Abstract
Desensitization of activated receptors is an important mechanism for terminating signal transduction. Here we show that phytochrome (phy) A, a predominant photoreceptor for seedling deetiolation, colocalizes in nuclear bodies with CONSTITUTIVELY PHOTOMORPHOGENIC (COP) 1, a RING motif-containing E3 ligase. The phyA PAS domain interacts with the COP1 WD40 domain. Both the Pr and the Pfr forms of phyA, as well as the PHYA apoprotein, are ubiquitinated by COP1 in vitro. The phyA destruction rate is decreased in cop1 mutants and by expression of a COP1 RING motif mutant. Our results indicate that COP1 acts as an E3 ligase to regulate phyA signaling by targeting elimination of the phyA photoreceptor itself.
Keywords: Photoreceptor, desensitization, COP1, phytochrome A signaling
Light provides a major source of information from the environment during plant growth and development. To perceive light, plants have evolved a large set of photoreceptors. Among these, the cryptochromes and phototropins are responsible for UV-A/blue light sensing, whereas the phytochromes predominantly regulate responses to red (R) and far-red (FR) light (Neff et al. 2000).
The Arabidopsis phytochrome gene family comprises five members, named PHYA-PHYE (Nagy and Schäfer 2002). Each forms a homodimer of ∼240 kD and light sensitivity is conferred by a tetrapyrrole chromophore covalently bound to the N-terminal half of each monomer (Montgomery and Lagarias 2002). Dimerization domains are located within the C-terminal halves of the proteins, as are other domains involved in the activation of signal transduction (Quail et al. 1995; Quail 2002a).
In dark-grown plants, phytochrome (phy) A is the most abundant photoreceptor and makes a major contribution to initiating photomorphogenesis (Casal et al. 2003). In darkness, phyA is in the stable Pr form, but on illumination it is converted to the active Pfr form, which is rapidly degraded (Clough and Vierstra 1997; Nagy and Schäfer 2002; Sharrock and Clack 2002). phyA ubiquitination was reported more than a decade ago (Jabben et al. 1989a,b) and the extreme photolability of phyA is well documented (Sharrock and Clack 2002). Nevertheless, the failure to identify the E3 ligase that mediates phyA ubiquitination has impeded further progress in elucidating the mechanism that underlies this signal termination event.
Mutant screens in Arabidopsis have identified many components of the signaling pathways that act downstream of photoreceptors (Quail 2002b). Among the gene products that are important for deetiolation are the pleio-tropic CONSTITUTIVELY PHOTOMORPHOGENIC (COP)/DET/FUS proteins, which are required for the repression of photomorphogenesis in darkness (Wei and Deng 1996). COP1 is a 76-kD repressor that contains single RING-finger, coiled-coil, and WD40 domains (Deng et al. 1992). RING-fingers are commonly conserved in a subclass of ubiquitin protein ligases (Pickart 2001). It has recently been shown that COP1 has E3 ubiquitin protein ligase activity toward the transcription factors LAF1 (Seo et al. 2003) and HY5 (Saijo et al. 2003), both of which transduce the phyA signal. COP1 thus regulates phyA signaling at least in part through proteasome-mediated degradation of these intermediates. Further in vitro analyses indicated that COP1 activity on both substrates is regulated by SPA1 (Saijo et al. 2003; Seo et al. 2003), a nuclear-localized repressor of phyA-mediated signaling (Hoecker et al. 1999).
PhyB, cryptochrome (cry) 1, and cry2 were found to interact with COP1 (Wang et al. 2001; Yang et al. 2001), but under the same condition phyA failed to interact with the latter (Yang et al. 2001). Although Shalitin et al. (2002) showed that cry2 accumulates in cop1-4 and cop1-6 mutants, it is not known whether this accumulation is due to the direct effect of COP1 E3 ligase activity. In general, there is still little information about the role of photoreceptor degradation in the desensitization of plant light signal transmission. Termination of signaling through a pathway ensures that responses to a single stimulatory event are not perpetuated indefinitely. This regulatory mechanism is particularly important for receptors that initiate signaling, and many eukaryotic receptors are down-regulated by proteolysis. We have thus focused on the requirement of COP1 activity for termination of phyA signaling by reassessing whether COP1 and phyA interact directly and whether this interaction might promote ubiquitination of phyA. Indeed, COP1 and phyA interact in vitro and colocalize in nuclear bodies (NBs) in vivo. Furthermore, cop1 mutant plants and transgenic plants expressing COP1 dominant-negative mutant protein accumulate phyA after exposure to light. Our results provide the first example of how plants use E3 ubiquitin protein ligase activity to desensitize photoreceptor signaling.
