Abstract
The differentiation of CD4+ T cells into different T helper lineages is driven by cytokine milieu in the priming site and the underlying transcriptional circuitry. Even though many positive regulators have been identified, it is not clear how this process is inhibited at transcriptional level. Here we report that the ETS transcription factor ELF4 suppresses the differentiation of Th17 cells both in vitro and in vivo. Culture of naive Elf4−/− CD4+ T cells in the presence of IL-6 and TGFβ (or IL-6, IL-23, and IL-1β) resulted in increased numbers of IL-17A positive cells compared to wild-type controls. In contrast, the differentiation to Th1, Th2, or Treg was largely unaffected by loss of ELF4. The increased expression of genes involved in Th17 differentiation observed in Elf4−/− CD4+ T cells suggested that ELF4 controls their programming into the Th17 lineage rather than only IL-17A gene expression. Despite normal proliferation of naïve CD4+ T cells, loss of ELF4 lowered the requirement of IL-6 and TGFβ signaling for IL-17A induction in each cell division. ELF4 did not inhibit Th17 differentiation by promoting IL-2 production as proposed for another ETS transcription factor, ETS1. Elf4−/− mice showed increased numbers of Th17 cells in the lamina propria at steady state, in lymph nodes after immunization, and most importantly in the CNS following EAE induction, contributing to the increased disease severity. Collectively, our findings suggest that ELF4 restrains Th17 differentiation in dividing CD4+ T cells by regulating commitment to the Th17 differentiation program.
Introduction
The main function of CD4+ T cells is to provide “help” to cells of the innate and adaptive immune system. There are different T helper cell lineages with unique features (i.e. Th1, Th2, and Th17 cells) that modulate immune responses against infections and tumor growth by promoting the effector functions and memory formation of CD8+ T cells, inducing antibody class switching of B cells, and enhancing the activity of phagocytes (1, 2). Lineage differentiation of CD4+ T cells is driven by TCR activation and a specific cytokine milieu. Alterations in the balanced generation of different T helper cells often lead to immunodeficiencies or autoimmune disorders (2). Although many studies have focused on identifying specific regulators of CD4+ T cell differentiation, the cell-intrinsic mechanisms downstream of the TCR and cytokine receptors are largely unknown.
APC activate naïve CD4+ T cells via TCR recognition, costimulation, and secretion of cytokines that induce differentiation into effector cells. These cytokines imprint unique molecular signatures on CD4+ T cells by creating a transcriptional program that controls lineage commitment and effector functions. In the case of Th17 cells, potent inducers of tissue inflammation, IL-6 (or IL-21) and TGFβ initiate the differentiation program in activated CD4+ T cells by inducing expression of the master regulator RORγt, which subsequently turns on IL-17A and other Th17-associated genes (3–6). Differentiating Th17 cells are not fully functional until receiving IL-23 signal, which stabilizes and expands in vivo Th17 responses in a STAT3-dependent manner (7). Consequently, transcription factors are important cell-intrinsic mediators that translate environmental cues into effector functions of Th17 cells. However, negative regulators that prevent excessive Th17 responses are ill-defined.
ELF4 is a member of the ETS family transcription factors with a highly conserved ETS domain that mediates protein-DNA and protein-protein interactions (8). Since ETS proteins generally work in concert with other co-regulatory proteins by forming supramolecular complexes, both protein-DNA and protein-protein interactions contribute to their transcriptional activity and specificity (8, 9). Some ETS family proteins have been linked to carcinogenesis because of their roles in cellular proliferation, differentiation, and apoptosis (8–11). Given that certain ETS transcription factors such as ETS1 and PU.1 are involved in T helper cell differentiation (12–16), we decided to investigate the role of ELF4 in this process. ELF4 is widely expressed in several tissues including bone marrow, thymus, and the spleen (17). ELF4 regulates cell cycle progression in hematopoietic stem cells and endothelial cells, and has both tumor suppressor and oncogenic activity (18–21). In the immune system, ELF4 plays important roles in both innate and adaptive immune cells, as embryonic deletion of ELF4 resulted in impaired lytic activity of NK cells as well as aberrant proliferation and trafficking of naïve CD8+ T cells (22, 23). Given that ELF4 is generally considered a transcriptional activator, its aforementioned effects on NK cells and CD8+ T cells were caused at least in part by direct regulation of the Prf1 and Klf4 genes, respectively (22, 23). We previously showed that TCR activation leads to rapid downregulation of ELF4 transcripts in naïve CD4+ T cells (24), suggesting a regulatory role of ELF4 in TCR-mediated biological processes such as T cell differentiation.
In this work, we report that loss of ELF4 specifically enhanced Th17 differentiation both in vitro and in vivo. ELF4 did not significantly affect the proliferation or survival of CD4+ T cells but instead regulated commitment to the Th17 differentiation program downstream of STAT1, STAT3, STAT5, and SMAD2/3 proteins, likely by suppressing Notch1 signaling. As a consequence of deregulated Th17 differentiation, Elf4−/− mice showed more severe symptoms after induction of EAE and increased numbers of Th17 cells in the CNS. Taken together, our results suggest that ELF4 is a novel lineage-specific regulator that inhibits the development of Th17 cells, emerging as an ideal therapeutic target in treating Th17-mediated immune disorders.
Materials and Methods
Mice
Elf4−/− mice were provided by Dr. S. Nimer (Memorial Sloan-Kettering Cancer Center, New York, NY) and backcrossed to the C57BL/6 (B6) background for additional 10 generations (23). B6 mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Il17f rfp mice were crossed with Elf4−/− mice to generate Elf4−/− Il17f rfp mice (25). All mice were bred and maintained in specific pathogen-free conditions at Baylor College of Medicine. The animal studies were approved by the Institutional Animal Care and Usage Committee of Baylor College of Medicine.
Flow cytometry
The following antibodies were purchased from eBioscience (San Diego, CA): αIL-4 Alexa 647 (11B11), αFoxp3 Alexa 488 (FJK-16s), and αIFNγ Alexa 488 (XMG1.2), and αNK1.1 PE (PK136). Antibodies for αCD4 PE-Cy7 (GK1.5), αGM-CSF FITC (MP1-22E9), αTCRγ/δ FITC (GL3), αCD45.1 FITC (A20), and αIL-17A Alexa 647 (TC11-18H10.1) were purchased from BioLegend (San Diego, CA). Samples were analyzed using the flow cytometer FACSCanto (BD Biosciences, San Jose, CA) and FlowJo software (Treestar, Ashland, OR). For intracellular staining of cytokines, cells were first stimulated with PMA (50 ng/ml) and Ionomycin (500 ng/ml) in the presence of GolgiPlug (1:1000; BD Biosciences) for 4–6 hours at 37°C, followed by cell surface staining with αCD4 PE-Cy7 and the viability dye eFluor 780 (eBioscience). Then, cells were fixed and permeabilized using BD Cytofix/Cytoperm kit (BD Biosciences), followed by intracellular staining with αIFNγ Alexa 488, αIL-4 Alexa 647, or αIL-17A Alexa 647. For Foxp3 staining, cells were stained with αCD4 PE-Cy7 and viability dye eFluor 780, and then fixed and permeabilized using Foxp3 Staining Buffer Set (eBioscience), followed by intracellular staining of αFoxp3 Alexa 488. All samples were analyzed on gated live cells (eFluor 780 negative).
CD4+ T cell proliferation and differentiation in vitro
CD4+ T cells were purified from the spleen of 2–3 month old mice using the negative selection BD IMag magnetic-bead separation system according to the manufacturer’s instructions (BD Biosciences). Biotinylated αCD25 antibody (PC61, eBioscience) was used in the enrichment process to deplete CD25+ T cells. Purity of enriched CD4+ T cells was routinely 90–95%. Of note, WT and Elf4−/− CD4+ T cells purified from young mice had a similar frequency of CD44hi cells (<5%).
For proliferation assays, CD4+ T cells were labeled with 2 μM CFSE according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). CFSE-labeled cells were then stimulated with 10 μg/ml plate-bound αCD3 antibodies (145-2C11; BD Biosciences) and 2 μg/ml αCD28 antibody (37.51; BD Biosciences) in X-VIVO medium (Lonza, Basel, Switzerland). After three days, proliferation was determined by CFSE dilution using flow cytometry. For differentiation assays, CD4+CD25− T cells were stimulated with 10 μg/ml plate-bound αCD3 and 2 μg/ml soluble αCD28 antibody in the presence of indicated cytokines and neutralizing antibody. Th0: 10 ng/ml IL-2 (PeproTech, Rocky Hill, NJ), 5 μg/ml αIFNγ (XMG1.2; BioLegend) and 5 μg/ml αIL-4 (11B11; BioLegend); Th1: 10 ng/ml IL-2, 40 ng/ml IL-12 (PeproTech) and 5 μg/ml αIL-4; Th2, 10 ng/ml IL-2, 50 ng/ml IL-4 (PeproTech) and 5 μg/ml αIFNγ; Treg, 10 ng/ml IL-2, 5 ng/ml TGFβ (PeproTech), 5 μg/ml αIFNγ and 5 μg/ml αIL-4; Th17, 30 ng/ml IL-6 (PeproTech), 0.1 ng/ml TGFβ, 5 μg/ml αIFNγ and 5 μg/ml αIL-4 unless otherwise indicated. Cells were analyzed for intracellular levels of IFNγ, IL-4, IL-17A or Foxp3 after three days. Inhibition of Notch1 signaling during Th17 differentiation was achieved using the γ-secretase inhibitor (GSI) compound E (Enzo Life Sciences, Farmingdale, NY).
