Abstract
The myelodysplastic syndromes (MDS) are characterized by ineffective hematopoiesis, dysplasia, and transformation to acute myeloid leukemia (AML). Although it has been suggested that additional mutations lead to progression of MDS to AML, the causative agent(s) for such mutations remains unclear. Oxidative stress is a potential cause, therefore, we evaluated levels of reactive oxygen species (ROS) in NUP98-HOXD13 (NHD13) transgenic mice, a murine model for MDS. Increased levels of ROS were detected in bone marrow nucleated cells (BMNC) that express CD71, a marker for cell proliferation, as well as immature, lineage negative bone marrow nucleated cells from NHD13 mice. In addition to the increase in ROS, increased DNA double strand breaks and activation of a G2/M phase cell cycle checkpoint were noted in NHD13 BMNC. Finally, using an in vivo assay for mutation frequency, we detected an increased mutation frequency in NHD13 BMNC. These results suggest that oxidative stress may contribute to disease progression of MDS to AML through ineffective repair of DNA damage and acquisition of oncogenic mutations.
Keywords: MDS, AML, ROS, Mutation, NHD13, Big Blue® mice
1. Introduction
The progression of a pre-malignant condition to a lethal malignancy is thought to involve an accumulation of mutations in genes that regulate cellular proliferation, survival, and differentiation [1, 2]. The myelodysplastic syndromes (MDSs) can be considered as a representative pre-malignant hematopoietic disorder that transforms to acute myeloid leukemia (AML) in approximately 35% of cases [3]. It has been proposed that AML requires a minimum of two complementary mutations, one leading to enhanced proliferation and a second leading to impaired differentiation [4]. Although the genetic basis of MDS is not completely understood, a significant percentage of MDS cases are characterized by chromosomal aberrations including deletions, amplifications, inversions, and translocations [5, 6]. The transformation of MDS to AML is often accompanied by additional mutations, such as cytogenetically detectable gross chromosomal rearrangements, or point mutations detectable only by DNA sequence analysis [7]. The causative agent(s) for these secondary events is poorly understood.
Reactive oxygen species (ROS) are normally produced in all aerobic cells, and are normally eliminated by antioxidants. Oxidative stress occurs when this critical balance is disrupted due to excess ROS production, antioxidant depletion, or both [8]. The excess ROS are also known to be a genotoxic stress that can induce DNA damage and mutation following ineffective repair of DNA damage [9–12].
Increased levels of ROS have been detected in both AML and chronic myeloid leukemia (CML) [1, 13]. In the case of CML, it has been speculated that the increased oxidative stress in cells carrying the BCR-ABL fusion results in genomic instability. Furthermore, this genomic instability is predicted to result in mutations of genes that collaborate with the BCR-ABL fusion protein, resulting in the transition from chronic phase to blast crisis [14, 15]. With regard to MDS and oxidative stress, several studies have reported that increased levels of ROS or oxidative DNA damage could be detected in hematopoietic cells from MDS patients [16–20].
The NUP98-HOXD13 (NHD13) fusion gene was initially identified in patients with therapy-related MDS [21]. Subsequently, several additional NUP98-HOX gene fusions were noted to be associated with MDS (for review see ref [22]). Transgenic mice that express an NHD13 transgene in the hematopoietic compartment display impaired hematopoietic differentiation and develop MDS, characterized by peripheral blood cytopenias, bone marrow dysplasia, and transformation to AML [23, 24]. Importantly, over one-third of the NHD13 mice that transform to AML spontaneously acquire a mutation in NRAS, KRAS, CBL, or PTPN11 [25] ; these mutations are often present in patients with either AML or advanced stage MDS [26]. In this study, we demonstrate that hematopoietic stem and precursor cells which express an NHD13 transgene show increased ROS and a G2/M arrest. Furthermore, we show that these cells have an increased mutation rate in vivo, providing a potential mechanism for mutations which accompany the transformation of MDS to AML in vivo.
2. Materials and methods
2.1. Cells and transfection
Bone marrow mononucleated cells (BMNC) were obtained from the femora and tibiae of C57Bl6 NHD13 mice and wild type (WT) mice at age 6 to 8 months. Lineage depletion of BMNC was performed by using StemSep (StemCell Technologies) for Lineage negative (LN) BMNC analysis. The mouse hematopoietic progenitor cell lines 32D and Ba/F3 were transfected with pEF1a-NHD13 and pEF1a-mock expression vectors using Amaxa nucleofection technology (www.Amaxa.com). Briefly, 1 × 106 cells were transfected with 2µg of plasmid vector using nucleofection (Solution V), and program E-32 for 32D or X-01 for Ba/F3 cells. Stable transfectants were established after 48 hours by transfer to media containing 400µg/mL of G418 (Geneticin, Invetrogen, CA).
2.2. Measurement of intracellular ROS level
Intracelluar ROS levels were determined by staining with 5-(and-6) chloromethyl-2’,7’-dichlorodihydro-fluorescein diacetate (CM-H2DCFDA; Invitrogen, OR). Cells were resuspended with Hanks Balanced Salt Solution supplemented with 2% FBS (HF2). 5µM of CM-H2DCFDA was added to the cell suspension in a final concentration, followed by incubation at 37°C with shaking at 150rpm for 20 minutes. The ROS level in cells was determined by flow cytometry (FACScan; Cytek, CA). In order to analyze specific subsets of BMNC, the cells were first stained with biotin-conjugated anti-mouse lineage antibody cocktail (StemCell Technologies) followed by washing with PBS once and stained with Alexa Flour 780-conjugated streptavidin, allophycocyanin (APC)-conjugated anti-mouse CD71, CD117, Sca-1 (eBioscience) along with CM-H2DCFDA.
2.3. Immunocytochemistry for γH2AX foci
Immunocytochemistry for γH2AX foci was performed as described previously [13].
2.4. Pathway-Focused gene expression profiling using real-time PCR
The RT2 Profiler™ PCR array system (SABiosciences, MD) was used to examine expression of 84 genes associated with oxidative stress using PAMM-065C plates. Total RNA was prepared from LN BM using Trizol (Invitrogen, CA) reagent. First strand cDNA was synthesized using the RT2 First strand kit (C-03; SABiosciences, MD ). Real-Time PCR was performed with a 7500 Fast real-time PCR system (Applied Biosystems, CA). Data analysis was carried out using software available at theSABioscience web site (http://sabioscience.com/pcrarraydataanalysis.php).