Results and Discussion
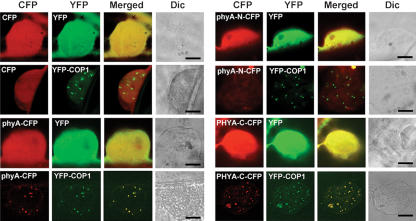
Previous studies revealed that phyA is exclusively cytosolic in darkness but that on illumination, a pool of phyA becomes sequestered in NBs (Kircher et al. 2002; Nagy and Schäfer 2002). Because the RING motif protein COP1 is also localized in NBs (Stacy and von Arnim 1999), we asked whether the two proteins colocalize. To this end, we transiently expressed in onion epidermal cells Arabidopsis PHYA tagged with cyan fluorescent protein (CFP) and COP1 tagged with yellow fluorescent protein (YFP). To what extent onion cells can recapitulate the situation in Arabidopsis cells is not known; nevertheless, we found that the two proteins were indeed localized in the same NBs, indicating interaction in vivo (Fig. 1). This subnuclear localization appears to be a property of the PHYA C-terminal half (amino acids 591-1121).
Figure 1.
Subcellular distribution and colocalization of COP1 and phyA. COP1 and phyA colocalize in nuclear bodies in onion epidermal cells. PHYA-N-CFP and PHYA-C-CFP refer to CFP fusions to the N-terminal (amino acids 1-600) and C-terminal (amino acids 591-1121) fragments of Arabidopsis PHYA, respectively. (Dic) Differential interference contrast. Bar, 20 μm.
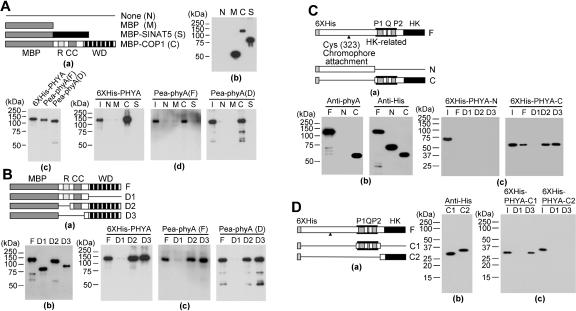
Either direct or indirect interactions could account for the colocalization of phyA and COP1 in NBs. Although previous work failed to detect an interaction between COP1 and a PHYA C-terminal fragment in yeast two-hybrid assays (Yang et al. 2001), we used in vitro pull-down assays to investigate potential interactions between pea phyA holoprotein, Arabidopsis PHYA apoprotein, and COP1. Maltose-binding protein (MBP) and MBP-SINAT5 (Xie et al. 2002) were used as negative controls. Figure 2A shows that MBP-COP1, but not MBP-SINAT5 or MBP alone, was able to pull down Arabidopsis PHYA, pea phyA, and N-terminal-deleted pea phyA (Tokutomi et al. 1988). Experiments using deletion mutants demonstrated that the COP1 WD40 domain interacts with the PAS domain region (amino acids 591-850) of Arabidopsis PHYA (Fig. 2B-D). These in vitro results suggest that the colocalization of phyA and COP1 in NBs likely reflects their direct interaction in vivo.
Figure 2.