RNA isolation and quantitative RT-PCR
Total RNA was extracted from CD4+ T cells using RNeasy Plus Mini kit (Qiagen, Valencia, CA), and cDNA was synthesized from 100–500 ng RNA using random hexamer primers and SuperScript III First-Strand Synthesis kit (Invitrogen). FastStart Universal SYBR Green Master was used for quantitative real-time PCR as specified by the manufacturer (Roche, Indianapolis, IN). Primer sequences for PCR were as follows: β-actin forward, 5′-CTGGGCCGCTCTAGGCACCA-3′, and reverse, 5′-CGGTTGGCCTTAGGGTTCAGGGG-3′; ELF4 forward, 5′-CGGA AGTGCTTTCAGACTCC-3′, and reverse, 5′-GGTCAGTGACAGGTGAGGTA-3′; IL-17A forward, 5′-ACTTTCAGGGTCGAGAAGA-3′, and reverse, 5′-TTCTGAATCTGCCTCTGAAT-3′; IL-17F forward, 5′-TGCTACTGTTGATGTTGGGAC-3′, and reverse, 5′-AATGCCCT GGTTTTGGTTGAA-3′; IL-21 forward, 5′-TGACATTGTTGAACAGCTGAAA-3′, and reverse, 5′-AAAACAGGCAAAAGCTGCAT-3′; IL-22 forward, 5′-GTGAGAAGCTAACGTCCATC-3′, and reverse, 5′-GTCTACCTCTGGTCTCATGG-3′; IL-10 forward, 5′-ATCGATTTCTCCCCTGTGAA-3′, and reverse, 5′-TGTCAAATTCATTCATGGCCT-3′; Foxp3 forward, 5′-CTCGTCTGAAGGCAGAGTCA-3′, and reverse, 5′-TGGCAGAGAGGTATTGAGGG-3′; GATA3 forward, 5′-AGGATGTCCCTGCTCTCCTT-3′, and reverse, 5′-GCCTG CGGACTCTACCATAA-3′; T-bet forward, 5′-CAATGTGACCCAGATGATCG-3′, and reverse, 5′-GCGTTCTGGTAGGCAGTCAC-3′;IRF4 forward, 5′-CAAAGCACAGAGTCACCTGG-3′, and reverse, 5′-TGCAAGCTCTTTGACACACA-3′; RORγt forward, 5′-AGCTTTGTGCAGATCTAAGG-3′, and reverse, 5′-TGTCCTCCTCAGTAGGGTAG-3′; RUNX1 forward, 5′-GGTGGACAGAGGAAGAGGTG-3′, and reverse, 5′-TTGCCACCTACCATAGAGCC-3′; IL-23R forward, 5′-TTCAGATGGGCATGAATGTTTCT-3′, and reverse, 5′-CCAAATCCGAGCTGTTGTTCTAT-3′; CCR6 forward, 5′-CTGGAACTCTGCAGAACGCT-3′, and reverse, 5′-TGGCCAGTCTACTTTGGAGC-3′; ETS1 forward, 5′-ATCTCGAGCTTTTCCCTTCC-3′, and reverse, 5′-TTTTCAAGGCTTGGGACATC-3′; Hes1 forward, 5′-ACACCGGACAAACCAAA GAC-3′, and reverse, 5′-ATGCCGGGAGCTATCTTTC-3′; c-Myc forward, 5′-TCAAGAGGCGAACACACAAC-3′, and reverse, 5′-GGAGGAAGTCCAGTGTCCAGCC-3′; HeyL forward, 5′-CAGTGGAACAACAGAGAATGAAC-3′, and reverse, 5′-ACCAGCAGTAGTGAGTAACCAG-3′. Reactions were performed in the Mx3005P instrument (Stratagene, La Jolla, CA). Expression was calculated relative to β-actin for each sample.
Induction of EAE
WT and Elf4−/− mice were immunized subcutaneously on the hind flanks with 200 μg MOG35-55 (EZBiolab, Carmel, IN) emulsified in CFA (Sigma, St. Louis, MO). Pertussis toxin (List Biological Laboratories, Campbell, CA) was administered intraperitoneally on day 0 and day 2 (500 ng in PBS). Mice were monitored daily for body weight and clinical symptoms; clinical scores were defined as previously described (26).
ELISA
WT and Elf4−/− CD4+ T cells were cultured under Th17 conditions and the supernatant was collected on day 3 (for IL-17A) or on day 1, 2, and 3 (for IL-2). IL-17A and IL-2 production was measured using Mouse IL-17A ELISA Ready-Set-Go kit and Mouse IL-2 ELISA Ready-SET-Go kit following manufacturer’s instructions (eBioscience).
Isolation of lamina propria lymphocytes (LPL)
Small intestine LPL were isolated as previously described (27). Briefly, small intestine was removed, cut into small pieces, and washed with CMF solution (Ca2+/Mg2+-free HBSS with 10 mM HEPES, 25 mM NaHCO3, and 2% FCS). After incubation with DTE solution (CMF with 1 mM dithioerythritol), intraepithelial lymphocytes were removed and the remaining tissues were incubated with HBSS containing 1.3 mM EDTA. After washing with RPMI, tissues were digested with 100 U/ml collagenase and subjected to centrifugation on a 44%/67% Percoll gradient. Finally, LPL were obtained from the interface.
Isolation of mononuclear cells from brain and spinal cord
Mice were perfused with cold PBS through left cardiac ventricle. Brain was dissected and spinal cord was flushed out by hydrostatic pressure using a 18G1.5 needle. CNS tissues were minced and digested with collagenase D (2.5 mg/ml) and DNase I (1 mg/ml) in DMEM at 37°C for 45 min. After passing through a 70 μm cell strainer, cells were subjected to a 37%/70% Percoll gradient centrifugation at 500g for 20 min. Mononuclear cells were collected from the interface.
Retroviral transduction of CD4+ T cells
293T cells were transfected with the retroviral vector Migr1 carrying ELF4 cDNA (or empty vector as control) and a Ψ ecotropic envelope using ProFection Mammalian Transfection System (Progema, Madison, WI). Two days after transfection, supernatant containing retrovirus was passed through a 0.45 μm filter. CD4+ T cells were activated by plate-bound αCD3 (10 μg/ml) and αCD28 (2 μg/ml) in X-VIVO medium for 24 hours and transduced with the viral supernatant by spinoculation at 1200g for 90 min in the presence of polybrene (8 μg/ml). After 24 hours, cells were transduced again and then cultured under Th17 polarizing condition for additional two days before intracellular cytokine staining.
Results
ELF4 selectively inhibits in vitro differentiation of Th17 cells
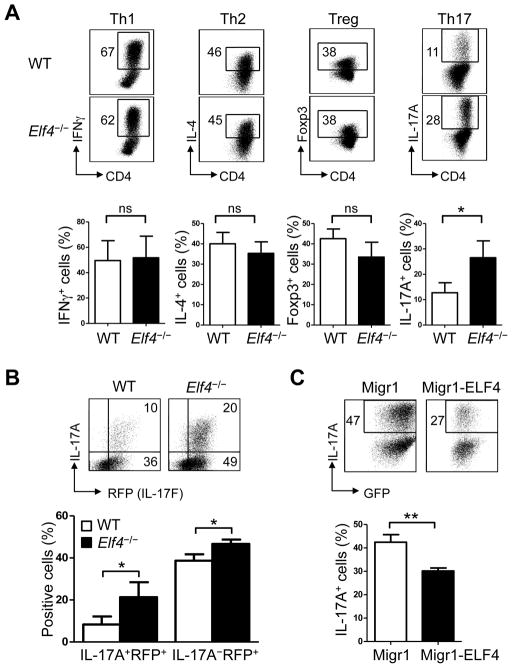
To examine whether ELF4 was involved in T helper cell differentiation, naïve CD4+CD25− T cells from the spleens of wild-type (WT) and Elf4−/− mice were cultured under Th1, Th2, Treg, or Th17 polarizing conditions. Elf4−/− CD4+ T cells showed similar percentages of IFNγ, IL-4, and Foxp3 positive cells compared to WT controls, indicating that the ability to differentiate into Th1, Th2, and Treg lineages was not significantly altered by loss of ELF4 (Fig 1A). In contrast, Elf4−/− CD4+ T cells showed a 2-fold increase in IL-17A+ cells compared to WT counterparts (Fig 1A). Since Elf4−/− CD4+ T cells showed skewed differentiation towards Th17 lineage despite normal Treg differentiation, this bias to the Th17 lineage was likely not due to a reciprocal imbalance between Treg and Th17 cells.
Fig. 1. ELF4 selectively inhibits in vitro differentiation of Th17 cells.
