Abstract
Tn916-like conjugative transposons carrying antibiotic resistance genes are found in a diverse range of bacteria. Orf14 within the conjugation module encodes a bifunctional cell-wall hydrolase CwlT that consists of an N-terminal bacterial lysozyme domain (N-acetylmuramidase, bLysG) and a C-terminal NlpC/P60 domain (γ-d-glutamyl-l-diamino acid endopeptidase) and is expected to play an important role in the spread of the transposons. We determined the crystal structures of two CwlT from pathogens Staphylococcus aureus mu50 (SaCwlT) and Clostridium difficile 630 (CdCwlT). These structures reveal that NlpC/P60 and LysG domains are compact and conserved modules, connected by a short flexible linker. The LysG domain represents a novel family of widely distributed bacterial lysozymes. The overall structure and the active site of bLysG bear significant similarity to other members of the glycoside hydrolase family 23 (GH23), such as the g-type lysozyme (LysG) and Escherichia coli lytic transglycosylase MltE. The active site of bLysG contains a unique structural and sequence signature (DxxQSSES+S) that is important for coordinating a catalytic water. Molecular modeling suggests that the bLysG domain may recognize glycan in a similar manner to MltE. The C-terminal NlpC/P60 domain contains a conserved active site (Cys-His-His-Tyr) that appears to be specific for tetrapeptide. Access to the active site is likely regulated by isomerism of a side chain atop the catalytic cysteine, allowing substrate entry or product release, or closing during catalysis.
Keywords: bifunctional cell-wall lysin, bacterial lysozyme, muramidase, NlpC/P60 endopeptidase, Tn916 family conjugative transposons
Introduction
The Tn916-like family of conjugative transposons consists of a diverse group of modular mobile genetic elements (MGEs) conferring antibiotic (mainly tetracycline) resistance 1; 2; 3. The Tn916 conjugative transposon was originally identified in Enterococcus faecalis strain DS16, and related transposons have since been found in many commensal and pathogenic bacteria, such as Streptococcus pneumoniae (Tn1545), Clostridium difficile (Tn5397), Staphylococcus aureus (Tn5801), Klebsiella pneumonia (Tn6009), and Bacillus subtilis (ICEBs1). These transposons contain genes organized in modules that encode all of the necessary functions for conjugation, recombination (excision and integration), transcriptional regulation and antibiotic resistance (accessory functions). The wide distribution of the Tn916 family of MGEs and their ability to acquire new accessory genes may contribute to the survival of bacteria in certain environments, and the emergence of multidrug resistance.
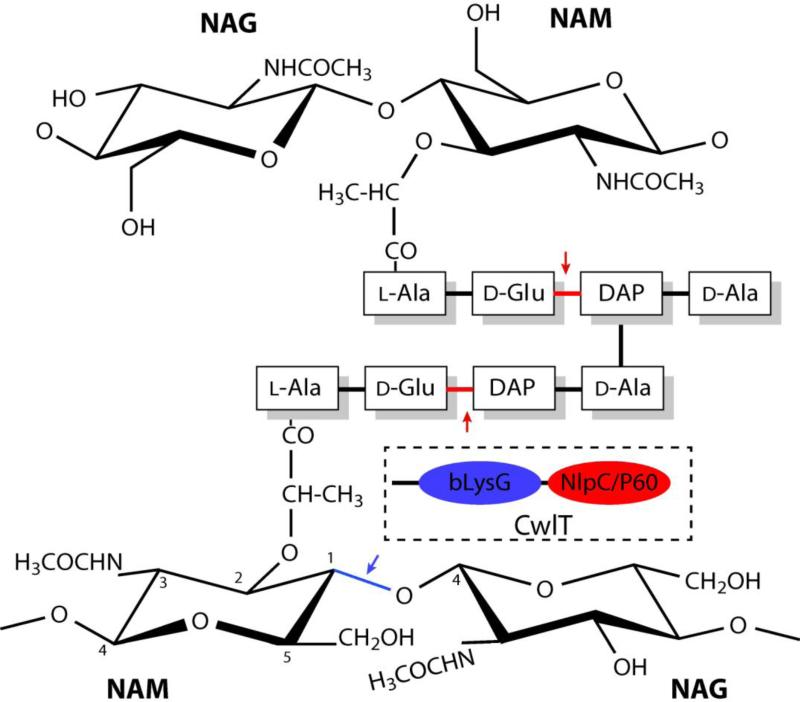
Nearly all Tn916-like MGEs contain Orf14, which encodes a two-domain protein with an N-terminal lysozyme-like domain and a C-terminal NlpC/P60 domain, both of which are involved in degrading cell-wall peptidoglycan. Thus, this protein might facilitate conjugative transfer of transposons by creating a localized opening in the peptidoglycan shield of recipient cells4. Peptidoglycans are composed of glycan strands, oligosaccharides with alternating N-acetylmuramic acid (NAM) and N-acetylglucosamine (NAG) residues, cross-linked by short muropeptides attached to NAM (Fig.1). Biochemical characterization of the B. subtilis Orf14 ortholog5, CwlT [cell wall lytic enzyme T (Two-catalytic domains)], showed that the NlpC/P60 domain is a dl-endopeptidase (or amidase) that hydrolyzes the linkage of d-γ-glutamyl-DAP (meso-diaminopimelic acid), while the g-type lysozyme-like domain (referred as bLysG hereafter, for bacterial lysozyme G) is an N-acetylmuramidase (lysozyme) that hydrolyzes the β(1-4) glycosidic bond between NAM and NAG (Fig.1) and is not a lytic transglycosylase (LT) as previously annotated.
Fig. 1.
Schematic representation of typical bacterial peptidoglycan. The putative cleavage sites by the N-terminal bLysG domain (blue arrow) and the C-terminal NlpC/P60 domain (red arrows) of CwlT are indicated. Muropeptides of B. subtilis and E. coli contain L-Ala-D-Glu-DAP-D-Ala cross-linked between D-Ala and DAP. The muropeptide of S. aureus peptidoglycan consist L-Ala-d-Glu-NH2-L-Lys-D-Ala cross-linked via penta-glycine between D-Ala and L-Lys. A schematic drawing of the domain architecture of CwlT is shown in a box.
However, the sequence of the N-terminal, bLysG domain of CwlT is very distant from g-type lysozyme or LTs sequences and represents a new family of widely distributed cell-wall hydrolases5, containing >1100 members (Pfam family PF13702; version 27.0, March 2013) that are almost exclusively found in Firmicutes (99%). In addition to CwlT orthologs, which constitute ~40% of the members of the PF13702 family, the bLysG domain is present in Pneumococcal vaccine antigen A6, B. subtilis YocA, and other proteins encoded by ORFs such as Lactobacillus plantarum bacteriophage LP657, plasmid pXO1 harboring the anthrax toxin genes in Bacillus cereus8, and vancomycin-resistant operons of E. faecalis9 and S. aureus10. PF13702 is part of the Pfam Lysozyme clan (CL0037), which currently includes 12 families, such as soluble LTs (SLT and SLT2) and several other glycoside hydrolase families (GH19, GH22, GH24, GH46, GH73, and GH108). Although, LTs share the same substrate specificity as lysozymes, they are not hydrolases. They instead cleave the glycosidic linkage between NAM and NAG residues with a concomitant intramolecular transglycosylation reaction, resulting in the formation of the 1,6-anhydro ring at the NAM residue of the product. The catalytic mechanism of the bLysG domain hydrolase activity of CwlT remains to be elucidated.
We have been interested in the structure and function of the NlpC/P60 family of papain-like cell-wall hydrolases11; 12; 13, particularly in enzymes that contain multiple functional modules. Orf14/CwlT homologs were selected for structural studies based on the presence of the novel bLysG domain in addition to the NlpC/P60 domain. Here, we report the crystal structures of two Orf14/CwlT homologs, SaCwlT and CdCwlT, from S. aureus and C. difficile respectively, which are two clinically important Gram-positive pathogens often involved in infections in hospitals or long-term care facilities. SaCwlT (SAV0400, 340aa) is encoded by the Tn5801 transposon14, while CdCwlT (CD0372, 335aa) is encoded by the CTn1 transposon whose accessory module harbors a novel ATP-binding cassette (ABC) transport system. SaCwlT shares 61% sequence identity with CdCwlT, and 41% with the previously characterized B. subtilis CwlT (BsCwlT, 328aa, Fig. 2). The structures indicate that the two catalytic domains of CwlT are coupled with a flexible linker, allowing each domain to function independently of the other. CwlT represents the first structural characterization of a bLysG domain, and reveals that its active site contains unique features distinctive from LTs and previously characterized lysozymes.