Interaction of COP1 and phyA in vitro. (A) In vitro pull down of Arabidopsis PHYA or pea phyA with full-length COP1. (Panel a) Schematic diagram of bait proteins (MBP, MBP-COP1, and MBP-SINAT5. (Panel b) Purified bait proteins detected by anti-MBP antibody (Santa Cruz Biotechnology). (Panel c) Purified prey proteins detected by anti-phyA antibody. (Panel d) 6×His-PHYA, full-length pea phyA [Pea-phyA (F)], and deleted pea phyA [Pea-phyA (D)] pulled down with the indicated bait protein were detected by anti-phyA antibody. (B) In vitro pull down of phyA with COP1 deletion mutants. (Panel a) Schematic diagram of full-length COP1 and deletion mutants. (MBP) Maltose-binding protein; (R) RING motif; (CC) coiled-coil motif; (WD) WD40 motif. (Panel b) Detection of purified deletion mutants by anti-MBP antibody. (Panel c) 6×His-PHYA, full-length pea phyA, and deleted pea phyA pulled down with the indicated COP1 deletion mutant were detected by anti-phyA antibody. (C) In vitro pull down of the 6×His-tagged full-length, N-terminal, or C-terminal fragments of Arabidopsis PHYA with full-length COP1 and COP1 deletion mutants. (Panel a) Schematic diagram of full-length Arabidopsis PHYA and deletion mutants. (P1) PAS1 motif; (Q) quail motif; (P2) PAS2 motif; (HK) histidine kinase motif. (Panel b) Purified N-terminal or C-terminal fragment of 6×His-tagged Arabidopsis PHYA detected by anti-phyA antibody or anti-His antibody. (Panel c) 6×His-tagged N-terminal or C-terminal fragments of Arabidopsis PHYA pulled down with COP1 or the indicated COP1 deletion mutant were detected by anti-His antibody. (I) Input. (D) In vitro pull down of 6×His-tagged C-terminal deletion mutants of Arabidopsis PHYA with COP1 deletion mutants. (Panel a) Schematic diagram of Arabidopsis PHYA C-terminal deletion mutants. (Panel b) Purified C-terminal deletion mutants of His6-tagged Arabidopsis PHYA detected by anti-His antibody. (panel c) 6×His-tagged C-terminal deletion mutants of Arabidopsis PHYA pulled down with the indicated COP1 deletion mutant were detected by anti-His antibody. (I) Input. D1 and D3 are COP1 deletion mutants shown in panel a of part B.
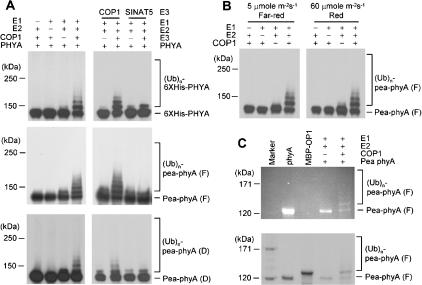
The colocalization of phyA with COP1 in vivo and their direct interaction in vitro suggest that COP1 may act as an E3 ligase for phyA. We produced Arabidopsis PHYA tagged with His6 in Escherichia coli and also purified photoreversible phyA from peas. Figure 3A shows that purified His6-PHYA was polyubiquitinated by COP1 in a reaction dependent on E1 and E2 activities. The integrity of the COP1 RING motif was essential for this reaction (data not shown). No PHYA polyubiquitination was seen with SINAT5, another Arabidopsis RING motif-containing E3 ligase (Xie et al. 2002), demonstrating specificity. Similar results were obtained with purified pea phyA, presented either in the Pr or the Pfr form (Fig. 3B). However, we cannot strictly rule out the possibility that the Pfr form, which is continuously generated under FR light, is the preferred substrate. Detection of phyA by Zn2+-induced fluorescence verified that polyubiquitination of the holoprotein indeed occurred (Fig. 3C).
Figure 3.
Ubiquitination of phyA by COP1. (A) COP1 E3 activity was assayed in the presence or absence of rabbit E1, UbcH5b (E2), COP1, 6×His-Ub, 6×His-PHYA, full-length pea phyA (F), or deleted pea phyA (D). (B) Ubiquitination of phyA by COP1 under R or FR light. Reaction mixtures without full-length pea phyA were preincubated under R or FR light for 90 min. Full-length pea phyA was then added and the reaction mixtures were further incubated under the same light conditions for 2 h. (C) Ubiquitination of bilin-bound phyA by COP1. The products of the same reactions performed in A were subjected to SDS-PAGE including 1 mM zinc acetate, before analysis of zinc-induced florescence (top) and staining with Coomassie blue (bottom).
