Abstract
Vibrio species are Gram-negative, rod-shaped bacteria that live in aqueous environments. Several species, such as V. harveyi, V. alginotyticus, and V. splendidus, are associated with diseases in fish or shellfish. In addition, a few species, such as V. cholerae and V. parahaemolyticus, are risky for humans due to infections from eating raw shellfish infected with these bacteria or from exposure of wounds to the marine environment. Bacterial flagella are not essential to live in a culture medium. However, most Vibrio species are motile and have rotating flagella which allow them to move into favorable environments or to escape from unfavorable environments. This review summarizes recent studies about the flagellar structure, function, and regulation of Vibrio species, especially focused on the Na+-driven polar flagella that are principally responsible for motility and sensing the surrounding environment, and discusses the relationship between flagella and pathogenicity.
Keywords: bacterial flagellum, ion-driven motor, motility, chemotaxis, pathogenicity
Introduction
Vibrio species are Gram-negative, rod-shaped bacteria that live in all types of aqueous environments, including marine, freshwater, and estuary (Blake et al., 1980; Joseph et al., 1982; Johnson et al., 2012). All Vibrio species can move using flagella, which are cell surface organelles that can propel them. Unlike eukaryotic flagella, each bacterial flagellum is driven by a rotary motor embedded in the cell envelope, and the flagellar rotation is harnessed by the ion-motive force across the cell membrane (Berg, 2003; Terashima et al., 2008). The basic structure and function of the flagellum produced by Vibrio species are the same as those in other species (Figure 1) (Aizawa, 1996; Chen et al., 2011). Each flagellum consists of a filament acting as a helical propeller, a hook functioning as a universal joint and a basal body working as a rotary motor (Figure 1) (Sowa and Berry, 2008; Li et al., 2011). More than 50 gene products are involved in flagellar synthesis (MacNab, 2003). Since flagella are relatively large motility organelles for the cell, the formation of the flagella and the expression of their components are tightly regulated. Their assembly process has been described in many reviews (Chilcott and Hughes, 2000; Kim and McCarter, 2000; Aldridge, 2002; Chevance and Hughes, 2008).
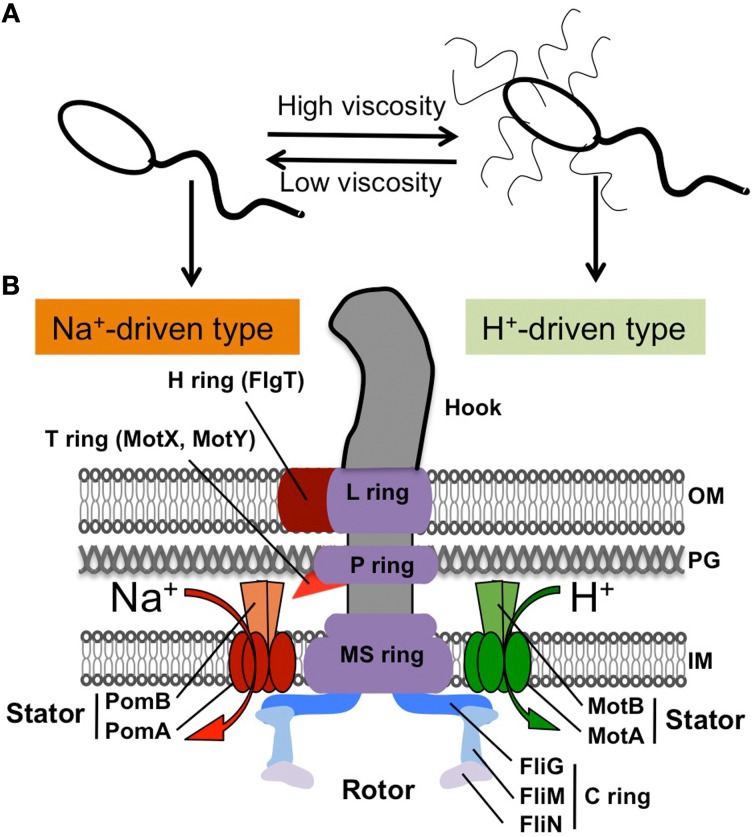
Figure 1.
Flagellar structure of Vibrio. (A) Polar flagellum and lateral flagella in some Vibrio species (not including Vibrio cholerae). (B) Schematic diagram of the hook basal body for a polar flagellum shown in the left half (based on EM images, (Terashima et al., 2006, 2013)) and the lateral flagellar structure shown in the right half, according to the peritrichous flagellum from Salmonella (MacNab, 2003). The characteristics of the polar flagellum are the H ring and the T ring, shown in red.
All Vibrio species have single or multiple flagella at the cell pole (called “polar flagellum”) and can swim freely in a liquid environment. With respect to the flagellum, V. cholerae has a single polar flagellum (monotrichous). However, some species, such as V. parahaemolyticus and V. alginolyticus, produce enormous numbers of flagella at lateral or peritrichous positions (called “lateral flagella”) in addition to the single polar flagellum when they are exposed to viscous environments (Figure 1A) (McCarter et al., 1988; Kawagishi et al., 1996). The polar and lateral flagella are structurally and functionally distinct from each other (McCarter, 2004). Each polar flagellum is covered with a sheath that is contiguous with the outer membrane, so that it is thick and can be observed using dark-field microscopy with a mercury lamp. Lateral flagella are not covered with a sheath, so they are difficult to observe using light microscopy. Although both types of flagella are driven by rotary motors embedded at their bases, the power source is different. Polar flagella are driven by the Na+-motive force, whereas lateral flagella are driven by the H+-motive force, as seen in the lateral flagella of Escherichia coli or Salmonella enterica (Figure 1) (Atsumi et al., 1992; Asai et al., 2000; Blair, 2003).
In terms of the pathogenicity of Vibrio, some species, such as V. cholerae, V. vulnificus, and V. parahaemolyticus, have been described extensively as human pathogens (Daniels and Shafaie, 2000; Yildiz and Visick, 2009), and Vibrios are also pathogenic to fishes or the other animals. In this review, we focus on flagellar function and assembly and on the relationship between flagellar motility and pathogenicity.
Flagellar basal body structure and motor
The overall structure of the flagellar base is shown schematically in Figure 1B, based on electron microsopic images of the purified hook-basal body from a peritrichous flagellum of Salmonella enterica (right side) and from a polar flagellum of Vibrio alginolyticus (left side) (Francis et al., 1994; Thomas et al., 2006; Terashima et al., 2008, 2013). Both kinds of basal bodies share common features even though they originate from different species of Gram-negative bacteria: the hook and basal body with several rings embedded in the cell envelope. The flagellar basal body functions as a rotary motor, and consists of two parts: the rotary part (rotor) and the stationary part (stator). The stator complex is composed of two proteins, MotA/MotB (LafT/LafU), for the H+-driven motor of lateral flagella from E. coli, Salmonella and Vibrio, and PomA/PomB, for the Na+-driven polar flagellar motor of Vibrio (Figure 1). The ion flux through the stator couples to the rotor-stator interaction that generates torque. The rotor contains several rings: from the cytoplasmic face, there is the C ring (also called the “switch complex”) composed of FliG, FliM, and FliN, and the MS ring embedded in the cytoplasmic membrane (made of at most 26 copies of FliF). These rings are connected by a rod whose tip connects to the hook. The basal body contains two other rings, the P ring (FlgI), which is associated with the peptidoglycan layer, and the L ring (FlgH), which is located in the outer membrane (Aizawa, 1996; Terashima et al., 2008). Thus, the LP ring does not rotate but functions as a bushing for the central rod.