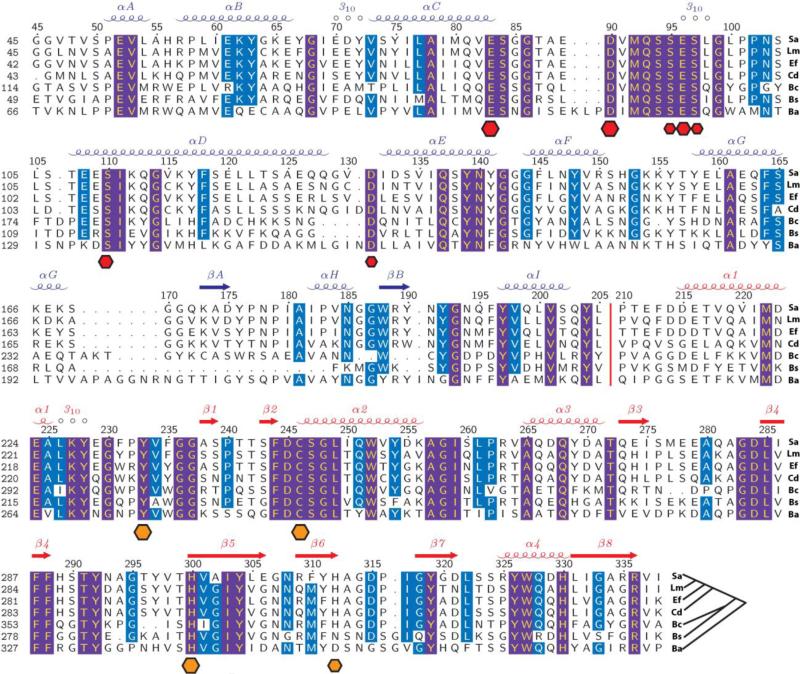
Fig. 2.
Sequence alignment of CwlT homologs from bacteria pathogens. Sa, S. aureus strain Mu50 (SAV0400, SaCwlT); Lm, Listeria monocytogenes serovar 1/2a strain ATCC BAA-679/EGD-e (LMO1104); Ef, E. faecalis (ORF14); Cd, C. difficile 630 (CD0372, CdCwlT); Bc, B. cereus Rock1-15 (BCERE0018_53500); Bs, B. subtilis subsp. subtilis str. 168 (YddH, CwlT); and Ba, B. anthracis strain Ames ‘Ancestor’ Plasmid pXO2 (GBAA_pXO2_0007). An unrooted phylogenetic tree is shown at the end of the alignment. The secondary structure elements of SaCwlT are shown at the top row, colored by domains (bLysG in blue and NlpC/P60 in red). A red vertical bar indicates the location of the flexible linker region between the domains. Sequence numbering is shown at the left column, and SaCwlT numbering is also shown on the top of the sequence. Effects of site-directed mutagenesis in the bLysG domain of BsCwlT5 are shown at the bottom as hexagonal dots (red), whose size correlates with the severity of the loss of activity (i.e. the largest dots denote a complete loss of activity when mutated). Similarly, the catalytic residues of the C-terminal NlpC/P60 domain are marked as orange dots with their putative importance reflected by size (larger means more important). Completely conserved residues in all sequences are shown in yellow on a purple background and highly conserved residues are shown in white on a cyan background.
Results and Discussion
Sequence analysis of CwlT orthologs or paralogs
CwlTs are commonly associated with the Tn916-like family of transposons. For example, the multidrug-resistant pathogen C. difficile strain 630 contains four closely related (sequence identity >65%) CwlT orthologs, CD0372 (CdCwlT), CD3336, CD3380 and CD0504 (Orf14 of Tn5397), each encoded by an ORF in a Tn916-like transposon. CwlT orthologs or paralogs are predominantly found in Firmicutes, and are generally highly conserved. Clustering of unique CwlT orthologs using a 50% sequence identity threshold results in only 12 clusters, with the largest cluster covering 83% of the members. A sequence alignment of SaCwlT, CdCwlT, BsCwlT and a few representatives from other pathogens is shown in Fig. 2. The previously identified5 putative active site residues are strictly conserved in these proteins, indicative of conserved enzymatic functions.
Some CwlT homologs, including BsCwlT, were previously predicted to contain an N-terminal lipoprotein signal peptide. However, sequence analyses using the Phoibus 15 server indicate that ~70% of CwlTs, including those from B. ceres, B. anthrax, B. subtilis and E. faecalis, likely carry an N-terminal trans-membrane helix, rather than a signal peptide. Both SaCwlT and CdCwlT contain predicted N-terminal signal peptides with putative cleavage sites located after residues 28 and 29 respectively. Therefore, for crystallization purposes, the first 28 residues of both proteins were omitted in the cloned constructs.
Structure determination
To increase the chances of obtaining a structure, we initially selected 10 CwlT orthologs or paralogs from S. aureus Mu50, C. difficile 630 (4), Eubacterium ventriosum ATCC 27560, E. faecalis V583 (2), Streptococcus agalactiae 2603V/R, and B. subtilis. These targets were processed using the semi-automated, high-throughput pipeline of the Joint Center for Structural Genomics (JCSG, http://www.jcsg.org)16, as part of the NIH National Institute of General Medical Sciences (NIGMS), Protein Structure Initiative (PSI). The crystal structures of two of these targets were determined. Selenomethionine derivatives of SaCwlT and CdCwlT were expressed in Escherichia coli with an N-terminal, TEV-cleavable, His-tag and purified by metal affinity chromatography. The purification tag was removed prior to crystallization trials. Crystals were harvested from several conditions and screened for diffraction to identify the best crystals for structure determination.
The crystal structure of SaCwlT was determined in space group P21 using the MAD method, and was refined using data up to a resolution of 2.23 Å with an Rcryst of 19.9% and an Rfree of 24.2%. The asymmetric unit (asu) contains two monomers (chains A and B, residues 46-340) and 201 water molecules. The final model (PDB code 4fdy) has good geometry based on Molprobity17 Ramachandran analysis, and all residues are within allowed backbone conformations with 97% in the most favorable regions. The electron density is continuous for the entire main chain and is also good for the majority of the side chains. Only 1.4% of the side-chain conformations are considered rotamer outliers by Molprobity and four surface side chains (A/325, B/70, B/73 and B/306) are partially modeled due to the lack of interpretable electron density. The N-terminal residues 28-45 and a residual glycine from the purification tag are disordered and not included in the final structure.
The structure of CdCwlT was determined in space group P1 by the molecular replacement (MR) method using the SaCwlT structure as the template. The MR search was initiated with the placement of the larger bLysG domain first, followed by the NlpC/P60 domain. The structure was refined using data up to a resolution of 2.38 Å with an Rcryst of 17.6% and an Rfree of 21.1%, and comparable geometrical quality to the SaCwlT model. The final model (PDB code 4hpe) contains six monomers in the asu (chains A-F, residues 43-335), 590 water molecules, four glycerol molecules and nine chloride ions. The N-terminal residues 29-42, the linker between the two domains (residues 206-209), and 28 surface side chains are disordered and not included in the final model. A summary of data collection, processing and refinement statistics of SaCwlT and CdCwlT is shown in Table 1.
Table 1.
Data collection and refinement statistics
| Crystal | SaCwlT (4fdy) | CdCwlT (4hpe) | ||
| Data collection | ||||
| Beamline | SSRL 11-1 | SSRL 12-2 | ||
| Space group | P21 | P1 | ||
| Data | λ1 MADSe | λ2 MADSe | λ3 MADSe | λ1 SADSe |
| Wavelength (Å) | 0.9790 | 0.9184 | 0.9785 | 0.9794 |
| Resolution range (Å) | 47.0-2.23 | 47.0-2.33 | 47.0-2.41 | 49.4-2.38 |
| No. of observations | 114,923 | 97,941 | 91,693 | 293,405 |
| No. of unique reflections | 33,428 | 28,768 | 26,611 | 76,631 |
| Completeness (%)a | 95.4 | 93.7 (83.5) | 95.8 (95.2) | 87.6 (84.5) |
| Mean I/σ (I)a | 12.9 (1.8) | 14.0 (2.2) | 19.2 (2.2) | 9.0 (2.3) |
| Rmerge on Ia (%) | 5.2 (68) | 5.0 (49) | 4.9 (56) | 10.9 (58) |
| Rmeas on Ia (%) | 6.2 (80) | 6.0 (59) | 5.8 (66) | 12.8 (68) |
| Rpim on Ia (%) | 3.3 (42) | 3.2 (31) | 3.1 (35) | 6.5 (34) |
| Highest resolution shell | 2.35-2.23 | 2.46-2.33 | 2.54-2.41 | 2.51-2.38 |
| Model and refinement statistics | ||||
| Data used in refinement | λ1 MADSe | λ1 SADSe | ||
| No. of reflections (total) | 33,396 | 76,620 | ||
| No. of reflections (test) | 1677 | 3897 | ||
| Cutoff criteria | |F|>0 | |F|>0 | ||
| Rcryst (%) | 19.9 | 17.6 | ||
| Rfree (%) | 24.2 | 21.1 | ||
| Stereochemical parameters | ||||
| Restraints (RMSD observed) | ||||
| Bond lengths (Å) | 0.010 | 0.010 | ||
| Bond angles (°) | 1.04 | 1.03 | ||
| MolProbity all atom clash score | 7.2 | 4.3 | ||
| Ramachandran plot (%)b | 97.1 (0) | 97.7 (0) | ||
| Rotamer outlier (%) | 1.4 | 0.6 | ||
| Average isotropic B-value (Å2)c | 65.5 (53.5) | 50.1 (48.9) | ||
| ESU based on Rfree (Å) | 0.21 | 0.22 | ||
| No. protein residues / chains | 590/2 | 1735/6 | ||
| Non-protein entities | 201 H2O | 590 H2O, 4 GOL, 9 CL | ||
Highest resolution shell in parentheses.
