasonable assessment of the efficiency of the system. A total of 34 unique lines were recovered, excluding apparent duplicates recovered from sibling F1 flies. Overall, the rate of new phenotypic hits was 1/500 flies, with 1.7% of all new hops yielding a phenotype. To date, we verified the insertion site in 22 of these lines, plus four more lines isolated from other GMR-GAL4 pilot screens (SMH, CXM, BRW, and WGL; screens not included in Table 1 since the hop rates were not determined). These 26 verified lines are described here.
Table 1. Eye screens for disruptive Hto inserts.
| Screen | F1 flies screeneda | “Rough-eyed” flies recovered | Unique lines established (rate) | Trans hop rateb | New hops screenedc | Unique lines per hop |
|---|---|---|---|---|---|---|
| #1 | 4,500 | 12 | 8 (1/560) | ND | 290 | 1/36 |
| #2 | 7,600 | 14 | 11 (1/690) | 5.6% (n = 695) | 490 | 1/45 |
| #3 (2 × HS) | 4,800 | 26 | 15 (1/320) | 20.0% (n = 501) | 1210 | 1/81 |
| Total, #1–3 | 16,900 | 52 | 34 (1/500) | NA | 1990 | 1/59 (1.7%) |
Summary of results from three F1 screens for rough or otherwise defective eyes. More stringent screening criteria were used in screen 3; F1 progeny with weak phenotypes were not retained, resulting in a lower rate of positives.
An estimate of the total number of offspring from the mosaic Starter males; all of these F1 were inspected for defects.
Percentage of female F1 with new trans hops as determined by RFP in the eye; the rate in males would be reduced since they cannot inherit X chromosome hops from the paternal side.
An estimate of the total number of new insertions present in the F1, extrapolated from the female hop rate (see Materials and Methods).
Hto inserts cause strong, diverse phenotypes and primarily target regulatory genes
The recovered Hto lines were tested for GAL4-dependent adult phenotypes. In Figure 2 and Figure 3, each row of three images represents a different Hto line, independently crossed to ms1096w-GAL4 (expresses in the developing wing blade), ey-GAL4 (expresses early in the eye disc), and GMR-GAL4 (expresses in the retina after the furrow). Importantly, each Hto insert produces a unique combination of phenotypes with these three drivers; the phenotypes do not repeat as they would if the Hto products acted through a common mechanism. Phenotype strength is summarized in the master table of line data (Table S1).
To identify the molecular basis of these phenotypes, the Hto insertion sites were amplified, sequenced, and mapped to the Drosophila genome (Table 2 and Table S1). The genome annotations were scanned to find the candidate target gene, which is generally the first downstream gene in the same orientation as Hto. Hto exon 1 is expected to splice to the first available exon with a 3′ ss, and in 23/26 cases this splice would be in-frame with Hto, allowing a functional fusion protein to be produced. Splicing models for 14 such lines are shown in Figure 4, Figure S4, and Figure S5. The three exceptions were inserts KPF, LNP, and SMH. KPF lies in the coding region for a 385-kDa Zn finger protein (CG11122); the next natural splice is out-of-frame, but we hypothesize that the Hto transcript uses an in-frame cryptic splice site(s), as suggested by large protein products seen on preliminary anti-FLAG Western blots. For LNP and SMH, there is no in-frame protein fusion, but the functional Hto product appears instead to be a noncoding (nc) RNA.
Table 2. Locations and target genes of verified Hto inserts.
| Hto insert name | Target gene | Gene product notes | Chromosome: insertion site (orientation) |
|---|---|---|---|
| Transcription factors and other nuclear proteins | |||
| BRO | cut | Homeobox transcription factor | X: 7,475,721 (+) |
| KPF | CG11122 | Zn-finger protein | X: 11,010,000 (−) |
| GND | mamo/CG11071 | Zn-finger protein similar to human ZNF121 | X: 13,760,086 (−) |
| GLO | CG34340 | Similar to human dorsal root ganglia homeobox | 2L: 3,669,091 (+) |
| DRE | elbow B | Similar to human Zn-finger protein 503 | 2L: 14,399,989 (−) |
| POV | vestigial | Transcription cofactor for Sd | 2R: 8,777,614 (+) |
| REM | mastermind | Transcription cofactor for Notch | 2R: 9,901,271 (+) |
| QSR | CG17181/kahuli | Snail family transcription factor | 3L: 594,766 (−) |
| XTC | klumpfuss | Zn-finger protein, similar to Hum. WT1 | 3L: 10,983,471 (−) |
| FAR | araucan (Iro-C) | homeobox transcription factor | 3L: 12,573,566 (+) |
| WGL | AP-2 | Transcription factor | 3L: 21,598,379 (+) |
| GTA | taranis | Trithorax group protein; SERTA domain | 3R: 12,052,275 (+) |
| QBP | taranis | Trithorax group protein; SERTA domain | 3R: 12,068,742 (+) |
| QYE | bonus | Tripartite motif-containing 24 (TRIM24) homolog | 3R: 16,430,713 (+) |
| HJF | Sox102F | HMG box transcription factor | 4: 824,130 (−) |
| OMD | shaven | D-Pax2, paired box transcription factor | 4: 1,099,612 (+) |
| Cytoplasmic proteins | |||
| CXM | l(1)G0232 | Nonreceptor protein tyrosine phosphatase (PTP) Meg2 | X: 9,535,463 (−) |
| BRW | nuclear fallout | Arfophilin/FIP3 (binds both Rab11 and ARF) | 3L: 14,212,366 (+) |
| GYB | Synaptotagmin 4 | Regulates membrane traffic | 3R: 3,079,839 (+) |
| DPQ | pumilio | RNA-binding protein | 3R: 4,943,989 (−) |
| GER | pumilio | RNA-binding protein | 3R: 4,971,465 (−) |
| WEB | pumilio | RNA-binding protein | 3R: 5,046,677 (−) |
| Membrane proteins | |||
| WRS | seizure | 6-TM K+ channel | 2R: 19,938,312 (+) |
| KCM | CG42235 | Multipass TM protein, similar to SLC5A8 solute transporter | 3R: 21,740,248 (+) |
| ncRNAs | |||
| SMH | mir-274 | microRNA | 3L: 11,649,595 (+) |
| LNP | iab-8, abd-A | iab-8 is an ncRNA in Bithorax Complex | 3R: 12,723,886 (−) |
The Hto inserts and their target genes are ordered by function and genome position. Orientation of the insert (plus or minus) is given relative to the standard genome sequence. Each target gene is in the same orientation as the Hto insert. See Table S1 for insertion site, splicing, and protein data.
