Abstract
The Drosophila external sensory organ forms in a lineage elaborating from a single precursor cell via a stereotypical series of asymmetric divisions. HAMLET transcription factor expression demarcates the lineage branch that generates two internal cell types, the external sensory neuron and thecogen. In HAMLET mutant organs, these internal cells are converted to external cells via an unprecedented cousin-cousin cell-fate respecification event. Conversely, ectopic HAMLET expression in the external cell branch leads to internal cell production. The fate-determining signals NOTCH and PAX2 act at multiple stages of lineage elaboration and HAMLET acts to modulate their activity in a branch-specific manner.
Keywords: Cell fate, respecification, cell lineage, Drosophila, external sensory organ, hamlet
Tissues that develop from progenitor cells, including the vertebrate hematopoietic system and the central and peripheral nervous systems, generate multiple cell types from a single precursor via iterative cell divisions. The Drosophila external sensory organ (ESO) also forms in this way. It consists of five different cell types descended from one ESO precursor cell (ESOP) via a stereotypical series of asymmetric divisions (Figs. 1A, 2A-B; Orgogozo et al. 2001; Roegiers et al. 2001a).
Figure 1.
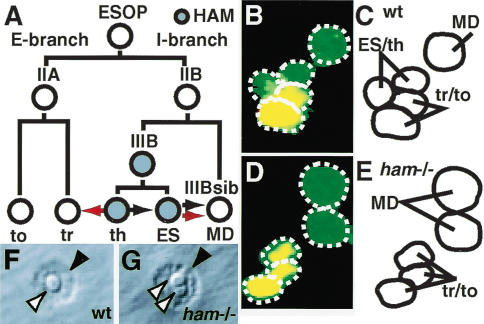
A thecogen-to-trichogen conversion in hamlet mutant embryos. (A) An illustration of ESO lineage elaboration; each cell is represented by a circle (filled is HAM-positive). The lines running from each cell indicate its daughters; the daughter with high N activity is on the left. The black arrows represent the potential sibling-sibling or uncle-nephew conversions available with the I-branch. The red arrows show the conversions that occur in the ham mutant. (B-E) The vp4a cluster at embryonic stage 13 labeled with fluorescent antibodies that detect CUT (green) and A1-2-29 (red). (B,C) A wild-type vp4a cluster contains five cells all expressing CUT; the two external cells also express A1-2-29. (D,E) A ham mutant vp4a cluster contains five cells all expressing CUT, three of which also express A1-2-29. (F,G) The ventral pore organ external cells on the cuticle of a second instar larvae. A wild-type organ (F) has one trichogen (white arrowhead) and one tormagen (black arrowhead); a ham mutant organ (G) has two trichogen and one tormagen. (ESOP) External sensory organ precursor; (to) tormagen; (tr) trichogen; (ES) external sensory neuron; (th) thecogen; (MD) multidendritic neuron; (I-branch) internal cell branch; (E-branch) external cell branch; (wt) wild type; (ham-/-) ham mutant.
Figure 2.
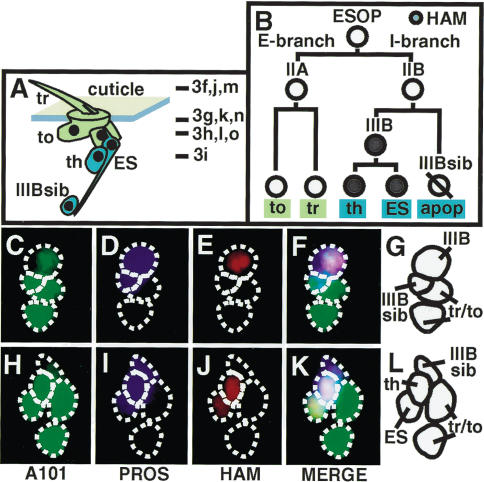
The expression pattern of HAMLET in the ESO lineage. (A) An illustration of the adult ESO with external cells in green and internal cells in blue; the lines indicate the planes of section through the ESO shown in Figure 3. (B) An illustration of the elaboration of the adult ESO lineage. (apop) Apoptotic cell. (C-L) The elaborating adult ESO labeled with fluorescent antibodies to detect A101 (C,H, green), PROS (D,I, blue), and HAM (E,J, red). F and K are overlays of C-E and H-J, respectively. G and L are illustrations of F and G, respectively.