Percentage of residues in favored regions of Ramachandran plot (No. outliers in parenthesis).
This value represents the total B that includes TLS and residual B components (Wilson B-value in parenthesis). The average B-values of the protein and the solvents included in the refinement are very similar.
ESU = Estimated Standard Uncertainty in coordinates.
Rmerge=ΣhklΣi|Ii(hkl)-<I(hkl)>|/ΣhklΣiIi(hkl), Rmeas(redundancy-independent Rmerge) = Σhkl[Nhkl/(Nhkl-1)]1/2Σi|Ii(hkl)-<I(hkl)>|/ΣhklΣiIi(hkl), and Rpim(precision-indicating Rmerge)=Σhkl[1/(Nhkl-1)]1/2Σi|Ii(hkl)-<I(hkl)>|/ΣhklΣiIi(hkl).
Rcryst =Σ| |Fobs|-|Fcalc| |/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rcryst, but for 5.0% of the total reflections chosen at random and omitted from refinement.
Two enzymes linked by a short flexible peptide
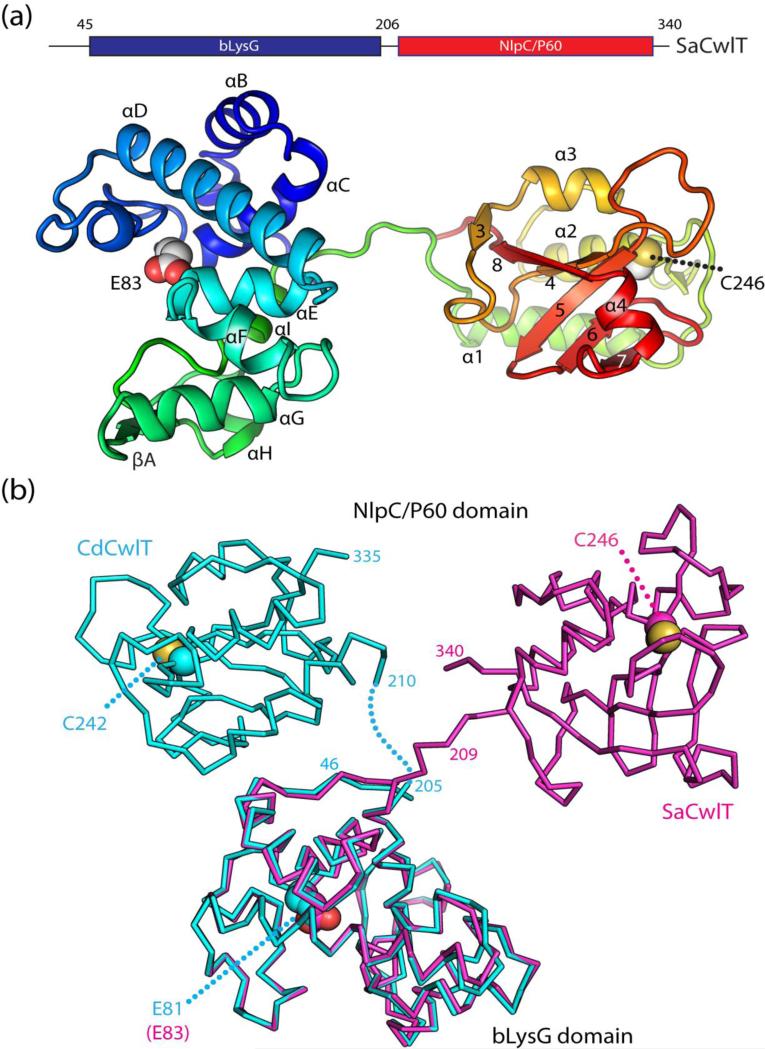
Analysis of the protein-protein interfaces in SaCwlT or CdCwlT crystal lattice using the PISA server18 indicates that there is no strong association between the monomers, suggesting each protein likely exists as a monomer in solution. In both cases, the monomers of SaCwlT (or CdCwlT) in the asu are almost identical to each other with RMSD of 0.72 Å for 295 Cα atoms (average pairwise RMSD of 0.39 Å for 289 Cα atoms for CdCwlT). The CwlT structures consist of two domains (Fig. 3a): an N-terminal helical bLysG domain (residues 46-206 for SaCwlT, 43-205 for CdCwlT) and a C-terminal NlpC/P60 domain with an α/β/α fold (residues 210-340 for SaCwlT, 210-335 for CdCwlT). The respective bLysG and NlpC/P60 domains are structurally very similar in SaCwlT and CdCwlT (RMSDs ~0.7 Å for both, Fig. S1). The two functional domains are coupled by a short flexible linker, which is ordered in SaCwlT, presumably due to crystal packing interactions, but is disordered in CdCwlT where it is not involved in packing. The putative general acid of bLysG is a glutamate (Glu83 in SaCwlT and Glu81 in CdCwlT). Two glycerol molecules, from the cryoprotectant, are found in the bLysG active site groove of one CdCwlT monomer. The catalytic cysteine of the NlpC/P60 domains, corresponding to Cys246 of SaCwlT and Cys242 of CdCwlT, are both oxidized based on the electron density (Fig. S2). The structure of CwlT is one of the few structurally characterized cell-wall lysins with two active catalytic subunits; the structure of another multi-subunit lysin, PlyC, was recently determined19. A combination of multiple cell-wall degrading modules may increase the potency against the bacterial cell wall19, which could be exploited to develop approaches to control pathogenic antibiotic resistant bacteria20.
Fig. 3.
Crystal structures of SaCwlT and CdCwlT. (a) Ribbon representation of SaCwlT colored as a gradient from the N-terminus (blue) to the C-terminus (red). The secondary structure elements are labeled as defined in Fig. 2. Catalytic residues (Glu83: general acid and Cys246: nucleophile) are shown as spheres. (b) Comparison of SaCwlT (magenta) and CdCwlT (cyan). SaCwlT and CdCwlT are superimposed based on the bLysG domain.
The relative orientation of the two domains in SaCwlT and CdCwlT differs significantly, most likely a result of differences in crystal packing (Fig. 3b). The two domains in SaCwlT are distal to each other without direct contacts, while the two domains in CdCwlT are proximal to each other with a total buried interface of 580 Å2. When their bLysG domains are superimposed, the distance between the two catalytic cysteines of the NlpC/P60 domains is ~52 Å. The active sites of the two catalytic domains are 52 Å apart in SaCwlT, compared to 32 Å in CdCwlT. Nonetheless, the active site of each catalytic domain is unaffected by the other domain or other molecules in the asu and is fully accessible in both SaCwlT and CdCwlT. Furthermore, the N- and C- termini are distal to the active sites in the bLysG and the NlpC/P60 domains, respectively (Fig. 3). It is geometrically impossible for the two active sites within a molecule to come into close proximity to each other due to the short length of the linker, which may prevent interference between the two catalytic domains. Thus, the two enzyme activities within a SaCwlT or CdCwlT monomer are expected to function independently of each other. Due to crystal packing, both active sites are accessible in the SaCwlT crystal, but inaccessible in the CdCwlT crystal (Fig. S3). Due to the highly conserved nature of the individual modules, hereafter we will focus our analysis on SaCwlT unless specified.
The bLysG domain
The bLysG domain (Figs. 3 and 4) is an α-helical protein (αA-αI) consisting of two lobes. The N-terminal lobe consists of four helices (αA-αD), while the C-terminal lobe consist of five helices (αE-αI), and a small two-stranded β-sheet (βA and βB) between helices αG and αI. The longest helix in the structure, αD, connects the two lobes. Notably, the bLysG domain has a large groove traversing one face of the molecule.
Fig. 4.
The bLysG domain has a g-type lysozyme fold. (a) Comparison SaCwlT-bLysG with MltE (PDB code 4hjv) and g-type lysozyme (PDB code 3gxk). The three proteins are shown in a similar orientation with common secondary structures colored in green, while the conserved residues in the active site are shown as sticks (pink). Two catalytically important residues in bLysG (Asp90 and Glu96), and their counterparts in the other two proteins are colored in blue. (b) Surface representation of the substrate binding groove on SaCwlT-bLysG, MltE and the g-type lysozyme. The penta-NAG ligand bound to MltE is shown as sticks.