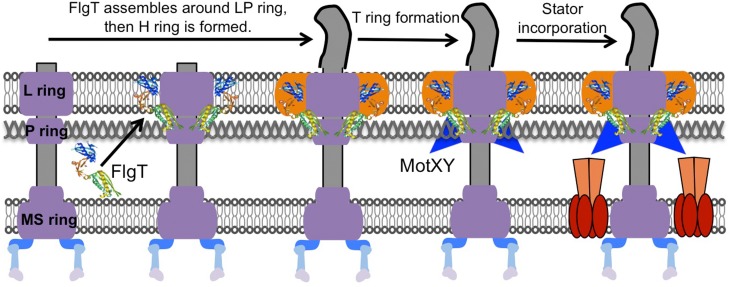
Although high resolution ultrastructural images have been reported for the basal body, intact images of the entire flagellar motor have remained ambiguous until recently due to the complexity of the stator units, which dynamically assemble around the rotor (Leake et al., 2006). However, the stator always dissociates from the detergent-solubilized flagellar basal body and no one has been able to isolate the basal body intact with the stator. In 2006, Murphy and co-workers (Murphy et al., 2006) first showed the structure of the complete flagellar motor in situ using the whole cell electron cryotomography method from the spirochete Treponema primitia. Their images captured electron densities corresponding to the stator units surrounding around the rotor. The resolution was still not sufficient to identify the detailed structure of the stator, but those images revealed the relative locations of the C-ring and the stator units for the first time. The structure of the polar flagellum of Vibrio contains two additional ring structures, named the T ring, which surrounds the periplasmic side of the P ring (Terashima et al., 2006), and the H ring, which is located at the outer rim of the L ring. The T ring is made of two proteins, MotX and MotY, that are essential components for the polar flagellar motor of Vibrio, and it is probably involved in stabilizing the stator surrounding the rotor (McCarter, 1994a,b; Okabe et al., 2001; Terashima et al., 2006). The crystal structure of MotY from V. alginolyticus in conjunction with biochemical analysis revealed that the C-terminal domain of MotY stabilizes the association with the peptidoglycan layer and that the N-terminal domain of MotY is involved in the association with the basal body (Kojima et al., 2008). The H ring is located at the outer rim of the LP ring, so that we initially thought that the LP ring was somehow bigger in this basal body (Figure 2). The loss of flgT function results in an almost non-motile phenotype and there is no formation of the T ring in addition to the lack of an outer rim structure of the LP ring. Therefore, FlgT is likely to be a component of the H ring, and is also important for assembly of the T ring. Our crystal structure and functional analyses of FlgT support this idea (Terashima et al., 2013). Perhaps these two unique extra ring structures in the polar flagellum are required to fix the stator units around the rotor (Figure 2). The Vibrio polar flagella can rotate extraordinarily up to 1700 Hz (Magariyama et al., 1994), while the rotational speed of the H+-driven flagellar motor is just around 300 Hz (Chen and Berg, 2000).
Figure 2.
Model for the basal body ring formation of the Na+-driven polar flagellum. The H ring is constructed dependent on FlgT proteins that assemble around the previously formed L ring and P ring. Next, MotX and MotY assemble around the basal body to stably form the T ring with the help of the middle domain of FlgT. As a result, the stator is incorporated properly and works in the presence of the T ring. The model is drawn based on a previous report (Terashima et al., 2013).
It is noteworthy that in the E. coli H+-driven motor, unexpectedly dynamic properties of the stator units were observed by fluorescent photobleaching experiments: the stator units are not really static components of the motor, but can dynamically associate with and dissociate from the rotor (Leake et al., 2006). Such a dynamic characteristic can be observed also for the PomA/PomB stators of the Vibrio polar flagellum. Their assembly around the rotor is dependent on the external Na+ concentration (Fukuoka et al., 2009). Removal of Na+ from the medium caused the dissociation of the stators from the rotor, and the subsequent addition of Na+ to the medium restored the stator incorporation to the motor. The physiological meaning of the dynamic property of the stator is not known, but the rapid and stable rotation of the motor may be compensated by the turnover of the units to components pooled on the membrane.
Flagellar gene regulation and assembly
About 50 gene products are involved in the construction of a functional flagellum. Since the flagellum is such a big organelle and its production and assembly requires a large commitment of resources, especially for filament formation, bacteria have developed a precise regulation system that controls flagellar construction (MacNab, 2003). The control mechanism has been well-characterized in the system of Salmonella flagella (Chilcott and Hughes, 2000). In brief, the flagellum assembles from the inner structure base to the outer ones, beginning with basal body construction followed by hook assembly, and finally by filament formation. Therefore, any defects in gene products that disrupt the basal body or the hook formation inhibit the filament assembly. The hook assembly step is the critical check point: if the hook subunits are polymerized to the proper length (about 55 nm), then genes required for filament formation are induced. This assembly-coupled flagellar gene regulation is achieved by a cascade of flagellar gene operons (called the flagellar regulon). In Salmonella, there are three classes of operons: early, middle, and late (Kutsukake et al., 1990). The master regulator for the flagellar regulon (FlhDC) belongs to the early operon that induces the expression of the middle operon. The middle operon contains genes encoding for basal body and hook proteins. It also contains regulator σ28 (FliA) that controls the expression of genes belonging to the late operon, which encodes filament and motor proteins (Ohnishi et al., 1990). After completion of the hook assembly, σ28 is released from anti-sigma factor FlgM binding, and so is able to induce the expression of genes in the late operon (Ohnishi et al., 1992).