ESOP cell division forms the IIA and IIB cells. The IIA gives rise to the external cells, the trichogen (hair), and the tormagen (the socket) that are visible on the surface of the cuticle (external cell [E]-branch; Figs. 1A, 2A-B). The IIB cell divides to give rise to neuronal and glial internal cell types that lie beneath the surface of the cuticle (internal cell [I]-branch; Figs. 1A, 2A-B). At each stage of elaboration, each cell can be clearly visualized and distinguished; hence, the ESO is an excellent model in which to examine the elaboration of multiple cell types from one precursor at the single-cell level in vivo.
Each division of the ESO lineage is asymmetric; one of the two cells formed inherits the NUMB protein, causing it to have a lower level of NOTCH (N) activity than its sibling. N-mediated signaling between the siblings then determines a difference in identity between them (Guo et al. 1996; Van De Bor and Giangrande 2001). This difference is expressed in terms of gene expression (for example, the IIB cell expresses the transcription factor PROSPERO [PROS], whereas the IIA does not) and in terms of cell behavior (for example, the IIB cell divides with a different plane of mitotic spindle orientation to the IIA).
The disruption of the function of genes that generate asymmetry between siblings (e.g., N) always leads to a sibling-sibling conversion. In many cases, the disruption of transcription factors required for cell differentiation also leads to sibling-sibling conversions; for example, loss of pros activity leads to IIB-to-IIA cell conversions at low frequency, and ectopic expression of PROS in the IIA cell converts this cell into a IIB (Manning and Doe 1999; Reddy and Rodrigues 1999b). A second type of conversion is a nephew-uncle conversion; for example, in the Drosophila embryo, the HAMLET (HAM) transcription factor is required for external sensory (ES) neuron fate, and in the absence of HAM the ES neuron is converted to the IIIBsib cell (Fig. 1A; Moore et al. 2002). This represents a conversion between internal cell types, with a shared ancestor (IIB cell) that is a direct parent of one of the two cell types.
To date the only type of cell-fate conversions described in the ESOP lineage are sibling-sibling or nephew-uncle conversions. This is unsurprising given the binary nature of each cell-fate decision. Here, we demonstrate that loss of HAM activity in the thecogen (and also the ES neuron in the adult ESO lineage) causes a fate switch not to that of an internal cell type, but to an external cell type. This switch represents a cousin-cousin conversion, which we show takes place by a respecification mechanism. In ham mutants, the ES neuron and thecogen first express markers associated with the I-branch of the lineage, but they fail to fully differentiate and later convert into external cell types without first reentering the cell cycle. We demonstrate that one role of HAM in the development of the ESO is to modulate the activity of N and PAX2 signals used at multiple points in the lineage in a branch-specific manner.
Results and Discussion
The thecogen cell is lost and an extra trichogen is present in the external sensory organ of hamlet mutant embryos
In the embryonic ESO lineage, the transcription factor HAM is expressed in the ES neuron but not in the multidendritic (MD) neuron (Fig. 1A). Loss of HAM function converts the ES to an (uncle) MD neuron fate, and indeed HAM acts as a binary genetic switch between these two internal cell fates (Fig. 1B). HAM is also expressed in the thecogen cell, which is the sibling of the ES neuron (Fig. 1A). If the thecogen cell undergoes a cell-fate conversion in the absence of HAM, however, it could not have switched fates to a sibling or uncle cell type, as both of these are neurons, and in ham mutant ES organs there is no increase in neuron number (Fig. 1A; Moore et al. 2002).
To investigate the fate of the “thecogen” in a ham mutant, we stained wild-type and ham mutant embryos with antibodies to detect expression of the transcription factors CUT, PROS, and PAX2, and the placW enhancer trap A1-2-29 (Table 1). CUT marks all cells descended from the ESOP (Blochlinger et al. 1990). On terminal differentiation of the ESO, PAX2 marks the thecogen and trichogen (Kavaler et al. 1999), PROS marks only the thecogen, and A1-2-29 drives β-galactosidase expression in the trichogen and tormagen (Kavaler et al. 1999). In the dorsal external sensory (des) and ventral pore (vp) organs of ham mutant embryos, the total number of cells remains unchanged (Fig. 1B-C), and the thecogen cell (expressing CUT, PAX2, and PROS) is replaced by a cell expressing CUT, PAX2, and A1-2-29; this combination of markers normally defines trichogen fate (Table 1; Fig. 1C-F). Given that this staining pattern implies that this cell is a trichogen, we examined the ESO structures on the cuticle surface of ham mutant second instar larvae. Sixty-five percent of organs showed a wild-type one-trichogen and one-tormagen phenotype, and 34% a two-trichogen and one-tormagen phenotype (Table 2; Fig. 1G-H).
Table 1.
