Abstract
The heritability of Borderline Personality (BP) features has been established in multiple twin and family studies. Using data from the borderline subscale of the Personality Assessment Inventory (PAI-BOR) collected in two Dutch cohorts (N total=7,125), the Netherlands Twin Register (NTR) and the Netherlands Study of Depression and Anxiety (NESDA), we show that heritability of the PAI-BOR total score using genome-wide SNPs is estimated at 23%, and that the genetic variance is substantially higher in affect instability items compared to the other three subscales of the PAI-BOR (42.7% vs non-significant estimates for self-harm, negative relations, and identity problems). We present results from a first GWAS of BP features, which shows a promising signal on chromosome 5 corresponding to SERINC5, a protein involved in myelination. Reduced myelination has been suggested as possibly playing a role in the development of psychiatric disorders characterized by lack of social interaction. The signal was confirmed in a third independent Dutch cohort drawn from the Erasmus Rucphen Family study (ERF, N=1301). Our analyses were complemented by investigating the heterogeneity that was implied by the differences in genetic variance components in the four subscales of the PAI-BOR. These analyses show that the association of SNPs tagging SERINC5 differs substantially across the 24 items of the PAI-BOR. Further, using reverse regression we showed that the effects were present only in subjects with higher scores on the PAI-BOR. Taken together, these results suggest that future genome-wide analyses can benefit substantially by taking into account the phenotypic and genetic heterogeneity of BP features.
Keywords: GCTA, GWAS, borderline personality disorder, PAI-BOR, heterogeneity
Introduction
Borderline personality disorder is a severe psychiatric disorder that is characterized by a pervasive dysregulation of emotion as well as behavior, including suicidal behaviors, instability of relations and self image, and impulse control. The disorder is highly comorbid with mood and anxiety disorders, substance use, and other personality disorders.1–3 Heritability estimates of borderline personality (BP) in twin and family studies vary between 30% and 70% depending on age group, the type of measurement (diagnosis vs. dimensional), and the statistical approach (latent trait vs observed measurements).4–7
This study is a first genome-wide association analysis focusing on borderline personality features (BP features). The study uses data from three cohorts, a Dutch population based-sample that originated from the Netherlands Twin Register (NTR, N=5802), a subsample of the Netherlands Study of Depression and Anxiety study with available borderline phenotypes (NESDA, N=1332), and a Dutch sample from the Erasmus Rucphen Family study (ERF, N=1301).8–10
In all participants the phenotype was assessed with the same 24 items of the borderline subscale of the Personality Assessment Inventory (PAI-BOR).11 By design the PAI-BOR scale is divided into four subscales each consisting of six items. The four subscales target affect instability (AI), identity problems (IP), negative relations (NR), and self harm (SH), respectively. Initial factor analytical studies supported the designed 4-factor structure, while more recent studies indicate that six factors, or an alternative 4-factor solution might provide a better fit to the data.11–13 The factors in these studies have moderate to high intercorrelations, underlining the fact the 24 PAI-BOR features have substantial common variability. This is in line with evidence of high clinical validity of the PAI-BOR total score in discriminating diagnosed vs non-diagnosed subjects (73%–80% correct assignment).14–16 The PAI-BOR expands on the nine DSM-IV borderline criteria. The DSM-IV criteria support a single factor, although a similarly well-fitting 3-factor structure has been advocated as providing more detailed information concerning the different aspects of borderline personality (i.e., disturbed relatedness, behavioral dysregulation, and affective dysregulation).17–19
BP has been described as phenotypically heterogeneous, and different biological pathways may be associated with the different aspects of BP.20,21 In the context of this paper we define phenotypic heterogeneity as differences in the phenotype profiles across subgroups in the population. Examples are gender and age differences especially of the impulsivity and affect instability symptoms, and gender differences in comorbidity patterns.22,23 We define genetic heterogeneity as differential genetic effects across subgroups. Phenotypic heterogeneity can indicate but does not imply genetic heterogeneity. In addition to phenotypic and genotypic heterogeneity, it is possible that genetic effects differ for different aspects of BP.24 Our main analyses focus on the common core of the 24 features assessed with the PAI-BOR by using an aggregate score, however, these analyses are complemented by a set of analyses that address phenotypic and genetic heterogeneity as well as differential genetic effects across PAI-BOR subscales and items.
The total sample size in the current study is relatively small for a genome-wide analysis of a psychiatric disorder (N=8426 for the three cohorts), however, the study benefits from the fact that the phenotype is measured with exactly the same instrument in all participants. Using different measures of a disorder across samples can prioritize different aspects of the disorders in the different samples, which dilutes power in the meta-analysis. Furthermore, the borderline phenotype in our association study is quantitative, which can provide considerably increased power compared to case/control phenotypes.25
We present analyses consisting of (1) a phenotype comparison of the NTR and NESDA samples, (2) estimation of SNP-heritability using the software package GCTA v1.11 in the NTR and combined NTR/NESDA samples,26 (3) a GWAS with NTR/NESDA as discovery sample, and ERF as replication samples, (4) an inverse variance meta-analysis of all three samples, and (5) a set of analyses addressing heterogeneity. For the GCTA and association analyses (i.e., (2), (3), and (4)) we aggregated the 24 PAI-BOR items to obtain a dimensional measure of BP features. Previous research has suggested that a dimensional conceptualization of BPD is preferable compared to a case/control approach because a dimensional approach does not necessitate a cutoff, and is more robust against fluctuations over time between sub-threshold and above threshold levels of the disorder.27 The GCTA analyses (2) were also done on the subscale level, and for parts (1) and (5) we assessed item level data.