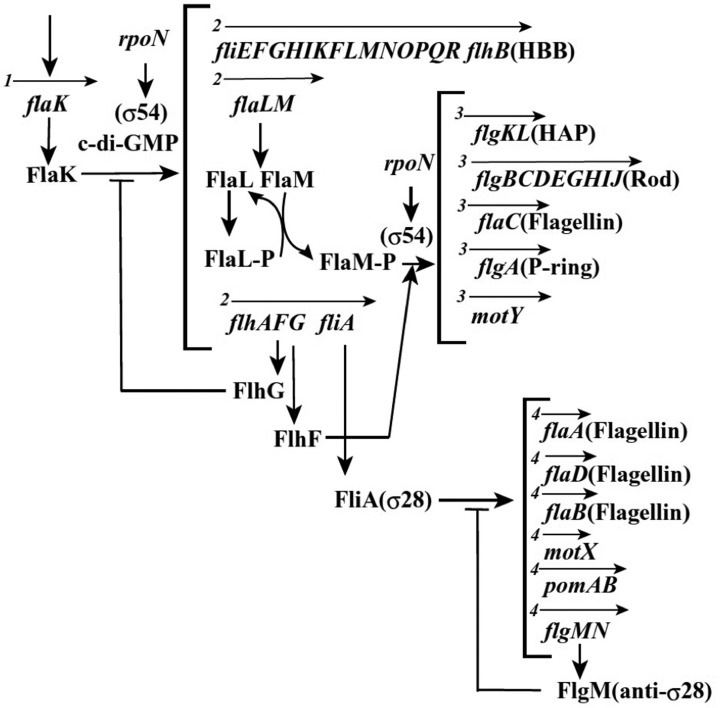
In Vibrio, the morphogenetic pathway for the polar and lateral flagella is quite likely to be the same as the one identified for Salmonella. However, the gene regulation for polar flagellar synthesis is more complex (Figure 3). Detailed analyses have been carried out for V. cholerae and V. parahaemolyticus and have found that the flagellar regulon is composed of a combination of both a RpoN (σ54) and a FlrA/FlaK (master regulator) dependent transcriptional hierarchy, organized into four classes (Klose and Mekalanos, 1998; Klose et al., 1998). Distinct from the Salmonella cascade, the middle operons are divided into two classes (class 2 and 3), and two sigma factors are involved in the regulation (Kim and McCarter, 2000). σ54 controls class 2 and 3 operons, which encode basal body and hook components, and σ28 (FliA) controls class 4 operons, which encode filament, chemotaxis, and motor components (Stewart and McCarter, 2003). Since V. alginolyticus is so closely related to V. parahaemolyticus, it is likely to have a similar mechanism that regulates the polar flagellar synthesis, and the putative organization of its polar flagellar regulon is shown in Figure 3 (McCarter, 2001; Correa et al., 2005; Kojima et al., 2011). It is noteworthy that Vibrio species have two chromosomes: the larger one contains all genes involved in the polar flagellar systems, whereas the smaller one contains all genes for the lateral flagellar systems (Makino et al., 2003). Recent work that the flagellar regulatory hierarchy facilitates the correct spatiotemporal expression patterns for optimal V. cholerae colonization and disease progression, is consistent with the idea that motility and the expression of specific virulence factors are inversely regulated (Syed et al., 2009).
Figure 3.
Plausible hierarchy of polar flagellum gene expression in Vibrio, based on previous reports (McCarter, 2001; Correa et al., 2005; Kojima et al., 2011). The arrows indicate the transcription unit and the number at the beginning of arrow indicates the class of transcriptional hierarchy.
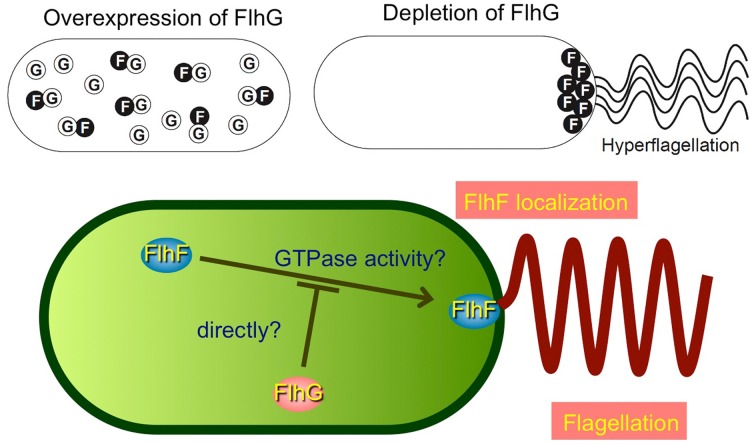
Another interesting topic regarding polar flagellar assembly is the regulation of their number and placement: V. alginolyticus cells have only a single polar flagellum at the cell pole. Genetic analyses identified two proteins, FlhF and FlhG, which are involved in this regulation (Kusumoto et al., 2008). FlhF is a GTPase and signal-recognition particle (SRP) homolog that positively regulates the number of polar flagella, and also determines the polar positioning (Salvetti et al., 2007; Balaban et al., 2009). FlhG is a MinD homolog and a putative ATPase that negatively regulates the flagellar number (Kusumoto et al., 2006). Biochemical and protein localization studies using GFP fusion derivatives revealed that FlhG binding to FlhF prevents the polar localization of FlhF, so that the proper number of FlhF is localized at the pole to initiate a single polar flagellum (Figure 4) (Correa et al., 2005; Kusumoto et al., 2008). From the flhF flhG double mutant, which mostly lacks flagella, a suppressor strain was isolated that forms peritrichous flagella in the majority of cells (Kojima et al., 2011). A mutation was recently found in a dnaJ family gene, named sflA (suppressor of ΔflhFG), which changed the monotrichous flagellar Vibrio cells into peritrichous flagellated cells (Kitaoka et al., 2013). SflA protein is proposed to prevent the initiation of flagellar construction.
Figure 4.
Model for regulation of the number of polar flagella in Vibrio. The function of FlhF is probably inhibited by an interaction with FlhG, a negative regulator of FlhF, and thereby the number of polar flagellum is limited to one. The schemes are drawn based on a previous report (Kusumoto et al., 2008).
Sensing the environmental conditions
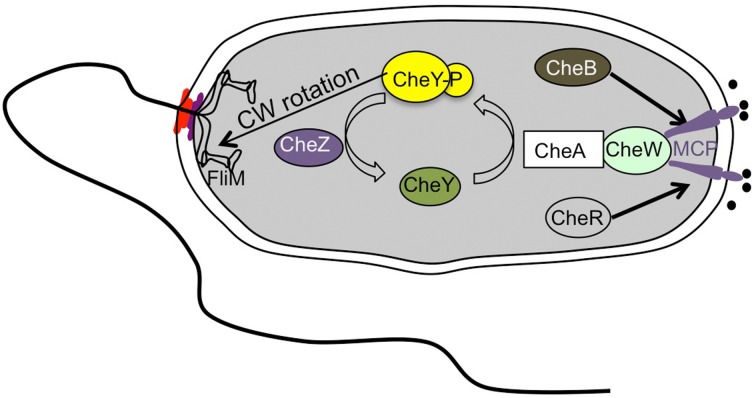
As seen in other bacteria like E. coli, Vibrio species also show chemotaxis, moving toward favorable conditions and avoiding unfavorable environments (Figure 5) (Szurmant and Ordal, 2004). The chemotaxis-related genes have been well-characterized for V. cholerae. Whole genome sequence analysis predicted three che gene clusters: cluster I and II on chromosome I and cluster III on chromosome II (Heidelberg et al., 2000). Although many homologs of various chemotaxis components found in E. coli have been identified, only one of three chemotaxis operons, cluster II, is required for chemotaxis in V. cholerae (Gosink et al., 2002). Current understanding of chemotactic behavior using a polar flagellar system is similar to that of E. coli (Figure 5). Once an attractant chemical binds to the periplasmic region of the methyl-accepting chemotaxis proteins (MCPs), a conformational change is generated that triggers autophosphorylation of the cytoplasmic kinase CheA, which is assumed to be associated with MCPs via adaptor protein CheW. Consequently, CheA donates the phosphate group to a response regulator CheY that triggers a rotational switch from counter-clockwise to clockwise by binding to FliM, a component of the switch complex. Meanwhile, some specific residues in the cytoplasmic domain of the MCP will be in a dynamic state of methylation with the help of CheR, a constitutively active methyltransferase, and of CheB, a methylesterase activated by phosphorylation. Methylated MCPs cease the signal output, so that cells are back to the original state (Boin et al., 2004).