Figure 4.
Genomic maps of Hto insertions BRO, GND, CXM, and DRE. Maps show the relevant transcripts and alternative splices; for complete transcript maps, see FlyBase.org (McQuilton et al. 2012). Lower scale bars indicate the genomic coordinates based on D. melanogaster Genome Release 5.47. Arrows, transcription starts; boxes, exons; gray boxes, coding regions; start codons indicated by “M”; position of Hto insert indicated by red triangle; angled lines indicate Hto splices or endogenous alternative splices.
To test the splicing models, RNA was isolated from Hto-expressing flies, and RT-PCR was used amplify across the junction between Hto exon 1 and the target gene. Eleven lines were tested, and usage of the next downstream exon was confirmed in eight cases; as an example, the RT-PCR sequence from XTC is shown in Figure S5. In two cases, BRO and BRW, the next downstream exon is a known alternative exon that was skipped over by the Hto transcripts that we recovered; these lines use other in-frame exons as shown (Figure 4 and Figure S4). The other exceptional line was LNP, which was found to splice into the bithorax complex ncRNA gene iab-8. The RT-PCR sequence from LNP helped to characterize iab-8 exon structure and demonstrated transcription from iab-8 into the adjacent abd-A gene, as detailed in Gummalla et al. (2012).
The 26 lines feature 23 different target genes, 2 of which have multiple hits (WEB, GER, and DPQ lie in pum; GTA and QBP lie in tara) (Figure S4). The genes fall into several functional classes, but two strong trends are apparent from this list: nearly all of the target genes make regulatory proteins (or RNAs), and, more specifically, there is a bias toward transcription factor genes. Eleven genes (48%) encode DNA-binding transcription factors such as cut, klu, and sv/Pax2 (compared to ∼2–5% transcription factor genes in the genome; Pfreundt et al. 2010); two encode transcriptional co-activators (vg and mam), and two encode other nuclear proteins (bonus/TRIM24/TRIM33 and tara/TRIP-Br2) for a total of 15 (65%) nuclear regulators. Four genes encode cytoplasmic regulatory proteins: synaptotagmin 4 (Syt4), PTP-Meg2 (l(1)G0232), Pumilio (pum), and Nuf/Rab11-FIP3/arfophilin (nuf). Two lines can express ncRNAs (iab-8/miR-iab-8 and miR-274) that are known or predicted to modulate the expression of other regulatory genes (http://miRBase.org). Overall, then, 21/23 (91%) of the target genes regulate cell biological processes or gene expression.
Some lines match known misexpression phenotypes and biological functions, as exemplified by XTC, which expresses a full-length Klu transcription factor (Figure S5, A and B). Loss of klu leads to bristle loss, while, conversely, both UAS-klu (Kaspar et al. 2008) and the XTC line (Figure S5) yield ectopic bristles. We find that XTC expression by appropriate drivers converts the eye disc, retina, or wing blade into dense beds of bristles, often with tufts of multiple bristle shafts, as seen by SEM (Figure S5, C–E). Dorsal expression of XTC via pnr-GAL4 strikingly leads to fusion of the head and thorax (Figure S5, F–I). XTC also inhibits the growth of salivary gland cells in a cell-autonomous manner (Figure S5, J and K).
Hto fusions typically include substantial portions of the target protein
In most lines, Hto lies 5′ of most of the coding region and thus can make a full-length or near full-length fusion protein (Table S1). For other lines, the Hto product includes just a C-terminal portion of the target protein, but most of these are predicted to include at least one functional domain. The five lines QSR and GYB (Figure S4), XTC (Figure S5A), GLO, and OMD splice to the target gene’s 5′ UTR and thus express full-length proteins fused to FLAG-RFP via an extended linker encoded by the 5′ UTR. For the nine lines BRO and DRE (Figure 4), GTA/QBP and HJF (Figure S4), POV, FAR, WGL, and QYE, the fusion transcripts bypass the target gene’s start codon, but still include >92% of its coding region, and so these products are considered “near full length.” Twelve of the 13 target genes in the full- and near-full-length categories encode transcription factors or other nuclear regulatory proteins.
For the nine lines CXM and GND (Figure 4), BRW and the pum lines WEB/GER/DPQ (Figure S4), REM (below), WRS, and KCM, significant portions of the endogenous coding region are bypassed, such that the fusion products include 50–85% of the full-length protein (Table S1). Seven of these fragments are expected to retain key functions; for example, the GND product includes a functional DNA-binding domain, since it localizes to discrete sites on polytene chromosomes. (REM, CXM, BRW, and the pum lines are detailed individually below.) The remaining two truncated products are multi-pass membrane transporters: WRS expresses a fragment of the Seizure K+ channel, and KCM expresses a fragment of the CG42235 product, a SLC-family sodium-solute cotransporter. The only phenotype found using the WRS line is a rough eye with GMR>WRS (Figure 3). KCM, however, shows several phenotypes suggestive of the Notch pathway: glazed eyes, thickened veins, and extra macrochaetae (Figure 2). The mechanisms underlying the WRS and KCM phenotypes are not clear.
The Hto system adds a biochemical tag, 3xFLAG, to the target protein. The tag allows us to test whether the size of the actual fusion product(s) matches the predictions from the splicing models described above. For eight lines, the fusion was expressed in adult flies and examined by anti-FLAG Western blots (Figure 5). These lines each express a single major fusion protein near the predicted size, with some less abundant smaller bands that may be breakdown products or smaller isoforms. The exceptions are two transcription factor lines, HJF and GND, whose fusions run ∼15 kDa larger than predicted; this may be due to protein modifications or other structural features that cause reduced mobility. The Starter2 line makes no fusion protein, and there are no endogenous proteins that cross-react with anti-FLAG M2 (Figure 5, “St.” lane). We note that the Starter element, and most other Hto lines, can express at varying levels a 32-kDa product corresponding to unfused FLAG-RFP. This may arise from polyadenylation of some Hto transcripts using signals within the Minos right IR.
Figure 5.