The overall structure and the active site of bLysG resemble that of E. coli LT MltE and LysGs (Fig. 4a and Fig. S4). The bLysG domain of SaCwlT can be superposed onto MltE (PDB code 3t36)21 with an RMSD of 3.0 Å and sequence identity of 24% for 130 aligned Cα atoms (Dali22 Z-score 12.4), and onto LysG from Atlantic cod (PDB code 3gxk)23 with an RMSD of 2.9 Å and sequence identity of 18% for 119 Cα atoms (Z-score 9.9). The common secondary structure elements include helices αB-αG and αI (Figs. 4a). The glutamate residue expected to function as the general acid during catalysis corresponds to Glu83 of SaCwlT, consistent with experimental evidence that showed the equivalent glutamate (Glu87) in BsCwlT (Fig. 2) is critical for enzymatic activity5. Glu83 of SaCwlT is located at the C-terminus of αC and adopts a conformation identical to that of Glu64 in MltE and Glu73 in LysG, and is stabilized by a hydrogen bond between the Oε2 atom and the Oη atom of a conserved tyrosine (Tyr197 of SaCwlT). In addition, a few residues near the catalytic glutamate (Ser84, Gln93, Tyr139, and Tyr197 of SaCwlT) are also strictly conserved among bLysG, MltE and LysG. Overall, bLysG appears to be phylogenetically closer to MltE (Fig. S5)24.
bLysG and LysG are muramidases, while MltE is a lytic transglycosylase. The difference in enzymatic activity can be attributed to structural variations in the active sites. Notably, an asparagine (Asn140 of SaCwlT) near the general acid (Glu83 of SaCwlT) is conserved in both bLysG and LysG. Also, Glu96 of SaCwlT is located in a similar location as Asp101 of LysG, which is essential for the enzymatic activity23, while a lysine is located the equivalent position of MltE (Figs. 4a). Lysozymes have a conserved β-hairpin at the N-terminal lobe that carries a family-specific sequence signature important for catalysis25. This hairpin is absent in bLysG, and the equivalent region (residues 90-103 in SaCwlT) adopts a conformation similar to that of MltE, which contains loops and helices. Nonetheless, SaCwlT also carries a sequence motif specific to PF13072 in this region that contributes a third carboxylate group (Asp90) to the active site (see below).
Active site groove and substrate recognition by bLysG
The active site groove of bLysG is fully accessible from both ends (Fig. 4b), suggestive of endo-acting activity. Active site residues that are conserved among bLysG, MltE and LysG are located at the bottom of the groove, while the walls of the groove differ more significantly. The C-terminal end of the groove is formed by conserved helices αE-αF, and αI, and is further extended by a βαβ insertion (βA-αH-βB) between αG and αI (Fig. 4a). Both MltE and LysG lack the βαβ insertion, but have other structural elements that occupy an equivalent space, and as a result, the C-terminal end of the groove of SaCwlT traverses a similar span compared to the groove in MltE or LysG. On the other hand, an insertion between αC and αD of SaCwlT (residues 84-106), which contributes to the opposite side of the active site groove, is shorter than that of MltE (residue 66-110) or LysG (residues 75-113). As a result, SaCwlT appears to be less restrictive at both ends of the groove (Fig. 4b). The two lysozymes, LysG and bLysG, possess a wider groove at one end compared to MltE (Fig. 4b).
Individual units of the polysaccharide substrate of lysozymes and other glycosyl hydrolases are commonly assigned as positions −i through +j (the reducing end), with the saccharide units flanking the scissile glycosidic bond designated as positions −1 and +1. Structural comparison between bLysG and MltE suggest that bLysG possesses similar binding sites to the six subsites (−4 to +2) identified in MltE, in particular at −2,−1 and +1. Thus, it is expected that the bLysG domain recognizes saccharide units in a similar manner as MltE. In order to further investigate substrate recognition by the bLysG domain, we computationally docked an eight-saccharide glycan into the active site of SaCwlT (Fig. 5). Interestingly, a glycerol molecule in the active site of CdCwlT interacts with the protein in a similar manner as predicted by our model (Fig. 6a), suggesting that it might mimic the substrate conformation in the productive binding state.
Fig. 5.
Substrate recognition by SaCwlT-bLysG. (a) A computational model of an eight-unit glycan (stick, green/gray) bound to the active site groove of the SaCwlT-bLysG. The protein surface is colored by a gradient based on sequence conservation ranging from non-conserved (blue) to strictly conserved residues (green). Ligand (shown as sticks) atoms that are within 4.5 Å of the protein are colored red. Individual saccharide positions are labeled from −4 to +4. (b) A schematic representation of the predicted protein-ligand interactions. Hydrogen bonds and salt bridges are shown as dashed lines (red).
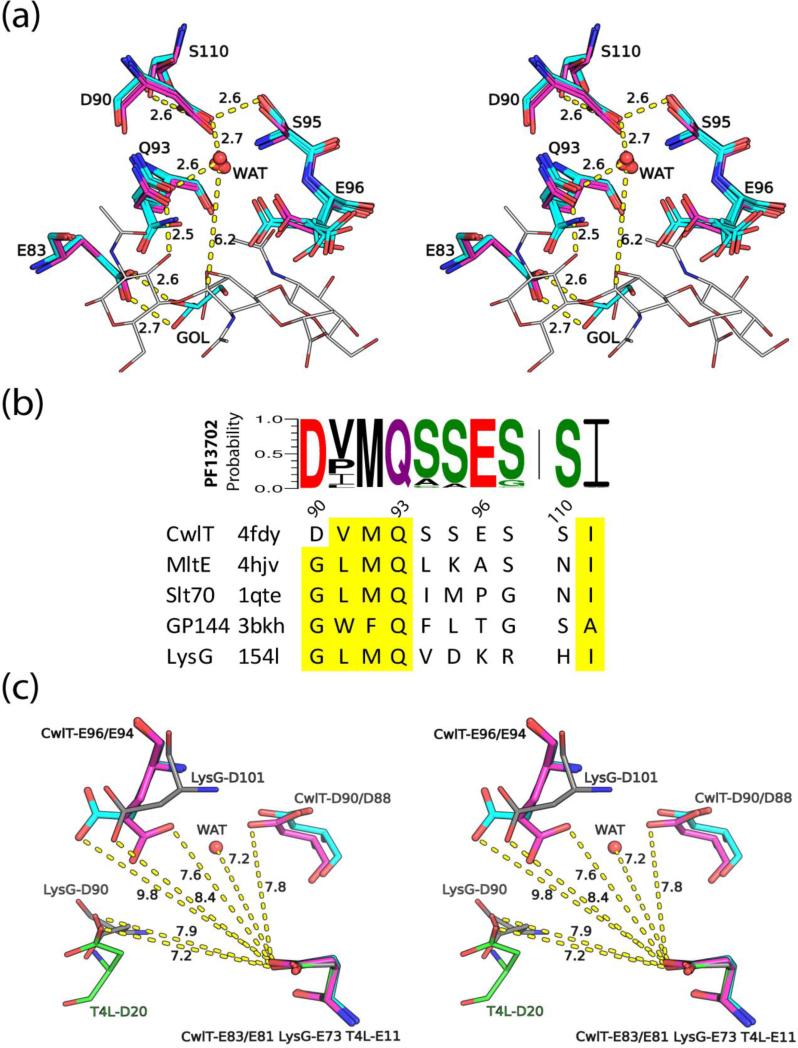
Fig. 6.
The active site and a proposed catalytic mechanism for bLysG. (a) A stereoview of the superposed bLysG domain active sites of SaCwlT (magenta/blue/red, two monomers) and CdCwlT (cyan/blue/red, six monomers in the asu). Residue numbering is based on SaCwlT. Distances are labeled near dashed lines. The putative catalytic water (WAT) and a glycerol molecule (GOL) in the active site of CdCwlT are also shown as spheres and sticks respectively. Part of the modeled glycan ligand is shown as thin sticks (gray). (b) Sequence motif for residues around Asp90, derived from sequence alignment of all family members of PF13702. The height of a stacked letter in a column is proportional to the probability of occurrence of that residue at that position. Equivalent residues of other GH23 members are shown below (conserved residues highlighted in yellow). (c) Stereoview of the arrangement of carboxylate groups in the active sites of SaCwlT (magenta), CdCwlT (cyan), T4L (green, PDB code 4lzm) and LysG (gray, PDB code 3gxk). All structures are superposed based on the common proton donor (Glu83 of SaCwlT).