(A) Flow cytometric analysis of intracellular IFNγ, IL-4, Foxp3, or IL-17A expression in wild-type (WT) and Elf4−/− CD4+ T cells cultured under Th1, Th2, Treg, or Th17 polarizing conditions. Percentages of positive cells are summarized in the lower panels (n=3; mean ± s.d.). (B) Flow cytometric analysis of intracellular IL-17A and expression of the reporter IL-17F-RFP in WT Il17frfp/+ and Elf4−/− Il17frfp/+ CD4+ T cells polarized under Th17 condition. Percentages of IL-17A+IL-17F+ and IL-17A−IL-17F+ cells are summarized in the lower panel (n=3; mean ± s.d.). (C) Flow cytometric analysis of intracellular IL-17A and GFP expression in WT CD4+ T cells transduced with either empty retroviral vector (Migr1) or retroviral carrying ELF4 (Migr1-ELF4) and cultured under Th17 condition. Percentages of IL-17A+ in CD4+GFP+ cells are summarized in the lower panel (n=3; mean ± s.d.). Data are representative of at least two independent experiments. ns: not significant, *P<0.05, **P<0.01 (Two-tailed Student’s t-test).
IL-17F, another Th17 signature cytokine closely related to IL-17A, is largely co-expressed with IL-17A but can also be expressed independently (25, 28). By crossing IL-17F-RFP reporter (Il17f rfp) mice (25) with Elf4−/− mice, we were able to examine the effect of ELF4 on the expression of the two cytokines on a per cell basis by monitoring RFP expression and the intracellular levels of IL-17A using flow cytometry. Both WT and Elf4−/− CD4+ T cells showed two populations of Th17 cells, IL-17A+RFP(IL-17F)+ and IL-17A−RFP(IL-17F)+ cells (Fig 1B). Deletion of ELF4 augmented the formation of not only IL-17A+RFP+ but also IL-17A−RFP+ cells (Fig 1B), suggesting that ELF4 regulated the lineage commitment of Th17 cells rather than expression of the Il17a gene. Conversely, we confirmed the inhibitory effect of ELF4 on Th17 differentiation using a gain-of-function model, where retroviral expression of ELF4 in WT CD4+ T cells significantly reduced the frequency of IL-17A+ cells (Fig 1C).
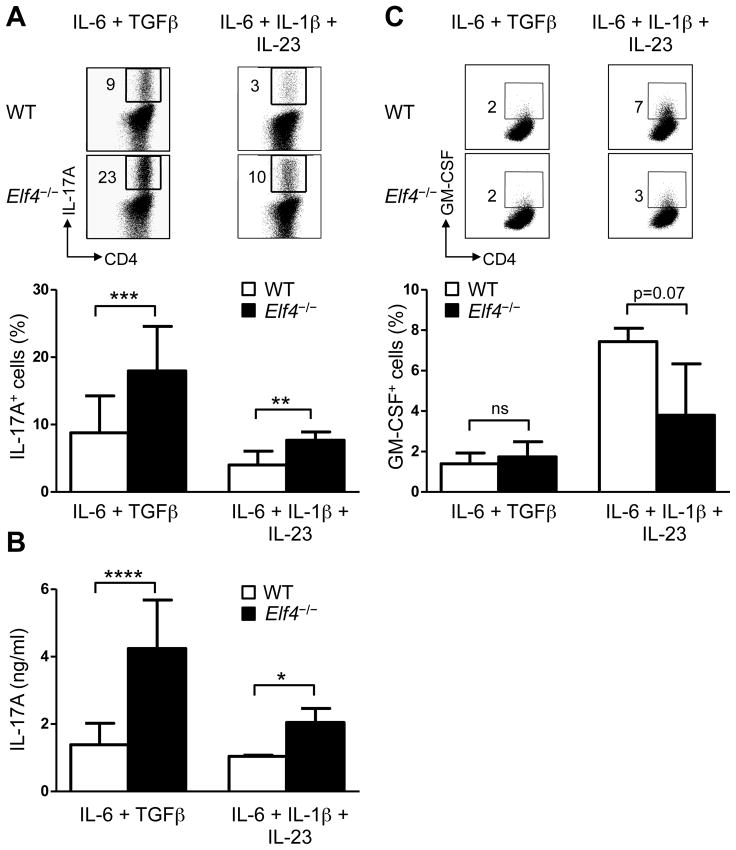
Despite a close association with inflammatory responses, not all in vitro-generated Th17 cells are endowed with the same capacity to induce inflammation in vivo, which depends on the cytokines present during differentiation (29–31). IL-6/TGFβ-induced Th17 cells are unable to cause EAE after adoptive transfer (29, 30, 32) whereas Th17 cells induced by IL-6/IL-1β/IL-23 are highly pathogenic (30). These findings suggested that pathogenicity of Th17 cells is regulated by specific differentiation programs. Elf4−/− CD4+ T cells showed greater frequency of Th17 cells induced by both IL-6/TGFβ (non-pathogenic) and IL-6/IL-1β/IL-23 (pathogenic) conditions (Fig 2A). The enhanced production of IL-17A was further confirmed by increased secretion of IL-17A protein measured by ELISA (Fig 2B). Given the role of GM-CSF in the pathogenesis of EAE (33, 34), we also examined whether ELF4 regulates its production in CD4+ T cells cultured in both Th17 conditions (Fig 2C). Even though ELF4 can activate the human GM-CSF promoter in vitro (17), ELF4 deletion did not significantly affect the production of GM-CSF in Th17 cells (Fig 2C). These data suggest that ELF4 selectively regulates the differentiation of Th17 cells and potentially their pathogenicity.
Fig. 2. ELF4 impairs Th17 differentiation induced by both IL-6 + TGFβ and IL-6 + IL-1β + IL-23.
(A) Flow cytometric analysis of IL-17A expression in WT and Elf4−/− CD4+ T cells cultured with IL-6 + TGFβ (n=15) or IL-6 + IL-1β + IL-23 (n=5). Percentages of IL-17A+ cells are summarized in the lower panel (mean ± s.d.). (B) The secretion of IL-17A was measured by ELISA in WT and Elf4−/− CD4+ T cells cultured with IL-6 + TGFβ (n=9) or IL-6 + IL-1β + IL-23 (n=3) (mean ± s.d.). (C) Flow cytometric analysis of GM-CSF expression in WT and Elf4−/− CD4+ T cells cultured with IL-6 + TGFβ (n=3) or IL-6 + IL-1β + IL-23 (n=3). Percentages of GM-CSF+ cells are summarized in the lower panel (mean ± s.d.). Data are representative of at least two independent experiments. ns: not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (Two-tailed Student’s t-test).
Naïve CD4+ T cells acquire effector functions during Th17 differentiation through the acquisition of a specific gene signature controlled by transcription factors such as RORγt (35). Therefore, we measured the transcript levels of additional Th17-associated genes to investigate the effect of ELF4 on a global Th17 gene signature. Real-time PCR analysis showed that the increased expression of IL-17A in Elf4−/− CD4+ T cells was accompanied by elevated levels of additional cytokines (IL-17F, IL-21, IL-22, and IL-10), transcription factors (RORγt and IRF4), and cytokine/chemokine receptors (IL-23R and CCR6) (Fig 3). This finding raises the possibility that ELF4 may regulate a transcriptional repressor of Irf4 and Rorc genes to control the differentiation of Th17 cells. Despite comparable levels of GATA3 (Th2) and lower levels of Foxp3 (Treg), Elf4−/− CD4+ T cells cultured under a Th17-polarizing condition expressed higher levels of T-bet (Fig 3), which were likely due to the occurrence of T-bet positive Th17 cells since few Th1 cells emerged in this experimental condition (29, 30). Collectively, our findings support a novel role of ELF4 as a lineage-specific inhibitor of the Th17 differentiation program.
Fig. 3. Loss of ELF4 activates the Th17 differentiation program in CD4+ T cells.
Quantitative real-time PCR analysis of Th17-associated genes was performed in Th17-polarized WT and Elf4−/− CD4+ T cells. Relative expression is expressed as log2 fold change of Elf4−/− over WT controls after normalization with β-actin. Data include two independent experiments (n=6; mean ± s.d.). ns: not significant, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (Two-tailed Student’s t-test).