The protein expression profile of external sensory organ lineage cells
| CUT | PROS | HAM | PAX2 | A1-2-29 | A101 | ELAV | Su(H) | |
|---|---|---|---|---|---|---|---|---|
| ESOP | ++ | − | − | + | − | ++ | − | − |
| (E) IIA | ++ | − | − | + | − | ++ | − | − |
| (E) tr | ++ | − | − | ++ | ++ | ++ | − | − |
| (E) to | ++ | − | − | − | ++ | ++ | − | ++ |
| (I) IIB | ++ | ++ | − | + | − | ++ | − | − |
| (I) IIIB | ++ | ++ | ++ | + | − | ++ | − | − |
| (I) IIIB sib | + | +/− | − | − | − | ++ | ++ or − | − |
| (I) th | ++ | ++ | +/− | ++ | − | ++ | − | − |
| (I) ES | + | +/− | +/− | − | − | ++ | ++ | − |
Each row represents a different ESO lineage cell; abbreviations are as in Figure 1. Each column shows the expression level of a specific protein: (++) high; (+) low, (+/−) transient; (−) none. The embryonic IIIBsib (MD neuron) expresses PROS transiently and ELAV permanently; the adult IIIBsib expresses PROS until apoptosis but never expresses ELAV.
Table 2.
The external cell number in hamlet mutants
| Phenotype: | 1 tr/1 to | 2 tr/1 to | 3 tr/1 to | 2 tr/2 to | 1 tr/2 to |
|---|---|---|---|---|---|
| Larvae | 89/137 | 46/137 | 1/137 | 0/137 | 1/137 |
| % | 65 | 34 | <1 | 0 | <1 |
| Adult | 84/304 | 21/304 | 0/304 | 70/304 | 129/304 |
| % | 28 | 7 | 0 | 23 | 42 |
The replacement of the thecogen by a trichogen in the ham mutant could have occurred by two different mechanisms. In the first, an alteration of the ESO lineage division pattern is responsible for the replacement of an internal cell type by an external one. In the second, the thecogen derived from the IIIB cell is converted not to another cell type derived from the I-branch of the lineage, but to a cell type that is representative of the E-branch. These two possibilities have very different implications for understanding how the ESO develops. The first is consistent with the idea that all cell-fate decisions within this lineage are essentially binary and the fates of the daughters are restricted by the identity of the precursors giving rise to them. The second, on the other hand, implies there is no such restriction.
In the Drosophila adult external sensory organ lineage, HAMLET is expressed in the IIIB and its daughters in the external sensory neuron and thecogen
In order to distinguish between these possibilities, we turned our analysis to the adult microcheate ESO lineage, in which we can follow the elaboration of each ESO derived from a single ESOP cell by live imaging (Roegiers et al. 2001a). The adult ESO lineage is considered analogous to that of the embryo, except that the IIIBsib in the embryo becomes an MD neuron, whereas in the adult it undergoes apoptosis (Table 1; Figs. 1A, 2A-B; Gho et al. 1999; Reddy and Rodrigues 1999a; Orgogozo et al. 2001).
The HAM expression pattern in the adult ESO lineage is analogous to that seen in the embryo. We dissected pupae at a stage in which the ESO lineage was elaborating (Materials and Methods) and stained them with antibodies to detect HAM, PROS, and the A101 enhancer trap. A101 is expressed in all cells of the ESO lineage (Kavaler et al. 1999; Table 1) and PROS at least transiently in all cells of the I-branch (Manning and Doe 1999; Orgogozo et al. 2001). HAM is first expressed in the IIIB cell (Fig. 2C-G) and then inherited by its daughters, the ES neuron and thecogen (Fig. 2H-L).
Duplication of both trichogen and tormagen cells occurs in hamlet mutant adult external sensory organ lineages
Next we investigated whether the supernumerary external cell seen in embryonic ham mutant ESOs was also present in the adult. We examined the external phenotype of ham mutant ESO clusters in nota mosaic for wild-type and ham mutant tissue (Materials and Methods). ham mutant ESOs could be identified by the loss of YELLOW (Y) in the thecogen and tormagen cells (Fig. 3A). Seven percent of y- sensory organs had a two-trichogen, one-tormagen phenotype similar to the ham mutant phenotype in the embryonic ESO. In addition, 42% of ham mutant ESOs had a one-trichogen, two-tormagen phenotype and 23% had a two-trichogen, two-tormagen phenotype (Table 2; Fig. 3A).
Figure 3.