2.5. Cell cycle, bromodeoxyuridine (BrdU), and western blot analysis
Cells were fixed with 70% cold EtOH, rinsed once with PBS, and incubated in 1mL of PBS containing RNase A (180µg/mL) for 30 minutes at RT. After washing with PBS, the cells were incubated for 15 minutes in 1mL propidium iodide (15ug/mL) solution. The histogram data were analyzed with FlowJo 7.5 software. For cell proliferation assessment, Mice were euthanized and BMNC were harvested at 4 or 16 hours after injecting 1 mg of BrdU intraperitoneally. Lineage and BrdU staining were performed using the manufacturer’s suggested protocol and reagents (BrdU Flow Kit; BD-Pharmingen).
For western blot, Cell lysate equivalent to 4X105 of LN BMNC or whole BM was separated on a 12% SDS-polyacrylamide gel and blotted onto nitrocellulose membrane. The antibodies (Abs) for Chk1 (2G1D5, Cell Signaling Technology), for Phospho-Chk1 (Ser345, Cell Signaling Technology), and for β-actin (4967, Cell Signaling Technology) were used at 1/1000 dilutions. Secondary Abs corresponding to protein detection antibodies were used at 1/10,000 dilutions. The ECL system (Thermo Scientific, IL) was used for the autoradiograms.
2.6. Determination of mutation frequency in vivo (The Big Blue® cII mutation detection assay)
To determine mutation rate changes in NHD13 MDS mice, Big Blue® mice (Homozygous C57BL/6 transgenic mice; Taconic, NY) were mated to NHD13 mice. F1 mice 6–7 months of age were used in this study. Genomic DNA was extracted from BMNC of Big Blue® NHD13 mice and littermate control mice (WT Big Blue®) using the RecoverEase™ DNA Isolation Kit (Stratagene, CA). The mutation detection assay was carried out using the Transpak® and the λ Select-cII™ protocol (Stratagene, CA). Verification of cII mutation was accomplished by direct sequencing of PCR products. Putative cII mutants were cored using sterile pipette tips and the each agar plug was inoculated into 400µL of G1250 culture, prepared with overnight culture and suspended to an OD600 of 0.1 with LB media. The inoculums were incubated under selective (24°C) condition for 24 hours, centrifuged, and the supernatants recovered. After boiling the supernatants for 5 minutes, an aliquot of the supernatant was used as a template for PCR. The cII gene was amplified using primers 5’-CCG CTC TTA CAC ATT CCA GC-3’ (asp-Fw) and 5’-CCT CTG CCG AAG TTG AGT AT-3’ (cII-Rw), and the profile consisted of incubation at 94°C for 3 min, followed by 35 cycles of 94°C for 30 seconds, 61°C for 30 seconds, and 72°C for 1 min. PCR products were purified using Qiagen (Valencia, CA) protocols, and were directly sequenced (DNA Sequencing and Gene Expression Core, NCI, Bethesda, MD) with primer 5’-CAT TTA TTT GCA TAC ATT CA-3’. Sequences were compared to the wild type cII Genbank reference sequence (J02459). Mutant frequency was calculated by dividing the number of mutant plaques by the total number of plaque forming units (pfu’s) as estimated from the number of plaques on titer plates incubated under control temperature conditions.
3. Results
3.1. Proliferative and primitive hematopoietic stem/progenitor cells show increase ROS in NHD13 mice
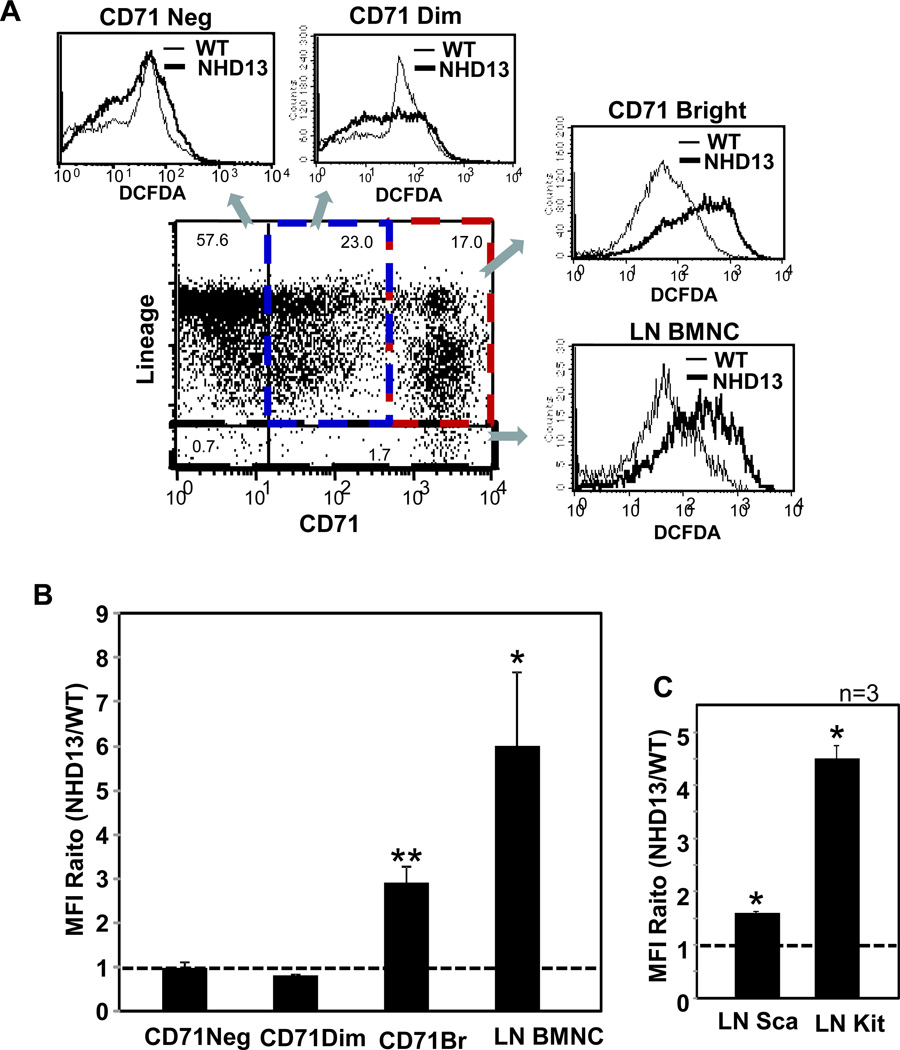
Increased ROS levels were detected in two distinct populations of NHD13 hematopoietic cells. Lineage negative (LN) BMNCs, which contain hematopoietic stem and progenitor cells from the NHD13 mice showed a 6-fold increase in ROS compared to WT controls (Fig. 1 A and B). In addition, CD71bright lineage positive (LP) cells from NHD13 mice showed a more modest 3-fold increase in ROS. CD71, or transferrin receptor, is an established marker for cell proliferation [27]. To investigate the possibility of proliferation-induced oxidative stress, we performed a cell proliferation assay using BrdU. Similar cell proliferation rates were detected in both genotypes (Suppl. Fig. S1) suggesting that the increased ROS level in NHD13 BM cells was not due to an increase in cell proliferation rates. In order to determine whether the increased level of ROS in NHD13 LN BM could be scavenged by an antioxidant, we treated LN BMNCs in vitro with N-Acetyl-L-cysteine (NAC). LN BMNCs from NHD13 mice treated with NAC showed decreased levels of ROS, in a dose-dependent manner (Suppl. Fig. S2). Using a high dose of NAC (50mM), ROS levels from the NHD 13 LNBMNCs were reduced to the level of WT LNBMNCs.