Materials and Methods
Participants
1. NTR sample
The sample used in the analyses described below consisted of N=5802 participants of the NTR.8 NTR participants are ascertained because of the presence of twins or triplets in the family and consist of multiples, their parents, siblings and spouses. Twins are born in all strata of society and NTR represents a general sample from the Dutch population. For all analysis we excluded one MZ twin per pair. Additional corrections for family resemblance are analysis specific, and described where appropriate. Age ranged between 13 and 90 (median 38), and 62.7% was female.
2. NESDA sample
We included N=1323 participants from NESDA with available phenotype data that passed quality control. NESDA is a longitudinal study focusing on the course and consequences of depression and anxiety disorders.9 Subjects for NESDA were recruited from three sources, namely the general population, mental health organizations, and general practices. The vast majority of NESDA subjects is selected for depression and anxiety, but the sample also includes healthy controls without lifetime psychiatric disorders.9 Age ranged between 18 and 65 in NESDA (median 43), and the proportion of females was 66.1%.
3. ERF sample
ERF is embedded in the Genetic Research in Isolated Population (GRIP) program. The ERF study selected 22 related (first /second degree, non-consanguineous) couples living in Rucphen, the Netherlands, between 1850–1900, and having at least six children. All living descendants of those couples were invited to participate in ERF.10 About 4,000 relatives included in the study form a very complex pedigree, which consists of more than 23,000 members. N=1,466 individuals from this population were phenotyped for borderline personality features, and N=1,301 that passed quality control were included in the current analyses. ERF has an age range of 18 to 80 with a mean of 47.9, and 56% females.
Phenotype
BP features were measured with the borderline subscale of the Personality Assessment Inventory (PAI-BOR).11 The PAI-BOR consists of 24 items that are grouped in parcels of six items focusing on affect instability (e.g., “mood shifts”, “little control over anger”), identity problems (e.g., “can’t handle separation”, “feel empty”), negative relations (e.g., “mistakes in picking friends”, “stormy relations”), and self-harm (e.g., “when upset hurt self”, “too impulsive”). Item level data were available for analyses in the NTR and NESDA samples. We excluded subjects with response rates per subscale <67%, resulting in a median missing proportion across items of 0.0027. The same aggregate scores were computed in the ERF sample, and were used for replication of association results and meta-analysis.
Genotype
Genotyping in the NTR and NESDA samples was done on multiple platforms for partly overlapping subsets of the total sample (Affymetrix Perlegen 5.0, Illumina 370, Illumina 660, Illumina Omni Express 1M, and Affymetrix 6.0). Quality control was done within as well as between platforms. Imputation was done in IMPUTEv2.1 using the 1000 Genomes panel as a reference set. SNP removal criteria included mismatching alleles, allele frequencies differing >0.20 with the reference set, MAF <1%, HWE p-value <.00001, and call rate <95%. Samples were excluded in case of sex mismatch, genotype missing rate > 10%, or Plink F inbreeding value was either >0.10 or <−0.10 (heterozygosity). On the merged data, the HWE and MAF SNP filters were re-applied, as well as the reference allele frequency difference <0.20 check, and an imputation quality cutoff R2 <0.30. Finally, all SNPs were removed that were by then known to show problems in the reference sequence data of the phase I Interim set.
Genotyping in the ERF sample was also performed on different platforms including Illumina 318K, Illumina 370K, Illumina 610K and Affymetrix 250K. QC was comparable to the protocol described above. Pre-imputations QC included snp call rate >95%, individual call rate >90%, HWE p-value=10−06, high autosomal heterozygosity (FDR <1%), high IBS (>=0.95), MAF >0.01, ethnic outliers and sex mismatch. Post imputation QC included removal of monomorphic variants. Genetic imputations were performed using minimac 2012.8.15 and the 1000 Genomes phase1 v3 as the reference set.
Software
SNP-heritability was calculated using the package GCTA v1.11.26 Plink was used for the association analyses.28 All other analyses were carried out in R.29
Results
1. Phenotype analysis in the NTR and NESDA samples
Most means of the individual items were elevated in the NESDA sample which is expected based on the comorbidity of BP features and mood and anxiety disorders, and the fact that the majority of NESDA subjects were selected for elevated levels of depression and anxiety disorders. Not surprisingly, the most pronounced difference concerned AI item “happy person”. However, the elevated pattern was not observed for all items. NR item “let people know they’ve hurt me”, IP item “can’t handle separation”, and SH item “too impulsive” had significantly higher means in NTR subjects, and SH items “do things impulsively” and “when upset hurt self” did not differ significantly across the two samples. Correcting for the level of depression using the 28-item Inventory of Depressive Symptomatology (IDS) score did not change the dissimilarity of the pattern of mean differences between the two samples.30 As expected, depression levels explained a large proportion of variance of the aggregate score of NESDA subjects (r2=46%), but the influence varied considerable across the individual items (0.4%–45.5%) and subscales (9.6%–46.1%).
Due to the item mean differences between the NTR and NESDA samples all subsequent analyses were done for NTR and NESDA samples separately as well as jointly. For all joint analyses we used a NTR/NESDA indicator as covariate to adjust for the overall elevated scores in the NESDA sample.