ELF4 is dispensable for the proliferation and survival of differentiating Th17 cells
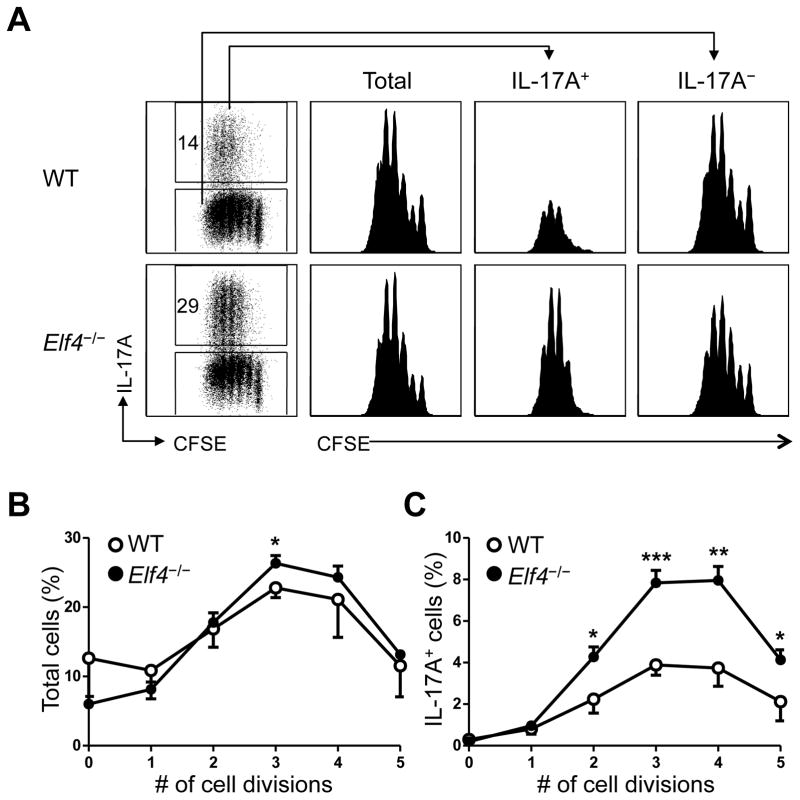
Cytokines are required but not sufficient to induce T helper differentiation, as naïve CD4+ T cells only undergo differentiation after receiving TCR stimulation. TCR-driven proliferation is known to be pre-requisite for the production of effector cytokines such as IFNγ and IL-4 (36). Since ELF4 inhibits proliferation of naive CD8+ T cells (22), we explored the possibility that ELF4 may suppress Th17 differentiation in CD4+ T cells by blocking their cell cycle progression. Two models of TCR-mediated induction of proliferation, in vitro TCR crosslink and adoptive transfer into lymphopenic mice, showed a normal proliferative capacity in Elf4−/− CD4+ T cells (data not shown). In addition, we examined the proliferation of CD4+ T cells cultured under Th17-polarizing condition by dual detection of intracellular IL-17A and dilution of the CFSE dye. Interestingly, Elf4−/− CD4+ T cells displayed increased frequency of cells producing IL-17A despite a normal dilution of the CFSE dye compared to WT controls (Fig 4A). The cell division of neither IL-17A+ nor IL-17A− populations was significantly affected by loss of ELF4 (Fig 4A). A more detailed analysis showed that loss of ELF4 did not substantially affect the overall cell cycle progression (Fig 4B) but rather increased the proportion of IL-17A+ cells after two or more cell divisions (Fig 4C), suggesting that the increased Th17 differentiation was not caused by augmented proliferation of Elf4−/− CD4+ T cells. A similar frequency of Annexin V+ cells in WT and Elf4−/− CD4+ T cells cultured under Th17 condition (27.7±1.4% for WT and 31.1±2.6% for Elf4−/− CD4+ T cells) ruled out the possibility that deletion of ELF4 promoted the survival of CD4+ T cells. Thus, the greater frequency of IL-17A+ cells caused by deletion of ELF4 was likely not due to defects in proliferation or survival but rather resulted from an enhanced Th17 differentiation program, indicating that ELF4 prevents proliferating cells from committing to the Th17 lineage.
Fig. 4. ELF4 modulates Th17 differentiation without affecting proliferation of CD4+ T cells.
(A) Flow cytometric analysis of intracellular IL-17A expression and dilution of the CFSE dye in WT and Elf4−/− CD4+ T cells cultured under Th17 condition. CFSE histograms are shown for total, IL-17A+, and IL-17A− cells. (B) Percentages of total (IL-17A+ and IL-17A−) cells that had undergone indicated cell divisions were calculated in WT and Elf4−/− CD4+ T cells (n=3; mean ± s.d.). (C) Percentages of IL-17A+ cells for each cell division were calculated in WT and Elf4−/− CD4+ T cells (n=3; mean ± s.d.). Data are representative of three independent experiments. *P<0.05, **P<0.01, ***P<0.001 (Two-tailed Student’s t-test).
ELF4 increases cytokine threshold for Th17 differentiation
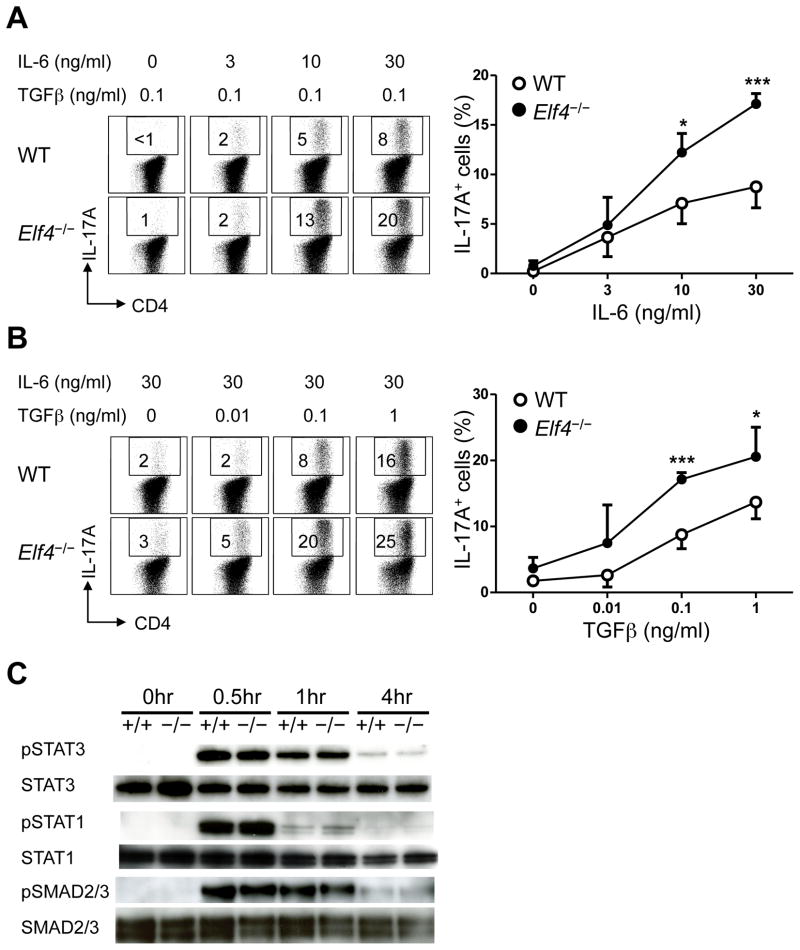
Since TCR-induced proliferation was normal in Elf4−/− CD4+ T cells (Fig 4), we next focused on cytokine signaling driving Th17 differentiation. A dose-dependent study revealed increased expression of IL-17A in Elf4−/− CD4+ T cells in response to intermediate (10 ng/ml) and high (30 ng/ml) concentrations of IL-6 (Fig 5A). However, this hyper-responsiveness to IL-6 was not caused by elevated levels of STAT3 phosphorylation (Fig 5C) or increased surface IL-6Rα expression (data not shown). IL-6 induced a similar level of phosphorylated STAT1 in WT and Elf4−/− CD4+ T cells (Fig. 5C), which is known to negatively regulate Th17 differentiation (37, 38). A parallel study of TGFβ showed increased frequency of IL-17A positive Elf4−/− CD4+ T cells at intermediate (0.1 ng/ml) and high (1 ng/ml) concentrations of TGFβ (Fig 5B) despite normal kinetics of SMAD2/3 phosphorylation (Fig 5C). Of note, a similar frequency of Foxp3+ cells was induced by TGFβ in the presence of IL-6 in Elf4−/− CD4+ T cells compared to WT controls, confirming that enhanced Th17 differentiation was not caused by impaired Treg differentiation (data not shown). Taken together, our findings suggest that ELF4 heightens cytokine requirements of differentiating CD4+ T cells to prevent exacerbated Th17 responses downstream of STAT1, STAT3 and SMAD2/3.
Fig. 5. Increased Th17 differentiation of Elf4−/− CD4+ T cells in response to IL-6 and TGFβ stimulation.
Flow cytometric analysis of intracellular IL-17A in WT and Elf4−/− CD4+ T cells cultured in the presence of either TGFβ (0.1 ng/ml) and increasing concentrations of IL-6 (0–30 ng/ml) (A) or IL-6 (30 ng/ml) and increasing concentrations of TGFβ (0–1 ng/ml) (B). Percentages of IL-17A+ cells are summarized on the right (n=4; mean ± s.d.). (C) Immunoblot analysis shows kinetics of STAT3, STAT1, SMAD2/3 phosphorylation (pSTAT3, pSTAT1, and pSMAD2/3) and total STAT3, STAT1, and SMAD2/3 levels in WT and Elf4−/− CD4+ T cells after activation with αCD3/αCD28 in the presence of IL-6 and TGFβ. Data are representative of two independent experiments. *P<0.05, ***P<0.001 (Two-tailed Student’s t-test).