Fig. 1.
Increased ROSs in proliferative cells and primitive hematopoietic progenitor cells of NHD13 mice. (A) Representative FACS profiles showing increased ROS in CD71 positive BMNC and LN BMNC from NHD13 mice. Histograms show DCFDA staining intensity from NHD13 BMNC (thick line) compared to WT littermate control BMNC (thin line). (B) Average mean fluorescence intensity (MFI) ratio of NHD13 versus age matched WT littermates was determined by FACS analysis from 3 independent experiments using CD71 and a lineage cocktail, and 9 independent experiments with lineage depleted cells. (C) MFI of DCFDA was analyzed in Lineage negative (LN) Sca-1 positive cells and LN cKit positive cells from 3 independent experiments. Error bars represent SEM. *, p<0.05; **, p<0.01
The elevated ROS levels from NHD13 LN BMNC was evident in both the LN cKit+ population, which includes the common myeloid progenitor (CMP), granulocyte monocyte progenitor (GMP), and megakaryocyte erythrocyte progenitor (MEP), and the LN Sca+ population, which include hematopoietic stem cell (HSC) and common lymphocyte progenitor (CLP) populations, although the effect was more dramatic in the LN cKit+ population (Fig. 1C). These data suggest that proliferative cells, including primitive progenitor cells from NHD13 mice, have increased oxidative stress compared to their WT counterparts.
3.2. Expression NHD13 leads to increased ROS in hematopoietic cell lines
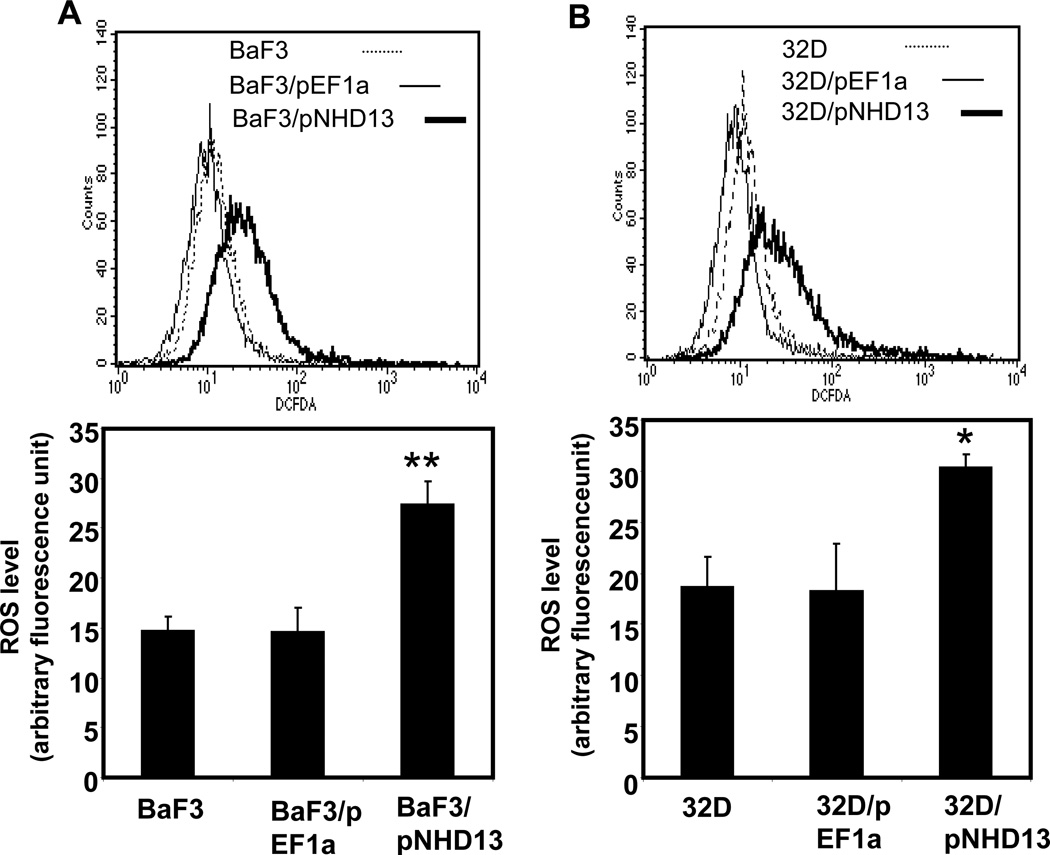
To determine whether the observed increase in ROS associated with expression of the NHD13 fusion gene was unique to the line of transgenic mice that we studied, we transfected two independent murine hematopoietic progenitor cell lines, 32D and BaF3, with an NHD13 expression vector or empty vector control, and selected stably transfected cells with G418. NHD13 expression was verified by RT-PCR of RNA extracted from the stable transfectants (Suppl. Fig. S3). ROS levels were significantly elevated in 32D/NHD13 and BaF3/NHD13 compared to controls transfected with an empty vector (EF1a) or parental cells (Fig. 2A and B). These data demonstrate that expression of an NHD13 fusion leads to increased production of ROS in established hematopoietic progenitor cell lines as well as populations of primary hematopoietic cells.
Fig. 2.
Increased ROS in hematopoietic cell lines expressing an NHD13 transgene. (A) Ba/F3 cells stably transfected with pEF1a-NHD13 construct (BaF3/pNHD13) showed higher ROS level than cells transfected with empty vector (BaF3/pEF1a) or Ba/F3 cells. (B) Elevated ROS level was observed in 32D/ pEF1a-NHD13 (32D/pNHD13) cells compared with 32D/pEF1a and 32D cells. Bar graphs and statistics were acquired from stable transfectants at 3 different passages. The error bars represent SEM. *, p<0.05; **, p<0.01
3.3. Expression of the NHD13 transgene is associated with decreased expression of peroxidase genes in hematopoietic stem and progenitor cells
To explore the mechanism(s) that might lead to increased ROS in LN BMNC from the NHD13 mice, we used the RT2 Profiler™ PCR system. This system uses quantitative real-time PCR to detect the expression of a panel of 84 genes associated with oxidative stress simultaneously. The three most significantly down regulated genes in LN BMNC from NHD13 mice compared to WT BM encoded peroxidase enzymes, including myeloperoxidase (Mpo; −9.9 fold), eosinophil peroxidase (Epx; −8.8 fold) and lactoperoxidase (Lpo; −5.2 fold) (Suppl. Fig. S4). Although the principal role of Mpo is to metabolize hydrogen peroxide (H2O2) to hypochlorous acid (HOCl) in phagosomes of mature neutrophils, the role of Mpo and other peroxidiases in hematopoietic stem and progenitor cells has not been elucidated in detail. However, H2O2 produced by intracellular metabolism is thought to be processed by peroxidases to prevent the accumulation of free oxygen radicals. Therefore, the absence of peroxidases would be expected to lead to an accumulation of H2O2 as well as other oxidants that depend on H2O2 for their formation [28].