SDS/PAGE–Western analysis shows that Hto lines primarily express a single major fusion protein of the expected size, as well as unfused FLAG-RFP (arrowheads). Each lane shows whole adult protein from heat-shock-induced Hsp70-GAL4>Hto flies, stained with anti-3xFLAG and AP-linked secondary antibody. Each line had a different optimal BCIP development time, suggesting that the efficiency of fusion protein expression varies significantly across lines. The first lane is Starter2 element (St.) only; no fusion is made. Remaining lanes show the designated Hto line. BRW was not heat-shocked; it expresses a constitutive fusion protein in the absence of GAL4 induction (likely due to the adjacent nested gene CG7768; Figure S3). For each line’s major Hto product, the expected/observed molecular weight (in kDa) is as follows: BRW, 64.4/62; POV, 72.9/71; QSR, 79.9/82; DRE, 83.5/79; GYB, 84.4/84; HJF, 93.6/110; GND, 116.1/131; XTC, 125.5/133; and unfused FLAG-RFP, 31.5/32.
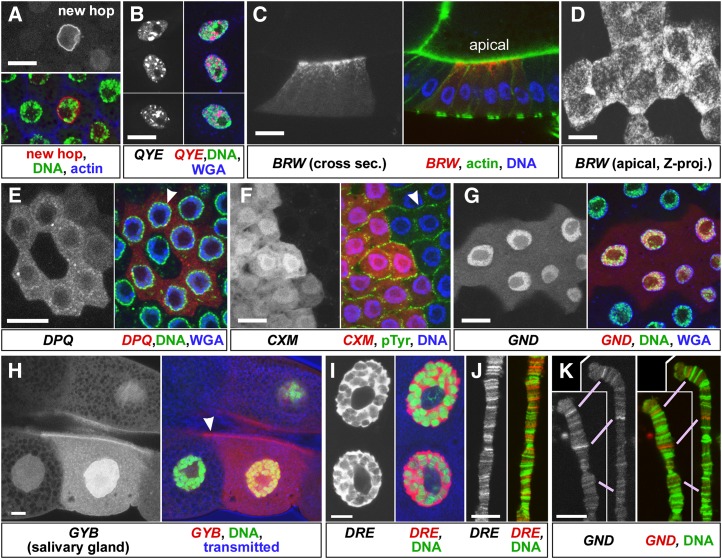
Imaging Hto protein traps in cells and on chromatin
The localization of Hto fusion proteins was documented by confocal microscopy (Figure 6 and Figure S3). We primarily used the ovary follicle cells and the larval salivary glands as model polarized epithelia to determine whether the fusions localize to specific subcellular regions. First, we tested whether the Starter element can transpose to new loci to produce new RFP localization patterns in response to Minos transposase. In Starter2 flies, Minos transposase and GAL4 were induced by heat shock, and numerous novel RFP patterns appeared in follicle cell clones, indicating that Hto hops efficiently in somatic cells and tags a variety of proteins. Examples include a highly specific labeling of the nuclear envelope (Figure 6A), and an RFP fusion protein that assembles into bars and filaments in the cytoplasm (Figure S3H). Those hops cannot be molecularly characterized since they are limited to small clones, but they highlight the range of localizations that are possible with the Hto system. Next, the Hto lines that tag cytoplasmic proteins were expressed in follicle cell clones using the FLP-out GAL4 system or in salivary glands. For each line, a different localization pattern was observed, and the patterns are consistent with the nature of the protein. Nuf/Rab11-FIP3/arfophilin fusion (BRW) is strongly polarized to the apical cortex (Figure 6, C and D) and the leading edge of migrating border cells approaching and contacting the oocyte (Figure S3E). The pattern matches that of anti-Nuf staining in egg chambers (Xu et al. 2011). The synaptotagmin 4 (GYB) fusion is broadly distributed, but accumulates at the apical membranes of the salivary gland cells (Figure 6H) and on follicle cell membranes (Figure S3C). The PTP-Meg2 fusion (CXM) lacks the protein’s known localization domain and accordingly has a fairly uniform distribution with no polarization. The fusion includes the PTP domain, but overexpression of the PTP does not affect phosphotyrosine accumulation at the adherens junction (Figure 6F and Figure S3D), suggesting that it retains some substrate specificity (i.e., is not acting as a general Tyr phosphatase). A Pumilio fusion accumulates at low levels and has a granular appearance with a few strong puncta appearing on the nuclear envelope (Figure 6E and Figure S3F), similar to the pattern of a standard protein trap in Pumilio (Harris et al. 2011).
Figure 6.
Subcellular localization patterns of Hto fusion proteins. Bars, 10 µm. Confocal analyses of egg-chamber follicle cells (A–G) or salivary glands (H–K) expressing Hto inserts. In each set, the grayscale image is the RFP signal shown with original contrast; the color image includes the RFP channel in red and structural markers in green and blue as indicated beneath the picture. (A) In tissues expressing Minos transposase, new hops of the Starter element can yield new localization patterns (Hsp70-MiT/Starter2; Hsp70-GAL4, heat-shocked twice prior to fixation). In this example, RFP fusion with an unknown target protein causes strong localization to the nuclear envelope. Blue, f-actin; green, SYBR Green. (B–G) Hto inserts expressed in clones of follicle cells using the Act5C-GAL4 FLP-out system. (B) Three examples of stretched nurse-cell follicle cells with relatively flat nuclei. QYE fusion protein forms very dense aggregates in the nucleus. Green, SYBR Green; blue, WGA. (C) Cross-section of a stage 9 BRW clone, apical up. The fusion with Nuf/FIP3 labels puncta or vesicular structures that accumulate toward the apical side. Green, f-actin; blue, SYBR Green. (D) Projection of a z-series through the apical region of a stage 10 BRW clone. (E) Stage 9 DPQ clone expresses a fusion to Pumilio that accumulates in the cytoplasm and especially near the nuclear envelope (arrowhead). Green, SYBR Green; blue, WGA. (F) A fusion to PTP-Meg2 in a CXM clone is relatively uniform in the cytoplasm. Accumulation of pTyr (green) at the adherens junction (arrowhead) is not reduced in the clone vs. the wild-type neighbors. Blue, SYBR Green. (G) GND clone shows nuclear localization of the fusion protein. Green, SYBR Green; blue, WGA; anterior follicle cells at stage 8/9. (H–K) Hto expression in salivary glands using Hsp70-GAL4 with no heat shock. Green, SYBR Green. (H) GYB expresses a fusion to Synaptotagmin 4, enriched on the apical membrane facing the lumen (arrowhead). Blue, transmitted light image. (I) The DRE fusion strongly concentrates in the nucleus. Blue, transmitted light image. (J) The DRE fusion accumulates at distinct sites on chromatin, seen as bands on spread polytene chromosomes. SYBR Green staining (green) gives a different banding pattern. (K) The GND fusion protein also shows a distinct banding pattern; two homologized chromosomes are shown; matching bands are indicated by purple lines.