Figure 5.
Simplified scheme of the chemotaxis signal transduction cascade in Vibrio species. The scheme is drawn based on a previous report (Boin et al., 2004).
The number of chemotaxis genes in V. cholerae is much greater than in E. coli, but only some of them are functional under experimental conditions (Banerjee et al., 2002). In V. cholerae, only deletion of cheA-2 in cluster II on chromosome I impairs chemotaxis and the che genes in this cluster are the most homologous to the che genes in other Vibrio species, suggesting that che cluster II is involved in chemotaxis. Moreover, only the Che-Y3 protein among the 5 cheY paralogs showed a relationship to flagellar rotation (Boin et al., 2004; Hyakutake et al., 2005). Furthermore, there are 45 proteins predicted to be potential chemoreceptors. However, only some chemoreceptors such as Mlp24 (methyl-accepting chemotaxis like protein, MCP homologs) recently reported were implicated in pathogenicity (Nishiyama et al., 2012). Thus, multiple chemotaxis-related genes and a large number of predicted MCP-like proteins imply a distinct chemotaxis mechanism that allows V. cholerae to adapt to various environmental conditions. However, Hyakutake et al. suggested that che clusters I and III are perhaps involved in controlling function(s) other than chemotaxis (Hyakutake et al., 2005). With regard to chemotaxis and pathogenicity, a non-chemotactic strain with counter-clockwise-biased flagellar rotation displays increased colonization and infectivity whereas reduced competition of a clockwise-biased strain was found (Butler and Camilli, 2004, 2005). A global transcription profile approach from stool samples showed that expression of many chemotaxis genes is at a low level, but some of them (cheA-2, cheY-3, cheW-1, and cheR-2) are not altered (Merrell et al., 2002). CheA-1 is highly expressed during human infection (Hang et al., 2003). Furthermore, it has been reported that human-shed V. cholerae have a 10-fold lower oral infectious, and have a transiently reduced chemotactic state in which the protein level of CheW was reduced (Butler et al., 2006). This shows that a pathogen alters its chemotactic state in response to human infection.
In addition to the chemotaxis, which mediates the direction of flagellar rotation, the polar flagellum seems to be a mechanosensor. As described above, V. alginolyticus and V. parahaemolyticus induce numerous lateral flagella in response to increases in external viscosity. The details of this sensing mechanism are still not understood. Besides an increase in viscosity, lateral flagellar expression is also induced by the addition of an anti-polar flagellum antiserum (McCarter et al., 1988). Since polar flagellar rotation is restricted in these two conditions, the polar flagellum also functions as a mechanosensor to induce lateral flagellar systems. Moreover, it was reported that inhibition of polar flagellar rotation by the specific inhibitor phenamil could induce lateral flagellar expression in media devoid of viscous agents, indicating the possibility that Vibrio cells can sense a decrease in the rotation rate of (or sodium influx through) the polar flagellar motor as a trigger for lateral gene expression (Kawagishi et al., 1996). Recently, it was reported that the mechanosensing mechanism by flagella is due to the dynamic assembly of the stator in E. coli (Lele et al., 2013; Tipping et al., 2013). Tipping and colleagues showed that the number of the stators bound to the flagellar motor is dependent on the external mechanical load, with more stators at higher viscosity and fewer stators at lower loads (Tipping et al., 2013). The flagellar motor of bacteria senses external load and regulates the strength of stator binding to the rest of the motor. Through this process the flagella can sense the external viscosity, suggesting that they function as a mechanosenseor as well as a locomotion component, and impact cellular functions (Lele et al., 2013). In this regard, it is suggested that the polar flagellum in Vibrio perhaps applies the same mechanosensing mechanism to sense and respond to its environment.
Pathogenesis and motility
Of particular interest to the pathogenesis of Vibrio is to determine the relationships between virulence factors, motility, and biofilm formation. V. cholerae, V. parahaemolyticus, and V. vulnificus, in addition to fish pathogens like V. alginolyticus have been established as important pathogens (DePaola et al., 2003; Xie et al., 2005; Jones and Oliver, 2009; Rasmussen et al., 2011; Kim et al., 2013; Ren et al., 2013; Sreelatha et al., 2013). In V. cholerae, the major virulence factors are cholera toxin (CT) and toxin co-regulated pilus (TCP), which are mediated in response to environmental stimuli through a hierarchical regulatory cascade called the ToxR regulon (Childers and Klose, 2007). ToxR and TcpP, two membrane-localized transcription factors, activate the expression of ToxT, another important transcription factor in the cytoplasm, which results in the activation of CT and TCP (DiRita et al., 1991; Krukonis and DiRita, 2003). CT can cause severe diarrhea in humans, and is encoded in the CTXΦ prophage (Hassan et al., 2010). A large Vibrio pathogenicity island (VPI), encoded in the VPIΦ prophage (Karaolis et al., 1998), is required for the TCP gene cluster, functioning both as an essential colonization factor and as a CTXΦ receptor (Taylor et al., 1987; Herrington et al., 1988; Lowden et al., 2010). However, recent research has shown that the ToxR cascade is also involved in repressing the production of virulence factors, suggesting that the ToxR regulon in V. cholerae may play a broader role in pathogenesis (Bina and Bina, 2010; Bina et al., 2013).
With regard to the pathogenesis of V. parahaemolyticus, a wide variety of virulence factors, including adhesins, thermostable direct hemolysin (TDH), TDH related hemolysin and Type 3 Secretion Systems, have been well-described by Zhang and Orth (2013). Interestingly, although hemolysin is thought to be a major virulence factor (Makino et al., 2003; Zhang and Austin, 2005), a deletion of the gene encoding both hemolysins does not drastically affect its cytotoxic effects on cultured cells (Zhang and Orth, 2013), suggesting that other virulence factors play important roles in the pathogenesis of V. parahaemolyticus. With regard to the pathogenesis of V. vulnificus, a recent review described that it possesses a variety of virulence factors, including capsular polysaccharide, acid neutralization, iron acquisition, acid neutralization and expression of proteins involving in motility, attachment and adhesion (Jones and Oliver, 2009).