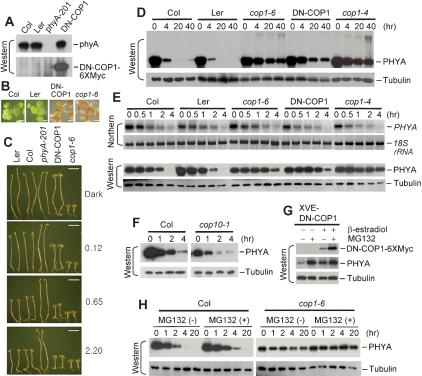
The in vitro results suggest that COP1 may target phyA for ubiquitin-mediated proteolysis in vivo. We previously showed that a COP1 RING motif mutant, which lacks E3 activity, can interfere with wild-type COP1 E3 activity in vitro (Seo et al. 2003). Such RING motif mutants function in a dominant negative manner, presumably by forming inactive dimers with wild-type COP1. To reduce COP1 E3 activity in vivo, we used a 35S promoter or XVE promoter (Kost et al. 1998; Zuo et al. 2000) to express a dominant negative version of COP1 (DNCOP1) with mutations in four cysteine residues (C52S, C55S, C86S, C89A) of the RING motif. Figure 4C shows that transgenic plants expressing 35S-DN-COP1 exhibited deetiolation in the dark, as did the cop1-6 mutant. In addition, these transgenic plants phenocopied the cop1-6 mutant when grown under white light or exposed to FR light of different intensities (Fig. 4B,C). In general, however, the transgenic phenotype is not as severe as the mutant phenotype with respect to inhibition of hypocotyl elongation. Similar results were obtained with transgenic plants expressing XVE-DN-COP1 in the presence of inducer (data not shown).
Figure 4.
Phenotypes of DN-COP1-overexpression plants and regulation of phyA levels by COP1 in vivo. (A) DN-COP1 expression. Extracts of wild-type (Col and Ler), phyA-201 (Reed et al. 1994), and 35S-DN-COP1-6×Myc (DN-COP1) transgenic seedlings grown in darkness for 4 d were analyzed by Western blotting using anti-phyA antibody or anti-Myc antibody. (B) Morphological comparison of 4-day-old wild-type, cop1-6 (McNellis et al. 1994), and 35S-DN-COP1-6×Myc transgenic seedlings grown in 16 h light/8 h dark with 1% sucrose. Bar, 0.5 mm. (C) Hypocotyl and cotyledon phenotypes of wild-type, phyA-201, cop1-6, 35S-DN-COP1-6×Myc transgenic seedlings germinated and grown under continuous FR light for 4 d. Numbers on the right indicate the FR light intensity (μmole m-2sec-1). Bar, 2.5 mm. (D) Western analyses of wild-type, cop1-4 (McNellis et al. 1994), cop1-6, and transgenic 35S-DN-COP1-6×Myc seedlings treated with R light (20 μmole m-2sec-1) for the indicated time. (E) Northern and Western analyses of seedlings treated as in D but for shorter time periods. (F) phyA degradation in cop10-1. Four-day-old dark-grown wild-type and cop10-1 (Suzuki et al. 2002) seedlings were treated with R light (20 μmole m-2sec-1) for the indicated time and phyA levels were assessed by Western blot using anti-phyA antibody. After exposure to light, only seedlings homozygous for the cop10-1 mutation were analyzed by Western blotting. Different exposure times were used for the wild-type and cop10-1 samples. In separate experiments, dark-grown seedlings of wild type and cop10-1 showed comparable phyA levels. (G) phyA degradation is inhibited by DN-COP1 overexpression. Four-day-old dark-grown transgenic seedlings expressing XVE-DN-COP1-6×Myc were induced by 10 μM β-estradiol under R light (20 μmole m-2sec-1) with or without MG132. After 15 h, harvested samples were analyzed by Western blotting using anti-phyA antibody. (H) phyA degradation is mediated by the 26S proteasome. Dark-grown wild-type and cop1-6 seedlings were treated with MG132 under R light (20 μmole m-2sec-1) for the indicated times. Extracts were analyzed by Western blotting using anti-phyA antibody with tubulin levels as loading controls. Comparable RNA levels (5 μg/lane) were determined by Northern blotting and detection of 18S rRNA abundance. Note that the exposure times of D-H were different.