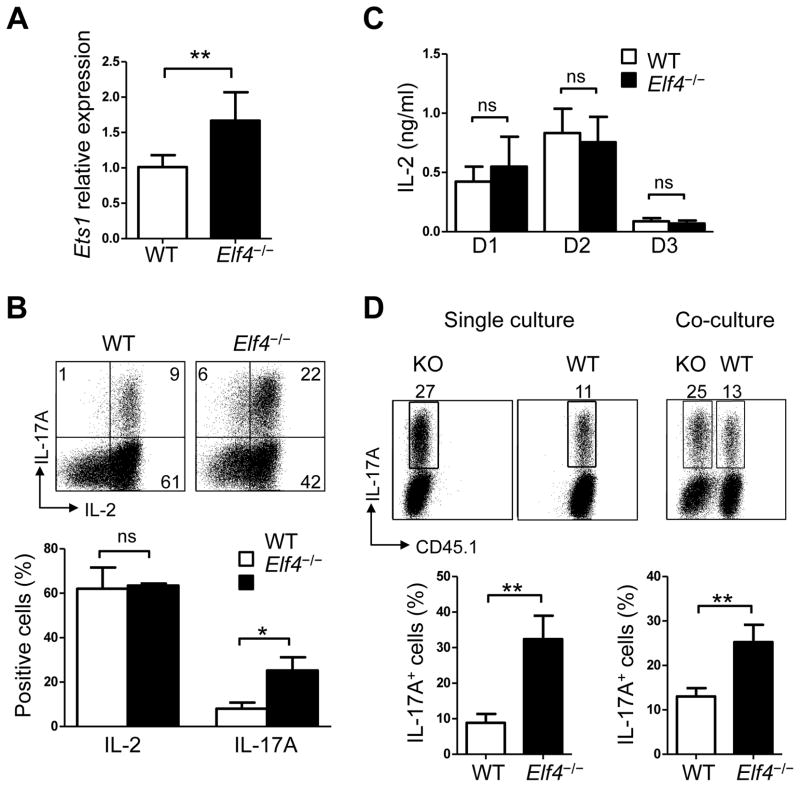
ELF4 inhibits Th17 differentiation in an IL-2-independent manner
Given the similar effect on Th17 cells, we sought to determine whether ELF4 inhibits Th17 differentiation by an IL-2-dependent mechanism similar to ETS1 (13). We first measured the transcript levels of Ets1 and found higher expression in Elf4−/− Th17 cells compared to WT controls (Fig 6A), suggesting that ELF4 does not drive Ets1 gene transcription and that ETS1 is unable to reverse the defects caused by ELF4 deletion. In contrast to the impaired IL-2 production reported for Ets1−/− CD4+ T cells (12), Elf4−/− CD4+ T cells showed normal frequency of IL-2-producing cells (Fig 6B) and secretion of IL-2 to the media (Fig 6C). Next, we co-cultured WT and Elf4−/− CD4+ T cells during Th17 differentiation to examine whether aberrant secretion of IL-2 or other cytokine causes increased Th17 differentiation of Elf4−/− CD4+ T cells. As shown in Fig 6D, Elf4−/− CD4+ T cells generated more Th17 cells even when co-cultured with WT CD4+ T cells, suggesting that the increased Th17 differentiation was caused by a cell intrinsic defect rather than secreted factors, which should have been normalized in a co-culture system. Of note, since IL-2 represses Th17 differentiation via STAT5 (39, 40), we also examined the kinetics of STAT5 phosphorylation under Th17 polarizing condition. STAT5 phosphorylation was not impaired in Elf4−/− CD4+ T cells (data not shown).
Fig. 6. ELF4 does not use the same mechanism as ETS1 to regulate Th17 differentiation.
(A) Quantitative real-time PCR analysis of Ets1 expression was performed in Th17-polarized WT and Elf4−/− CD4+ T cells (n=6; mean ± s.d.). (B) Flow cytometric analysis of intracellular IL-2 and IL-17A expression in Th17-polarized WT and Elf4−/− CD4+ T cells. Percentages of IL-2+ and IL-17A+ cells are summarized in the lower panel (n=3; mean ± s.d.). (C) The secretion of IL-2 on day 1, 2, and 3 was measured by ELISA in WT and Elf4−/− CD4+ T cells cultured under Th17 condition (n=3) (mean ± s.d.). (D) WT (CD45.1+) and Elf4−/− (CD45.1−) CD4+ T cells were cultured separately or co-cultured under Th17-polarizing condition and analyzed for intracellular expression of IL-17A. The percentage of IL-17A+ cells for single and co-culture are summarized in the lower panel (n=3; mean ± s.d.). Data are representative of two independent experiments. ns: not significant, *P<0.05, **P<0.01 (Two-tailed Student’s t-test).
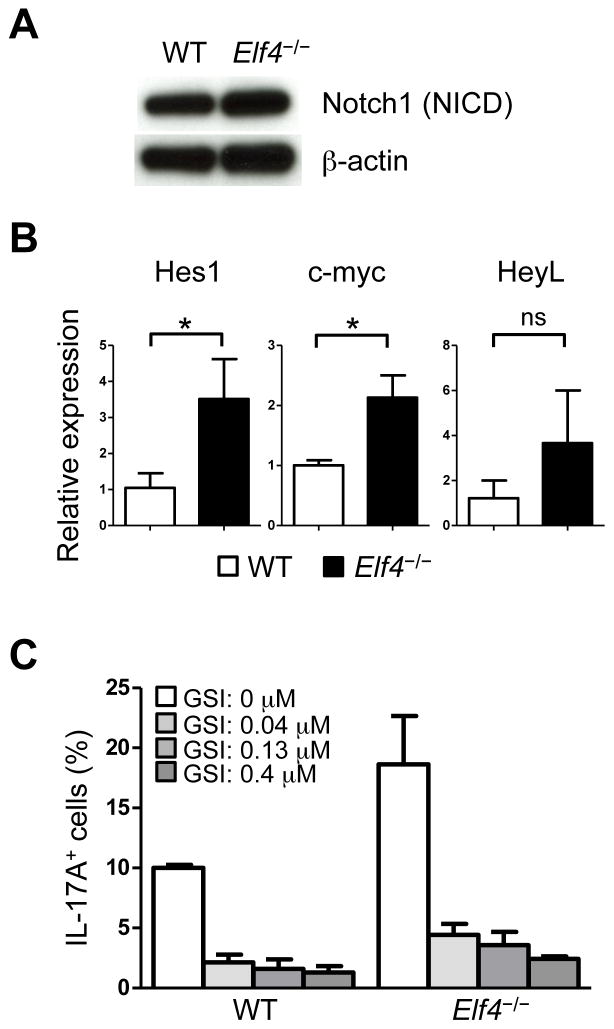
Finally, Notch1 signaling has been shown to promote Th17 differentiation of murine and human CD4+ T cells by directly regulating the expression of the Il17a and Rorc genes (41). Since the expression of both genes was upregulated in the absence of ELF4 (Fig 3), we investigated whether Notch1 pathway was augmented in Elf4−/− CD4+ T cells. Although the protein level of Notch1 intracellular domain (NICD) was similar (Fig 7A), the transcript levels of the Notch1 target genes Hes1 and Myc, and to a lesser extent HeyL, were significantly increased in Elf4−/− Th17 cells (Fig 7B). This finding suggests that ELF4 likely inhibits Notch1 signaling by modulating not the levels but the transcriptional activity of NICD. Chemical disruption of Notch1 pathway by the γ-secretase inhibitor compound E brought the frequency of IL-17A+ cells in Elf4−/− CD4+ T cells down to that of WT controls (Fig 7C), suggesting that increased Th17 differentiation in Elf4−/− CD4+ T cells may be caused by augmented Notch1 signaling. Collectively, these data suggest that ELF4 controls Th17 differentiation using at least in part an ETS1-independent and Notch1-dependent mechanism in differentiating CD4+ T cells.
Fig. 7. Notch1 pathway is associated with ELF4-mediated inhibition on Th17 differentiation.
(A) Immunoblot analysis shows Notch1 intracellular domain (NICD) and β-actin levels in WT and Elf4−/− CD4+ T cells cultured under Th17-polarizing condition for three days. (B) Quantitative real-time PCR analysis of Hes1, Myc, and Heyl expression was performed in Th17-polarized WT and Elf4−/− CD4+ T cells (n=3; mean ± s.d.). (C) Summary of percentages of IL-17A+ fraction in WT and Elf4−/− CD4+ T cells cultured under Th17 polarizing condition in the presence of 0, 0.04, 0.13, or 0.4 μM of the γ-secretase inhibitor (GSI) compound E. Data are representative of two independent experiments. ns: not significant, *P<0.05 (Two-tailed Student’s t-test).
Loss of ELF4 leads to stronger in vivo Th17 responses
To further dissect the selective role of ELF4 in Th17 cell responses in vivo, we examined the composition of lamina propria (LP) T helper cells in the small intestine, a site where Th1/Th17 differentiation is constitutively driven by commensal microbiota (42, 43). Consistent with our in vitro findings, we found a significant increase in Th17, but not Th1, cells in the LP of unchallenged Elf4−/− mice compared to WT controls (Fig 8A), suggesting increased homeostatic differentiation of Th17 cells in Elf4−/− mice. Next, we induced Th1 and Th17 differentiation in vivo by immunizing mice with the MHC-II-restricted peptide myelin oligodendrocyte glycoprotein 35–55 (MOG35-55) emulsified in CFA (44–46). Elf4−/− mice displayed a stronger induction of Th17 cells in the draining LNs, whereas the magnitude of Th1 response was largely unaffected (Fig 8B). A normal Th1 response along with normal expression of CTLA-4 and GITR in CD4+CD25+Foxp3+ cells in the thymus and the spleen of Elf4−/− mice (data not shown) suggest that the Th17-skewed response was likely not due to impaired function of Treg cells. These data demonstrate that ELF4 selectively modulates Th17 differentiation in vivo both at steady state and after immunization.