Several studies have probed the relationship between the motility and the pathogenicity of V. cholerae in vivo (Gardel and Mekalanos, 1996; Krukonis and DiRita, 2003; Silva et al., 2006; Syed et al., 2009). It was initially thought that the expression of motility genes may repress the virulence genes (Gardel and Mekalanos, 1996). Mutational analyses have shown that motility is a virulence determinant, since non-motile mutants do not adhere to isolated rabbit brush borders and non-motile strains seem to have attenuated virulence for humans (Richardson, 1991; Silva et al., 2006). However, non-motile mutants have no significant defect in the ability to colonize suckling mice, implying that motility does not affect colonization dependent on TCP (Silva et al., 2006). The expression of motility genes is likely to repress virulence genes and vice-versa (Gardel and Mekalanos, 1996). Subsequently, Häse and coworkers showed that reducing the motility of V. cholerae by increasing the medium viscosity or disrupting the Na+-motive force results in an increase in the expression of ToxT (Häse and Mekalanos, 1999). It is noteworthy that the flagellar stator as a mechanosensor responds to assembly around the flagellar rotor in the case of high viscosity and consequently conducts massive Na+ ions influx. Meanwhile, a large number of Na+ ions are inside of cell in the case of disrupting the Na+-motive force. Thus, we can speculate that the influx of Na+ ions through the flagellar stator regulates the expression of ToxT.
With regard to the motility and the pathogenicity, Silva et al. found a remarkable increase in CT and TCP major subunit in a non-motile strain (motY), but a decrease in CT production in both wild-type and mutant strains when flagellar motility was inhibited (Silva et al., 2006). Recent work revealed a clear contribution of the flagellar regulatory hierarchy to the virulence of V. cholerae (Syed et al., 2009). Virulence factors such as toxin and hemolysin, which are up-regulated in flagellar regulatory mutants, were confirmed by quantitative reverse transcription PCR. The flagellar regulatory system positively mediates the transcription of diguanylate cyclase, named CdgD which results in the transcription of a hemagglutinin that enhances intestinal colonization (Syed et al., 2009). Thus, this whole-genome expression analysis supports the concept that motility and virulence gene expression are inversely regulated. Meanwhile, it has been found that TCP is expressed before the generation of CT during infection (Lee et al., 1999). These findings specify another transitional phase from motility to pathogenicity: colonization prior to release of CT (Figure 6A).
Figure 6.
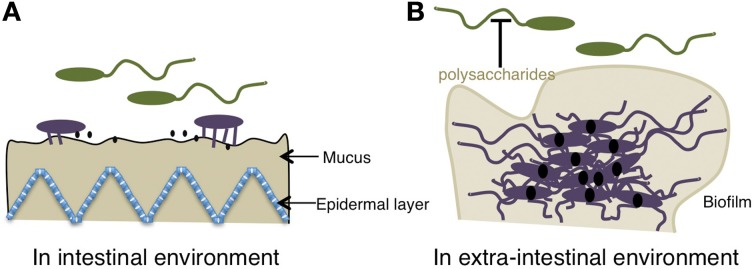
Relationship between the motility of the polar flagellum of Vibrio and pathogenicity. (A) In an intestinal environment, because of the presence of bile, the gene expression of virulence is “OFF.” When the polar flagellum senses the high viscosity of the mucus gel, the motility is blocked, lateral flagella or pili are induced, and pathogenic gene expression, including toxins (black solid dots) is kept “ON” presumably because of the reduced bile concentration. (B) In an extra-intestinal environment, c-di-GMP (solid black dots) facilitates Vibrio from the motile-to-sessile transition by decreasing migration due to the binding of the YcgR homolog to the flagellar motor, which causes a reduction of motility and induces the biofilm formation and the suppression on the motility by releasing a lots of polysaccharides. The schemes are drawn based on previous reports (Krasteva et al., 2010; Boyd and O'Toole, 2012).
A similar relationship between motility and pathogenicity can be found between motility and biofilm formation in the extra-intestinal environment (Figure 6B). Once Vibrio cells are colonized, they form a biofilm and are encapsulated and thereby protected (Costerton et al., 1999). Biofilm formation is an important lifestyle of pathogenic Vibrio and renders bacteria resistant to environmental stresses, such as antimicrobial compounds or drugs (Yildiz and Visick, 2009; Lasarre and Federle, 2013). Biofilm formation in V. cholerae is controlled by quorum sensing (QS) through the modulation of cyclic di-guanosine monophosphate (c-di-GMP) that is used by bacterial pathogens to regulate the expression of genes involved in defense and invasion (Hammer and Bassler, 2003; Zhu and Mekalanos, 2003; Waters et al., 2008). A role of c-di-GMP in biofilm and motility is becoming clear. A flaA motX double mutant formed smooth colonies, indicating that flagella are involved in the initial stages of biofilm formation (Lauriano et al., 2004). For biofilm formation in V. cholerae, VpsT, a transcriptional regulator, has been reported to control Vibrio polysaccharide gene expression and to inversely regulate biofilm formation and motility, via c-di-GMP (Krasteva et al., 2010). Polysaccharide is a very important factor regarding the pathogenicity of Vibrio in the case of immune evasion and biofilm formation (Figure 6B) (Waldor et al., 1994; Naka et al., 2011; Johnson et al., 2012).
Regarding motility, several groups have shown that in E. coli and Salmonella, c-di-GMP directly bound to YcgR interacts with components of the flagellar motor to disrupt flagellar rotation, thereby leading to decreased motility (Boehm et al., 2010; Fang and Gomelsky, 2010; Paul et al., 2010). Since homologs of the YcgR protein that contains a binding domain of c-di-GMP extensively exist in Vibrio, the same impacting mechanism for the transition from the motile to the sessile phenotype by regulating c-di-GMP also exists in Vibrio (Pratt et al., 2007). Interestingly, on one hand, flagellar motility is involved in biofilm formation in the initial stage, but on the other hand, it is repressed when the biofilm is formed. This perhaps implies much broader functions for flagella in addition to motility and mechanosensing. Further, QS also influences motility and polar flagellar biogenesis via a master regulator for QS, SmcR that down-regulates flhF expression at the transcriptional level (Kim et al., 2012). Following the loss of motility and the formation of bacterial community, the responses induced by QS signals are usually involved as virulence determinants, such as toxins, proteases, polysaccharides and other relative fitness factors (Rutherford and Bassler, 2012).