We also identified genes that were overexpressed >1.5 fold in LN BMNC from the NHD13 mice. These genes included several that are associated with increases in oxidative stress, such as Ctsb (Cathepsin B) and Prdx2 (Peroxiredoxin). Prdx2 (increased 1.9 fold, p=0.016367) is produced in response to oxidative stress and is known to reduce intracellular H2O2 [29–31]; an increase in expression of this gene is not surprising given the decrease in peroxidases seen in the LN BMNC from the NHD13 mice. Ctsb (1.8 fold, p=0.01396) was also increased; Ctsb has been linked to oxidative stress as its release from lysosomes is thought to contribute to oxidative stress induced by H2O2 [32]. In addition, we noted Hbq1 to be increased 6.1-fold (p value = 0.049627) in the LN BMNC from the NHD13 mice. Hbq1 is a member of the alpha globin cluster, and, although Hbq1 has not been noted to be increased in response to oxidative stress, expression of other members of this cluster, including HBA have recently been reported to be increased following oxidative stress [33].
3.4. Increased ROS are associated with increased DNA double strand breaks in NHD13-expressing cells
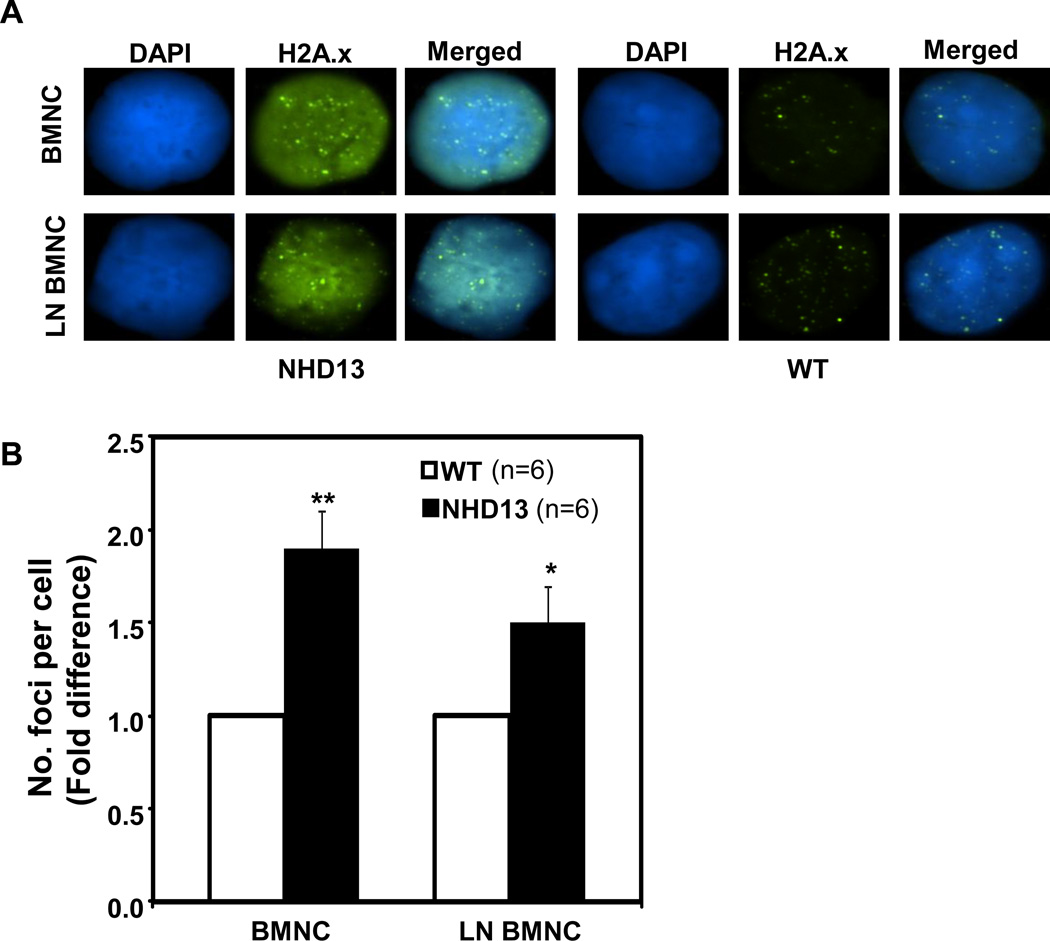
ROS are well known genotoxins that have been associated with DNA double strand breaks (DSBs) in mammalian cells [9]. We examined γH2AX foci, an established measure of DNA DSBs, in BMNC and LN BMNC from NHD13 and WT mice (Fig. 3A). The number of γH2AX foci per cell were 1.9 and 1.5 fold higher in NHD13 BMNCs and LN BMNC, respectively, compared with WT counterparts (Fig. 3B). These results demonstrate that the increased levels of ROS in NHD13 LN BMNC can lead to DSBs and the DSBs accumulate in mature BMNC as primitive progenitor cells differentiate.
Fig. 3.