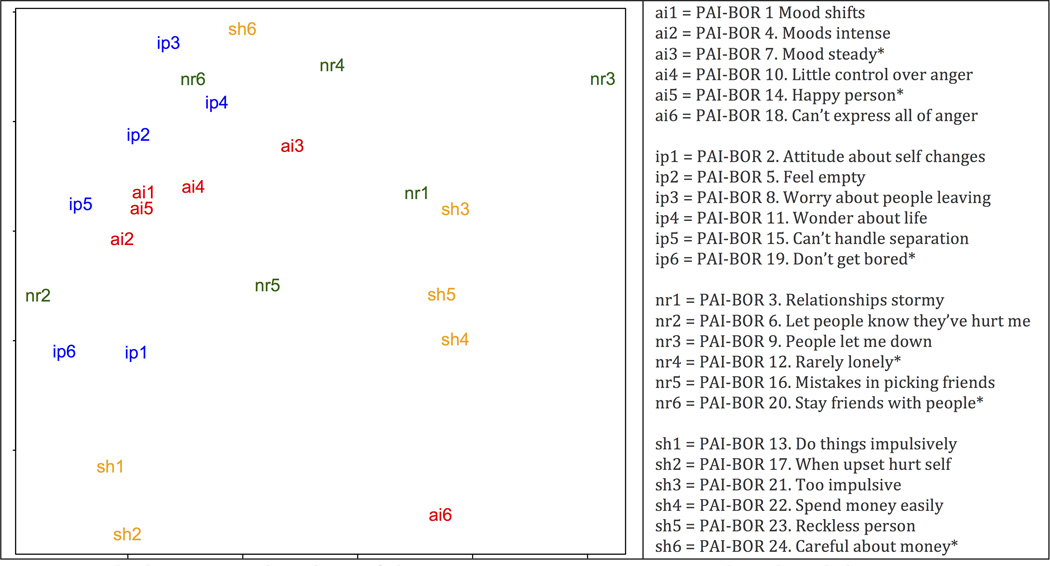
The PAI-BOR is designed to measure four interrelated aspects of the borderline phenotype (e.g., affect instability, AI, self-harm, SH, negative relationships, NR, and identity problems, IP). We explored the relations between the 24 PAI-BOR items in our sample using multidimensional scaling, which confirmed the common content of 5 of the 6 AI items, but also showed a less clear grouping of the items of the other 3 scales.31 A projection of the 24 items onto two dimensions is shown in Figure 1. Using three dimensions did not increase the clarity of the grouping of the items.
Figure 1.
Note: Multidemensional scaling of the 24 PAI-BOR items. Items indexed with * are reverse scored.
For the association study we aggregated item level data by averaging the items. This aggregate score represents the common content of the PAI-BOR items, and has been shown to have good validity in discriminating between cases and controls.14 Averaging cancels out item-specific content. Item-specific analyses are presented in part 5 of our analyses.
Based on previous studies, we assessed the effect of age, gender, and their interaction on the total aggregate score as well as the four subscale scores. In the NRT sample the interaction between age and gender significantly predicted total and subscale scores with younger males having relatively more elevated scores compared to older females although females had generally higher scores. The interaction was most pronounced in the IP subscale (p= 1.15e-09) explaining 4.7% of the variance. Interestingly, we found no significant age or gender effects in the NESDA sample. The age/gender effects were included as covariates in the association and SNP-heritability analyses.
2. Heritability estimated from SNP data
We used the software package GCTA v1.11 to estimate the heritability of the aggregate measure of BP features in the NTR and combined NTR/NESDA samples that is due to all SNPs.26 We also estimated the variance in the four subscales AI, NR, ID, and SH that is due to SNPs. We used likelihood ratio tests to establish significance. In all analyses we used a cutoff of 0.025 to limit genetic relatedness. Results presented in Table 1 show that in the combined NTR/NESDA sample the SNP-heritability of the total score is significant (23.1%, pLRT=0.003). The estimate in the NTR sample was somewhat lower, 20.7%, and had larger standard errors due to the smaller sample size. The proportion of phenotypic variance due to SNPs was still significant with pLRT=0.03. Note that affect instability had a substantially higher genetic component (42.7% in the combined sample) compared to negative relations (11.5%, not significant) and identity problems and self-harm, which had essentially zero estimates. This result is consistent with the multidimensional scaling results (see Figure 1), which showed that AI items are closely grouped, reflecting their common content. The items of the SH, IP, and NR subscales are more spread out, indicating comparatively more item-specific content. The higher within subscale diversity of the SH, IP, and NR subscales explains at least to some extent the lower genetic variance estimates for these subscales since item-specific variance cancels out when aggregating items in a subscale. The results are also consistent with a twin study showing larger subscale specific genetic variance components for those subscales compared to AI.24 Differential genetic effects across the 24 individual BP features are further explored in part 5 of our analyses.
Table 1.
Estimating SNP-heritability with GCTA v1.11
| Total score | AI | NR | IP | SH | |
|---|---|---|---|---|---|
| NTR/NESDA N=3951 | |||||
| V(G)/Vp | 0.231 | 0.427 | 0.115 | 0.083 | 0.0599 |
| SE | 0.088 | 0.087 | 0.088 | 0.087 | 0.0868 |
| LRT | 7.437 | 26.575 | 1.765 | 0.941 | 0.484 |
| p-value | 0.003 | 1e-07 | 0.09 | 0.2 | 0.2 |
| NTR N=3300 | |||||
| V(G)/Vp | 0.207 | 0.257 | 0.174 | 0.119 | 0.089 |
| SE | 0.106 | 0.105 | 0.106 | 0.105 | 0.104 |
| LRT | 3.773 | 6.349 | 2.738 | 1.291 | 0.757 |
| p-value | 0.03 | 0.006 | 0.05 | 0.1 | 0.2 |
Note: We used a cutoff of 0.025 of genetic resemblance resulting approximately in 1 member per family in all analyses. In the analyses of the combined NTR/NESDA sample an ntr/nesda indicator was used as a covariate. Other covariates were age, gender, their interaction, and a PC controlling for stratification within the Dutch population.