Summary
Overall, this Review has focused on the structure, gene regulation and sensing of the polar flagellum in Vibrio spp., and emphasizes the inverse relationship between motility and pathogenicity. However, it is undeniable that motility induced by the polar flagellum in Vibrio spp. contributes to the virulence of pathogenic Vibrio through adhesion or biofilm formation regardless of the environment. We are interested in understanding how the Vibrio flagellar structure and function are involved in the pathogenicity. We also want to know why and how this very complicated structure has evolved and been maintained in Vibrio.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Aizawa S.-I. (1996). Flagellar assembly in Salmonella typhimurium. Mol. Microbiol. 19, 1–5 10.1046/j.1365-2958.1996.344874.x [DOI] [PubMed] [Google Scholar]
- Aldridge P. (2002). Regulation of flagellar assembly. Curr. Opin. Microbiol. 5, 160–165 10.1016/S1369-5274(02)00302-8 [DOI] [PubMed] [Google Scholar]
- Asai Y., Kawagishi I., Sockett R. E., Homma M. (2000). Coupling ion specificity of chimeras between H+- and Na+-driven motor proteins, MotB and PomB, in Vibrio polar flagella. EMBO J. 19, 3639–3648 10.1093/emboj/19.14.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi T., McCarter L. L., Imae Y. (1992). Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature 355, 182–184 10.1038/355182a0 [DOI] [PubMed] [Google Scholar]
- Balaban M., Joslin S. N., Hendrixson D. R. (2009). FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J. Bacteriol. 191, 6602–6611 10.1128/JB.00884-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee R., Das S., Mukhopadhyay K., Nag S., Chakrabortty A., Chaudhuri K. (2002). Involvement of in vivo induced cheY-4 gene of Vibrio cholerae in motility, early adherence to intestinal epithelial cells and regulation of virulence factors. FEBS Lett. 532, 221–226 10.1016/S0014-5793(02)03678-5 [DOI] [PubMed] [Google Scholar]
- Berg H. C. (2003). The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 10.1146/annurev.biochem.72.121801.161737 [DOI] [PubMed] [Google Scholar]
- Bina X. R., Bina J. E. (2010). The cyclic dipeptide cyclo(Phe-Pro) inhibits cholera toxin and toxin-coregulated pilus production in O1 El Tor Vibrio cholerae. J. Bacteriol. 192, 3829–3832 10.1128/JB.00191-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bina X. R., Taylor D. L., Vikram A., Ante V. M., Bina J. E. (2013). Vibrio cholerae ToxR downregulates virulence factor production in response to cyclo(Phe-Pro). MBio 4, 5 10.1128/mBio.00366-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. F. (2003). Flagellar movement driven by proton translocation. FEBS. Lett. 545, 86–95 10.1016/S0014-5793(03)00397-1 [DOI] [PubMed] [Google Scholar]
- Blake P. A., Weaver R. E., Hollis D. G. (1980). Diseases of humans (other than cholera) caused by vibrios. Ann. Rev. Microbiol. 34, 341–367 10.1146/annurev.mi.34.100180.002013 [DOI] [PubMed] [Google Scholar]
- Boehm A., Kaiser M., Li H., Spangler C., Kasper C. A., Ackermann M., et al. (2010). Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141, 107–116 10.1016/j.cell.2010.01.018 [DOI] [PubMed] [Google Scholar]
- Boin M. A., Austin M. J., Häse C. C. (2004). Chemotaxis in Vibrio cholerae. FEMS Microbiol. Lett. 239, 1–8 10.1016/j.femsle.2004.08.039 [DOI] [PubMed] [Google Scholar]
- Boyd C. D., O'Toole G. A. (2012). Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28, 439–462 10.1146/annurev-cellbio-101011-155705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler S. M., Camilli A. (2004). Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 101, 5018–5023 10.1073/pnas.0308052101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler S. M., Camilli A. (2005). Going against the grain, chemotaxis and infection in Vibrio cholerae. Nat. Rev. Microbiol. 3, 611–620 10.1038/nrmicro1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler S. M., Nelson E. J., Chowdhury N., Faruque S. M., Calderwood S. B., Camilli A. (2006). Cholera stool bacteria repress chemotaxis to increase infectivity. Mol. Microbiol. 60, 417–426 10.1111/j.1365-2958.2006.05096.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Beeby M., Murphy G. E., Leadbetter J. R., Hendrixson D. R., Briegel A., et al. (2011). Structural diversity of bacterial flagellar motors. EMBO J. 30, 2972–2981 10.1038/emboj.2011.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Berg H. C. (2000). Solvent-isotope and pH effects on flagellar rotation in Escherichia coli. Biophys. J. 78, 2280–2284 10.1016/S0006-3495(00)76774-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance F. F., Hughes K. T. (2008). Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6, 455–465 10.1038/nrmicro1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott G. S., Hughes K. T. (2000). Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708 10.1128/MMBR.64.4.694-708.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childers B. M., Klose K. E. (2007). Regulation of virulence in Vibrio cholerae: the ToxR regulon. Future Microbiol. 2, 335–344 10.2217/17460913.2.3.335 [DOI] [PubMed] [Google Scholar]
- Correa N. E., Peng F., Klose K. E. (2005). Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J. Bacteriol. 187, 6324–6332 10.1128/JB.187.18.6324-6332.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284, 1318–1322 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Daniels N. A., Shafaie A. (2000). A review of pathogenic Vibrio infections for clinicians. Infect. Med. 17, 665–685 12364378 [Google Scholar]
- DePaola A., Ulaszek J., Kaysner C. A., Tenge B. J., Nordstrom J. L., Wells J., et al. (2003). Molecular, serological, and virulence characteristics of Vibrio parahaemolyticus isolated from environmental, food, and clinical sources in North America and Asia. Appl. Environ. Microb. 69, 3999–4005 10.1128/AEM.69.7.3999-4005.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRita V. J., Parsot C., Jander G., Mekalanos J. J. (1991). Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 88, 5403–5407 10.1073/pnas.88.12.5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X., Gomelsky M. (2010). A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76, 1295–1305 10.1111/j.1365-2958.2010.07179.x [DOI] [PubMed] [Google Scholar]
- Francis N. R., Sosinsky G. E., Thomas D., Derosier D. J. (1994). Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235, 1261–1270 10.1006/jmbi.1994.1079 [DOI] [PubMed] [Google Scholar]
- Fukuoka H., Wada T., Kojima S., Ishijima A., Homma M. (2009). Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol. Microbiol. 71, 825–835 10.1111/j.1365-2958.2008.06569.x [DOI] [PubMed] [Google Scholar]
- Gardel C. L., Mekalanos J. J. (1996). Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64, 2246–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosink K. K., Kobayashi R., Kawagishi I., Häse C. C. (2002). Analyses of the roles of the three cheA homologs in chemotaxis of Vibrio cholerae. J. Bacteriol. 184, 1767–1771 10.1128/JB.184.6.1767-1771.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer B. K., Bassler B. L. (2003). Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50, 101–104 10.1046/j.1365-2958.2003.03688.x [DOI] [PubMed] [Google Scholar]