The presumed nuclear proteins tagged by Hto generally show the expected nuclear localization in vivo. Examples are GND, which expresses a Zn-finger-containing fragment from the mamo gene (Figure 6G), and DRE, which makes a near-full-length fusion to Zn-finger protein ElbowB. The DRE product is almost entirely nuclear, but excluded from the nucleolus, in the salivary gland (Figure 6I). The QYE product, a near full-length fusion to Bonus/TRIM24/33, displays an unusual behavior in follicle cells. At lower expression levels it has a granular localization, with the brighter regions mostly lying adjacent to SYBR-Green-stained chromatin (Figure S3). This matches the distribution of TRIM24 in HeLa cells (Herquel et al. 2011). At higher levels, the fusion accumulates into large, very bright (densely packed with RFP) nuclear aggregates (Figure 6B). This is the only case that we have found where the localization pattern changes notably based on expression levels.
Since the Hto system tagged numerous transcription factors, we asked whether the transcription factor fusions retain sequence-specific binding. If so, they should show discrete and reproducible RFP-positive bands on polytene chromosomes. Indeed, the lines DRE, GND, and QSR each show different patterns of polytene banding. DRE produces numerous strong RFP bands, which sometimes complement and sometimes overlap with SYBR-Green-rich bands (Figure 6J). GND and QSR each produce fewer and weaker bands, but the banding pattern is reproducible within each line across multiple chromosome spreads (Figure 6K and Figure S3B). There were no lines found to yield just a few strong bands; this may indicate that these transcription factors normally bind numerous sites (not all of which may be functionally important) (Li et al. 2008; Fisher et al. 2012) and/or that overexpression with the Hsp70-GAL4 driver was forcing the fusion protein onto weak binding sites. The latter explanation is not consistent with results from neighboring cells that express very different levels of fusion (an artifact of the GAL4 system). Both strongly and weakly expressing cells from a given line show the same relative distribution of RFP from band to band (not shown).
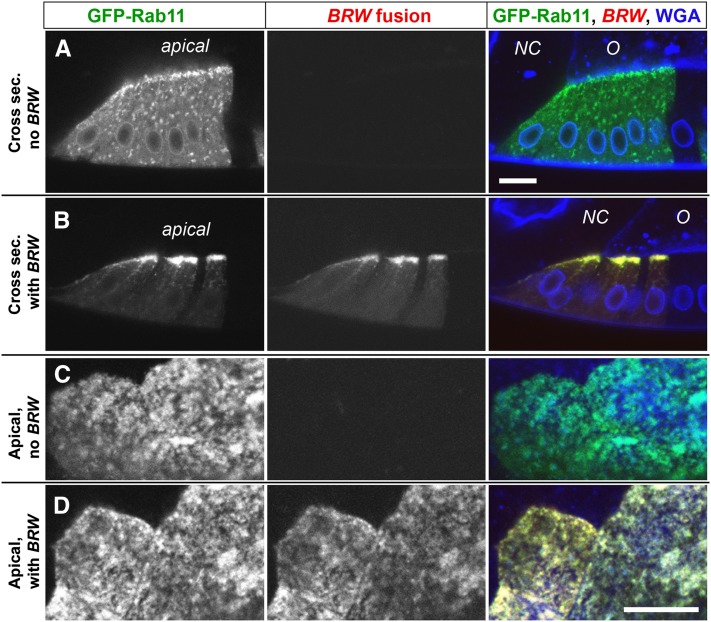
Colocalization of the Nuf fusion with its binding partner, Rab11
To assess whether Hto lines are suited to characterizing in vivo protein interactions, we investigated the BRW insert in the nuf gene. The resulting Nuf fusion protein is mostly apical (Figure 6, C and D); much of the pattern is reminiscent of transport vesicles, but does not match the pattern observed for early endosomes (e.g., Belenkaya et al. 2008; Velichkova et al. 2010). Nuf normally interacts with Rab11 on the recycling endosome (Riggs et al. 2003; Horgan and McCaffrey 2009; Baetz and Goldenring 2013), and disruption of recycling endosomes during retinal differentiation could account for BRW’s very strong GMR-GAL4 phenotype (Figure 2) (Alone et al. 2005). The Nuf fusion is predicted to dimerize via an extended coiled-coil region, whose C-terminal ∼30 aa forms the Rab11-binding domain (Eathiraj et al. 2006; Shiba et al. 2006). These structural data imply that Nuf fusion protein dimers, and heterodimers between the fusion protein and endogenous Nuf, should interact with Rab11 properly. However, such dimers would have missing or disrupted N-terminal regions and thus may not interact properly with other effectors, such as cytoskeletal motors (Horgan et al. 2010).
We predicted that the Nuf fusion either would be recruited to recycling endosomes by Rab11 or would recruit Rab11 to new sites. To test this, we coexpressed UAS-GFP-Rab11 with BRW in follicle cell clones. The Nuf fusion showed nearly complete colocalization with GFP-Rab11 (Figure 7, B and D). Furthermore, in co-expressing cells, the localization of Nuf fusion and especially of GFP-Rab11 became more compact. On its own, GFP-Rab11 was seen throughout the cell on vesicle-like structures with a variable concentration at the apical cortex (Figure 7, A and C). Addition of BRW greatly diminished the cytoplasmic signal, and a dense accumulation of Nuf fusion and GFP-Rab11 appeared apically (Figure 7, B and D). The result indicates that the Nuf fusion retains the Rab11-binding function, but alters Rab11 behavior, likely through overexpression (outcompeting other binding partners for Rab11) and/or the absence of the Nuf N terminus in the fusion protein.
Figure 7.
Interaction of an Hto fusion protein with its predicted partner. Bar in A, 10 µm for rows A and B. Bar in D, 10 µm for rows C and D. Confocal sections of egg chambers from sibs with FLP-out clones that express UAS-GFP-Rab11 (green in right column), either without (A and C) or with (B and D) the BRW insert expressing the RFP-Nuf fusion (red in right column). The left column is GFP (original contrast); the middle column is RFP (original contrast); the right column is the overlay, with WGA in blue. Note the lack of signal in the RFP channel in the absence of BRW. (A and B) Cross-sections of follicle cells at the anterior of the oocyte (O) adjacent to the nurse cells (NC) at late stage 9–early stage 10; apical is up. Much of the GFP-Rab11 moves from the cytoplasm (A) into dense apical aggregates when the RFP-Nuf fusion is also present (B). (C and D) Sections along the apical cortex of the oocyte follicular epithelium at clone borders in stage 10. Colocalization of GFP-Rab11 and the RFP-Nuf fusion is very strong; note yellow in the B and D overlays. Pearson’s correlation coefficient was 0.92 for GFP vs. RFP signal (excluding background; summed over clones from seven different egg chambers), but 0.00 for GFP vs. a randomized version of the RFP channel.