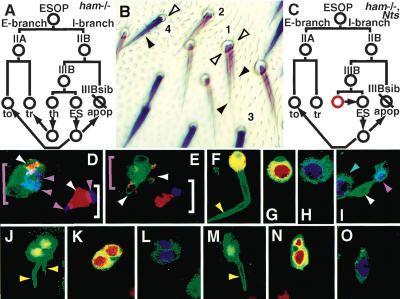
A conversion of internal-to-external cell types in adult hamlet mutant external sensory organs by cell-fate respecification. (A) An illustration of the elaboration of a ham mutant adult ESO lineage. Arrows represent the stages of cell-fate conversion. The daughters of the IIIB cell first begin to differentiate normally, then undergo respecification; the thecogen becomes a trichogen and the ES neuron either undergoes apoptosis (IIIBsib fate) or converts to a tormagen. (B) Adult ESOs. ham mutant trichogen and tormagen cells are y- and hence light brown in color. ESOs 1 and 2 are ham-/- and have two trichogen (black arrowheads) and two tormagen (open arrowheads). Wild-type trichogen and tormagen are y+ and hence darker: ESO 3 (ham+/-) and 4 (ham+/+ in ck-/- twinspot) have one trichogen and one tormagen. (C) An illustration of the effect of removing N activity in both IIIB cell daughters in a ham mutant. The daughter that normally has high N (red circle) now becomes the same as the low-N daughter (normally the ES neuron) and no supernumerary trichogen forms. (D,E) ESOs stained with fluorescent antibodies that detect PROS (blue) and ELAV (red). Green mCD8GFP fluorescence highlights the ham mutant ESO (pink bracket), whereas the wild-type ESO is mCD8GFP-negative (white bracket). (D) Twenty-two to twenty-four hours APF. Soon after IIIB cell division, both the wild-type and the ham mutant ESO contain one ELAV-positive neuron (white arrowhead), and two PROS-positive cells (the IIIBsib and “thecogen”, pink arrowheads). (E) Twenty-eight to thirty hours APF. The wild-type cluster contains one ES neuron (ELAV-positive) and one thecogen (PROS-positive). The ham mutant cluster contains apoptotic ES neuron fragments expressing ELAV (white arrowheads) and no PROS-positive “thecogen” cell. (F-O) Thirty-six to forty hours APF. Successive apical-to-basal sections (see Fig. 2A) through adult ESOs stained with fluorescent antibodies that detect mCD8GFP (green), PAX2 (blue), and Su(H) (red). (F-I) A wild-type ESO consists of one trichogen cell (F, yellow arrowhead); the nucleus of this cell contains PAX2 (H). (G) The trichogen projects through an Su(H)-positive tormagen cell. (I) Basal to the external cells are the ES neuron (white arrowhead) and PAX2-positive thecogen (pink arrowhead). The PAX2-positive trichogen of a neighboring ESO is also in this panel (blue arrowhead). (J-L) A ham mutant ESO has two PAX2-positive (L) trichogen cells (J, yellow arrowheads) projecting through two Su(H)-positive tormagen cells (K) and no internal cells. (M-O) When N activity is selectively reduced in the IIIB cell daughters in a ham mutant ESO, the supernumerary trichogen is lost (M, yellow arrowhead; O, PAX2 expression) but the Su(H)-positive supernumerary tormagen remain (N).
The IIIB cell daughters are converted to external cell-branch fates via cell-fate respecification in both embryonic and adult hamlet mutant external sensory organs
To distinguish whether these supernumerary external cells have arisen from either an altered division pattern in the ESO lineage or conversion of terminal cell fates, we carried out live imaging of ESO lineage elaboration in ham mutant clones between 18 and 38h after pupal formation (APF). ham mutant mosaic analysis via a repressible cell marker (MARCM; Lee and Luo 1999) clones were marked by Partner of Numb-GFP (PON-GFP) fusion protein expression (Materials and Methods), which is asymmetrically localized and inherited by only one daughter at each cell division of the ESO lineage (Lu et al. 1999; Roegiers et al. 2001a). In ESO lineages in which the IIB cell has been converted to the IIA, for example, by ectopic expression of activated N in the IIB, the timing and orientation of the division of the cells of the I-branch are altered to resemble those of the E-branch (Le Borgne et al. 2002). In contrast, the elaboration of ham mutant ESO lineages was indistinguishable from that in wild-type in the timing, orientation, and number of divisions in both the E-branch and the I-branch (n = 50; data not shown). Therefore, the supernumerary external cells in ham mutant clones are not due to a conversion of I-branch precursors into E-branch precursors or extra divisions within the E-branch itself; they are due to the conversion of the IIIB cell daughters to external cell fates.