DNA double strand breaks assessed by γH2AX foci. (A) γH2AX foci are increased in the BMNC and LN BMNC of NHD13 mice compared with wild type (WT) BMNC. (B) Bar graphs represent the fold differences of number of γH2AX foci per individual cells from 6 independent experiments. The error bars represent SEM. *, p<0.05; **, p<0.01
3.5. Increased ROS correlates with a partial G2/M cell cycle arrest in NDH13 LN BMNC
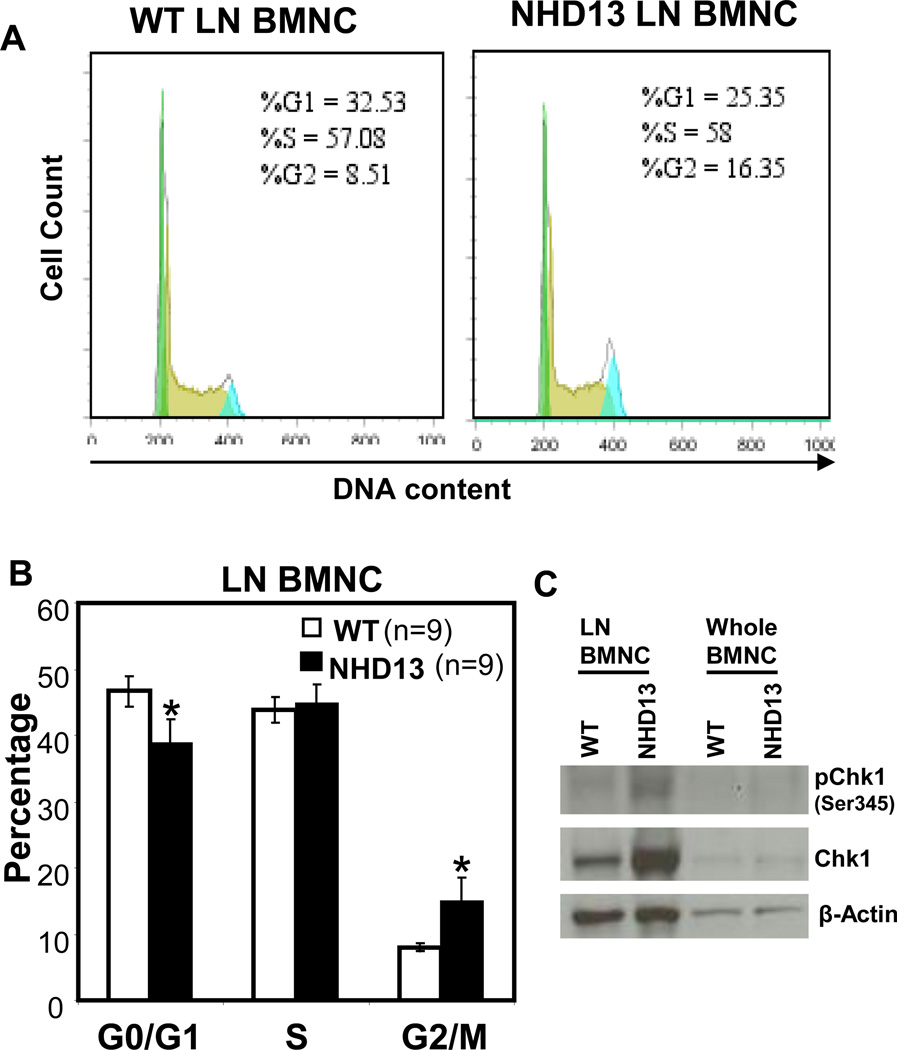
Given that increased ROS is known to interfere with normal cell cycle progression [34, 35], we analyzed DNA content in BMNC and LN BMNC from NHD13 and WT mice. Although the BMNC showed no difference in G0/G1, S, or G2/M (Suppl. Fig. S5), a two-fold increase of G2/M, with a corresponding decrease in G1 was detected in the LN BMNC from the NHD13 mice (Fig. 4A and B). This G2/M arrest seen in the NHD13 LN BMNC was associated with overexpression and activation of Chk1, a protein that plays a key role in the G2/M checkpoint response [36] (Fig. 4C). A similar result was detected in stable transfectants of 32D cells that expressed NHD13 (Suppl. Fig. S6). In addition, the G2/M arrest was accompanied by a marked increase in the proportion of binucleate cells (Suppl. Fig. S6B and C), which has previously been associated with some forms of G2/M arrest [37]. Taken together, these observations suggest that expression of NHD13 leads to increased ROS and DNA damage followed by a partial G2/M arrest, accompanied by an increase in Chk1 levels and increase in binucleated cells.
Fig. 4.
Association of cell growth arrest with increased level of ROS in lineage negative BM. (A) Representative histograms show the FACS results of cells stained with propidium iodide (PI) after fixation with 70% cold ethanol. Histogram data was analyzed with Flow Jo 7.5 software. (B) Bar graphs represent the mean percentage of each cell cycle phase from 9 independent experiments. (C) Western blot analysis of Chk1 and phospho-Chk1 expression in lineage negative BM (LN BMNC) or whole BM from NHD13 or WT mice. LN BMNCs were pooled from 3 mice to obtain sufficient protein for analysis. β-actin is used as a loading control *, p<0.05.
3.6. Increased level of ROS is accompanied by an increase in mutation frequency in BMNC from NHD13 mice
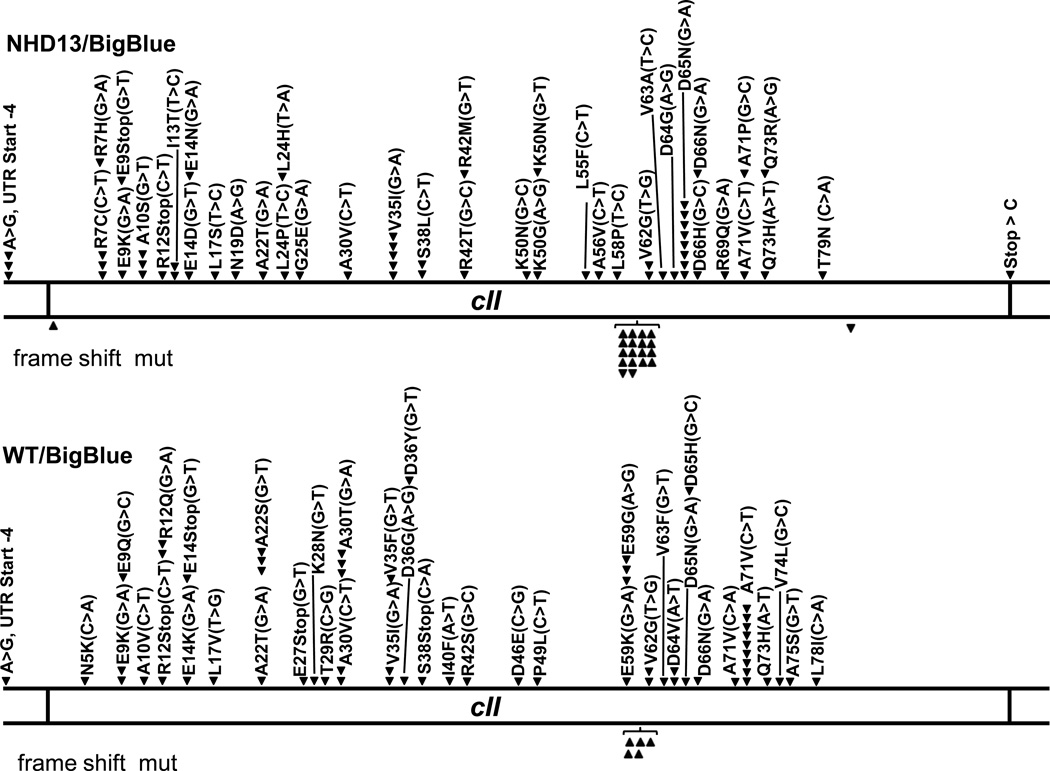
In order to determine if the ROS-induced DNA damage led to mutations of genomic DNA in vivowe crossed the NHD13 mice to ”Big Blue®” mice. The Big Blue mice have integrated a λLIZ shuttle vector derived from the coliphage lambda into their genomic DNA, and have been used to detect mutation frequencies in vivo [38]. Mutations of the lambda phage cII gene can be identified by virtue of λ phage replication via the lytic or lysogenic cycle in E. coli host cells. If the cII gene product is produced, the phage replicates via lysogeny; if the phage is mutant, it replicates by a lytic cycle. Thus, upon rescue of the λLIZ shuttle vector from mouse genomic DNA, the mutation frequency can be calculated by comparison of lytic to lysogenic plaques. We compared the mutation rates of the cII gene in mice that were positive for the NHD13 and Big Blue transgenes to littermate control mice that were positive for only the Big Blue transgene. Mutations were identified by the appearance of lytic plaques, and confirmed by PCR amplification and direct sequencing of the cII gene from lytic plaques. As shown in Table 1, there was a significant (1.8-fold) increase in the mutation frequency in BMC from the NHD13/BigBlue mice compared to the BigBlue only mice in four independent experiments. As shown in Fig. 5, there was no obvious “hotspot” for the single nucleotide substitution mutations, as the mutations were distributed throughout the entire cII gene in both the BigBlue/NHD13 mice as well as the BigBlue mice controls. All of the single nucleotide substitutions led to missense or nonsense mutations, which, combined with the appearance of lytic plaques, made it likely that these missense mutations were indeed loss of function mutations. There were no silent mutations identified. The frequency of single nucleotide transition or transversion mutations was similar between the NHD13 and control groups (Suppl. Table S1). In addition, although 7.7% of the mutations identified in the BigBlue mice were frameshift mutations, a mutant frequency that is similar to previous reports[38], 25% of the mutations identified in the NHD13/BigBlue mice were frameshift mutations (Suppl. Table S2), indicating that not only was the frequency of cII gene mutations altered by the presence of the NHD13 transgene, but the type of mutation was altered as well. Most of the frameshift mutations were single nucleotide insertions at a GGGGGG mononucleotide repeat; this was true for both the NHD13/BigBlue mice as well as the BigBlue control mice (Fig. 5).