Previous reports indicated that the Pfr form of phyA is light labile (Clough and Vierstra 1997; Sharrock and Clack 2002; Nagy and Schäfer 2002). We therefore compared the rate of phyA degradation in wild-type Arabidopsis and two cop1 mutants after irradiation with red (R) light. Under our conditions, irradiation of wild-type plants with R light for 4 h reduced phyA levels to ∼5% of the level in dark-grown seedlings (Fig. 4E), and little or no phyA was detectable after 20 h of irradiation (Fig. 4D). In contrast, in cop1-4 and cop1-6 mutants, phyA levels remained high (∼40% of the level in darkness) even after 40 h of irradiation with R light. This effect was specific for cop1 mutations, as the cop10-1 mutant displayed similar phyA degradation kinetics as wild type (Fig. 4F). The delayed degradation kinetics of phyA was phenocopied in transgenic plants overexpressing DN-COP1. In a shorter time course study, we confirmed that phyA transcript abundance was reduced by R light in wild-type seedlings (Col and Ler; Cantón and Quail 1999) and comparable kinetics were seen in cop1-6 and DN-COP1 transgenic plants (Fig. 4E). In contrast, the rate of phyA disappearance was delayed in cop1-6 and the transgenic line compared with wild type (Fig. 4D,E). The ability of MG132 to further block phyA disappearance in wild type and cop1-6 implicates proteasome-mediated degradation of the photoreceptor (Fig. 4H).
We used an inducible system to express the DN-COP1 mutant in transgenic plants and examine the effect of inhibiting endogenous COP1 E3 activity on phyA levels. Figure 4G shows that phyA levels were increased three- to fourfold on induced expression of DN-COP1, which was expected to block endogenous COP1 E3 activity. Addition of the proteasome inhibitor MG132 increased phyA levels a further twofold, suggesting incomplete inhibition of endogenous COP1 E3 activity by the DN-COP1. However, in the presence of MG132, phyA levels were comparable in transgenic plants with or without induced expression of DNCOP1. The expressed DN-COP1 was also labile and can be stabilized by MG132.
The results with mutants and transgenic plants indicate that compromising the endogenous COP1 E3 activity by mutation (cop1-4, cop1-6) or overexpression of DN-COP1 greatly reduced the phyA degradation rate in vivo. This suggests that COP1 indeed acts as an E3 ligase for phyA in vivo.
Light-induced degradation of phyA appears to constitute a primary regulatory mechanism, although phyA phosphorylation in the photoreceptor N-terminal domain (Stockhaus et al. 1992; Park et al. 2000) and degradation of downstream effectors (Saijo et al. 2003; Seo et al. 2003) have also been suggested to attenuate phyA signaling. Our findings suggest that COP1 acts in concert with the 26S proteasome to degrade phyA. In vitro experiments did not uncover any preference of COP1 for the Pr or Pfr forms of phyA, nor any effect of the presence or absence of the chromophore conjugated to PHYA apoprotein. Nonetheless, the Pfr form is likely the in vivo substrate because the rapid nuclear translocation of phyA occurs only after irradiation, at a time when COP1 is still nuclear. It is possible that Pfr phosphorylation or accessory factors might be required to increase the interaction of phyA with COP1, and these are lacking in our in vitro assay. Our results also show that polyubiquitination of phyA can proceed before disassembly of the chromophore.
Pull-down and localization results (Figs. 1, 2) suggest that the C-terminal domain of phyA interacts with COP1 in NBs, which might represent sites of phyA polyubiquitination. Subsequent degradation of the polyubiquitinated phyA may occur either in the nucleus or the cytosol. Recent results also suggest that the phyB C-terminal region, which interacts with COP1 (Stacy and von Arnim 1999), negatively regulates light signaling. Transgenic plants expressing a phyB N-terminal fragment (amino acids 625-1172) display a hyper-sensitive light response (Matsushita et al. 2003). The E3 ligase responsible for desensitization of phyB remains to be identified.