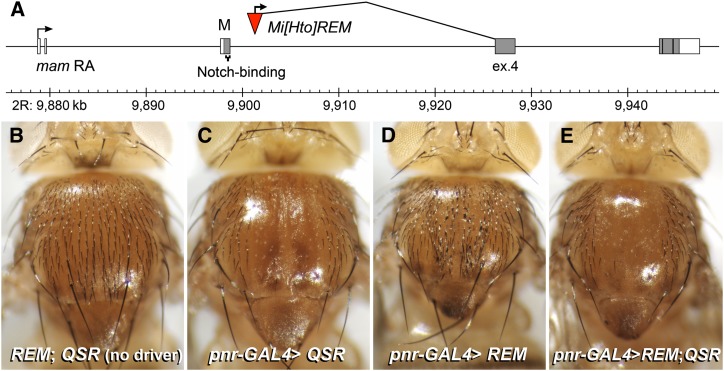
Hto lines are useful for genetic interaction analysis
The insert REM lies in mastermind (mam), which encodes a key transcriptional coactivator for the Notch intracellular domain/CBF1, Suppressor of Hairless, Lag-1 (CSL) complex. Mam binds to Notch/CSL via an α-helix at the Mam N terminus and recruits transcriptional and chromatin effectors to the complex (Wallberg et al. 2002; Nam et al. 2006). The REM fusion includes the C-terminal 85% of Mam protein, but lacks the crucial Notch/CSL-binding helix (Figure 8A), suggesting that the fusion has the capacity to bind effectors, but not to recruit them to Notch/CSL. We observed that several REM phenotypes are consistent with Notch loss-of-function effects. Most specifically, the wing veins expand to a much larger but uniform width (Figure 3), indicating that cells of the vein-competent region are not properly partitioned between vein and intervein fate by Notch (De Celis 2003). Also, pnr>REM gives a strong excess bristle phenotype on the thorax (Figure 8D). Thus, the molecular and phenotypic evidence indicates that REM’s fusion protein is a dominant negative, antagonizing Notch signaling. Interestingly, a fragment complementary to this, based on the Notch/CSL-binding helix, is also a strong dominant negative (Moellering et al. 2009).
Figure 8.
Use of Hto system for genetic interaction analysis: an epistasis experiment with a Notch pathway antagonist (product of REM) and a Snail family transcription factor (product of QSR). (A) Map of the REM insert in mastermind. The resulting Hto fusion protein lacks the entire Notch/CSL-binding region. (B–E), Siblings raised at 25°. (B) Wild-type thorax: the fly carries both REM and QSR inserts but no driver. (C) pnr-GAL4 driving QSR results in a loss of macrochaetae in the zone of pnr-GAL4 expression. (D) pnr-GAL4 driving REM results in an excess of macrochaetae, suggesting that the response to Notch signaling is blocked. (E) pnr-GAL4 driving both REM and QSR results in a loss of macrochaetae, as seen with QSR alone. Thus the expression of QSR eliminates bristles regardless of the presence or absence of Notch signaling.
The line QSR targets the Snail family transcription factor gene CG17181/kahuli (see Materials and Methods, Nomenclature of tagged genes). Expression of QSR in the thorax eliminates bristles leaving a stripe of naked cuticle, just as observed with excess Notch signaling (Chandra et al. 2003). To determine if Hto lines can yield informative epistasis results, we crossed REM and QSR with pnr-GAL4 in the background; the offspring included each Hto line alone or together (Figure 8, B–E). While REM alone gives extra bristles due to loss of Notch-mediated transcription, the REM; QSR flies have naked cuticle; thus the bristle program is halted by ectopic kahuli expression from QSR regardless of the Notch signal. The pnr>QSR bristle loss, then, does not arise due to ectopic Notch activation upstream of Mam.
Discussion
The goal of the project was to implement and test a novel protein-trapping system, Hto, that can simultaneously misexpress and tag endogenous target proteins and fragments. An Hto transposon insert is intended to provide a single reagent that can be used for multiple downstream analyses. We established that the Hto vector can mobilize to chromosomes X, 2, 3, and 4 and that Hto transcripts can splice cleanly to downstream target gene exons (Figure 1) across distances ranging from <0.5 to >50 kb (Table S1). Expression leads to diverse, GAL4-dependent dominant phenotypes (Figure 2, Figure 3, and Figure S5) that are useful for genetic interaction studies (Figure 8). The resulting Hto fusion proteins have dual FLAG-mCherry RFP tags that are effective for both microscopy and immunodetection; the RFP tag reports an array of informative subcellular localizations (Figure 6 and Figure S3), while the FLAG epitope can be used to verify the size of the fusion protein (Figure 5). RFP-tagged proteins from Hto can complement the many existing GFP protein traps and constructs, allowing for colocalization studies in fixed or live tissues (Figure 7). Unlike a typical cDNA expression construct, the target gene is endogenous and thus retains features such as alternative splices and poly(A) sites; this is especially useful for expressing large and complex genes. Genetic analysis is further facilitated by the inclusion of an I-SceI genomic restriction site. If an Hto insert lies near an exon or regulatory region of interest, then an I-SceI expression construct can be crossed with Hto to trigger double-stranded breaks and targeted chromosomal deletions (Bellaiche et al. 1999). We expect other uses to arise with further deployment of the system, including isolation of in vivo protein complexes or chromatin immunoprecipitation of transcription factors using the FLAG tag and suppressor screens to obtain loss-of-function alleles of target genes (described below).
We employed an F1 phenotypic screen for eye defects using the retinal GMR-GAL4 driver to collect only those inserts that can disrupt development. It is reasonable to assume that phenotype-causing inserts must make biologically active, and therefore properly folded, fusion proteins. On the other hand, it is also possible that strong overexpression of an RFP-tagged but otherwise misfolded protein could yield nonspecific phenotypes, perhaps as a general stress response. However, we conclude that the large majority of lines recovered from the screen make fusions that are properly folded and act via distinct developmental pathways rather than a common stress pathway, based on the following four arguments.