Table 1.
Spontaneous mutant frequencies from BMNC of 6 month old mice
| Mouse ID | Plaques | Mutants (Lytic plaques) |
Mutants (Verified by sequence) |
MF (per 105 plaques) |
|---|---|---|---|---|
| NHD13 | ||||
| C434 | 381500 | 16 | 15 | 3.93 |
| C446 | 435000 | 18 | 17 | 3.91 |
| C448 | 630000 | 22 | 19 | 3.02 |
| C453 | 463300 | 33 | 29 | 6.26 |
| Mean ± S. E. M. | 4.28 ± 0.69 | |||
| WT | ||||
| C435 | 478500 | 12 | 10 | 2.09 |
| C447 | 665000 | 17 | 14 | 2.11 |
| C440 | 456500 | 8 | 6 | 1.31 |
| C452 | 890000 | 38 | 35 | 3.93 |
| Mean ± S. E. M. | 2.36 ± 0.56 | |||
| MF, mutation frequency | p=0.0371 | |||
Fig. 5.
λ cII mutational spectra. Linear map of the cII gene. Numerical co-ordinates refer to amino acid (aa) residue. Downward arrowheads above the diagram indicate single nucleotide substitutions. Upward and downward arrows below the diagram indicate single nucleotide frameshift insertions or deletions, respectively. Specific nucleotide changes are indicated in brackets. The mutations marked on the figure represent combined results from 4 independent experiments. UTR, untranslated region.
4. Discussion
Several studies have demonstrated that markers of oxidative stress, such as elevated levels of ROS and DNA damage, can be detected in MDS patients [17–20]. In this report, we show an increase in ROS can be detected in both the proliferative, CD71+ BMNC as well as the LN BMNC from NHD13 mice. The markedly elevated levels of ROS in LN BMNC from NHD13 mice is consistent with the observation that primitive hematopoietic (CD34+) cells from MDS patients have higher level of intracellular ROS than normal CD34+ cells [20].
Although the mechanisms are not completely elucidated, numerous reports have shown that activation or overexpression of proto-oncogenes can lead to over-production of ROS [1, 13, 39]. We have shown a direct correlation between expression of an NHD13 fusion gene and increased level of ROS by introduction of the NHD13 fusion into two distinct hematopoietic progenitor cell lines. Although we have not yet established a direct link, expression of the NHD13 fusion was associated with decreased production of three different genes encoding peroxidases (Mpo, Lpoand Epx), providing a potential molecular mechanism leading to increased ROS production in the lineage negative BMNC from NHD13 mice.
All mammalian cells have cell cycle “checkpoints” that allow repair of DNA damage caused by either environmental or endogenous genotoxic insults. These cell cycle checkpoints slow or arrest cell cycle progression, allowing the cell to repair damaged DNA, to prevent the transmission of damaged or incompletely replicated chromosomes. Three checkpoints (G1/S, S and G2/M) have been identified [40]. The G2/M checkpoint blocks the initiation of mitosis when DNA damage has been incompletely repaired [41, 42]. Thus, we suspect that the G2/M phase growth arrest observed in the lineage negative stem and progenitor BMNC from NHD13 mice may be triggered by ROS-induced DNA damage. This hypothesis is supported by the increased numbers of γH2AX foci and elevated levels of Chk1 protein expression in the lineage negative BMNC from the NHD13 mice (Fig. 4). Alternatively, it has recently been suggested that cells which express an NHD13 fusion have an impaired ability to repair DNA damage caused by class switch recombination; this defect may also contribute to the G2/M growth arrest seen in BMNC from the NHD13 mice [43].
A large fraction of MDS patients will progress to AML [3]. This progression is often accompanied by additional cytogenetic changes, as well as more subtle mutations, leading to the suspicion that the transformation of MDS to AML can be triggered by genotoxic stress, including ROS, and the subsequent accumulation of mutated DNA [25, 44]. Using the ”BigBlue” strain of reporter mice, we show that increased ROS are associated with an increased rate of DNA mutation in the BMNC of NHD13 mice. The BMNC from NHD13 mice showed a 3-fold increase in frameshift mutations compared to BMNC from WT mice. These findings are consistent with reports of increased frameshift mutations associated with increased ROS [12, 45]. Moreover, several reports have linked increased levels of ROS with single nucleotide insertions or deletions at mono or di-nucleotide repeat sequences [46–48]. Although the “BigBlue” assay detects frameshift mutations that may result from increased ROS, it is unable to detect larger DNA sequence changes that have also been linked to increased ROS levels and result from aberrant DNA repair by pathways such as alternative non-homologous end-joining [39, 49, 50].