Whereas in vivo data with mutants and transgenic plants (Fig. 4) indicate that the COP1 E3 ligase regulates phyA abundance, two lines of evidence suggest that normal rates of phyA degradation require other pathways as well. First, a pool of phyA remains cytosolic even on illumination (Nagy and Schäfer 2002). Second, cop1-4 (which lacks the WD40 domain) and cop1-6 mutants display only reduced rates of phyA degradation (Fig. 4D,E). However, these mutant alleles are not null. In darkness, phyA and COP1 are located in the cytosol and nucleus, respectively. On illumination, a pool of phyA translocates to the nucleus (Nagy and Schäfer 2002) and presumably becomes polyubiquitinated by nuclear COP1. At the same time, COP1 gradually disappears from the nucleus and accumulates in the cytosol (von Arnim and Deng 1994; von Arnim 1997), where it may modify cytosolic phyA as well. We expect future studies on factors that regulate phyA polyubiquitination by COP1 and the effects on intracellular transport of these two proteins to further our understanding of the feedback regulation of phyA signaling.
Materials and methods
Preparation of recombinant proteins
Full-length Arabidopsis PHYA cDNA and its deletion mutants were cloned by PCR and inserted into pRSETA (Invitrogen) to generate 6xHis-PHYA (encoding full length), 6×His-PHYA-N (encoding amino acids 1-600), 6×His-PHYA-C (encoding amino acids 591-1121), 6×His-PHYA-C1 (encoding amino acids 591-850), and 6×His-PHYA-C2 (encoding amino acids 851-1121). Proteins were expressed in E. coli strain BL21 and purified (Seo et al. 2003). cDNA encoding full-length COP1 and deletion mutants fused with MBP were prepared as described (Seo et al. 2003). Full-length pea phyA and N-terminal deleted pea phyA (amino acids 53-1112) were purified according to Tokutomi et al. (1988).
Subcellular localization of phyA and COP1
The YFP coding sequences were fused to the 3′ end of full-length Arabidopsis PHYA cDNA, PHYA cDNA 5′ fragment, and PHYA cDNA 3′ fragment to generate phyA-YFP, phyA-N-YFP, and PHYA-C-YFP constructs, respectively. COP1-CFP was described previously (Seo et al. 2003). All fusion genes or genes for YFP and CFP were expressed from the 35S promoter. Onion epidermal cells were bombarded with different combinations of plasmids using a helium biolistic gun (Xie et al. 2000) and incubated in the dark for 14 h and treated with white light for 3 h before analysis by confocal microscopy.
In vitro binding and in vitro ubiquitination assays
For in vitro binding, 2 μg of bait (full-length COP1 or deletion mutants) and 2 μg of prey (6×His-PHYA or pea phyA) were added to 1 mL of binding buffer (50 mM Tris at pH 7.5, 100 mM NaCl, 0.2% glycerol, 0.6% Triton X-100, 0.5 mM β-mercaptoethanol). After incubation at 25°C for 2 h, the reaction mixture was further incubated with amylose resin beads for 2 h before being washed six times with the washing buffer (50 mM Tris at pH 7.5, 100 mM NaCl, 0.6% Triton X-100). Pulled-down proteins were analyzed by SDS-PAGE and detected by Western blotting using anti-phyA monoclonal antibody (Shinomura et al. 1996) or anti-His antibody (Santa Cruz Biotechnology). In vitro ubiquitination (Seo et al. 2003) was performed using 100 ng of purified 6×His-PHYA or pea phyA. After incubation at 30°C for 2 h, reaction mixtures were separated on 6% SDS-PAGE gels. Ubiquitinated 6×His-PHYA or pea phyA was detected by Western blotting using anti-phyA antibody. Covalently bound bilins were visualized by Zn2+-induced florescence of the complexes after resolution on 6% SDS-PAGE gels (Berkelman and Lagarias 1986).