- Hang L., John M., Asaduzzaman M., Bridges E. A., Vanderspurt C., Kirn T. J., et al. (2003). Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 100, 8508–8513 10.1073/pnas.1431769100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häse C. C., Mekalanos J. J. (1999). Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 96, 3183–3187 10.1073/pnas.96.6.3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan F., Kamruzzaman M., Mekalanos J. J., Faruque S. M. (2010). Satellite phage TLCϕ enables toxigenic conversion by CTX phage through dif site alteration. Nature 467, 982–985 10.1038/nature09469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg J. F., Eisen J. A., Nelson W. C., Clayton R. A., Gwinn M. L., Dodson R. J., et al. (2000). DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406, 477–483 10.1038/35020000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrington D. A., Hall R. H., Losonsky G. G., Mekalanos J. J., Taylor R. K., Levine M. M. (1988). Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 16, 1487–1492 10.1084/jem.168.4.1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyakutake A., Homma M., Austin M. J., Boin M. A., Häse C. C., Kawagishi I. (2005). Only one of the five CheY homologs in Vibrio cholerae directly switches flagellar rotation. J. Bacteriol. 187, 8403–8410 10.1128/JB.187.24.8403-8410.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. N., Bowers J. C., Griffitt K. J., Molina V., Clostio R. W., Pei S., et al. (2012). Ecology of Vibrio parahaemolyticus and Vibrio vulnificus in the coastal and estuarine waters of Louisiana, Maryland, Mississippi, and Washington (United States). Appl. Environ. Microb. 78, 7249–7257 10.1128/AEM.01296-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. K., Oliver J. D. (2009). Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77, 1723–1733 10.1128/IAI.01046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. W., Colwell R. R., Kaper J. B. (1982). Vibrio parahaemolyticus and related halophilic Vibrios. Crit. Rev. Microbiol. 10, 77–124 10.3109/10408418209113506 [DOI] [PubMed] [Google Scholar]
- Karaolis D. K., Johnson J. A., Bailey C. C., Boedeker E. C., Kaper J. B., Reeves P. R. (1998). A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. U.S.A. 95, 3134–3139 10.1073/pnas.95.6.3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi I., Imagawa M., Imae Y., McCarter L. L., Homma M. (1996). The sodium-driven polar flagellar motor of marine Vibrio as the mechanosensor that regulates lateral flagellar expression. Mol. Microbiol. 20, 693–699 10.1111/j.1365-2958.1996.tb02509.x [DOI] [PubMed] [Google Scholar]
- Kim S. M., Lee D. H., Choi S. H. (2012). Evidence that the Vibrio vulnificus flagellar regulator FlhF is regulated by a quorum sensing master regulator SmcR. Microbiology 158, 2017–2025 10.1099/mic.0.059071-0 [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Thanh X. T. T., Jeong K., Kim S. B., Pan S. O., Jung C. H., et al. (2013). Contribution of six flagellin genes to the flagellum biogenesis of Vibrio vulnificus and in vivo invasion. Infect. Immun. [Epub ahead of print]. 10.1128/IAI.00654-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. K., McCarter L. L. (2000). Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 182, 3693–3704 10.1128/JB.182.13.3693-3704.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka M., Nishigaki T., Ihara K., Nishioka N., Kojima S., Homma M. (2013). A novel dnaJ family gene, sflA, encodes an inhibitor of flagellation in marine Vibrio species. J. Bacteriol. 195, 816–822 10.1128/JB.01850-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose K. E., Mekalanos J. J. (1998). Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28, 501–520 10.1046/j.1365-2958.1998.00809.x [DOI] [PubMed] [Google Scholar]
- Klose K. E., Novik V., Mekalanos J. J. (1998). Identification of multiple sigma54-dependent transcriptional activators in Vibrio cholerae. J. Bacteriol. 180, 5256–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M., Nishioka N., Kusumoto A., Yagasaki J., Fukuda T., Homma M. (2011). Conversion of mono-polar to peritrichous flagellation in Vibrio alginolyticus. Microbiol. Immunol. 55, 76–83 10.1111/j.1348-0421.2010.00290.x [DOI] [PubMed] [Google Scholar]
- Kojima S., Shinohara A., Terashima H., Yakushi T., Sakuma M., Homma M., et al. (2008). Insights into the stator assembly of the Vibrio flagellar motor from the crystal structure of MotY. Proc. Natl. Acad. Sci. U.S.A. 105, 7696–7701 10.1073/pnas.0800308105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasteva P. V., Fong J. C. N., Shikuma N. J., Beyhan S., Navarro M. V. A. S., Yildiz F. H., et al. (2010). Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327, 866–868 10.1126/science.1181185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukonis E. S., DiRita V. J. (2003). From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6, 186–190 10.1016/S1369-5274(03)00032-8 [DOI] [PubMed] [Google Scholar]
- Kusumoto A., Kamisaka K., Yakushi T., Terashima H., Shinohara A., Homma M. (2006). Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J. Biochem. 139, 113–121 10.1093/jb/mvj010 [DOI] [PubMed] [Google Scholar]
- Kusumoto A., Shinohara A., Terashima H., Kojima S., Yakushi T., Homma M. (2008). Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology 154, 1390–1399 10.1099/mic.0.2007/012641-0 [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Ohya Y., Iino T. (1990). Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasarre B., Federle M. J. (2013). Exploiting quorum sensing to confuse bacterial pathogens. Microbiol. Mol. Biol. Rev. 77, 73–111 10.1128/MMBR.00046-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauriano C. M., Ghosh C., Correa N. E., Klose K. E. (2004). The sodium-driven flagellar motor controls exopolysaccharide expression in Vibrio cholerae. J. Bacteriol. 186, 4864–4874 10.1128/JB.186.15.4864-4874.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake M. C., Chandler J. H., Wadhams G. H., Bai F., Berry R. M., Armitage J. P. (2006). Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 10.1038/nature05135 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Hava D. L., Waldor M. K., Camilli A. (1999). Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99, 625–634 10.1016/S0092-8674(00)81551-2 [DOI] [PubMed] [Google Scholar]
- Lele P. P., Hosu B. G., Berg H. C. (2013). Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 110, 11839–11844 10.1073/pnas.1305885110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Kojima S., Homma M. (2011). Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes Cells. 16, 985–999 10.1111/j.1365-2443.2011.01545.x [DOI] [PubMed] [Google Scholar]
- Lowden M. J., Skorupski K., Pellegrini M., Chiorazzo M. G., Taylor R. K., Kull F. J. (2010). Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. U.S.A. 107, 2860–2865 10.1073/pnas.0915021107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNab R. M. (2003). How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100 10.1146/annurev.micro.57.030502.090832 [DOI] [PubMed] [Google Scholar]
- Magariyama Y., Sugiyama S., Muramoto K., Maekawa Y., Kawagishi I., Imae Y., et al. (1994). Very fast flagellar rotation. Nature 371, 752–752 10.1038/371752b0 [DOI] [PubMed] [Google Scholar]
- Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., et al. (2003). Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361, 743–749 10.1016/S0140-6736(03)12659-1 [DOI] [PubMed] [Google Scholar]
- McCarter L. L. (1994a). MotX, the channel component of the sodium-type flagellar motor. J. Bacteriol. 176, 5988–5998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L. (1994b). MotY, a component of the sodium-type flagellar motor. J. Bacteriol. 176, 4219–4225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L. (2001). Polar flagellar motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 65, 445–462 10.1128/MMBR.65.3.445-462.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarter L. L. (2004). Dual flagellar systems enable motility under different circumstances. J. Mol. Microbiol. Biotechnol. 7, 18–29 10.1159/000077866 [DOI] [PubMed] [Google Scholar]
- McCarter L. L., Hilmen M., Silverman M. (1988). Flagellar dynamometer controls swarmer cell differentiation of V. parahaemolyticus. Cell 54, 345–351 10.1016/0092-8674(88)90197-3 [DOI] [PubMed] [Google Scholar]