Taken together, these findings indicate that BMNC from NHD13 mice have increased levels of ROS, increased levels of DNA damage, a partial G2/M cell cycle arrest and defect in DNA repair. We hypothesize that rare errors in the repair of ROS-induced DNA damage result in single nucleotide substitution and frameshift mutations. These mutant proteins then collaborate with expression of the NHD13 protein and result in the progression of MDS to AML.
Supplementary Material
Acknowledgments
We are grateful to our colleagues Sarah Beachy, Masahiro Onozawa, Deborah Silverman, and Helder Paiva for insightful discussion. We thank the NCI intramural mini-sequencing core for Sanger sequence generation and Maria Jorge for excellent animal husbandry.
Funding
This research was supported by the Intramural Research Program of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors’ contributions: Y.J.C. designed and performed research, analyzed data and wrote the first draft of the manuscript; C.R. performed γ-H2AX staining and analyzed data; S.G. participated in the design of the study and data analysis; F.V.R. designed research and analyzed data; P.D.A. designed research, analyzed data and wrote the final draft of the manuscript.
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Rassool FV, Gaymes TJ, Omidvar N, Brady N, Beurlet S, Pla M, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer research. 2007;67:8762–8771. doi: 10.1158/0008-5472.CAN-06-4807. [DOI] [PubMed] [Google Scholar]
- 2.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mijovic A, Mufti GJ. The myelodysplastic syndromes: towards a functional classification. Blood reviews. 1998;12:73–83. doi: 10.1016/s0268-960x(98)90018-2. [DOI] [PubMed] [Google Scholar]
- 4.Gilliland DG, Tallman MS. Focus on acute leukemias. Cancer cell. 2002;1:417–420. doi: 10.1016/s1535-6108(02)00081-8. [DOI] [PubMed] [Google Scholar]
- 5.Heaney ML, Golde DW. Myelodysplasia. The New England journal of medicine. 1999;340:1649–1660. doi: 10.1056/NEJM199905273402107. [DOI] [PubMed] [Google Scholar]
- 6.Corey SJ, Minden MD, Barber DL, Kantarjian H, Wang JC, Schimmer AD. Myelodysplastic syndromes: the complexity of stem-cell diseases. Nat Rev Cancer. 2007;7:118–129. doi: 10.1038/nrc2047. [DOI] [PubMed] [Google Scholar]
- 7.Walter MJ, Shen D, Ding L, Shao J, Koboldt DC, Chen K, et al. Clonal architecture of secondary acute myeloid leukemia. The New England journal of medicine. 2012;366:1090–1098. doi: 10.1056/NEJMoa1106968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waris G, Ahsan H. Reactive oxygen species: role in the development of cancer and various chronic conditions. J Carcinog. 2006;5:14. doi: 10.1186/1477-3163-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sedelnikova OA, Redon CE, Dickey JS, Nakamura AJ, Georgakilas AG, Bonner WM. Role of oxidatively induced DNA lesions in human pathogenesis. Mutation research. 2010;704:152–159. doi: 10.1016/j.mrrev.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slupphaug G, Kavli B, Krokan HE. The interacting pathways for prevention and repair of oxidative DNA damage. Mutation research. 2003;531:231–251. doi: 10.1016/j.mrfmmm.2003.06.002. [DOI] [PubMed] [Google Scholar]
- 11.Steinboeck F, Hubmann M, Bogusch A, Dorninger P, Lengheimer T, Heidenreich E. The relevance of oxidative stress and cytotoxic DNA lesions for spontaneous mutagenesis in non-replicating yeast cells. Mutation research. 2010;688:47–52. doi: 10.1016/j.mrfmmm.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Themeli M, Petrikkos L, Waterhouse M, Bertz H, Lagadinou E, Zoumbos N, et al. Alloreactive microenvironment after human hematopoietic cell transplantation induces genomic alterations in epithelium through an ROS-mediated mechanism: in vivo and in vitro study and implications to secondary neoplasia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24:536–543. doi: 10.1038/leu.2009.284. [DOI] [PubMed] [Google Scholar]
- 13.Sallmyr A, Fan J, Datta K, Kim KT, Grosu D, Shapiro P, et al. Internal tandem duplication of FLT3 (FLT3/ITD) induces increased ROS production, DNA damage, and misrepair: implications for poor prognosis in AML. Blood. 2008;111:3173–3182. doi: 10.1182/blood-2007-05-092510. [DOI] [PubMed] [Google Scholar]
- 14.Koptyra M, Cramer K, Slupianek A, Richardson C, Skorski T. BCR/ABL promotes accumulation of chromosomal aberrations induced by oxidative and genotoxic stress. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008;22:1969–1972. doi: 10.1038/leu.2008.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sattler M, Verma S, Shrikhande G, Byrne CH, Pride YB, Winkler T, et al. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. The Journal of biological chemistry. 2000;275:24273–24278. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- 16.Farquhar MJ, Bowen DT. Oxidative stress and the myelodysplastic syndromes. Int J Hematol. 2003;77:342–350. doi: 10.1007/BF02982641. [DOI] [PubMed] [Google Scholar]
- 17.Ghoti H, Amer J, Winder A, Rachmilewitz E, Fibach E. Oxidative stress in red blood cells, platelets and polymorphonuclear leukocytes from patients with myelodysplastic syndrome. Eur J Haematol. 2007;79:463–467. doi: 10.1111/j.1600-0609.2007.00972.x. [DOI] [PubMed] [Google Scholar]
- 18.Novotna B, Bagryantseva Y, Siskova M, Neuwirtova R. Oxidative DNA damage in bone marrow cells of patients with low-risk myelodysplastic syndrome. Leukemia research. 2009;33:340–343. doi: 10.1016/j.leukres.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Peddie CM, Wolf CR, McLellan LI, Collins AR, Bowen DT. Oxidative DNA damage in CD34+ myelodysplastic cells is associated with intracellular redox changes and elevated plasma tumour necrosis factor-alpha concentration. Br J Haematol. 1997;99:625–631. doi: 10.1046/j.1365-2141.1997.4373247.x. [DOI] [PubMed] [Google Scholar]
- 20.Voukelatou G, Thanopoulou E, Dallas K, Fertakis V, Dimopoulou A, Micheva I, et al. CD34+marrow cells from MDS patients are characterized by higher levels of intracellular reactive oxygen species and inadequate antioxidant defences. Leukemia research. 2009;33:S93–SS4. [Google Scholar]
- 21.Raza-Egilmez SZ, Jani-Sait SN, Grossi M, Higgins MJ, Shows TB, Aplan PD. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer research. 1998;58:4269–4273. [PubMed] [Google Scholar]