Most fusion proteins initially produced by randomly mobilizing the Hto element are predicted to be inactive, but those inserts are effectively culled by the use of a phenotypic screen. Specifically, Hto is in reading frame 0, leaving most random fusion events out-of-frame. Of the in-frame fusions, many can be expected to express only part of a folded domain and thus to misfold. Fusions to secreted or transmembrane proteins will also fail to be processed correctly if they require an N-terminal signal sequence, since one is not provided by Hto. But, of the 23 targets, only two fusions are predicted to be significantly misfolded (WRS and KCM), and only KPF is out-of-frame with the next exon, although it is a strong candidate to make an in-frame cryptic splice.
This analysis is also supported by the yield of the screen. It is difficult to estimate how many new hops result in the production of some kind of fusion protein, but we expect this to be a majority, since gene density is 1 per ∼9 kb, and Hto can splice across at least 50 kb to its target. Only 1.7% of new hops or (therefore) ∼2–3% of fusion proteins cause a phenotype. This is much less than expected if random/misfolded polypeptides often caused phenotypes with GMR-GAL4, but on par with the number of developmental genes that Hto could legitimately target.
Each line yields a unique combination of GAL4 phenotypes in the eye and wing (Figure 2 and Figure 3). This demonstrates a high degree of biological specificity for each fusion protein, incompatible with general toxicity of misfolded products.
Of 23 target genes, 21 are primarily regulatory in nature, and 13 of these encode transcription factors/cofactors (Table 2). This strong functional bias indicates that the phenotypes are due to misexpression of biologically active proteins/domains.
Many of the targets are previously named and well characterized, as expected given the enrichment for important regulatory genes. The misexpression phenotypes shown here generally match well with previous reports, where available (e.g., Kaspar et al. 2008), but nonetheless some are quite striking, such as the ability of klu to fuse the head to the thorax under pnr-GAL4 control (Figure S5), and the ability of bon, a regulator of p53 and nuclear receptors (Beckstead et al. 2001, 2005; Allton et al. 2009), to drive production of hairs on the retina (see SEM data in Tardi et al. 2012). The Hto system has also provided functional information for several little-characterized CG genes. For example, the QSR line expresses a full-length fusion to the product of CG17181/kahuli, a Snail superfamily transcription factor, and revealed that it can specifically block thoracic bristle development (Figure 8).
Use of Minos facilitates expression of C-terminal protein fragments
N-terminal fragments of proteins are often sampled in mutagenesis screens, arising either from gene truncations or premature stop codons. However, C-terminal fragments are rarely generated by random mutation, since they require intact 5′ regulatory sequences to direct transcription and translation, followed by an in-frame deletion of just the N-terminal portion of the ORF. It appears that this part of the mutational spectrum has never been systematically mined for phenotypes. Unlike P elements, Minos has no preference for inserting near promoter regions and routinely inserts within coding spans of genes (Bellen et al. 2011; Venken et al. 2011). Hto insertions should randomly sample nearly all exons with a 3′ ss, yielding a variety of C-terminal fragments for phenotypic screening. The RFP tag that Hto adds to the N terminus likely makes these fragments more stable in the cell, especially if they are small. For modular proteins (common in signaling, the cytoskeleton, etc.), tagged C-terminal fragments can yield a wealth of structure–function information, tying specific domains to phenotypes, cellular localizations, and binding partners. Some C-terminal fragments can act as dominant negatives (e.g., the Mam fragment produced by REM; Figure 8) or can sequester interacting proteins (e.g., Nuf and Rab11; Figure 7) and thus may prove generally useful as pathway-disrupting reagents.
For three of the cytoplasmic target proteins, Hto express C-terminal fragments with well-defined functional domains. BRW expresses just 58% of Nuf/FIP3, but this contains the ARF- and Rab11-binding sites (Shiba et al. 2006). For CXM, the product includes the full PTP domain of PTP-Meg2, but deletes part of the N-terminal Sec14 localization domain (Saito et al. 2007). The pumilio inserts all make fusions that include the Puf RNA-binding domain of Pum. Interestingly, for all three genes, there are endogenous alternatively spliced species that closely match the Hto-derived fragments; that is, they express the binding or enzymatic domain and remove much of the N-terminal region of the protein: isoforms l(1)G0232-RE (Figure 4), nuf-RB, and pum-RH (Figure S4) (modENCODE Consortium et al. 2010). This indicates that the screen can highlight biologically relevant subregions of proteins.
Using a Minos-based vector expands the range of genes that can be targeted. Minos has very little insertion-site bias and thus provides more thorough coverage of the genome than P-element-based vectors (Bellen et al. 2011). Even though our sample of Hto targets is biased toward well-studied genes, 9 of the 23 target genes are P-element coldspots based on FlyBase map data, with either zero or one reported P insert (anywhere in the transcription unit or the 5′ flanking intergenic region; Table S1). The three loci vg, sv, and mir-274 (including its surrounding gene CG32085) all lack any reported P-element constructs. Also, only about half the target genes have publicly available insertions of P-element expression vectors (of the types EP, EPgy2, Mae-UAS.6.11, or GSV7; Table S1).
Recently, a large-scale collection of inserts of a Minos element called MiMIC was released (Drosophila Gene Disruption Project) (Venken et al. 2011; Venken and Bellen 2012). MiMIC is extremely powerful, providing a means to alter the local target gene in nearly any manner using recombinase-mediated cassette exchange (RMCE) to add a desired sequence at the insertion site (Bateman et al. 2006). MiMIC differs from Hto in function and in the approach to site selection. Unless RMCE is performed, a MiMIC insert does not tag or control the target gene, but rather acts as a gene disruption tool. MiMIC lines are not phenotypically selected, but generated randomly and retained based on the potential usefulness of the insertion site. Hto insertion sites (on a much smaller scale) are preselected as being “functional” in the sense that misexpression from that site will cause a useful phenotype. Given the large selection of mapped MiMIC inserts, it will be quite useful to make an RMCE donor containing the Hto core sequence to convert appropriate MiMIC lines to Hto lines.