3. GWAS
After QC a total of 6,670,475 SNPs and N=7125 and N=5802 were included in the association studies for NTR/NESDA and NTR, respectively. The association study was carried out using the average PAI-BOR score described above as an outcome in a linear additive genetic model implemented in Plink, with covariates sex, age, interaction between sex and age, a study indicator (NTR vs. NESDA), and a principal component to correct for population stratification within the Dutch population. To minimize computation time we carried out the association analysis first using all subjects but excluding one MZ twin per pair. This step provided a crude ranking of all SNPs although p-values and summary statistics are biased due remaining family relatedness (e.g., DZ twins). Next, we selected the top 100,000 SNPs, and carried out an ANOVA to control for differential associations across platforms, and eliminated 8 SNPs that had platform effects at a conservative level p<5e-5. Seven SNPs that emerged as promising association signals (defined as p<5e-6 in the ANOVA) were tested using Generalized Estimating Equations (GEE) to control for non-independence of observations due to family relatedness.32
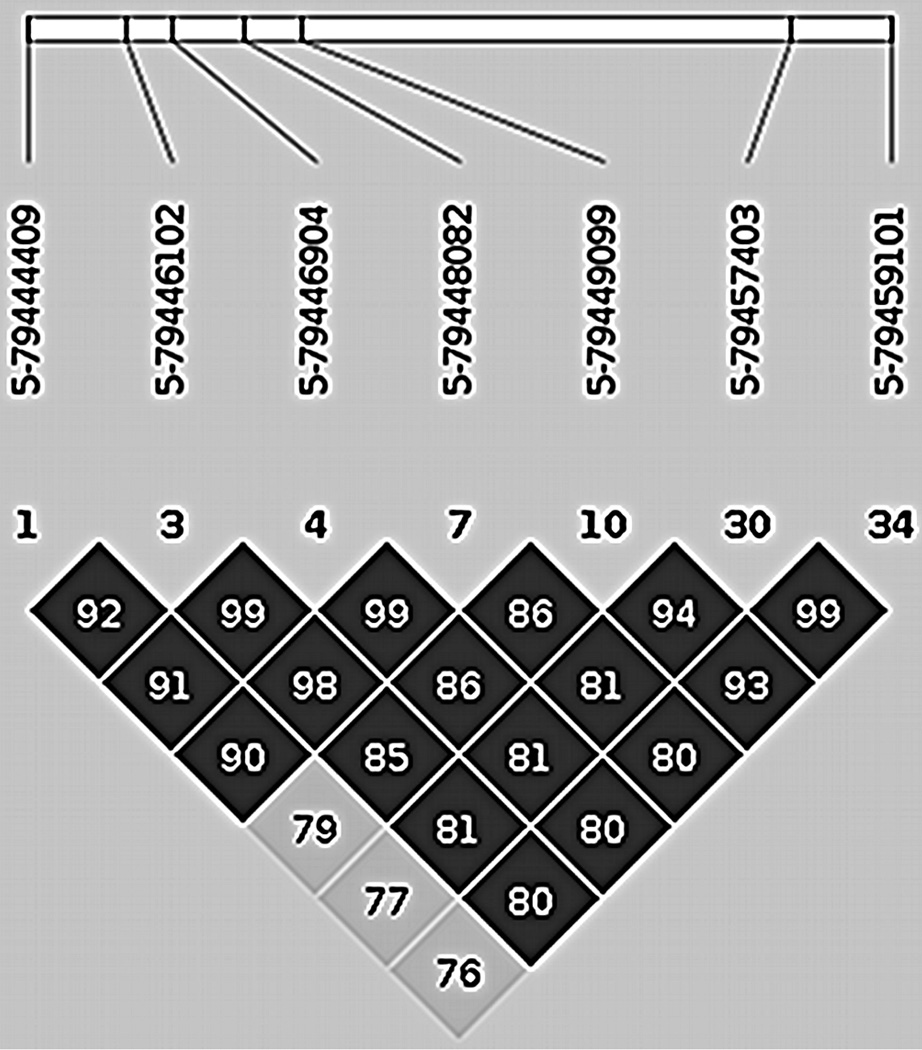
The seven SNPs were all located in a small region on chromosome 5. Detailed results concerning these SNPs are shown in Table 2. The p-values of these SNPs from fitting GEE were between 3.28e-6 and 8.22e-7 in the NTR sample, and between 4.89e-5 and 3.02e-6 in the combined NTR/NESDA sample. LD of the top 7 SNPs is shown in Figure 2.
Table 2.
Promising SNPs on chromosome 5 tested using GEE in the NTR sample N=5802
| rs id | location | alleles | MAF | coefficient | SE | p-value |
|---|---|---|---|---|---|---|
| rs6882423 | 79444409 | G/t | 0.1987 | −0.0378 | 0.00800 | 2.29e-06 |
| rs6866910 | 79446102 | A/G | 0.1963 | −0.0389 | 0.00803 | 1.28e-06 |
| rs73772260 | 79446904 | A/G | 0.1955 | −0.0397 | 0.00806 | 8.55e-07 |
| rs6888413 | 79448082 | T/G | 0.1953 | −0.0392 | 0.00808 | 1.24e-06 |
| rs6894288 | 79449099 | T/G | 0.2166 | −0.0377 | 0.00764 | 8.22e-07 |
| rs73125991 | 79457403 | A/T | 0.2203 | −0.0366 | 0.00759 | 1.40e-06 |
| rs11951568 | 79459101 | T/C | 0.2200 | −0.0352 | 0.00757 | 3.28e-06 |
Note: In the GEE analyses the covariates were age, gender, their interaction, and a PC controlling for stratification within the Dutch population.