Effects of COP1 on phyA levels in vivo
DN-COP1-6×Myc (C52S, C55S, C86S, C89A) was cloned into pBA002 (Kost et al. 1998) or the β-estradiol inducible vector, pER8 (Zuo et al. 2000). Constructs were transformed into Arabidopsis thaliana (Landsberg erecta) by vacuum infiltration (Clough and Bent 1998). Wild-type (Col and Ler), cop1 mutants (cop1-4 and cop1-6 in Col background), and transgenic seedlings (in Ler background) expressing 35S-DN-COP1-6×Myc were germinated and grown in darkness for 4 d before transfer to R light (20 μmole m-2sec-1) for the designated time, so that all seedlings were of the same age when protein or RNA was extracted. For examination of phyA level in XVE-DN-COP1-6×Myc transgenic seedlings, dark-grown 4-day-old seedlings were exposed to R light (20 μmole m-2 sec-1) with or without 10 μM β-estradiol (Sigma) for 15 h and then with or without 50 μM MG132 (Calbiochem) for 15 h. Samples were extracted and analyzed by Western blotting using anti-phyA antibody or anti-Myc antibody (Santa Cruz Biotechnology).
Acknowledgments
We thank Peter Hare for thoughtful comments. This work was supported by NIH grant GM 44640.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1187804.
References
- Berkelman T.R. and Lagarias, J.C. 1986. Visualization of bilin-liked peptides and proteins in polyacrylamide gels. Anal. Biochem. 156: 194-201. [DOI] [PubMed] [Google Scholar]
- Cantón F.R. and Quail, P.H. 1999. Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol. 121: 1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal J.J., Luccioni, L.G., Oliverio, K.A., and Boccalandro, H.E. 2003. Light, phytochrome signalling and photomophogenesis in Arabidopsis. Photochem. Photobiol. Sci. 2: 1-13. [DOI] [PubMed] [Google Scholar]
- Clough S.J. and Bent, A.F. 1998. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735-743. [DOI] [PubMed] [Google Scholar]
- Clough R.C. and Vierstra, R.D. 1997. Phytochrome degradation. Plant Cell Environ. 20: 713-721. [Google Scholar]
- Deng X.W., Minami, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. 1992. COP1, an Arabidoipsis regulatory gene, encodes a protein with both a zinc-binding and a Gβ homologous domain. Cell 71: 791-801. [DOI] [PubMed] [Google Scholar]
- Hoecker U., Tepperman, J.M., and Quail, P.H. 1999. SPA1: A WD-repeat protein specific to phytochrome A signal transduction. Science 284: 496-499. [DOI] [PubMed] [Google Scholar]
- Jabben M., Shanklin, J., and Vierstra, R.D. 1989a. Red light-induced accumulation of ubiquitin-phytochrome conjugates in both monocots and dicots. Plant Physiol. 90: 380-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1989b. Ubiquitin-phytochrome conjugates: Pool dynamics during in vivo phytochrome degradation. J. Biol. Chem. 264: 4998-5005. [PubMed] [Google Scholar]
- Kircher S., Gil, P., Kozma-Bognar, L., Fejes, E., Speth, V., Husselstein-Muller, T., Bauer, D., Adam, E., Schäfer, E., and Nagy, F. 2002. Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits diurnal rhythm. Plant Cell 14: 1541-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B., Spielhofer, P., and Chua, N.H. 1998. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16: 393-401. [DOI] [PubMed] [Google Scholar]
- Matsushita T., Mochzuki, N., and Nagatani, A. 2003. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature 424: 571-574. [DOI] [PubMed] [Google Scholar]
- McNellis T., von Arnim, A.G., Araki, T., Komeda, Y., Misera, S., and Deng, X.W. 1994. Genetic and molecular analysis of an allelic series of cop1 mutants suggests functional roles for the multiple protein domains. Plant Cell 6: 487-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery B.L. and Lagarias, J.C. 2002. Phytochrome ancestry: Sensors of bilins and light. Trends Plant Sci. 7: 357-366. [DOI] [PubMed] [Google Scholar]
- Nagy F. and Schäfer, E. 2002. Phytochromes control photomorphogenesis by differentially regulated, interacting signalling pathways in higher plants. Annu. Rev. Plant Biol. 53: 329-355. [DOI] [PubMed] [Google Scholar]
- Neff M.M., Frankhauser, C., and Chory, J. 2000. Light: An indicator of time and place. Genes & Dev. 14: 257-271. [PubMed] [Google Scholar]