Finally, we note that in some cases an Hto line could form the basis for a very efficient F1 suppressor screen to recover many loss-of-function missense mutations in the target protein. Some full-length Hto fusions result from long-distance tagging (e.g., BRO/cut, Figure 4), which leaves the target allele essentially wild type. Even intronic insertions may behave as wild-type alleles, since Hto is minimally disruptive to the gene (only 2 kb long, with no 3′ ss). If one were to chemically mutagenize an Hto chromosome and then express it with the appropriate driver, any F1 flies that lack the Hto phenotype should carry a new mutation that inactivates either the target gene or Hto itself. Those two classes could be quickly distinguished by the presence of the RFP marker or by FLAG Western blots. If the screen is performed on flies that are otherwise lethal due to a strong GAL4 driver, then it becomes an F1 viability screen, which could test hundreds of thousands of chromosomes. This strategy would essentially turn recessive mutations into dominant, scorable ones and work even for redundant genes with little or no recessive loss-of-function phenotype. The resulting bank of tagged missense mutants could be used for fine structure–function analysis, linking specific amino acids to in vivo localization patterns and binding interactions.
Supplementary Material
Acknowledgments
We thank Katie Molohon, Christopher Greulich, Troy Larson, and Craig Gatto for technical assistance and advice with the project and the Bloomington Drosophila Stock Center at Indiana University and the H. Bellen/Gene Disruption Project for fly stocks. This work was supported by National Institutes of Health grant GM62185 and by Illinois State University.
Footnotes
Communicating editor: W. Sullivan
Literature Cited
- Aleksic J., Lazic R., Müller I., Russell S. R., Adryan B., 2009. Biases in Drosophila melanogaster protein trap screens. BMC Genomics 10: 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allton K., Jain A. K., Herz H. M., Tsai W. W., Jung S. Y., et al. , 2009. Trim24 targets endogenous p53 for degradation. Proc. Natl. Acad. Sci. USA 106: 11612–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alone D. P., Tiwari A. K., Mandal L., Li M., Mechler B. M., et al. , 2005. Rab11 is required during Drosophila eye development. Int. J. Dev. Biol. 49: 873–879. [DOI] [PubMed] [Google Scholar]
- Ashburner M., Bergman C. M., 2005. Drosophila melanogaster: a case study of a model genomic sequence and its consequences. Genome Res. 15: 1661–1667. [DOI] [PubMed] [Google Scholar]
- Baetz N. W., Goldenring J. R., 2013. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol. Biol. Cell 24: 643–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J. R., Lee A. M., Wu C. T., 2006. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics 173: 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead R., Ortiz J. A., Sanchez C., Prokopenko S. N., Chambon P., et al. , 2001. Bonus, a Drosophila homolog of TIF1 proteins, interacts with nuclear receptors and can inhibit betaFTZ-F1-dependent transcription. Mol. Cell 7: 753–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckstead R. B., Ner S. S., Hales K. G., Grigliatti T. A., Baker B. S., et al. , 2005. Bonus, a Drosophila TIF1 homolog, is a chromatin-associated protein that acts as a modifier of position-effect variegation. Genetics 169: 783–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenkaya T. Y., Wu Y., Tang X., Zhou B., Cheng L., et al. , 2008. The retromer complex influences Wnt secretion by recycling wntless from endosomes to the trans-Golgi network. Dev. Cell 14: 120–131. [DOI] [PubMed] [Google Scholar]
- Bellaiche Y., Mogila V., Perrimon N., 1999. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics 152: 1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellen H. J., Levis R. W., He Y., Carlson J. W., Evans-Holm M., et al. , 2011. The Drosophila gene disruption project: progress using transposons with distinctive site specificities. Genetics 188: 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H., Perrimon N., 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. [DOI] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., et al. , 2007. The Carnegie protein trap library: a versatile tool for Drosophila developmental studies. Genetics 175: 1505–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S., Ahmed A., Vaessin H., 2003. The Drosophila IgC2 domain protein Friend-of-Echinoid, a paralogue of Echinoid, limits the number of sensory organ precursors in the wing disc and interacts with the Notch signaling pathway. Dev. Biol. 256: 302–316. [DOI] [PubMed] [Google Scholar]
- Clyne P. J., Brotman J. S., Sweeney S. T., Davis G., 2003. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics 165: 1433–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Celis J. F., 2003. Pattern formation in the Drosophila wing: the development of the veins. Bioessays 25: 443–451. [DOI] [PubMed] [Google Scholar]
- del Valle Rodríguez A., Didiano D., Desplan C., 2011. Power tools for gene expression and clonal analysis in Drosophila. Nat. Methods 9: 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit T., Dekker S., Maas A., Breedveld G., Knoch T. A., et al. , 2010. Tagged mutagenesis by efficient Minos-based germ line transposition. Mol. Cell. Biol. 30: 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMario P., Rosby R., Cui Z., 2006. Direct visualization of GFP-fusion proteins on polytene chromosomes. Drosoph. Inf. Serv. 89: 115–118. [Google Scholar]
- Dow J. A., 2003. The Drosophila phenotype gap—and how to close it. Brief. Funct. Genomics Proteomics 2: 121–127. [DOI] [PubMed] [Google Scholar]
- Duffy J. B., 2002. GAL4 system in Drosophila: a fly geneticist’s Swiss army knife. Genesis 34: 1–15. [DOI] [PubMed] [Google Scholar]
- Eathiraj S., Mishra A., Prekeris R., Lambright D. G., 2006. Structural basis for Rab11-mediated recruitment of FIP3 to recycling endosomes. J. Mol. Biol. 364: 121–135. [DOI] [PubMed] [Google Scholar]
- Edwards K., Doescher L., Kaneshiro K., Yamamoto D., 2007. A database of wing diversity in the Hawaiian Drosophila. PLoS ONE 2(5): e487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D. A., Brand A. H., 2008. The GAL4 system: a versatile system for the expression of genes. Methods Mol. Biol. 420: 79–95. [DOI] [PubMed] [Google Scholar]
- Fisher W. W., Li J. J., Hammonds A. S., Brown J. B., Pfeiffer B. D., et al. , 2012. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc. Natl. Acad. Sci. USA 109: 21330–21335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummalla M., Maeda R. K., Castro Alvarez J. J., Gyurkovics H., Singari S., et al. , 2012. abd-A regulation by the iab-8 noncoding RNA. PLoS Genet. 8: e1002720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. E., Pargett M., Sutcliffe C., Umulis D., Ashe H. L., 2011. Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell 20: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herquel B., Ouararhni K., Khetchoumian K., Ignat M., Teletin M., et al. , 2011. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 108: 8212–8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horgan C. P., McCaffrey M., 2009. The dynamic Rab11-FIPs. Biochem. Soc. Trans. 37: 1032–1036. [DOI] [PubMed] [Google Scholar]