Figure 2.
Replication of the 7 SNPs was carried out in the ERF sample. Correction for family structure in the ERF sample was done using the Grammar-Gamma method in GenABEL v1.7-4.33 To correct for multiple testing of correlated SNPs we used the program matSpD to obtain a conservative significance level, which for the seven SNPs was 0.027.34 Using this significance level two of the seven SNPs were significantly associated with the total aggregate phenotype in the ERF sample, and hence replicated the results in the discovery sample. Interestingly, when testing the association using the subscales as phenotype in the ERF sample, only the affect instability subscale (AI) was significantly related to some of these SNPs. All other subscales had insignificant associations. The results of the replication are presented in Table 3.
Table 3.
Replication in the ERF sample and meta-analysis NTR/NESDA/ERF
| ERF replication N=1301 | meta-analysis NTR/NESDA/ERF |
|||
|---|---|---|---|---|
| rs id | Coefficient | SE | p-value | p-value |
| rs6882423 | −1.60132 | 0.87256 | 0.06648 | 9.36e-07 |
| rs6866910 | −1.59989 | 0.88724 | 0.07135 | 5.03e-07 |
| rs73772260 | −1.72634 | 0.91117 | 0.05814 | 3.61e-07 |
| rs6888413 | −1.74865 | 0.90877 | 0.05433 | 4.10e-07 |
| rs6894288 | −1.43317 | 0.76133 | 0.05977 | 7.41e-07 |
| rs73125991 | −1.64513 | 0.61909 | 0.00788 | 1.27e-06 |
| rs11951568 | −1.67161 | 0.61933 | 0.00695 | 2.85e-06 |
4. Meta-analysis
We combined the NTR, NESDA, and ERF samples as three cohorts in an inverse variance meta-analysis to assess the SNPs on chr5.35 Results presented in Table 3 show that p-values of the 7 SNPs range between 2.85e-6 and 3.61e-7.
The genomic location of these SNPs is 5:79444409-79459289, which corresponds to serine incorportator 5 (SERINC5, synonyms: TPO1, C5orf12). The C5orf12 protein belongs to a protein family that facilitates the biosynthesis of lipids, an important component of the nervous system. More specifically, the protein incorporates serine into newly forming membrane lipids.36 Myelin in the brain is enriched with SERINC5. A recent mouse study established a relation between social isolation and decreased myelination in the brain, leading the authors to hypothesize a relation between myelination and psychiatric disorders in humans characterized by lack of social interaction.37 A different study showed that N-acetylaspartate (NAA), which supplies acetyl groups for myelin lipid synthesis, is decreased in the prefrontal region in borderline patients.38,39 SERINC5 has not been reported in any GWAS, however, SNPs tagging SERINC5 were included in a genotype score that was developed to predict the chance of success of smoking cessation.40
5. Heterogeneity
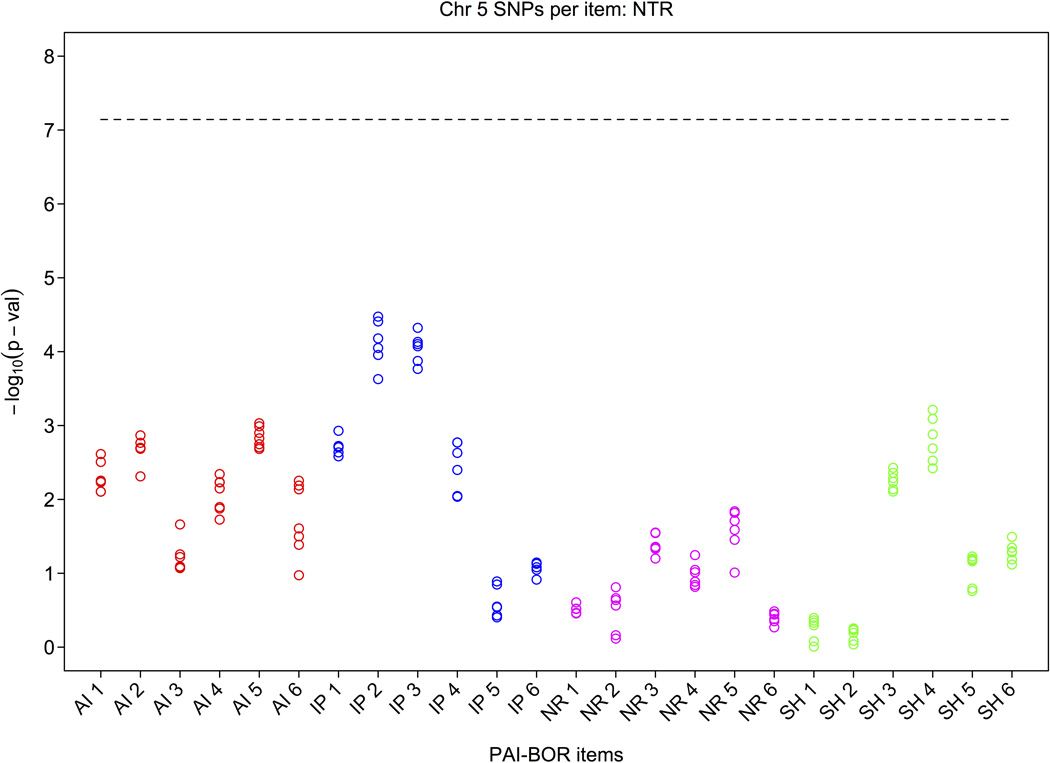
Given the evidence concerning the differential heritability of the four BP subscales that emerged from the heritability analyses carried out in Part 1, we conducted post-hoc tests of the top 7 SNPs for each of the 24 PAI-BOR items separately. Figure 3 shows lower p-values especially for items of the affect instability and identity problems subscales whereas items of the negative relation and self-harm subscales (with one exception) seem to have much weaker or no associations. Note that the reliability of the BP aggregate score is higher than that of individual items because when summing items, error in individual items cancels out. This explains the larger p-values of the individual items. The items with the smallest p-values were IP items “feel empty” and “worry about people leaving”, and AI item “happy person”.