Next we investigated the expression of I-branch specific markers in ham mutant ESO lineages during elaboration. We made ham mutant MARCM clones positively marked with mCD8GFP fusion protein expression in all ESOP-derived cells (Materials and Methods). We stained ham mutant clones at all stages of ESO lineage elaboration with antibodies that detect the following markers: PROS, PAX2, ELAV (EMBRYONIC LETHAL ABNORMAL VISION), which labels all differentiated neurons (Robinow and White 1991; Table 1), and SUPPRESSOR OF HAIRLESS [Su(H)], which labels differentiated tormagen (Gho et al. 1996; Table 1). In both wild-type and ham mutant lineages, the IIB, IIIB, and IIIBsib cells expressed PROS, thus confirming the live imaging findings that showed no differences between the elaborating I-branch in wild-type and ham mutant lineages (data not shown).
The IIIB progeny in ham mutant clusters, however, shifted their patterns of differentiation over time. Shortly after division of the IIIB cell (22-24 h APF), one daughter in ham mutant clones continued to express PROS, similar to a wild-type thecogen, and the other began to express ELAV, a marker of neuron fate (Fig. 3D). By 28-30 h APF, the ham mutant ESO cell clearly differed from the wild-type control: expression of the I-branch-specific maker PROS was lost (Fig. 3E). Moreover, ELAV-positive neurons were no longer present, but several ham mutant ESO showed small ELAV-positive apoptotic cell fragments (Roegiers et al. 2001b), indicative of the ES neuron undergoing cell death. Live confocal imaging of ham mutant clones showed the frequency of this event to be 15% (n = 33). By 36-40 h, wild-type ESO clearly contained one trichogen and one thecogen (both PAX2-positive), one tormagen [Su(H)-positive], and one ES neuron (ELAV-positive; Table 1; Fig. 3F-I). In contrast, ham mutant ESOs contained no ES neurons or thecogen, two trichogen (PAX2-positive), and either one or two tormagen [Su(H)-positive; Fig. 4J-L]. Whereas mCD8GFP activity and PAX2 staining revealed the two-trichogen phenotype with 100% frequency (n > 100), the appearance of two trichogens on the adult cuticle was less frequent (Table 2), indicating that one supernumerary trichogen must have failed to grow out onto the cuticle surface. Costaining of ham mutant embryos with antibodies to detect CUT, PROS, and ELAV revealed that PROS is also transiently expressed in the thecogen cell before it undergoes conversion to a trichogen (data not shown). Therefore, in both the adult and embryonic ham mutant ESO lineage, the daughters of the IIIB cell first express markers specific to internal cells, but expression of such markers ceases as these cells undergo respecification to an external cell fate.
Figure 4.
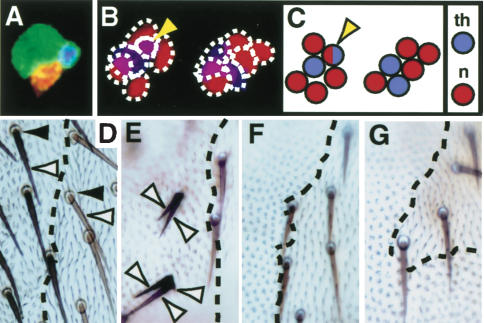
Ectopic expression of HAMLET converts external branch cells to internal fates and represses PAX2 hair-shaft-promoting activity. (A-C) Adult ESOs labeled with fluorescent antibodies to detect ELAV (red) and PROS (blue). (A) gal4109-68 × UAS-mCD8GFP. All ESO cells express mCD8GFP; the ESO cluster contains one ELAV-positive ES neuron, one PROS-positive thecogen, and two external cells. (B) gal4109-68 × UAS-mCD8GFP, UAS-ham. All cells express mCD8GFP and HAM (not shown). Six cells are present; four to five are neurons (ELAV-positive), and one to two are thecogen (PROS-positive). Occasionally a cell expresses both ELAV and PROS (yellow arrowhead). (C) A cartoon representing panel B; blue circles represent thecogen and red circles represent neurons. (D-G) Clones on the cuticle surface showing the effect of PAX2 and/or HAM ectopic expression in all cells of the ESO lineage. The region in which ectopic expression is occurring is y+,ck-; the clone boundary is marked by a broken line. (D) Control. Wild-type organs have one trichogen (white arrowhead) and one tormagen (black arrowhead). (E) neu-gal4 × UAS-pax2. Organs have multiple shaft structures (white arrowhead) and no tormagen. (F) neu-gal4 × UAS-ham. Both external cells are lost. (G) neu-gal4 × (UAS-pax2 + UAS-ham). Both external cells are lost.