Fig. 8. Elf4−/− mice show increased Th17 responses in vivo.
(A) Percentages of CD4+IL-17A+ and CD4+IFNγ+ cells isolated from lamina propria of small intestine of non-manipulated WT and Elf4−/− mice (n=3; mean ± s.d.). (B) Percentages of CD4+IL-17A+ and CD4+IFNγ+ cells isolated from the draining lymph nodes of WT and Elf4−/− mice ten days after immunization with MOG35-55 peptide emulsified in CFA. (C) Percentages of IL-17A+ fraction in NK cells (NK1.1+TCRβ−), γδ T cells (TCRγδ+), and LTi-like cells (CD4+TCRβ−NK1.1−B220−CD11b−CD11c−) isolated from the draining lymph nodes as in (B) (n=3 or 5; mean ± s.d.). Data are representative of two independent experiments. ns: not significant, *P<0.05, **P<0.01, ***P<0.001 (Two-tailed Student’s t-test).
Given that innate cells such as NK cells, γδ T cells, and lymphoid tissue inducer-like (LTi-like) cells are also important sources of IL-17A (47–49), we examined whether ELF4 regulates IL-17A production in these cell types. Ten days after MOG35-55/CFA immunization, very few NK cells were found to produce IL-17A regardless of ELF4 expression (Fig 8C). The frequency of IL-17A+ γδ T cells was also comparable between WT and Elf4−/− mice (Fig 8C). Interestingly, loss of ELF4 led to significant reduction in IL-17A-producing population in LTi-like cells (Fig 8C), suggesting that IL-17A is differentially regulated in different cell types.
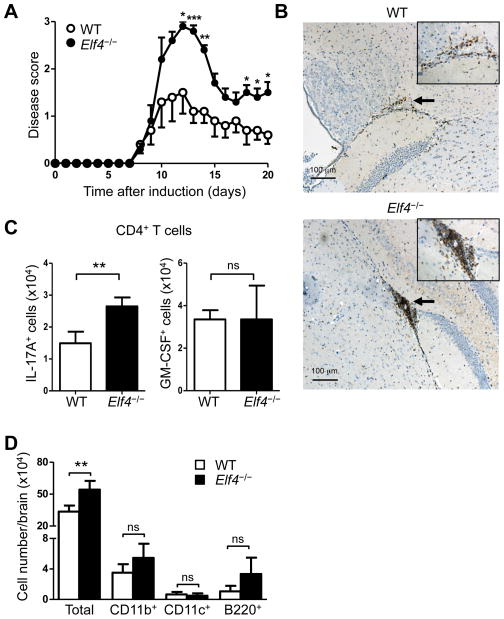
A growing body of evidence highlighted the importance of Th17 cells in the pathogenesis of multiple sclerosis, a neurodegenerative disease caused by autoimmune inflammatory responses in the central nervous system (50–52). Since ELF4 inhibited Th17 differentiation both in vitro and in vivo, we sought to determine the role of ELF4 in the pathogenesis of EAE, the most widely used animal model to study multiple sclerosis. As expected, the disease severity was worsened in Elf4−/− mice, which showed more severe paralysis (Fig 9A) and increased weight loss (data not shown) during the disease course. In addition, we detected larger perivascular infiltrates of CD3+ T cells in brain sections from diseased Elf4−/− mice (Fig 9B). In accord with these findings, brains of Elf4−/− mice collected 14 days after EAE induction contained increased numbers of IL-17A+ CD4+ T cells (Fig 9C), while no significant differences were found in the content of GM-CSF+ CD4+ T cells (Fig 9C) or IFNγ+ CD4+ T cells (data not shown). Finally, the increased numbers of cells infiltrated to the brain was not composed of more myeloid (CD11b+), dendritic cells (CD11c+), or B cells (B220+) (Fig 9D). In summary, expression of ELF4 plays a unique role in the differentiation of Th17 cells and attenuation of EAE.
Fig 9. Increased severity of EAE in Elf4−/− mice.
(A) EAE disease score of WT and Elf4−/− mice immunized with MOG35-55 peptide (n=5; mean ± s.e.m.). (B) Immunohistochemistry of CD3+ cells in brain sections from WT and Elf4−/− mice (fourteen days after EAE induction). Insets show higher magnification of areas indicated by arrows. Infiltrated mononuclear cells were isolated from brains of WT and Elf4−/− mice fourteen days after EAE induction. (C) The numbers of CD4+TCRβ+IL-17A+ and CD4+TCRβ+GM-CSF+ cells (n=3; mean ± s.d.). (D) The numbers of total infiltrating cells, CD11b+ cells, CD11c+ cells, and B220+ cells (n=3; mean ± s.d.). Data are representative of three independent experiments. ns: not significant, *P<0.05, **P<0.01, ***P<0.001 (Two-tailed Student’s t-test).
Discussion
Th17 cells are implicated in both pathogen clearance and development of autoimmune diseases owing to their proinflammatory properties, and thus extensive studies have focused on their generation and maintenance, aiming at developing novel strategies to control immune deregulation. Th17 cells share the same precursor – naïve CD4+ T cells– with other T helper lineages, making it difficult to specifically manipulate Th17 responses in patients. Therefore, the identification of lineage-specific regulators is essential for future clinical applications.
This is the first study identifying a novel function of ELF4 in CD4+ T cells as a cell-intrinsic negative regulator of Th17 differentiation. In this work, we found increased differentiation of Elf4−/− CD4+ T cells toward the Th17 lineage both in vitro and in vivo. Despite normal TCR-driven proliferation and activation of IL-6R and TGFβR proximal signaling, dividing Elf4−/− CD4+ T cells exhibited increased percentages of IL-17A positive cells in the second cell division and onward, suggesting that ELF4 represses Th17 lineage commitment in activated CD4+ T cells downstream of STAT3, STAT1, and SMAD2/3. Furthermore, the finding that the Th17 differentiation program was upregulated in Elf4−/− CD4+ T cells indicates that ELF4 acts earlier than the master regulators IRF4 and RORγt during lineage commitment. Elf4−/− mice exhibited worsened disease progression than WT mice in a model of experimental autoimmune encephalomyelitis, supporting a novel role of ELF4 in the pathobiology of autoimmune diseases. Interestingly, ELF4 expression was found elevated in patients suffering from multiple sclerosis after IFNβ treatment (53), and ELF4 is regulated by ligand-bound vitamin D receptor, which has a protective role against multiple sclerosis (54). The fact that ELF4 selectively modulates Th17 differentiation with no significant effect on the development of Th1, Th2, and Treg cells makes ELF4 an interesting transcriptional target in Th17-mediated immune disorders.
STAT3 mediates signal transduction downstream of the IL-6, IL-21, and IL-23 receptors and therefore is crucial for both initiation and maintenance of Th17 cells (3, 5, 55). Since Elf4−/− CD4+ T cells showed increased Th17 differentiation induced by two different combinations of cytokines that mediate STAT3 activation, we hypothesized that ELF4 abrogates STAT3 activation to prevent Th17 differentiation. However, immunoblots and flow cytometric analysis showed a normal kinetic of STAT3 phosphorylation in Elf4−/− CD4+ T cells. In addition to STAT3, we did not observe reduced STAT1 phosphorylation that would explain the increased Th17 differentiation of Elf4−/− CD4+ T cells. Similarly, Elf4−/− CD4+ T cells showed normal levels of phosphorylated SMAD2/3 in response to TGFβ. The increased Th17 differentiation of Elf4−/− CD4+ T cells driven by IL-6 and TGFβ stimuli with normal activation of STAT3 and SMAD2/3 implies that ELF4 may either suppress their transcriptional activity or interfere with their co-factors/downstream targets such as IRF4 and RORγt via protein-to-protein interactions.
The increased production of IL-17A in Elf4−/− CD4+ T cells could be due to a direct regulation of the Il17a gene by ELF4. However, promoter reporter assays showed that ELF4 did not suppress Il17a promoter activity in both COS7 and 293T cells (data not shown), consistent with the current understanding of ELF4 as a transcriptional activator (17, 22, 23). Therefore, ELF4-mediated repression requires proteins absent in the cell lines or ELF4 has no direct control in the Il17a gene. The fact that Th17 program is upregulated in Elf4−/− CD4+ T cells supports the latter model. In addition to binding DNA, the ETS family proteins can interact with other transcription factors or co-regulators to modulate gene expression, leading to gene activation or silencing in a cell context-dependent manner. For example, binding of ELF4 to RUNX1 results in a synergistic activation of Il3 promoter (56) while the interaction between ELF4 and RUNX2 inhibits RUNX2 transcriptional activity (57). Thus, it is plausible that ELF4 inhibits Th17 differentiation by forming a complex with RUNX1 that disrupts its function. This hypothesis is supported by two findings: RUNX1 has been shown to promote Th17 differentiation (58, 59), and RUNX1 physically interacts with ELF4 in t(8;21)-positive acute myeloid leukemia cells (56).