- Horgan C. P., Hanscom S. R., Jolly R. S., Futter C. E., McCaffrey M. W., 2010. Rab11–FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J. Cell Sci. 123: 181–191. [DOI] [PubMed] [Google Scholar]
- Hozumi A., Kawai N., Yoshida R., Ogura Y., Ohta N., et al. , 2010. Efficient transposition of a single Minos transposon copy in the genome of the ascidian Ciona intestinalis with a transgenic line expressing transposase in eggs. Dev. Dyn. 239: 1076–1088. [DOI] [PubMed] [Google Scholar]
- Kaspar M., Schneider M., Chia W., Klein T., 2008. Klumpfuss is involved in the determination of sensory organ precursors in Drosophila. Dev. Biol. 324: 177–191. [DOI] [PubMed] [Google Scholar]
- Kelso R. J., Buszczak M., Quiñones A. T., Castiblanco C., Mazzalupo S., et al. , 2004. Flytrap, a database documenting a GFP protein-trap insertion screen in Drosophila melanogaster. Nucleic Acids Res. 32: D418–D420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner P., Hung J., Béhague J., Le Gouar M., Balavoine G., et al. , 2009. Insights into the evolution of the snail superfamily from metazoan wide molecular phylogenies and expression data in annelids. BMC Evol. Biol. 9: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y., MacArthur S., Bourgon R., Nix D., Pollard D. A., et al. , 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6(2): e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. G., Chen Y., 2007. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques 43: 649–654. [DOI] [PubMed] [Google Scholar]
- McQuilton P., St Pierre S. E., Thurmond J.; FlyBase Consortium, 2012. FlyBase 101: the basics of navigating FlyBase. Nucleic Acids Res. 40: D706–D714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metaxakis A., Oehler S., Klinakis A., Savakis C., 2005. Minos as a genetic and genomic tool in Drosophila melanogaster. Genetics 171: 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium et al, 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering R. E., Cornejo M., Davis T. N., Del Bianco C., Aster J. C., et al. , 2009. Direct inhibition of the NOTCH transcription factor complex. Nature 462: 182–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X., Daneman R., Zavortink M., Chia W., 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y., Sliz P., Song L., Aster J. C., Blacklow S. C., 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124: 973–983. [DOI] [PubMed] [Google Scholar]
- Neumüller R. A., Wirtz-Peitz F., Lee S., Kwon Y., Buckner M., et al. , 2012. Stringent analysis of gene function and protein-protein interactions using fluorescently tagged genes. Genetics 190: 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Edwards K., 2004. “Marker removal” screen to generate an improved wing disc GAL4 driver. Drosoph. Inf. Serv. 87: 96–99. [Google Scholar]
- Pavlopoulos A., Oehler S., Kapetanaki M. G., Savakis C., 2007. The DNA transposon Minos as a tool for transgenesis and functional genomic analysis in vertebrates and invertebrates. Genome Biol. 8(Suppl 1): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfreundt U., James D. P., Tweedie S., Wilson D., Teichmann S. A., et al. , 2010. FlyTF: improved annotation and enhanced functionality of the Drosophila transcription factor database. Nucleic Acids Res. 38: D443–D447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignoni F., Zipursky S. L., 1997. Induction of Drosophila eye development by decapentaplegic. Development 124: 271–278. [DOI] [PubMed] [Google Scholar]
- Prelich G., 2012. Gene overexpression: uses, mechanisms, and interpretation. Genetics 190: 841–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones-Coello A. T., Petrella L. N., Ayers K., Melillo A., Mazzalupo S., et al. , 2007. Exploring strategies for protein trapping in Drosophila. Genetics 175: 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs B., Rothwell W., Mische S., Hickson G. R., Matheson J., et al. , 2003. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components Nuclear-fallout and Rab11. J. Cell Biol. 163: 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., 1996. A modular misexpression screen in Drosophila detecting tissue-specific phenotypes. Proc. Natl. Acad. Sci. USA 93: 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., Szabo K., Bailey A., Laverty T., Rehm J., et al. , 1998. Systematic gain-of-function genetics in Drosophila. Development 125: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Saito K., Williams S., Bulankina A., Höning S., Mustelin T., 2007. Association of protein-tyrosine phosphatase MEG2 via its Sec14p homology domain with vesicle-trafficking proteins. J. Biol. Chem. 282: 15170–15178. [DOI] [PubMed] [Google Scholar]
- Sasakura Y., Yaguchi J., Yaguchi S., Yajima M., 2010. Excision and transposition activity of Tc1/mariner superfamily transposons in sea urchin embryos. Zoolog. Sci. 27: 256–262. [DOI] [PubMed] [Google Scholar]
- Shiba T., Koga H., Shin H. W., Kawasaki M., Kato R., et al. , 2006. Structural basis for Rab11-dependent membrane recruitment of a family of Rab11-interacting protein 3 (FIP3)/Arfophilin-1. Proc. Natl. Acad. Sci. USA 103: 15416–15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., Bellen H. J., Hoskins R. A., 2011. Drosophila P elements preferentially transpose to replication origins. Proc. Natl. Acad. Sci. USA 108: 15948–15953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardi N. J., Cook M. E., Edwards K. A., 2012. Rapid phenotypic analysis of uncoated Drosophila samples with low-vacuum scanning electron microscopy. Fly (Austin) 6: 184–192. [DOI] [PubMed] [Google Scholar]
- Velichkova M., Juan J., Kadandale P., Jean S., Ribeiro I., et al. , 2010. Drosophila Mtm and class II PI3K coregulate a PI(3)P pool with cortical and endolysosomal functions. J. Cell Biol. 190: 407–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K. J., Bellen H. J., 2012. Genome-wide manipulations of Drosophila melanogaster with transposons, Flp recombinase, and ΦC31 integrase. Methods Mol. Biol. 859: 203–228. [DOI] [PubMed] [Google Scholar]
- Venken K. J., Schulze K. L., Haelterman N. A., Pan H., He Y., et al. , 2011. MiMIC: a highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods 8: 737–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A. E., Pedersen K., Lendahl U., Roeder R. G., 2002. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 22: 7812–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Lan L., Bogard N., Mattione C., Cohen R. S., 2011. Rab11 is required for epithelial cell viability, terminal differentiation, and suppression of tumor-like growth in the Drosophila egg chamber. PLoS ONE 6(5): e20180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.