In the ham mutant embryo, the ES neuron is transformed into a IIIBsib/MD neuron fate and the thecogen into a trichogen. On the basis of these embryo data, we propose that in the ham mutant adult, the thecogen is also converted into a trichogen and the ES neuron into either a IIIBsib (apoptotic cell) or a tormagen. A possible reason that the conversion properties of the ES neuron in the embryo are different from those of the adult could be that, as neuron-specific markers are already expressed in the ES neuron before it undergoes fate conversion, in the embryo, where a neuron-to-neuron respecification event can occur, it is favored over a neuron-to-nonneuron one.
The IIIB cell daughter with high NOTCH activity is transformed into the supernumary trichogen in a hamlet mutant external sensory organ
If the cell-fate conversions we propose take place, then it would mean that the thecogen (high N) becomes a trichogen (low N) and the ES neuron (low N) can become a tormagen (high N). To confirm this, we investigated whether the high-N or low-N IIIB daughter cell becomes the supernumerary trichogen in a ham mutant ESO. We made ham MARCM clones in an Nts (temperature sensitive) background (Materials and Methods) and inactivated N in these clones at the stage where N-mediated signaling is generating asymmetry between the IIIB daughter cells. We stained the resulting nota with antibodies to detect mCD8, Su(H), and PAX2 and examined the fate of the IIIB daughter cells. In the ham1, Nts ESOs 28/28 had a one-trichogen/multiple-tormagen phenotype (Fig. 3M-O), whereas in the ham1, N+ control 37/37 ESOs had a two-trichogen/multiple-tormagen phenotype.
This experiment clearly demonstrates that it is the high-N IIIB cell daughter that becomes the supernumerary trichogen, as reducing N activity resulted in a reduction in the number of trichogen in the ham mutant organ (to one; Fig. 3C). The conclusion to be drawn from this experiment is, therefore, that whereas N is acting to determine the difference between daughters of an asymmetric division, of either IIA or IIIB, a second signal is acting that makes the trichogen similar to the thecogen and the tormagen similar to the ES neuron.
Expression of HAMLET in cells of the external cell branch converts them to internal-cell-branch fates
In this study we show that in both the embryo and adult, ESO HAM is expressed solely in the IIIB cell and its daughters, the ES neuron and thecogen. We show that loss of HAM causes the conversion of the internal-cell-branch thecogen into an external-cell-branch trichogen by cell-fate respecification. In addition, loss of HAM in the ES neuron leads either to its conversion to an internal-cell-branch IIIBsib cell or an external-cell-branch tormagen. Therefore, HAM appears to act to determine the fate of the IIIB daughters with respect to all other terminally differentiated cell types in the ESO lineage. In other words, it acts as an intrinsic transcription factor determinant of IIIB cell-derived identity.
We wished to test whether HAM expression alone determines the difference between IIIB-derived and non-IIIB-derived fate. Ectopic expression of HAM in the embryonic IIB and IIIBsib cell (MD neuron) converts the IIIBsib into an ES neuron (Moore et al. 2002). We extended this analysis to the E-branch by using gal4109-68 (Abdelilah-Seyfried et al. 2000) to drive UAS-ham and UAS-mCD8GFP in all cells of the adult lineage. HAM expression in all ESO lineage cells led to the loss of both the E-branch-derived trichogen and tormagen from the surface of the cuticle (data not shown). Antibody staining of the ESOs showed that I-branch-specific cell types had replaced these external cells. When HAM was ectopically expressed in the entire ESO lineage, we saw five or six cells, all of which were expressing PROS or ELAV or (rarely) both PROS and ELAV (Fig. 4B-C). On the other hand, in a wild-type cluster, there are four cells including only one thecogen (PROS-positive) and one ES neuron (ELAV-positive; Fig. 4A). These ectopic expression experiments confirm that HAM determines IIIB-derived versus non-IIIB-derived fate.
HAMLET can repress the hair-shaft-promoting activity of PAX2
These ectopic expression experiments confirm that HAM determines IIIB-derived versus non-IIIB-derived fate. However, why in ham mutant ESOs are the transformations that occur thecogen (high N) to tricogen (low N) and ES neuron (low N) to tormagen (high N)? PAX2 expression in the ESO highlights a connection between these pairs of cell types; the thecogen and trichogen both express PAX2, whereas the tormagen and ES neuron do not. Moreover, PAX2 itself is required for hair-shaft differentiation in the trichogen, and ectopic PAX2 expression in the tormagen leads to ectopic hair-shaft development (Kavaler et al. 1999). In the thecogen, where PAX2 and HAM are coexpressed, PAX2 expression does not lead to hair-shaft development; however, in the absence of HAM, this cell now takes on a trichogen fate including the development of a hair shaft. We wondered therefore if one role of HAM in the thecgoen cell may be to suppress the ability of PAX2 to promote hair-shaft formation.