Although APC-derived cytokines are the main driving force during Th17 differentiation, factors secreted by CD4+ T cells also regulate development of Th17 cells in an autocrine/paracrine manner. For example, IL-6 induces CD4+ T cells to secret IL-21 (60) that promotes Th17 differentiation in cooperation with TGFβ1 (61, 62). Similarly, developing Th17 cells can secret TGFβ3 in response to IL-23, which induces the formation of pathogenic Th17 cells in concert with IL-6 (31). On the other hand, IL-2 produced by CD4+ T cells inhibits Th17 differentiation in a STAT5-dependent manner (39, 40). It has been demonstrated that Ets1−/− CD4+ T cells showed augmented Th17 differentiation due to impaired production of IL-2 and increased resistance to the inhibitory effects of IL-2 (13). However, the production of IL-2 in Elf4−/− CD4+ T cells was comparable to WT controls. Furthermore, co-culture experiments also suggested that autocrine/paracrine cytokines such as IL-2 and IL-21, whose transcript level was higher in Elf4−/− CD4+ T cells, are unlikely to account for the inhibitory effect of ELF4 on Th17 differentiation.
The immunosuppressive drug rapamycin has been found effective in the treatment of EAE in part by suppressing Th17 responses via inhibition of mTORC1 activity (63-66). The mTOR pathway appeared to be intact in Elf4−/− CD4+ T cells as evidenced by normal protein levels of phosphorylated mTOR, mTOR, RACTOR, RICTOR, and GβL (data not shown). Since mTOR represses the transcription of ELF4 downstream of TCR activation (24), it is possible that rapamycin inhibits Th17 differentiation by relieving mTOR-mediated repression of ELF4. Since mTOR is widely involved in many aspects of T cell biology (67), its potential downstream molecule ELF4 is an alternative molecular target for treating Th17-mediated autoimmune disorders.
Notch1 signaling pathway, a highly conserved cell signaling, participates in many aspects of the immune system including T helper cell differentiation (68). Different Notch1 ligands expressed on APC instruct CD4+ T cells towards Th1 or Th2 lineage by engaging the Notch1 receptor (68, 69). Since the Notch1 pathway is also activated upon TCR stimulation (70, 71) and that Notch1 is essential for Th17 differentiation (41), we decided to investigate whether ELF4 modulates Notch1 signaling in an APC-free in vitro culture system. The normal protein level of NICD in Elf4−/− CD4+ T cells indicates that the cleavage of Notch1 and subsequent release of NICD is not regulated by ELF4. However, Elf4−/− CD4+ T cells showed elevated expression of certain Notch1 target genes and their increased Th17 differentiation was normalized when Notch1 signaling was disrupted. These findings suggest that ELF4 either lowers the transcriptional activity of NICD through direct binding, which was suggested for GABPα and GABPβ via ETS domain and Notch-related structural motif (72), or directly suppresses the gene signature of Notch1 pathway to regulate Th17 differentiation.
Taken together, our data demonstrate that ELF4 selectively inhibits the differentiation program of Th17 cells with no major effects on the proliferation and survival of CD4+ T cells. Hence, loss of ELF4 worsened clinical symptoms of EAE at least in part by increasing infiltration of Th17 cells to the brain. The specificity on the Th17 lineage makes ELF4 an ideal therapeutic target to control Th17 responses. Future studies should target ELF4, or its upstream/downstream targets, to control aberrant immune response more specifically and efficiently than cytokine-based and immunosuppressive therapies.
Acknowledgments
This work was supported by National Institutes of Health Grants R01AI077536 and R01AI077536-02S1
We thank Dr. Dotti G for critically reading the manuscript and Goltsova T for cell sorting assistance.
Abbreviations
- ELF4
E74-like factor 4
- EAE
Experimental autoimmune encephalomyelitis
- RORγt
RAR-related orphan receptor gamma t
- RUNX
Runt-related transcription factor
- ETS
E-twenty six
- mTOR
Mammalian target of rapamycin
- LN
Lymph node
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Luckheeram RV, Zhou R, Verma AD, Xia B. CD4(+)T cells: differentiation and functions. Clin Dev Immunol. 20122012:925135. doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei L, Laurence A, Elias KM, O’Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–34610. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 5.Zhou L, Ivanov, Spolski RR, Shenderov K, Egawa T, Levy DE, Leonard WJ, Littman DR. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–974. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- 6.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 7.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 9.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 10.Dittmer J, Nordheim A. Ets transcription factors and human disease. Biochim Biophys Acta. 1998;1377:F1–11. doi: 10.1016/s0304-419x(97)00039-5. [DOI] [PubMed] [Google Scholar]
- 11.Lacorazza HD, Nimer SD. The emerging role of the myeloid Elf-1 like transcription factor in hematopoiesis. Blood Cells Mol Dis. 2003;31:342–350. doi: 10.1016/s1079-9796(03)00162-1. [DOI] [PubMed] [Google Scholar]
- 12.Grenningloh R, Kang BY, Ho IC. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J Exp Med. 2005;201:615–626. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moisan J, Grenningloh R, Bettelli E, Oukka M, Ho IC. Ets-1 is a negative regulator of Th17 differentiation. J Exp Med. 2007;204:2825–2835. doi: 10.1084/jem.20070994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouly E, Chemin K, Nguyen VH, Chopin M, Mesnard L, Leite-de-Moraes M, Burlen-defranoux O, Bandeira A, Bories JC. The Ets-1 transcription factor controls the development and function of natural regulatory T cells. J Exp Med. 2010;207:2113–2125. doi: 10.1084/jem.20092153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, Kaplan MH. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–534. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang HC, Zhang S, Thieu VT, Slee RB, Bruns HA, Laribee RN, Klemsz MJ, Kaplan MH. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 17.Miyazaki Y, Sun X, Uchida H, Zhang J, Nimer S. MEF, a novel transcription factor with an Elf-1 like DNA binding domain but distinct transcriptional activating properties. Oncogene. 1996;13:1721–1729. [PubMed] [Google Scholar]
- 18.Seki Y, Suico MA, Uto A, Hisatsune A, Shuto T, Isohama Y, Kai H. The ETS transcription factor MEF is a candidate tumor suppressor gene on the X chromosome. Cancer Res. 2002;62:6579–6586. [PubMed] [Google Scholar]
- 19.Yao JJ, Liu Y, Lacorazza HD, Soslow RA, Scandura JM, Nimer SD, Hedvat CV. Tumor promoting properties of the ETS protein MEF in ovarian cancer. Oncogene. 2007;26:4032–4037. doi: 10.1038/sj.onc.1210170. [DOI] [PubMed] [Google Scholar]
- 20.Sivina M, Yamada T, Park CS, Puppi M, Coskun S, Hirschi K, Lacorazza HD. The transcription factor E74-like factor controls quiescence of endothelial cells and their resistance to myeloablative treatments in bone marrow. Arterioscler Thromb Vasc Biol. 2011;31:1185–1191. doi: 10.1161/ATVBAHA.111.224436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacorazza HD, Yamada T, Liu Y, Miyata Y, Sivina M, Nunes J, Nimer SD. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9:175–187. doi: 10.1016/j.ccr.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Kruppel-like factors KLF4 and KLF2. Nat Immunol. 2009;10:618–626. doi: 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, Cordon-Cardo C, Mao S, Pandolfi PP, Nimer SD. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17:437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 24.Yamada T, Gierach K, Lee PH, Wang X, Lacorazza HD. Cutting edge: Expression of the transcription factor E74-like factor 4 is regulated by the mammalian target of rapamycin pathway in CD8+ T cells. J Immunol. 2010;185:3824–3828. doi: 10.4049/jimmunol.1000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XO, Chang SH, Park H, Nurieva R, Shah B, Acero L, Wang YH, Schluns KS, Broaddus RR, Zhu Z, Dong C. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008;205:1063–1075. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stromnes IM, Goverman JM. Active induction of experimental allergic encephalomyelitis. Nat Protoc. 2006;1:1810–1819. doi: 10.1038/nprot.2006.285. [DOI] [PubMed] [Google Scholar]
- 27.Laky K, Lefrancois L, Puddington L. Age-dependent intestinal lymphoproliferative disorder due to stem cell factor receptor deficiency: parameters in small and large intestine. J Immunol. 1997;158:1417–1427. [PubMed] [Google Scholar]