The active site groove of SaCwlT can accommodate a glycan of up to eight saccharide units (−4 to +4). The modeled glycan fits well into the groove and interacts favorably with the protein, through hydrophobic contacts as well as hydrogen bonds (Fig. 5). The protein residues involved in substrate binding are highly conserved among CwlT homologs, particularly from position −3 to +3 (Fig. 5a). The glycan makes most contacts with the protein at positions −3 to +2, while additional weaker interactions are also predicted for −4 and +3 positions. The −2 NAG interacts most extensively with the protein, where the groove is most restrictive. The N-acetyl group of the −2 NAG of the modeled glycan is inserted in a hole at the bottom of the groove, where it forms two hydrogen bonds with Ser97N and Tyr139O (carbonyl oxygen) (Fig. 5b), as well as hydrophobic interactions with the side chain of Tyr139. Overall, these interactions define a NAG-specific subsite, which is required for the correct placement of the cleavable bond. The subsite for binding the saccharide at the +1 position is also specific to NAG. The saccharide ring here is rotated by ~90° relative to the previous position, facilitated by the wider groove here. The 3-hydroxyl group is close to Gln93Oε1, leaving no space to accommodate the lactic acid moiety on the NAM. The lactic acid group of NAM at the +2 position interacts with the guanidinium group of Arg189, suggesting this subsite may have greater affinity for binding NAM.
The modeled glycan stayed bound to the enzyme during the entire molecular dynamics simulation (MD). The middle section of the ligand displays much smaller movements compared to the moieties at either end, correlated with ligand-protein interactions at each subsite and steric constraints imposed by the protein. The −1 saccharide always quickly converts to the low energy chair conformation (~50ps) during MD, which is accompanied by a shift in positions of sugars from +1 to +4 positions (Fig. S6). The larger shifts at the + positions between non-productive and productive state was also previously observed in MltE (Fig. S6)21. These results suggest that ligand-protein interactions at the downstream (+) sites play a critical role in maintaining the productive conformation at the −1 position.
Catalytic center and putative catalytic mechanism of bLysG
A site-directed mutagenesis study of BsCwlT identified Glu87 and Asp94 (corresponding to Glu83 and Asp90 of SaCwlT, Fig. 2) as most critical to the enzymatic activity5. Mutation of either of these residues into glutamine and asparagine respectively rendered BsCwlT inactive. Mutation of three other residues, Glu100 (E100Q), Ser99 (S99A) and Ser115 (S110A) (corresponding to Glu96, Ser95 and Ser110 of SaCwlT) also reduced the activity of BsCwlT by 90%, 70% and 85%, respectively suggesting that these residues also play an important role in catalysis. Interestingly, these residues are mapped to a single location near the general acid in the crystal structures of SaCwlT (Glu83) and CdCwlT (Glu81, Fig. 6a) with most of the residues located in a U-loop between αC and αD that harbors a short 310 helix (Fig. 2). The Oδ1 atom of Asp90 of SaCwlT forms hydrogen bonds with the side chains of Ser95 and Ser110, while its Oδ2 atom is exposed to solvent (Fig. 6a). Asp90 and nearby residues Gln93 and Glu96 form a small empty cavity (Fig. 5a). Interestingly, a water molecule is found in this cavity in CdCwlT, stabilized by hydrogen bonds with the side chains of three equivalent residues. Modeling studies indicated that this water is aligned to attack the anomeric carbon C1 atom from the α-side of the −1 NAM (distance ~6.0 Å, Fig. 6a), and thus could play a catalytic role. Two other nearby residues Gln93 and Glu96 could also interact with the water. Crystal structures of SaCwlT and CdCwlT show that both side chains can adopt alternate conformers, indicating they are mobile in the absence of ligand. The residues around Asp90 are highly conserved, in particular the sequence motif 90DvmQsses97+S110 (residues in upper case are strictly conserved, while residues in lower case are highly conserved) is identified in members of Pfam PF13072 (Fig. 6b). In contrast, a similar motif GhhQ (h-hydrophobic) is present in other structural homologs25, replacing the first aspartate in CwlT with a conserved glycine (Fig. 6b).
Two reaction mechanisms are most commonly suggested for lysozymes. Inverting enzymes employ a single-displacement mechanism that results in a net inversion of an anomeric carbon configuration, while retaining enzymes use a double-displacement mechanism with a net retention of the substrate configuration26. Both mechanisms involve two carboxyl groups in the active site, typically from glutamate or aspartate residues. The distance between the two carboxylates is typically ~5 Å in retaining enzymes26. The two carboxylates are further apart in inverting enzymes (6-11 Å), where one carboxyl acid acts as an acid to deprotonate the leaving group and the other as a base to activate a water molecule (nucleophile). While the general acid is strictly conserved in inverting enzymes, the general base residues that are involved in activating the hydrolyzing water molecule are not well conserved structurally. The distance between Glu83 and Asp90 is 7.8 Å, and 7.6 Å, between Glu83 and Glu96 (Fig. 6c). The arrangement of carboxylate groups in the active site of SaCwlT bears some similarity to LysG (Glu83-Glu96 of SaCwlT and Glu73-Asp101 of LysG, Fig. 6c). However, the third carboxylate group is different in SaCwlT from that of LysG 23 and T4 lysozyme T4L27, whereas Asp90 of SaCwlT is ~10 Å away from that of Asp20 of T4L or Asp90 of LysG. We speculate that SaCwlT may be an inverting enzyme where Glu83 functions as the general acid and Asp90 as the general base (Fig. S7), although further experiments are necessary to assign a reaction mechanism.
Lysozymes are one of the best-studied families of enzymes, and are found in all kingdoms of life. The bLysG domain characterized here is distinctive from other bacterial lysozymes, such as Chalaropsis lysozyme that have a β/α-barrel fold28 or pesticin T4L29. The overall structure and active site of bLysG share similarity with other members of the SLT family, such as LysG23, MltE21 and Slt7030. These results support the assignment of bLysG as a member of the glycoside hydrolase family 23 (GH23) in the Carbohydrate Active enZyme (CAZy) database (http://www.cazy.org/), which contains both Pfam SLT and bLysG families. However, bLysG contains a family-specific sequence motif that defines a site likely involved in the coordination of a nucleophilic water molecule, which is essential for the lysozyme activity of CwlT. Thus, bLysG represents a novel bacterial lysozyme.
Gated access to the active site of the NlpC/P60 endopeptidase
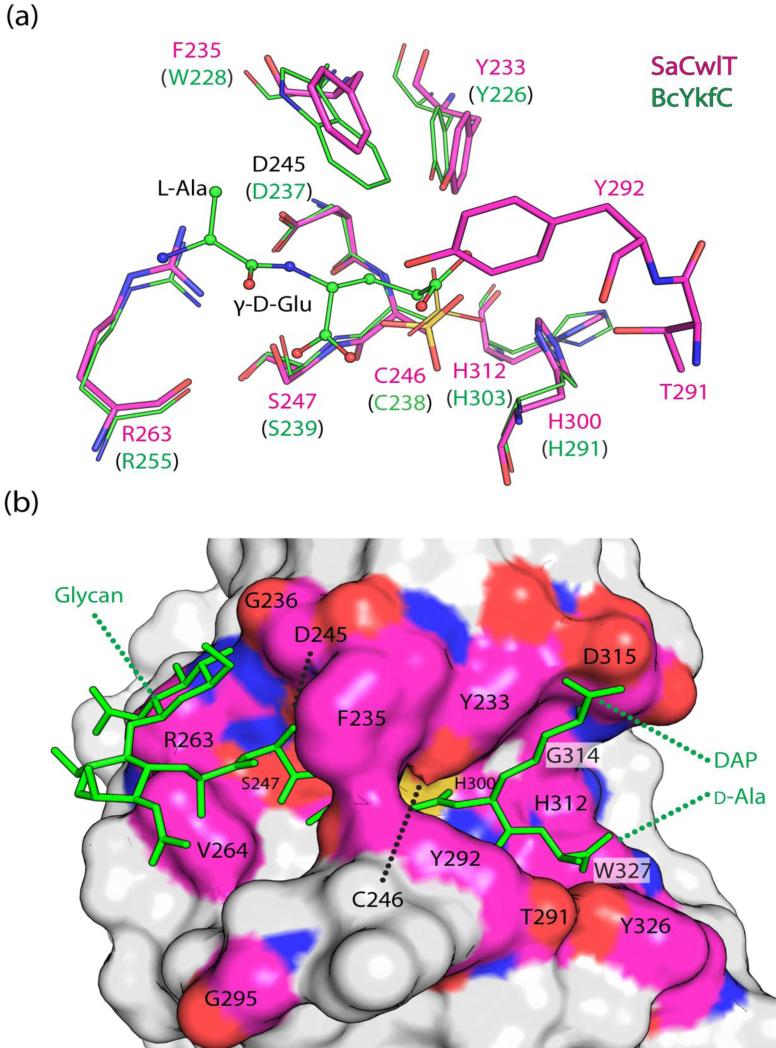
The C-terminal NlpC/P60 domain of BsCwlT can hydrolyze the γ-d-Glu-DAP linkage of peptidoglycan5. The NlpC/P60 domains of SaCwlT and CdCwlT are expected to have similar activity due to strictly conserved nature of the active site residues (Fig. 2). The NlpC/P60 domain of SaCwlT has a three-layered α/β/α fold consisting of a central six-stranded β-sheet (β3, β8, β4, β5, β6, and β7) protected on both sides by α-helices, with α1-α3 on one side and α4 on the other. The catalytic cysteine is located at the N-terminus of α2 (Fig. 3). The catalytic triad consists of Cys246, His300 and His312 (Fig. 7a). Sequence and structural studies have revealed that similar NlpC/P60 domains are prototypical papain-like cysteine peptidases or amidases 11; 31; 32. Both the overall structure and the NlpC/P60 active site of SaCwlT are similar to previously characterized NlpC/P60 domains (Fig. S8), most notably to the catalytic domain of BcYkfC, a γ-d-Glu-DAP(Lys) endopeptidase that is specific for muropeptides with a free N-terminal amino l-Ala 12. These two domains can be superposed with an RMSD of 1.5 Å and sequence identity of 30% for 118 aligned Cα atoms.