To investigate whether HAM can repress the hair-shaft-promoting activity of PAX2, we expressed HAM or PAX2 or coexpressed HAM and PAX2 at high levels in all cells of the ESO lineage. We used neu-gal4 to drive expression of UAS-ham and/or UAS-PAX2; however, this causes embryonic lethality. To get around this problem, we drove ectopic gene expression only in notum clones (Materials and Methods). Ectopic expression of PAX2 led to organs with multiple hair shafts, some organs with misshapen external cells, and in some cases loss of external cells. On the other hand, ectopic HAM or HAM and PAX2 caused the loss of external cells in almost 100% of ESO clones and never the formation of multiple hair shafts. These experiments demonstrate that HAM has the ability to modulate a PAX2-driven differentiation program, in this case, hair-shaft growth. Therefore, loss of HAM from the “thecogen” could lead to the depression of a program at least in part controlled by PAX2, which drives this cell to a trichogen fate.
Conclusion
The PAX2 cell-fate-determining signal is used at multiple points during the development of the ESO lineage, as is N. The presence or absence of HAM provides a branch-specific background state against which differentiating cells of the organ can interpret these signals. It is likely that a similar modulation of iterated signals by branch-specific factors occurs in vertebrate systems. For example, N signaling occurs at multiple points during the development of the hematopoietic lineage (Ohishi et al. 2003) and could be modulated by the presence of branch-specific transcription factors. We suggest that the analysis of HAM function in ESO lineage elaboration presented in this study provides useful insight into how cell fate is determined in many invertebrate and vertebrate lineages in which a single stem/precursor cell gives rise to multiple cell types via iterative cell divisions.
Materials and methods
Fly stocks and genetics
ham1 homozygous mutant embryos were made by crossing ham1, A1-2-29/CyO, Ftz-lacZ stocks. In the adult, mosaic clones of ham1 were made by using yw, Ubx-flp; ham1, E7-2-36, FRT40A/CyO virgins. These were crossed to yw; y+, ck, FRT40A/CyO to yield marked external notum clones, or yw; tubP-gal80, FRT40A; neu-gal4, Kgv, UAS-PON-GFP/Tm6, y+ or yw; tubP-gal80, FRT40A; neu-gal4, Kgv, UAS-mCD8GFP/Tm6, y+ to generate MARCM clones positively marked by 8GFP in the SOP lineage (Justice et al. 2003).
To inactivate N in a ham mutant background, Nts1 Ubx-flp; ham1, E7-2-36, FRT40A/CyO virgins were crossed to yw; tubP-gal80, FRT40A; neu-gal4, Kgv, UAS-mCD8GFP/Tm6, y+. Flies were grown at 25°C, then at 22 h APF the pupal case was removed and ham mutant clones at the four-cell stage, that is, hair, socket, IIIB, and IIIB sibling cell, were identified by live fluorescence microscopy for mCD8GFP activity. Pupae with ESO lineages at this stage were shifted to 29°C for 2 h and then back to a 25°C-temperature were allowed to develop overnight and then fixed for immunohistochemistry.
To get nonlethal neu-gal4 driven gene expression, we carried out the following self-crosses: for a control yw, Ubx-flp; y+, ck, tubP-gal80, FRT40A/FRT40A; neu-gal4, Kgv, UAS-mCD8-GFP/+; for PAX2 the “+” third chromosome was replaced by UAS-pax2; for HAM it was replaced by UAS-ham; and for PAX2 and HAM together by UAS-pax2, UAS-ham.
Live imaging and immunohistochemistry
Live imaging, immunohistochemistry and analysis of adult nota was carried out by standard protocols as described by Justice et al. (2003). The primary antibodies used in this study were guinea pig anti-HAM (1/100; Moore et al. 2002), rabbit anti-β-GAL (1/5000, ICN Biomedicals), rabbit anti-PROS (1/1000), rat anti-CUT F2 (1/2000), rabbit anti-PAX2 (1/5 gift of E. Frei, University of Zurich, Switzerland), rat anti-mCD8 (1/100, Caltag), rat anti-Su(H) (1/1500 gift of F. Schweisguth, Ecole Normale Superieur, Paris, France), and rat anti-ELAV 7E8A10 (1/200), mouse anti-CUT (1/100) and mouse anti-β-GAL (1/200), all from Developmental Studies Hybridoma Bank (DSHB), University of Iowa.