- Merrell D. S., Butler S. M., Qadri F., Dolganov N. A., Alam A., Cohen M. B., et al. (2002). Host-induced epidemic spread of the cholera bacterium. Nature 417, 642–645 10.1038/nature00778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy G. E., Leadbetter J. R., Jensen G. J. (2006). In situ structure of the complete Treponema primitia flagellar motor. Nature 442, 1062–1064 10.1038/nature05015 [DOI] [PubMed] [Google Scholar]
- Naka H., Dias G. M., Thompson C. C., Dubay C., Thompson F. L., Crosa J. H. (2011). Complete genome sequence of the marine fish pathogen Vibrio anguillarum harboring the pJM1 virulence plasmid and genomic comparison with other virulent strains of V. anguillarum and V. ordalii. Infect. Immun. 79, 2889–2900 10.1128/IAI.05138-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama S.-I., Suzuki D., Itoh Y., Suzuki K., Tajima H., Hyakutake A., et al. (2012). Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect. Immun. 80, 3170–3178 10.1128/IAI.00039-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Kutsukake K., Suzuki H., Iino T. (1990). Gene fliA encodes an alternative sigma factor specific for flagellar operons in Salmonella typhimurium. Mol. Gen. Genet. 221, 139–147 10.1007/BF00261713 [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Kutsukake K., Suzuki H., Lino T. (1992). A novel transcriptional regulation mechanism in the flagellar regulon of Salmonella typhimurium: an antisigma factor inhibits the activity of the flagellum−specific Sigma factor, sigma F. Mol. Microbiol. 6, 3149–3157 10.1111/j.1365-2958.1992.tb01771.x [DOI] [PubMed] [Google Scholar]
- Okabe M., Yakushi T., Asai Y., Homma M. (2001). Cloning and characterization of motX, a Vibrio alginolyticus sodium-driven flagellar motor gene. J. Biochem. 130, 879–884 10.1093/oxfordjournals.jbchem.a003061 [DOI] [PubMed] [Google Scholar]
- Paul K., Nieto V., Carlquist W. C., Blair D. F., Harshey R. M. (2010). The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38, 128–139 10.1016/j.molcel.2010.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt J. T., Tamayo R., Tischler A. D., Camilli A. (2007). PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282, 12860–12870 10.1074/jbc.M611593200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen L., White E. L., Pathak A., Ayala J. C., Wang H., Wu J.-H., et al. (2011). A high-throughput screening assay for inhibitors of bacterial motility identifies a novel inhibitor of the Na+-driven flagellar motor and virulence gene expression in Vibrio cholerae. Antimicrob. Agents Chemother. 55, 4134–4143 10.1128/AAC.00482-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Hu C., Jiang X., Sun H., Zhao Z., Chen C., et al. (2013). Distribution and pathogenic relationship of virulence associated genes among Vibrio alginolyticus from the mariculture systems. Mol. Cell Probe. 27, 164–168 10.1016/j.mcp.2013.01.004 [DOI] [PubMed] [Google Scholar]
- Richardson K. K. (1991). Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59, 2727–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S. T., Bassler B. L. (2012). Bacterial quorum sensing, its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2, 11 10.1101/cshperspect.a012427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvetti S., Ghelardi E., Celandroni F., Ceragioli M., Giannessi F., Senesi S. (2007). FlhF, a signal recognition particle-like GTPase, is involved in the regulation of flagellar arrangement, motility behaviour and protein secretion in Bacillus cereus. Microbiology 153, 2541–2552 10.1099/mic.0.2006/005553-0 [DOI] [PubMed] [Google Scholar]
- Silva A. J., Leitch G. J., Camilli A., Benitez J. A. (2006). Contribution of hemagglutinin/protease and motility to the pathogenesis of El Tor biotype cholera. Infect. Immun. 74, 2072–2079 10.1128/IAI.74.4.2072-2079.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa Y., Berry R. M. (2008). Bacterial flagellar motor. Q. Rev. Biophys. 41, 103–132 10.1017/S0033583508004691 [DOI] [PubMed] [Google Scholar]
- Sreelatha A., Bennett T. L., Zheng H., Jiang Q.-X., Orth K., Starai V. J. (2013). Vibrio effector protein, VopQ, forms a lysosomal gated channel that disrupts host ion homeostasis and autophagic flux. Proc. Natl. Acad. Sci. U.S.A. 110, 11559–11564 10.1073/pnas.1307032110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart B. J., McCarter L. L. (2003). Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 185, 4508–4518 10.1128/JB.185.15.4508-4518.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed K. A., Beyhan S., Correa N., Queen J., Liu J., Peng F., et al. (2009). The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J. Bacteriol. 191, 6555–6570 10.1128/JB.00949-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H., Ordal G. W. (2004). Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol. Mol. Biol. Rev. 68, 301–319 10.1128/MMBR.68.2.301-319.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. K., Miller V. L., Furlong D. B., Mekalanos J. J. (1987). Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. U.S.A. 84, 2833–2837 10.1073/pnas.84.9.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima H., Fukuoka H., Yakushi T., Kojima S., Homma M. (2006). The Vibrio motor proteins, MotX and MotY, are associated with the basal body of Na+-driven flagella and required for stator formation. Mol. Microbiol. 62, 1170–1180 10.1111/j.1365-2958.2006.05435.x [DOI] [PubMed] [Google Scholar]
- Terashima H., Kojima S., Homma M. (2008). Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 270, 39–85 10.1016/S1937-6448(08)01402-0 [DOI] [PubMed] [Google Scholar]
- Terashima H., Li N., Sakuma M., Koike M., Kojima S., Homma M., et al. (2013). Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc. Natl. Acad. Sci. U.S.A. 110, 6133–6138 10.1073/pnas.1222655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. R., Francis N. R., Xu C., Derosier D. J. (2006). The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188, 7039–7048 10.1128/JB.00552-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping M. J., Delalez N. J., Lim R., Berry R. M., Armitage J. P. (2013). Load-dependent assembly of the bacterial flagellar motor. mBio 4:e00551-13 10.1128/mBio.00551-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor M. K., Colwell R., Mekalanos J. J. (1994). The Vibrio cholerae O139 serogroup antigen includes an O-antigen capsule and lipopolysaccharide virulence determinants. Proc. Natl. Acad. Sci. U.S.A. 91, 11388–11392 10.1073/pnas.91.24.11388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters C. M., Lu W., Rabinowitz J. D., Bassler B. L. (2008). Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190, 2527–2536 10.1128/JB.01756-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. Y., Hu C. Q., Chen C., Zhang L. P., Ren C. H. (2005). Investigation of seven Vibrio virulence genes among Vibrio alginolyticus and Vibrio parahaemolyticus strains from the coastal mariculture systems in Guangdong, China. Lett. Appl. Microbiol. 41, 202–207 10.1111/j.1472-765X.2005.01688.x [DOI] [PubMed] [Google Scholar]
- Yildiz F. H., Visick K. L. (2009). Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17, 109–118 10.1016/j.tim.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Orth K. (2013). Virulence determinants for Vibrio parahaemolyticus infection. Curr. Opin. Microbiol. 16, 70–77 10.1016/j.mib.2013.02.002 [DOI] [PubMed] [Google Scholar]
- Zhang X. H., Austin B. (2005). Haemolysins in Vibrio species. J. Appl. Microbiol. 98, 1011–1019 10.1111/j.1365-2672.2005.02583.x [DOI] [PubMed] [Google Scholar]
- Zhu J., Mekalanos J. J. (2003). Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5, 10 10.1016/S1534-5807(03)00295-8 [DOI] [PubMed] [Google Scholar]