Fig. 7.
Active site of the NlpC/P60 domain. (a) Comparison of the active sites of SaCwlT-NlpC/P60 (magenta/red/blue) and BcYkfC (PDB code 3h41, green, labels in parentheses). The reaction product Ala-γ-D-Glu bound to BcYkfC is shown as ball-and-sticks. (b) Surface representation of the SaCwlT-NlpC/P60 active site. A modeled NAG-NAM-tetrapeptide is shown as green stick.
NlpC/P60 domains can specifically recognize l-Ala and γ-d-Glu involving conserved residues around the catalytic cysteine12. An aspartate, a serine and a tyrosine are involved in hydrogen bond interactions with the NH group, the carboxyl, and the carbonyl groups of γ-d-Glu respectively (Fig. 7a). These residues (Tyr233, Asp245, and Ser247) of SaCwlT, as well as the catalytic triad, are strictly conserved between SaCwlT and BcYkfC. A tryptophan side chain of BcYkfC (Trp228) interacts with Cβ atom of l-Ala, thus providing substrate side-chain specificity at this location. Interestingly, the equivalent Phe235 in SaCwlT (Tyr231 in CdCwT) has the same conserved side-chain orientation as Trp228 of YkfC (Fig. 7a). A large channel with a wide opening at either end runs across the catalytic triad (Fig. 7b). To further investigate the substrate binding specificity of the NlpC/P60 domain, we docked an NAG-NAM-tetrapeptide (Fig 1) into the active site of SaCwlT-NlpC/P60 assuming the substrate recognition for l-Ala-γ-d-Glu is conserved (Fig 7b). The docked substrate fits the active site groove reasonably well, and is stabilized by both hydrogen bonding and hydrophobic interactions (Fig. S9). Conserved residues Asp315, Tyr233, Thr291, Tyr292, and Asp315 interact with d-Ala and DAP, while the glycan strand is accommodated by the flat surface at the other end of the active site groove with no specific interactions (Fig. 7b). These results support our hypotheses that the CwlT-NlpC/P60 domain recognizes tetrapeptides and hydrolyze cross-linked peptidoglycan.
Phe235 and Tyr292 of SaCwlT, from the α1-α2 loop and the β4-β5 loop respectively (Fig. 7), define the narrowest part of the active site channel, which in the crystal structures is too narrow for the substrate to enter, indicating that conformational flexibility is necessary for the enzyme to function. A structural comparison between SaCwlT and CdCwlT indicates that these two loops, most notably the β4-β5 loop, exhibit significant structural differences (Fig. S10), which suggests that they are flexible. In particular, Phe235 likely has multiple side-chain conformations as it has weak density. A more accessible catalytic center is likely achieved through side-chain isomerism of the solvent-exposed Phe235, which likely switches to a different rotamer to expose the catalytic center for substrate binding or product release11.
Concluding comments
CwlT is a bifunctional cell-wall lysin encoded by transposons conferring antibiotic resistance in many bacteria. The crystal structures of CwlT from two pathogens reveal modular domain architecture that allow each domain with a conserved active site to function independently of each other. bLysG shares structural similarities to both lytic transglycosylases and lysozyme G despite very low sequence identities. More importantly, the structures reveal novel active-site features not present in other enzymes. bLysG defines a large, widespread bacterial protein family with >1100 members, and may play an important role in bacterial pathogenesis.
Material and Methods
Protein production
Clones were generated using the Polymerase Incomplete Primer Extension (PIPE) cloning method 33. The gene encoding CdCwlT (GenBank; YP_001086839, UniProt: Q18DB2) was amplified by polymerase chain reaction (PCR) from Clostridium difficile genomic DNA using PfuTurbo DNA polymerase (Stratagene) and I-PIPE (Insert) primers (forward primer, 5’-ctgtacttccagggcGCGGACAGCGACGACGAGAACAGCAAC-3’; reverse primer, 5’-aattaagtcgcgttaTTGTTTAACTCGTCCTGCACCGATTAAG-3’, target sequence in upper case) that included sequences for the predicted 5′ and 3′ ends. The gene encoding SaCwlT was amplified by polymerase chain reaction (PCR) from Staphylococcus aureus genomic DNA using PfuTurbo DNA polymerase (Stratagene) and I-PIPE (Insert) primers (forward primer, 5’-ctgtacttccagggcGATGATACGGACAGTGGCGAAAACAAC-3’; reverse primer, 5’-aattaagtcgcgttaGTTATGAATAACACGTCTAGCCCCAATC-3’, target sequence in upper case) that included sequences for the predicted 5′ and 3′ ends. The expression vector, pSpeedET, which encodes an amino-terminal tobacco etch virus (TEV) protease-cleavable expression and purification tag (MGSDKIHHHHHHENLYFQ/G), was PCR amplified with V-PIPE (Vector) primers (forward primer: 5’-taacgcgacttaattaactcgtttaaacggtctccagc-3’, reverse primer: 5’-gccctggaagtacaggttttcgtgatgatgatgatgatg-3’). V-PIPE and I-PIPE PCR products were mixed to anneal the amplified DNA fragments together. Escherichia coli GeneHogs (Invitrogen) competent cells were transformed with the I-PIPE / V-PIPE mixture and dispensed on selective LB-agar plates. The cloning junctions were confirmed by DNA sequencing. Using the PIPE method, the gene segment encoding residues M1-S28 for CdCwlT and M1-A28 for SaCwlT were deleted for expression of soluble protein because they were predicted to correspond to a signal peptide using SignalP and Phobius 15; 34. Expression was performed in a selenomethionine-containing medium at 37°C. Selenomethionine was incorporated via inhibition of methionine biosynthesis35, which does not require a methionine auxotrophic strain. At the end of fermentation, lysozyme was added to the culture to a final concentration of 250 μg/ml, and the cells were harvested and frozen. After one freeze/thaw cycle, the cells were homogenized and sonicated in lysis buffer [40 mM Tris, 300 mM NaCl, 10 mM imidazole, 1 mM Tris(2-carboxyethyl)phosphine-HCl (TCEP), pH 8.0]. Any remaining nucleic acids were digested with the addition of 0.4 mM MgCl2 and 1 μL of 250 U/μL Benzonase (Sigma) to the lysate. The lysate was clarified by centrifugation at 32,500 × g for 25 minutes. The soluble fraction was passed over nickel-chelating resin (GE Healthcare) pre-equilibrated with lysis buffer, the resin washed with wash buffer [40 mM Tris, 300 mM NaCl, 40 mM imidazole, 10% (v/v) glycerol, 1 mM TCEP, pH 8.0], and the protein was eluted with elution buffer [20 mM Tris, 300 mM imidazole, 10% (v/v) glycerol, 150 mM NaCl, 1 mM TCEP, pH 8.0]. The eluate was buffer exchanged with TEV buffer [20 mM Tris, 150 mM NaCl, 30 mM imidazole, 1 mM TCEP, pH 8.0] using a PD-10 column (GE Healthcare), and incubated with 1mg of TEV protease per 15 mg of eluted protein for 2 hours at ambient temperature followed by overnight at 4°C. The protease-treated eluate passed over nickel-chelating resin (GE Healthcare) pre-equilibrated with crystallization buffer [20 mM Tris, 150 mM NaCl, 30 mM imidazole, 1 mM TCEP, pH 8.0] and the resin was washed with the same buffer. The flow-through and wash fractions were combined and concentrated to 19 mg/ml for CdCwlT and 21 mg/ml for SaCwlT by centrifugal ultrafiltration (Millipore) for crystallization trials.