- Park C.M., Bhoo, S.H., and Song, P.S. 2000. Inter-domain crosstalk in the phytochrome molecules. Semin. Cell Dev. Biol. 11: 449-456. [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70: 503-533. [DOI] [PubMed] [Google Scholar]
- Quail P.H. 2002a. Photosensory perception and signaling in plant cells: New paradigms? Curr. Opin. Cell Biol. 14: 180-188. [DOI] [PubMed] [Google Scholar]
- ____. 2002b. Phytochrome photosensory signaling networks. Nat. Rev. Mol. Cell Biol. 3: 85-93. [DOI] [PubMed] [Google Scholar]
- Quail P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. 1995. Phytochromes: Photosensory perception and signal transduction. Science 268: 675-680. [DOI] [PubMed] [Google Scholar]
- Reed J.W., Nagatani, A., Elich, T.D., Fagan, M., and Chory, J. 1994. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol. 104: 1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y., Sullivan, J.A., Wang, H., Yang, J., Shen, Y., Rubio, V., Ma, L., Hoecker, U., and Deng, X.W. 2003. The COP1-SPA1 interaction defines a critical step in phytochrome A-mediated regulation of HY5 activity. Genes & Dev. 17: 2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H.S., Yang, J.Y., Ishikawa, M., Bolle, C., Ballesteros, M.L., and Chua, N.H. 2003. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423: 995-999. [DOI] [PubMed] [Google Scholar]
- Shalitin D., Yang, H., Mockler, T.C., Maymon, M., Guo, H., Whitelam, G.C., and Lin, C. 2002. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature 417: 763-767. [DOI] [PubMed] [Google Scholar]
- Sharrock R.A. and Clack, T. 2002. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 130: 442-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., and Furuya, M. 1996. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 93: 8129-8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey M.G. and von Arnim, A.G. 1999. A novel motif mediates the targeting of the Arabidopsis COP1 protein to subnuclear foci. J. Biol. Chem. 274: 27231-27236. [DOI] [PubMed] [Google Scholar]
- Stockhaus J., Nagatani, A., Halfter, U., Kay, S., Furuya, M., and Chua, N.H. 1992. Serine-to-alinine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes & Dev. 6: 2364-2372. [DOI] [PubMed] [Google Scholar]
- Suzuki G., Yanagawa, Y., Kwok, S.F., Matsui, M., and Deng, X.W. 2002. Arabidopsis COP10 is a ubiquitin-conjugating enzyme variant that acts together with COP1 and the COP9 signalosome in repressing photomorphogenesis. Genes & Dev. 16: 554-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokutomi S., Kataoka, M., Sakai, J., Nakasako, M., Tokunaka, F., Tasumi, M., and Furuya, M. 1988. Small-angle X-ray scattering studies on the macromolecular structure of the red-light-absorbing form of 121 kDa pea phytochrome and its 114 kDa chromopeptide. Biochim. Biophys. Acta 953: 297-305. [Google Scholar]
- von Arnim A.G. 1997. Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis. Plant Physiol. 114: 779-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim A.G. and Deng, X.W. 1994. Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79: 1035-1045. [DOI] [PubMed] [Google Scholar]
- Wang H., Ma, L.G., Li, J.M., Zhao, H.Y., and Deng, X.W. 2001. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154-158. [DOI] [PubMed] [Google Scholar]
- Wei N. and Deng, X.W. 1996. The role of the COP/DET/FUS genes in light control of Arabidopsis seedling development. Plant Physiol. 112: 871-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Frugis, G., Colgan, D., and Chua, N.H. 2000. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes & Dev. 14: 3024-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q., Guo, H.S., Dallman, G., Fang, S., Weissman, A.M., and Chua, N.H. 2002. SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167-170. [DOI] [PubMed] [Google Scholar]
- Yang H.Q., Tang, R.H., and Cashmore, A.R. 2001. The signaling mechanism of Arabidopsis CRY1 involves direct interaction with COP1. Plant Cell 13: 2573-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu, Q.W., and Chua, N.H. 2000. An estrogen receptor-based transactivator XVE mediates highly inducible gene expression in transgenic plants. Plant J. 24: 265-273. [DOI] [PubMed] [Google Scholar]