Figure 3.
Next we analyzed whether the relation between the top SNP and the quantitative BP phenotype was the same for all subjects, or whether the effect was more pronounced depending on the level of BP, in other words, whether the basic assumption in GWAS that SNPs have a fixed effect is correct. This question was addressed by dividing the aggregate BP phenotype into high and low groups using a median split, and carrying out a reverse regression. Importantly, the variance of the BP aggregate score was very similar in the higher and lower scoring groups, showing that the variability in BP features is comparable across the two groups, and that only the means differ. We then estimated the regression coefficients when regressing the SNPs on the phenotype in each of the two groups. In the low group none of the seven SNPs were related to BP features (all p-values >0.05) whereas in the higher scoring group p-values ranged between 0.00033 and 0.00108. Detailed results are shown in Table 4. The results show that for these SNPs the assumption of a fixed effect is not correct, and suggest genetic heterogeneity.
Table 4.
Reverse regression of SNPs on total score in high vs low scoring subjects derived by median split of the NTR sample
| rs6882423 | rs6866910 | rs73772260 | rs6888413 | rs6894288 | rs73125991 | rs11951568 | |
|---|---|---|---|---|---|---|---|
| high | 0.00108 | 0.00058 | 0.00093 | 0.00090 | 0.00033 | 0.00059 | 0.00092 |
| low | 0.49323 | 0.78759 | 0.85842 | 0.89423 | 0.49091 | 0.45804 | 0.56495 |
Note: For this analysis we selected 1 subject per family, total N=3142, high N=1605, low N=1537
Discussion
This first genome-wide analysis of borderline personality features as measured by the PAI-BOR in three Dutch cohorts (NTR, NESDA, ERF) showed a number of interesting results. First, SNP-heritability of the total aggregate score of all 24 items estimated using the package GCTA v1.1 was 23.1% in the NTR/NESDA sample, which, taking into account imperfect LD of imputed SNP with causal variants, is consistent with estimates in twin and family studies.6,24 Interestingly, this result is driven almost entirely by the affect instability subscale with an estimated 42.7% of variance due to SNPs while none of the other subscales had significant variance components in our sample. This result suggests that differentiating between aspects of a disorder can substantially increase the statistical power to detect genetic effects.
The GWAS showed a promising signal in a small region on chromosome 5. Seven SNPs in this region had p-values between 3.28e-6 and 8.22e-7, and effects of two of these SNPs were successfully replicated in an independent sample (ERF). An inverse variance meta-analysis combining the three cohorts also confirmed the associations. The SNPs tag the SERINC5 gene (synonyms TPO1, C5orf1). SNPs in this region have previously been included in genotype score in a smoking cessation.40 Furthermore, cDNA TPO1 recognizes mRNA that is highly expressed in human brain tissue during myelination.36 A recent study using a mouse model showed that social isolation is related to decreased myelination in the brain, which might provide a basis to generalize to a role of myelination in psychiatric disorders that are characterized by a lack of social interaction.37 A study focusing on borderline patients showed that N-acetylaspartate (NAA), which supplies acetyl groups for myelin lipid synthesis in the brain, is decreased in the prefrontal region in borderline patients.38,39 Further research is clearly needed to confirm the potential association of SERINC5 with BP features.
Our study provided suggestive evidence concerning phenotypic and genetic heterogeneity, as well as evidence for differential effect sizes across the different borderline features. In addition to the differences in the proportion of genetic variance in the different subscales, we showed that especially identity and affect dysregulation items seemed to be more strongly related to the SNPs tagging SERINC5. Moreover, there was a distinct difference in effect size for high and low scoring subjects concerning SNPs tagging SERINC5. These results indicate that increasing sample size is not necessarily the only option to improve statistical power in gene finding studies. Addressing phenotypic heterogeneity by using multivariate outcomes can substantially increase effect sizes and therefore statistical power.41 Similarly, studies that reveal differences in biological correlates between individuals belonging to different subtypes of a disorder can provide the basis to take into account potential genetic heterogeneity.42 Heterogeneity across subgroups requires statistical methods focusing on clustering, and further research concerning suitable methods seems advantageous especially for investigations of the genetic basis of phenotypes such as BP features. Taken together, the results of this study suggest that phenotypic and genetic heterogeneity play an important role in the heritability of BP features.