Crystallization
CdCwlT and SaCwlT were crystallized using the nanodroplet vapor diffusion method 36 with standard JCSG crystallization protocols37. Sitting drops composed of 100 nl protein solution mixed with 100 nl crystallization solution in a sitting drop format were equilibrated against a 50 μl reservoir at 277 K. The CdCwlT crystallization reagent consisted of 20% 2-propanol, 24% polyethylene glycol 4000, 0.1 M sodium citrate - citric acid pH 5.1. Glycerol was added to a final concentration of 10% (v/v) as a cryoprotectant. The SaCwlT crystallization reagent consisted of 0.2M calcium acetate, 20% polyethylene glycol 8000, 0.1M MES pH 6.0. Glycerol was added to a final concentration of 15% (v/v) as a cryoprotectant. Initial screening for diffraction was carried out using the Stanford Automated Mounting system (SAM)38 at the Stanford Synchrotron Radiation Lightsource (SSRL, Menlo Park, CA). CdCwlT diffraction data were indexed in triclinic space group P1 with unit cell dimensions as a=65.5, b=101.2, c=101.4 Å, α=109.7°, β=108.9°, and γ=101.4°. SaCwlT diffraction data were indexed in monoclinic space group P21 with unit cell a=40.7, b=72.0, c=124.9 Å, and β=95.9°.
Data collection, structure solution, and refinement
For SaCwlT MAD data were collected at wavelengths corresponding to the peak, inflection, and high energy remote of a selenium MAD experiment at 100 K using a Pilatus 6M detector (DECTRIS) at SSRL beamline BL11-1. For CdCwlT, SAD data were collected at the selenium edge using a Pilatus 6M detector (DECTRIS) at SSRL beamline BL12-2. Data processing and structure solution were carried out using an automated structure determination protocols developed at the JCSG39. In summary, the data were processed using XDS40. The structure of SaCwlT was determined using the MAD method, where the location of selenium sites, initial phasing, and identification of the space group were carried out using SHELXD 41. Phase refinement and initial model building were performed using autoSHARP42 and BUCCANEER43. Attempts at experimental phasing of CdCwlT by the SAD method were not successful, likely due to radiation decay of the anomalous signal over the long course of the data collection in space group P1. The structure of CdCwlT was therefore determined by molecular replacement (MR) as implemented in PHASER44 using the SaCwlT structure as the MR template. Anomalous difference maps calculated with the MR model phases clearly indicated the presence of Se sites (highest peak ~27 σ); however, the quality of the experimental SAD phases was very poor and the resulting map was uninterpretable. Model completion and refinement were performed manually with COOT45 and BUSTER46. The refinement included experimental phase restraints in the form of Hendrickson-Lattman coefficients (SaCwlT), TLS refinement with one TLS group per domain, and NCS LSSR restraints (Local Structure Similarity Restraints, BUSTER). Data and refinement statistics are summarized in Table 1. Analysis of the stereochemical quality of the model was accomplished using MolProbity47. Molecular graphics were prepared with PyMOL (http://www.pymol.org/).
Molecular modeling and molecular dynamics simulation
A (NAG-NAM)4-bound SaCwlT model was obtained as follows. An initial glycan model was created in CS ChemOffice Suite 12.0 (Cambridge Science Computing Inc). Ligand geometry restraints and topology then were generated with the PRODRG server48. The glycan ligand was initially modeled into the active site of MltE such that the saccharide residues at subsites −4 to +2 adopted very similar conformations as known ligands (chitopentaose, bulgecin and murodipeptide) bound to MltE 21 (PDB codes 4hjv and 4hjz). SaCwlT-bLysG was then superposed onto MltE creating the initial complex between the glycan ligand and CwlT. The saccharide residues at these subsites form favorable interactions with the protein with no significant steric clashes, consistent with a conserved mode of substrate recognition at these subsites in CwlT and MltE. Next, the NAG and NAM residues at position +3 and +4 were added to fill the SaCwlT-bLysG active site channel and adjusted manually using COOT to optimize protein/ligand interactions (hydrogen bonds and van der waals interactions). All saccharides were modeled as the low-energy chair conformation, except for position −1 that was modeled as a distorted sofa conformation similar to the conformation of the glycopeptide bulgecin A in the MltE structure (PDB code 4hjv), which is needed to facilitate the formation of an oxocarbeniumion-like transition state in an productive binding mode26. Molecular dynamics (MD) simulations were performed using GROMACS 4.5.5 with the GROMOS96 43A1 force field49. The modeled complex was placed in a rhombic dodecahedron box with the edges at least 10 Å away from the protein surface. The system was solvated with water molecules (SPC216) supplemented with sufficient counterions to neutralize the system. After energy minimization, position restrained MD simulations were carried out in the NVT and NPT ensembles (100 ps) to equilibrate the system at 300 K and 1 atm. The system was then subjected to unconstrained production MD simulation for 10 ns.
A NAG-NAM-tetrapeptide was docked onto the SaCwlT-NlpC/P60 domain using a similar protocol and restraining the middle section of the ligand (l-Ala and γ-d-Glu) to closely match the pose of the ligand observed in BcYkfC12 (PDB code 3h41).
Accession numbers
The structure factors and atomic coordinates are deposited in the RCSB Protein Data Bank (http://www.rcsb.org) with PDB codes 4fdy (SaCwlT) and 4hpe (CdCwlT). The plasmids for producing recombinant were deposited in PSI:Biology-Materials Repository (http://dnasu.asu.edu) with clone IDs SaCD00328273 (SaCwlT) and CdCD00085319 (CdCwlT).
Supplementary Material
Highlights.
CwlT is a bifunctional cell-wall lysin encoded by Tn916-like transposons
Crystal structures of CwlT from two pathogens were determined
Modular architecture of CwlT contains independent lysozyme and dl-endopeptidase catalytic domains.
The lysozyme represents a novel enzyme in the glycoside hydrolase family 23
Acknowledgements
We thank the members of the JCSG high-throughput structural biology pipeline for their contribution to this work. This work was supported by the NIH, National Institute of General Medical Sciences (NIGMS), Protein Structure Initiative [U54 GM094586]. Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209). Genomic DNA from Clostridium difficile (ATCC Number BAA-1382D-5) and Staphylococcus aureus Mu50 (ATCC Number 700699D) were obtained from the American Type Culture Collection (ATCC). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS, NCRR or NIH.
Abbreviations
- bLysG
bacterial lysozyme G
- CwlT
cell wall lytic enzyme T
- asu
asymmetric unit
- LT
lytic transglycosylase
- RMSD
Root-mean-square deviation
- GH
glycoside hydrolase family
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Data
Supplementary data to this article can be found online at http://dx.doi.org/...