- 28.Liang SC, Long AJ, Bennett F, Whitters MJ, Karim R, Collins M, Goldman SJ, Dunussi-Joannopoulos K, Williams CM, Wright JF, Fouser LA. An IL-17F/A heterodimer protein is produced by mouse Th17 cells and induces airway neutrophil recruitment. J Immunol. 2007;179:7791–7799. doi: 10.4049/jimmunol.179.11.7791. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Weiner J, Liu Y, Smith AJ, Huss DJ, Winger R, Peng H, Cravens PD, Racke MK, Lovett-Racke AE. T-bet is essential for encephalitogenicity of both Th1 and Th17 cells. J Exp Med. 2009;206:1549–1564. doi: 10.1084/jem.20082584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W, O’Shea JJ. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, Wu C, Kleinewietfeld M, Kunder S, Hafler DA, Sobel RA, Regev A, Kuchroo VK. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeachy MJ, Bak-Jensen KS, Chen Y, Tato CM, Blumenschein W, McClanahan T, Cua DJ. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 33.El-Behi M, Ciric B, Dai H, Yan Y, Cullimore M, Safavi F, Zhang GX, Dittel BN, Rostami A. The encephalitogenicity of T(H)17 cells is dependent on IL-1- and IL-23-induced production of the cytokine GM-CSF. Nat Immunol. 2011;12:568–575. doi: 10.1038/ni.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Codarri L, Gyulveszi G, Tosevski V, Hesske L, Fontana A, Magnenat L, Suter T, Becher B. RORgammat drives production of the cytokine GM-CSF in helper T cells, which is essential for the effector phase of autoimmune neuroinflammation. Nat Immunol. 2011;12:560–567. doi: 10.1038/ni.2027. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 36.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 37.Villarino AV, Gallo E, Abbas AK. STAT1-activating cytokines limit Th17 responses through both T-bet-dependent and -independent mechanisms. J Immunol. 2010;185:6461–6471. doi: 10.4049/jimmunol.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura A, Naka T, Nohara K, Fujii-Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci U S A. 2008;105:9721–9726. doi: 10.1073/pnas.0804231105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea J. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 40.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keerthivasan S, Suleiman R, Lawlor R, Roderick J, Bates T, Minter L, Anguita J, Juncadella I, Nickoloff BJ, Le Poole IC, Miele L, Osborne BA. Notch signaling regulates mouse and human Th17 differentiation. J Immunol. 2011;187:692–701. doi: 10.4049/jimmunol.1003658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanov, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Atarashi K, Tanoue T, Honda K. Induction of lamina propria Th17 cells by intestinal commensal bacteria. Vaccine. 2010;28:8036–8038. doi: 10.1016/j.vaccine.2010.09.026. [DOI] [PubMed] [Google Scholar]
- 44.Umemura M, Yahagi A, Hamada S, Begum MD, Watanabe H, Kawakami K, Suda T, Sudo K, Nakae S, Iwakura Y, Matsuzaki G. IL-17-mediated regulation of innate and acquired immune response against pulmonary Mycobacterium bovis bacille Calmette-Guerin infection. J Immunol. 2007;178:3786–3796. doi: 10.4049/jimmunol.178.6.3786. [DOI] [PubMed] [Google Scholar]
- 45.Sano K, Haneda K, Tamura G, Shirato K. Ovalbumin (OVA) and Mycobacterium tuberculosis bacilli cooperatively polarize anti-OVA T-helper (Th) cells toward a Th1-dominant phenotype and ameliorate murine tracheal eosinophilia. Am J Respir Cell Mol Biol. 1999;20:1260–1267. doi: 10.1165/ajrcmb.20.6.3546. [DOI] [PubMed] [Google Scholar]
- 46.Linnington C, Webb M, Woodhams PL. A novel myelin-associated glycoprotein defined by a mouse monoclonal antibody. J Neuroimmunol. 1984;6:387–396. doi: 10.1016/0165-5728(84)90064-x. [DOI] [PubMed] [Google Scholar]
- 47.Passos ST, Silver JS, O’Hara AC, Sehy D, Stumhofer JS, Hunter CA. IL-6 promotes NK cell production of IL-17 during toxoplasmosis. J Immunol. 2010;184:1776–1783. doi: 10.4049/jimmunol.0901843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takatori H, Kanno Y, Watford WT, Tato CM, Weiss Ivanov G, II, Littman DR, O’Shea JJ. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med. 2009;206:35–41. doi: 10.1084/jem.20072713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin B, Hirota K, Cua DJ, Stockinger B, Veldhoen M. Interleukin-17-producing gammadelta T cells selectively expand in response to pathogen products and environmental signals. Immunity. 2009;31:321–330. doi: 10.1016/j.immuni.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 50.Lock C, Hermans G, Pedotti R, Brendolan A, Schadt E, Garren H, Langer-Gould A, Strober S, Cannella B, Allard J, Klonowski P, Austin A, Lad N, Kaminski N, Galli SJ, Oksenberg JR, Raine CS, Heller R, Steinman L. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat Med. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- 51.Kebir H, Kreymborg K, Ifergan I, Dodelet-Devillers A, Cayrol R, Bernard M, Giuliani F, Arbour N, Becher B, Prat A. Human TH17 lymphocytes promote blood-brain barrier disruption and central nervous system inflammation. Nat Med. 2007;13:1173–1175. doi: 10.1038/nm1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishizu T, Osoegawa M, Mei FJ, Kikuchi H, Tanaka M, Takakura Y, Minohara M, Murai H, Mihara F, Taniwaki T, Kira J. Intrathecal activation of the IL-17/IL-8 axis in opticospinal multiple sclerosis. Brain. 2005;128:988–1002. doi: 10.1093/brain/awh453. [DOI] [PubMed] [Google Scholar]
- 53.Singh MK, Scott TF, LaFramboise WA, Hu FZ, Post JC, Ehrlich GD. Gene expression changes in peripheral blood mononuclear cells from multiple sclerosis patients undergoing beta-interferon therapy. J Neurol Sci. 2007;258:52–59. doi: 10.1016/j.jns.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 54.Satoh JI, Tabunoki H. Molecular network of chromatin immunoprecipitation followed by deep sequencing-based vitamin D receptor target genes. Mult Scler. 2013 doi: 10.1177/1352458512471873. [DOI] [PubMed] [Google Scholar]
- 55.Chirica CM, Timans J, Vaisberg E, Travis M, Cheung J, Pflanz S, Zhang R, Singh KP, Vega F, To W, Wagner J, O’Farrell AM, McClanahan T, Zurawski S, Hannum C, Gorman D, Rennick DM, Kastelein RA, de Waal Malefyt R, Moore KW. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 56.Mao S, Frank RC, Zhang J, Miyazaki Y, Nimer SD. Functional and physical interactions between AML1 proteins and an ETS protein, MEF: implications for the pathogenesis of t(8;21)-positive leukemias. Mol Cell Biol. 1999;19:3635–3644. doi: 10.1128/mcb.19.5.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim YJ, Kim BG, Lee SJ, Lee HK, Lee SH, Ryoo HM, Cho JY. The suppressive effect of myeloid Elf-1-like factor (MEF) in osteogenic differentiation. J Cell Physiol. 2007;211:253–260. doi: 10.1002/jcp.20933. [DOI] [PubMed] [Google Scholar]
- 58.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–1306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lazarevic V, Chen X, Shim JH, Hwang ES, Jang E, Bolm AN, Oukka M, Kuchroo VK, Glimcher LH. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORgammat. Nat Immunol. 2011;12:96–104. doi: 10.1038/ni.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suto A, Kashiwakuma D, Kagami S, Hirose K, Watanabe N, Yokote K, Saito Y, Nakayama T, Grusby MJ, Iwamoto I, Nakajima H. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med. 2008;205:1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nurieva R, Yang XO, Martinez G, Zhang Y, Panopoulos AD, Ma L, Schluns K, Tian Q, Watowich SS, Jetten AM, Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–483. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- 62.Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007;448:484–487. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurebayashi Y, Nagai S, Ikejiri A, Ohtani M, Ichiyama K, Baba Y, Yamada T, Egami S, Hoshii T, Hirao A, Matsuda S, Koyasu S. PI3K-Akt-mTORC1-S6K1/2 Axis Controls Th17 Differentiation by Regulating Gfi1 Expression and Nuclear Translocation of ROR3. Cell Reports. 2012;1:360–373. doi: 10.1016/j.celrep.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 65.Esposito M, Ruffini F, Bellone M, Gagliani N, Battaglia M, Martino G, Furlan R. Rapamycin inhibits relapsing experimental autoimmune encephalomyelitis by both effector and regulatory T cells modulation. J Neuroimmunol. 2010;220:52–63. doi: 10.1016/j.jneuroim.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 66.Donia M, Mangano K, Amoroso A, Mazzarino MC, Imbesi R, Castrogiovanni P, Coco M, Meroni P, Nicoletti F. Treatment with rapamycin ameliorates clinical and histological signs of protracted relapsing experimental allergic encephalomyelitis in Dark Agouti rats and induces expansion of peripheral CD4+CD25+Foxp3+ regulatory T cells. J Autoimmun. 2009;33:135–140. doi: 10.1016/j.jaut.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 67.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Yamane H, Paul WE. Early signaling events that underlie fate decisions of naive CD4(+) T cells toward distinct T-helper cell subsets. Immunol Rev. 2013;252:12–23. doi: 10.1111/imr.12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Palaga T, Miele L, Golde TE, Osborne BA. TCR-mediated Notch signaling regulates proliferation and IFN-gamma production in peripheral T cells. J Immunol. 2003;171:3019–3024. doi: 10.4049/jimmunol.171.6.3019. [DOI] [PubMed] [Google Scholar]
- 71.Guy CS, Vignali KM, Temirov J, Bettini ML, Overacre AE, Smeltzer M, Zhang H, Huppa JB, Tsai YH, Lobry C, Xie J, Dempsey PJ, Crawford HC, Aifantis I, Davis MM, Vignali DA. Distinct TCR signaling pathways drive proliferation and cytokine production in T cells. Nat Immunol. 2013;14:262–270. doi: 10.1038/ni.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson CC, Brown TA, McKnight SL. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]