References
- 1.Grant BF, Goldstein RB, Chou SP, Huang B, Stinson FS, Dawson DA, et al. Sociodemographic and psychopathologic predictors of first incidence of DSM-IV substance use, mood and anxiety disorders: results from the wave 2 national epidemiologic survey on alcohol and related conditions. Mol Psychiatry. 2008;14:1051–1066. doi: 10.1038/mp.2008.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friborg O, Martinussen M, Kaiser S, Øvergård KT, Rosenvinge JH. Comorbidity of personality disorders in anxiety disorders: A meta-analysis of 30 years of research. J Affect Disord. 2012;145:143–155. doi: 10.1016/j.jad.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Distel MA, Carlier A, Middeldorp CM, Derom CA, Lubke GH, Boomsma DI. Borderline personality traits and adult attention-deficit hyperactivity disorder symptoms: A genetic analysis of comorbidity. Am J Med Genet Part B. 2011;156:817–825. doi: 10.1002/ajmg.b.31226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornovalova MA, Hicks BM, Iacono WG, McGue M. Stability, change, and heritability of borderline personality disorder traits from adolescence to adulthood: a longitudinal twin study. Dev Psychopathol. 2009;21:1335–1353. doi: 10.1017/S0954579409990186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark LA. Stability and change in personality disorder. Current directions in psychological science. 2009;18:27–31. [Google Scholar]
- 6.Distel MA, Trull TJ, Derom CA, Thiery EW, Grimmer MA, Martin NG, et al. Heritability of borderline personality disorder features is similar across three countries. Psychol Med. 2008;38:1219–1229. doi: 10.1017/S0033291707002024. [DOI] [PubMed] [Google Scholar]
- 7.Distel MA, Trull TJ, Willemsen G, Vink JM, Derom CA, Lynskey M, et al. The five-factor model of personality and borderline personality disorder: a genetic analysis of comorbidity. Biol Psychiatry. 2009;66:1131–1138. doi: 10.1016/j.biopsych.2009.07.017. [DOI] [PubMed] [Google Scholar]
- 8.Boomsma DI, de Geus EJ, Vink JM, Stubbe JH, Distel MA, Hottenga JJ, et al. Netherlands Twin Register: from twins to twin families. Twin Res Hum Genet. 2006;9:849–857. doi: 10.1375/183242706779462426. [DOI] [PubMed] [Google Scholar]
- 9.Penninx BW, Beekman AT, Smit JH, Zitman FG, Nolen WA, Spinhoven P, et al. The Netherlands Study of Depression and Anxiety (NESDA): rationale, objectives and methods. Int J Meth Psych Res. 2008;17:121–140. doi: 10.1002/mpr.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pardo LM, MacKay I, Oostra B, van Duijn CM, Aulchenko YS. The effect of genetic drift in a young genetically isolated population. Ann Hum Genet. 2005;69:288–295. doi: 10.1046/j.1529-8817.2005.00162.x. [DOI] [PubMed] [Google Scholar]
- 11.Morey LC. The Personality Assessment Inventory: Professional manual. Odessa, FL: Psychological Assessment Resources; 1991. [Google Scholar]
- 12.Jackson KM, Trull TJ. The Factor Structure of the Personality Assessment Inventory-Borderline Features (PAI-BOR) Scale in a Nonclinical Sample. J Personal Disord. 2001;15:536–545. doi: 10.1521/pedi.15.6.536.19187. [DOI] [PubMed] [Google Scholar]
- 13.Gardner K, Qualter P. Reliability and validity of three screening measures of borderline personality disorder in a nonclinical population. Pers Individ Differ. 2009;46:636–641. [Google Scholar]
- 14.Stein MB, Pinsker-Aspen JH, Hilsenroth MJ. Borderline pathology and the personality assessment inventory (PAI): An evaluation of criterion and concurrent validity. J Pers Assess. 2007;88:81–89. doi: 10.1080/00223890709336838. [DOI] [PubMed] [Google Scholar]
- 15.Bell-Pringle VJ, Pate JL, Brown RC. Assessment of borderline personality disorder using the MMPI-2 and the Personality Assessment Inventory. Assessment. 1997;4:131–139. [Google Scholar]
- 16.Kurtz J, Morey L. Use of structured self-report assessment to diagnose borderline personality disorder during major depressive episodes. Assessment. 2001;8:291–300. doi: 10.1177/107319110100800305. [DOI] [PubMed] [Google Scholar]
- 17.Johansen M, Karterud S, Pedersen G, Gude T, Falkum E. An investigation of the prototype validity of the borderline DSM-IV construct. Acta Psychiatr Scand. 2004;109:289–298. doi: 10.1046/j.1600-0447.2003.00268.x. [DOI] [PubMed] [Google Scholar]
- 18.Sanislow CA, Grilo CM, Morey LC, Bender SD, Skodol AE, Gunderson JG, et al. Confirmatory factor analysis of the DSM-IV criteria for borderline personality disorder: Findings from the Collaborative Longitudinal Personality Disorders Study. American Journal of Psychiatry. 2002;159:284–290. doi: 10.1176/appi.ajp.159.2.284. [DOI] [PubMed] [Google Scholar]
- 19.Andión Ó, Ferrer M, Gancedo B, Calvo N, Barral C, Torrubia R, et al. Confirmatory factor analysis of borderline personality disorder symptoms based on two different interviews: the structured clinical interview for DSM-IV Axis II disorder and the revised diagnostic interview for borderlines. Psychiatry Res. 2011;190:304–308. doi: 10.1016/j.psychres.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Skodol AE, Gunderson JG, Pfohl B, Widiger TA, Livesley WJ, Siever LJ. The borderline diagnosis I: psychopathology, comorbidity, and personality structure. Biol Psychiatry. 2002;51:936–950. doi: 10.1016/s0006-3223(02)01324-0. [DOI] [PubMed] [Google Scholar]