References
- 1.Roberts AP, Mullany P. A modular master on the move: the Tn916 family of mobile genetic elements. Trends Microbiol. 2009;17:251–8. doi: 10.1016/j.tim.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Roberts AP, Mullany P. Tn916-like genetic elements: a diverse group of modular mobile elements conferring antibiotic resistance. FEMS Microbiol. Rev. 2011;35:856–71. doi: 10.1111/j.1574-6976.2011.00283.x. [DOI] [PubMed] [Google Scholar]
- 3.Ciric L, Jasni A, Elvira de Vries L, Agersø Y, Mullany P, Roberts AP. The Tn916/Tn1545 Family of Conjugative Transposons. In: Roberts AP, editor. Bacterial Integrative Mobile Genetic Elements. Landes Bioscience; Austin, USA: 2012. [Google Scholar]
- 4.Grohmann E. Conjugative transfer of the integrative and conjugative element ICEBs1 from Bacillus subtilis likely initiates at the donor cell pole. J. Bacteriol. 2010;192:23–5. doi: 10.1128/JB.01305-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukushima T, Kitajima T, Yamaguchi H, Ouyang Q, Furuhata K, Yamamoto H, Shida T, Sekiguchi J. Identification and characterization of novel cell wall hydrolase CwlT: a two-domain autolysin exhibiting N-acetylmuramidase and DL-endopeptidase activities. J. Biol. Chem. 2008;283:11117–25. doi: 10.1074/jbc.M706626200. [DOI] [PubMed] [Google Scholar]
- 6.Wizemann TM, Heinrichs JH, Adamou JE, Erwin AL, Kunsch C, Choi GH, Barash SC, Rosen CA, Masure HR, Tuomanen E, Gayle A, Brewah YA, Walsh W, Barren P, Lathigra R, Hanson M, Langermann S, Johnson S, Koenig S. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 2001;69:1593–8. doi: 10.1128/IAI.69.3.1593-1598.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chibani-Chennoufi S, Dillmann ML, Marvin-Guy L, Rami-Shojaei S, Brussow H. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 2004;186:7069–83. doi: 10.1128/JB.186.21.7069-7083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmaster AR, Ravel J, Rasko DA, Chapman GD, Chute MD, Marston CK, De BK, Sacchi CT, Fitzgerald C, Mayer LW, Maiden MC, Priest FG, Barker M, Jiang L, Cer RZ, Rilstone J, Peterson SN, Weyant RS, Galloway DR, Read TD, Popovic T, Fraser CM. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl. Acad. Sci. USA. 2004;101:8449–54. doi: 10.1073/pnas.0402414101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depardieu F, Bonora MG, Reynolds PE, Courvalin P. The vanG glycopeptide resistance operon from Enterococcus faecalis revisited. Mol. Microbiol. 2003;50:931–48. doi: 10.1046/j.1365-2958.2003.03737.x. [DOI] [PubMed] [Google Scholar]
- 10.Zhu W, Murray PR, Huskins WC, Jernigan JA, McDonald LC, Clark NC, Anderson KF, McDougal LK, Hageman JC, Olsen-Rasmussen M, Frace M, Alangaden GJ, Chenoweth C, Zervos MJ, Robinson-Dunn B, Schreckenberger PC, Reller LB, Rudrik JT, Patel JB. Dissemination of an Enterococcus Inc18-like vanA plasmid associated with vancomycin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2010;54:4314–20. doi: 10.1128/AAC.00185-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Q, Sudek S, McMullan D, Miller MD, Geierstanger B, Jones DH, Krishna SS, Spraggon G, Bursalay B, Abdubek P, Acosta C, Ambing E, Astakhova T, Axelrod HL, Carlton D, Caruthers J, Chiu HJ, Clayton T, Deller MC, Duan L, Elias Y, Elsliger MA, Feuerhelm J, Grzechnik SK, Hale J, Han GW, Haugen J, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Kumar A, Marciano D, Morse AT, Nigoghossian E, Okach L, Oommachen S, Paulsen J, Reyes R, Rife CL, Trout CV, van den Bedem H, Weekes D, White A, Wolf G, Zubieta C, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA. Structural basis of murein peptide specificity of a gamma-D- glutamyl-l-diamino acid endopeptidase. Structure. 2009;17:303–13. doi: 10.1016/j.str.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, Carlton D, Chen C, Chiu HJ, Chiu M, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Lam WW, Marciano D, Miller MD, Morse AT, Nigoghossian E, Nopakun A, Okach L, Puckett C, Reyes R, Tien HJ, Trame CB, van den Bedem H, Weekes D, Wooten T, Yeh A, Hodgson KO, Wooley J, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wilson IA. Structure of the gamma-D-glutamyl-L-diamino acid endopeptidase YkfC from Bacillus cereus in complex with L-Ala-gamma-D-Glu: insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta Crystallogr., Sect. F. 2010;66:1354–64. doi: 10.1107/S1744309110021214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, Rawlings ND, Chiu HJ, Jaroszewski L, Klock HE, Knuth MW, Miller MD, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wilson IA. Structural analysis of papain-like NlpC/P60 superfamily enzymes with a circularly permuted topology reveals potential lipid binding sites. PLoS ONE. 2011;6:e22013. doi: 10.1371/journal.pone.0022013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi NK, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–40. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 15.Kall L, Krogh A, Sonnhammer EL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wooley J, Wuthrich K, Wilson IA. The JCSG high-throughput structural biology pipeline. Acta Crystallogr., Sect. F. 2010;66:1137–42. doi: 10.1107/S1744309110038212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 19.McGowan S, Buckle AM, Mitchell MS, Hoopes JT, Gallagher DT, Heselpoth RD, Shen Y, Reboul CF, Law RH, Fischetti VA, Whisstock JC, Nelson DC. X-ray crystal structure of the streptococcal specific phage lysin PlyC. Proc. Natl. Acad. Sci. USA. 2012;109:12752–7. doi: 10.1073/pnas.1208424109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischetti VA. Bacteriophage lysins as effective antibacterials. Curr. Opin. Microbiol. 2008;11:393–400. doi: 10.1016/j.mib.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fibriansah G, Gliubich FI, Thunnissen AM. On the mechanism of peptidoglycan binding and cleavage by the endo-specific lytic transglycosylase MltE from Escherichia coli. Biochemistry. 2012;51:9164–77. doi: 10.1021/bi300900t. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helland R, Larsen RL, Finstad S, Kyomuhendo P, Larsen AN. Crystal structures of g-type lysozyme from Atlantic cod shed new light on substrate binding and the catalytic mechanism. Cell. Mol. Life Sci. 2009;66:2585–98. doi: 10.1007/s00018-009-0063-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlkonig A, Huet J, Looze Y, Wintjens R. Structural relationships in the lysozyme superfamily: significant evidence for glycoside hydrolase signature motifs. PLoS ONE. 2010;5:e15388. doi: 10.1371/journal.pone.0015388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zechel DL, Withers SG. Glycosidase mechanisms: anatomy of a finely tuned catalyst. Acc. Chem. Res. 2000;33:11–8. doi: 10.1021/ar970172+. [DOI] [PubMed] [Google Scholar]
- 27.Kuroki R, Weaver LH, Matthews BW. Structural basis of the conversion of T4 lysozyme into a transglycosidase by reengineering the active site. Proc. Natl. Acad. Sci. USA. 1999;96:8949–54. doi: 10.1073/pnas.96.16.8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rau A, Hogg T, Marquardt R, Hilgenfeld R. A new lysozyme fold. Crystal structure of the muramidase from Streptomyces coelicolor at 1.65 Å resolution. J. Biol. Chem. 2001;276:31994–9. doi: 10.1074/jbc.M102591200. [DOI] [PubMed] [Google Scholar]
- 29.Patzer SI, Albrecht R, Braun V, Zeth K. Structural and mechanistic studies of pesticin, a bacterial homolog of phage lysozymes. J. Biol. Chem. 2012;287:23381–96. doi: 10.1074/jbc.M112.362913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Asselt EJ, Thunnissen AM, Dijkstra BW. High resolution crystal structures of the Escherichia coli lytic transglycosylase Slt70 and its complex with a peptidoglycan fragment. J. Mol. Biol. 1999;291:877–98. doi: 10.1006/jmbi.1999.3013. [DOI] [PubMed] [Google Scholar]
- 31.Aramini JM, Rossi P, Huang YJ, Zhao L, Jiang M, Maglaqui M, Xiao R, Locke J, Nair R, Rost B, Acton TB, Inouye M, Montelione GT. Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli: structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry. 2008;47:9715–7. doi: 10.1021/bi8010779. [DOI] [PubMed] [Google Scholar]
- 32.Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klock HE, Koesema EJ, Knuth MW, Lesley SA. Combining the polymerase incomplete primer extension method for cloning and mutagenesis with microscreening to accelerate structural genomics efforts. Proteins. 2008;71:982–94. doi: 10.1002/prot.21786. [DOI] [PubMed] [Google Scholar]
- 34.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 35.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 1993;229:105–24. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 36.Santarsiero BD, Yegian DT, Lee CC, Spraggon G, Gu J, Scheibe D, Uber DC, Cornell EW, Nordmeyer RA, Kolbe WF, Jin J, Jones AL, Jaklevic JM, Schultz PG, Stevens RC. An approach to rapid protein crystallization using nanodroplets. J. Appl. Crystallogr. 2002;35:278–281. [Google Scholar]
- 37.Lesley SA, Kuhn P, Godzik A, Deacon AM, Mathews I, Kreusch A, Spraggon G, Klock HE, McMullan D, Shin T, Vincent J, Robb A, Brinen LS, Miller MD, McPhillips TM, Miller MA, Scheibe D, Canaves JM, Guda C, Jaroszewski L, Selby TL, Elsliger MA, Wooley J, Taylor SS, Hodgson KO, Wilson IA, Schultz PG, Stevens RC. Structural genomics of the Thermotoga maritima proteome implemented in a high-throughput structure determination pipeline. Proc. Natl. Acad. Sci. USA. 2002;99:11664–9. doi: 10.1073/pnas.142413399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen AE, Ellis PJ, Miller MD, Deacon AM, Phizackerley RP. An automated system to mount cryo-cooled protein crystals on a synchrotron beamline, using compact samples cassettes and a small-scale robot. J. Appl. Cryst. 2002;35:720–726. doi: 10.1107/s0021889802016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Bedem H, Wolf G, Xu Q, Deacon AM. Distributed structure determination at the JCSG. Acta Crystallogr., Sect. D. 2011;67:368–75. doi: 10.1107/S0907444910039934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabsch W. XDS. Acta Crystallogr., Sect. D. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheldrick GM. A short history of SHELX. Acta Crystallogr. Sect. A. 2008;64:112–22. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 42.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr., Sect. D. 2003;59:2023–30. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 43.Cowtan K. Fitting molecular fragments into electron density. Acta Crystallogr., Sect. D. 2008;64:83–9. doi: 10.1107/S0907444907033938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr., Sect. D. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr., Sect. D. 2004;60:2210–21. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 47.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–9. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuttelkopf AW, van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr., Sect. D. 2004;60:1355–63. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 49.Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–18. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.