- 22.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118:6247–6257. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YW, Slape C, Zhang Z, Aplan PD. NUP98-HOXD13 transgenic mice develop a highly penetrant, severe myelodysplastic syndrome that progresses to acute leukemia. Blood. 2005;106:287–295. doi: 10.1182/blood-2004-12-4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi CW, Chung YJ, Slape C, Aplan PD. Impaired differentiation and apoptosis of hematopoietic precursors in a mouse model of myelodysplastic syndrome. Haematologica. 2008;93:1394–1397. doi: 10.3324/haematol.13042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slape C, Liu LY, Beachy S, Aplan PD. Leukemic transformation in mice expressing a NUP98-HOXD13 transgene is accompanied by spontaneous mutations in Nras, Kras, and Cbl. Blood. 2008;112:2017–2019. doi: 10.1182/blood-2008-01-135186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bejar R, Levine R, Ebert BL. Unraveling the molecular pathophysiology of myelodysplastic syndromes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:504–515. doi: 10.1200/JCO.2010.31.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chitambar CR, Massey EJ, Seligman PA. Regulation of transferrin receptor expression on human leukemic cells during proliferation and induction of differentiation. Effects of gallium and dimethylsulfoxide. The Journal of clinical investigation. 1983;72:1314–1325. doi: 10.1172/JCI111087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klebanoff SJ. Myeloperoxidase: friend and foe. J Leukoc Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- 29.Low FM, Hampton MB, Peskin AV, Winterbourn CC. Peroxiredoxin 2 functions as a noncatalytic scavenger of low-level hydrogen peroxide in the erythrocyte. Blood. 2007;109:2611–2617. doi: 10.1182/blood-2006-09-048728. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, Lee TH, Park ES, Suh JM, Park SJ, Chung HK, et al. Role of peroxiredoxins in regulating intracellular hydrogen peroxide and hydrogen peroxide-induced apoptosis in thyroid cells. The Journal of biological chemistry. 2000;275:18266–18270. doi: 10.1074/jbc.275.24.18266. [DOI] [PubMed] [Google Scholar]
- 31.Stresing V, Baltziskueta E, Rubio N, Blanco J, Arriba M, Valls J, et al. Peroxiredoxin 2 specifically regulates the oxidative and metabolic stress response of human metastatic breast cancer cells in lungs. Oncogene. 2012 doi: 10.1038/onc.2012.93. [DOI] [PubMed] [Google Scholar]
- 32.Lee DC, Mason CW, Goodman CB, Holder MS, Kirksey OW, Womble TA, et al. Hydrogen peroxide induces lysosomal protease alterations in PC12 cells. Neurochemical research. 2007;32:1499–1510. doi: 10.1007/s11064-007-9338-5. [DOI] [PubMed] [Google Scholar]
- 33.Liu WS, Baker SS, Baker RD, Nowak NJ, Zhu LX. Upregulation of Hemoglobin Expression by Oxidative Stress in Hepatocytes and Its Implication in Nonalcoholic Steatohepatitis. Plos One. 2011:6. doi: 10.1371/journal.pone.0024363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clopton DA, Saltman P. Low-level oxidative stress causes cell-cycle specific arrest in cultured cells. Biochem Biophys Res Commun. 1995;210:189–196. doi: 10.1006/bbrc.1995.1645. [DOI] [PubMed] [Google Scholar]
- 35.Barzilai A, Yamamoto K. DNA damage responses to oxidative stress. DNA repair. 2004;3:1109–1115. doi: 10.1016/j.dnarep.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Cuddihy AR, O'Connell MJ. Cell-cycle responses to DNA damage in G2. International review of cytology. 2003;222:99–140. doi: 10.1016/s0074-7696(02)22013-6. [DOI] [PubMed] [Google Scholar]
- 37.Huang X, Tran T, Zhang L, Hatcher R, Zhang P. DNA damage-induced mitotic catastrophe is mediated by the Chk1-dependent mitotic exit DNA damage checkpoint. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1065–1070. doi: 10.1073/pnas.0409130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakubczak JL, Merlino G, French JE, Muller WJ, Paul B, Adhya S, et al. Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowicki MO, Falinski R, Koptyra M, Slupianek A, Stoklosa T, Gloc E, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104:3746–3753. doi: 10.1182/blood-2004-05-1941. [DOI] [PubMed] [Google Scholar]
- 40.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 41.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 42.Ishikawa K, Ishii H, Saito T. DNA damage-dependent cell cycle checkpoints and genomic stability. DNA Cell Biol. 2006;25:406–411. doi: 10.1089/dna.2006.25.406. [DOI] [PubMed] [Google Scholar]
- 43.Puthiyaveetil AG, Reilly CM, Pardee TS, Caudell DL. Non-homologous end joining mediated DNA repair is impaired in the NUP98-HOXD13 mouse model for myelodysplastic syndrome. Leukemia research. 2013;37:112–116. doi: 10.1016/j.leukres.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding L, Ley TJ, Larson DE, Miller CA, Koboldt DC, Welch JS, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pinto AV, Deodato EL, Cardoso JS, Oliveira EF, Machado SL, Toma HK, et al. Enzymatic recognition of DNA damage induced by UVB-photosensitized titanium dioxide and biological consequences in Saccharomyces cerevisiae: evidence for oxidatively DNA damage generation. Mutation research. 2010;688:3–11. doi: 10.1016/j.mrfmmm.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Cespedes M, Parrella P, Nomoto S, Cohen D, Xiao Y, Esteller M, et al. Identification of a mononucleotide repeat as a major target for mitochondrial DNA alterations in human tumors. Cancer research. 2001;61:7015–7019. [PubMed] [Google Scholar]
- 47.Jackson AL, Chen R, Loeb LA. Induction of microsatellite instability by oxidative DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12468–12473. doi: 10.1073/pnas.95.21.12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zienolddiny S, Ryberg D, Haugen A. Induction of microsatellite mutations by oxidative agents in human lung cancer cell lines. Carcinogenesis. 2000;21:1521–1526. [PubMed] [Google Scholar]
- 49.Fan X, Kavelaars A, Heijnen CJ, Groenendaal F, van Bel F. Pharmacological neuroprotection after perinatal hypoxic-ischemic brain injury. Current neuropharmacology. 2010;8:324–334. doi: 10.2174/157015910793358150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muvarak N, Nagaria P, Rassool FV. Genomic instability in chronic myeloid leukemia: targets for therapy? Current hematologic malignancy reports. 2012;7:94–102. doi: 10.1007/s11899-012-0119-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.