- 21.Skodol AE, Siever LJ, Livesley WJ, Gunderson JG, Pfohl B, Widiger TA. The borderline diagnosis II: biology, genetics, and clinical course. Biol Psychiatry. 2002;51:951–963. doi: 10.1016/s0006-3223(02)01325-2. [DOI] [PubMed] [Google Scholar]
- 22.Aggen SH, Neale MC, Røysamb E, Reichborn-Kjennerud T, Kendler A psychometric evaluation of the DSM-IV borderline personality disorder criteria: age and sex moderation of criterion functioning. Psychol Med. 2009;39:1967–1978. doi: 10.1017/S0033291709005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banzhaf A, Ritter K, Merkl A, Schulte-Herbrüggen O, Lammers CH, Roepke S. Gender differences in a clinical sample of patients with borderline personality disorder. J Personal Disord. 2012;26:368–380. doi: 10.1521/pedi.2012.26.3.368. [DOI] [PubMed] [Google Scholar]
- 24.Distel MA, Willemsen G, Ligthart L, Derom CA, Martin NG, Neale MC, et al. Genetic covariance structure of the four main features of borderline personality disorder. J Personal Disord. 2010;24:427–444. doi: 10.1521/pedi.2010.24.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Wray NR, Visscher PM. Comparing apples and oranges: equating the power of case-control and quantitative trait association studies. Genet Epidemiol. 2010;34:254–257. doi: 10.1002/gepi.20456. [DOI] [PubMed] [Google Scholar]
- 26.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torgersen S, Czajkowski N, Jacobson K, Reichborn-Kjennerud T, Røysamb E, Neale MC, Kendler KS. Dimensional representations of DSM-IV cluster B personality disorders in a population-based sample of Norwegian twins: a multivariate study. Psychol Med. 2008;38:1617–1625. doi: 10.1017/S0033291708002924. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2011. [Google Scholar]
- 30.Rush AJ, Giles DE, Schlesser MA, Fulton CI, Weissenburger JE, Burns CT. The Inventory of Depressive Symptomatology (IDS): Preliminary findings. Psychiatry Res. 1986;18:65–87. doi: 10.1016/0165-1781(86)90060-0. [DOI] [PubMed] [Google Scholar]
- 31.Kruskal JB. Nonmetric multidimensional scaling: a numerical method. Psychometrika. 1964;29:115–129. [Google Scholar]
- 32.Zeger SL, Liang KY, Albert PS. Models for longitudinal data: a generalized estimating equation approach. Biometrics. 1988;44:1049–1060. [PubMed] [Google Scholar]
- 33.Svishcheva GR, Axenovich TI, Belonogova NM, van Duijn CM, Aulchenko YS. Rapid variance components-based method for whole-genome association analysis. Nat Genet. 2012;44:1166–1170. doi: 10.1038/ng.2410. [DOI] [PubMed] [Google Scholar]
- 34.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hunter JE, Schmidt FL, Jackson GB. Meta-analysis. Beverly Hills, California: Sage publications; 1982. [Google Scholar]
- 36.Krueger WHH, Gonye GE, Madison DL, Murray KE, Kumar M, Spoerel N, et al. TPO1, a member of a novel protein family, is developmentally regulated in cultured oligodendrocytes. J Neurochem. 1997;69:1343–1355. doi: 10.1046/j.1471-4159.1997.69041343.x. [DOI] [PubMed] [Google Scholar]
- 37.Liu Jia, Dietz Karen, DeLoyht Jacqueline M, Pedre Xiomara, Kelkar Dipti, Kaur Jasbir, et al. Impaired adult myelination in the prefrontal cortex of socially isolated mice. Nat Neurosci. 2012;15:1621–1623. doi: 10.1038/nn.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Elst LT, Thiel T, Hesslinger B, Lieb K, Bohus M, Hennig J, et al. Subtle prefrontal neuropathology in a pilot magnetic resonance spectroscopy study in patients with borderline personality disorder. J Neuropsychiatry Clin Neurosci. 2001;13:511–514. doi: 10.1176/jnp.13.4.511. [DOI] [PubMed] [Google Scholar]
- 39.Chakraborty G, Mekala P, Yahya D, Wu G, Ledeen RW. Intraneuronal N-acetylaspartate supplies acetyl groups for myelin lipid synthesis: evidence for myelin-associated aspartoacylase. J Neurochem. 2001;78:736–745. doi: 10.1046/j.1471-4159.2001.00456.x. [DOI] [PubMed] [Google Scholar]
- 40.Rose JE, Behm FM, Drgon T, Johnson C, Uhl GR. Personalized smoking cessation: interactions between nicotine dose, dependence and quit-success genotype score. Mol Med. 2010;16:247–253. doi: 10.2119/molmed.2009.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Sluis S, Posthuma D, Dolan CV. TATES: Efficient multivariate Genotype-Phenotype analysis for Genome-Wide association studies. PLoS Genet. 2013;9(1) doi: 10.1371/journal.pgen.1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamers F, Vogelzangs N, Merikangas KR, de Jonge P, Beekman AT, Penninx BW. Evidence for a differential role of HPA-axis function, inflammation and metabolic syndrome in melancholic versus atypical depression. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.144. epub ahead of print, Oct 23 2012. [DOI] [PubMed] [Google Scholar]