Acknowledgments
We thank D. Anderson, N. Justice, S. Albin, and members of our lab for useful discussions, and F. Schweisguth, E. Frei, J. Posakony, and J. Knoblich for kindly providing reagents. This work was supported by NIH RO1 NS40929. A Wellcome Trust International Prize Traveling Research Fellowship supported A.W.M. F.R. is an associate and Y.N.J and L.Y.J. are investigators of the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1170904.
References
- Abdelilah-Seyfried S., Chan, Y.M., Zeng, C., Justice, N.J., Younger-Shepherd, S., Sharp, L.E., Barbel, S., Meadows, S.A., Jan, L.Y., and Jan, Y.N. 2000. A gain-of-function screen for genes that affect the development of the Drosophila adult external sensory organ. Genetics 155: 733-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blochlinger K., Bodmer, R., Jan, L.Y., and Jan, Y.N. 1990. Patterns of expression of cut, a protein required for external sensory organ development in wild-type and cut mutant Drosophila embryos. Genes & Dev. 4: 1322-1331. [DOI] [PubMed] [Google Scholar]
- Gho M., Lecourtois, M., Geraud, G., Posakony, J.W., and Schweisguth, F. 1996. Subcellular localization of Suppressor of Hairless in Drosophila sense organ cells during Notch signalling. Development 122: 1673-1682. [DOI] [PubMed] [Google Scholar]
- Gho M., Bellaiche, Y., and Schweisguth, F. 1999. Revisiting the Drosophila microchaete lineage: A novel intrinsically asymmetric cell division generates a glial cell. Development 126: 3573-3584. [DOI] [PubMed] [Google Scholar]
- Guo M., Jan, L.Y., and Jan, Y.N. 1996. Control of daughter cell fates during asymmetric division: Interaction of Numb and Notch. Neuron 17: 27-41. [DOI] [PubMed] [Google Scholar]
- Justice N., Roegiers, F., Jan, L.Y., and Jan, Y.N. 2003. Lethal giant larvae acts together with numb in notch inhibition and cell fate specification in the Drosophila adult sensory organ precursor lineage. Curr. Biol. 13: 778-783. [DOI] [PubMed] [Google Scholar]
- Kavaler J., Fu, W., Duan, H., Noll, M., and Posakony, J.W. 1999. An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development 126: 2261-2272. [DOI] [PubMed] [Google Scholar]
- Le Borgne R., Bellaiche, Y., and Schweisguth, F. 2002. Drosophila E-cadherin regulates the orientation of asymmetric cell division in the sensory organ lineage. Curr. Biol. 12: 95-104. [DOI] [PubMed] [Google Scholar]
- Lee T. and Luo, L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451-461. [DOI] [PubMed] [Google Scholar]
- Lu B., Ackerman, L., Jan, L.Y., and Jan, Y.N. 1999. Modes of protein movement that lead to the asymmetric localization of partner of Numb during Drosophila neuroblast division. Mol. Cell 4: 883-891. [DOI] [PubMed] [Google Scholar]
- Manning L. and Doe, C.Q. 1999. Prospero distinguishes sibling cell fate without asymmetric localization in the Drosophila adult external sense organ lineage. Development 126: 2063-2071. [DOI] [PubMed] [Google Scholar]
- Moore A.W., Jan, L.Y., and Jan, Y.N. 2002. hamlet, a binary genetic switch between single- and multiple- dendrite neuron morphology. Science 297: 1355-1358. [DOI] [PubMed] [Google Scholar]
- Ohishi K., Katayama, N., Shiku, H., Varnum-Finney, B., and Bernstein, I.D. 2003. Notch signalling in hematopoiesis. Semin. Cell Dev. Biol. 14: 143-150. [DOI] [PubMed] [Google Scholar]
- Orgogozo V., Schweisguth, F., and Bellaiche, Y. 2001. Lineage, cell polarity and inscuteable function in the peripheral nervous system of the Drosophila embryo. Development 128: 631-643. [DOI] [PubMed] [Google Scholar]
- Reddy G.V. and Rodrigues, V. 1999a. A glial cell arises from an additional division within the lineage during development of the microchaete on the Drosophila notum. Development 126: 4617-4622. [DOI] [PubMed] [Google Scholar]
- ____. 1999b. Sibling cell fate in the Drosophila adult external sense organ lineage is specified by prospero function, which is regulated by Numb and Notch. Development 126: 2083-2092. [DOI] [PubMed] [Google Scholar]
- Robinow S. and White, K. 1991. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22: 443-461. [DOI] [PubMed] [Google Scholar]
- Roegiers F., Younger-Shepherd, S., Jan, L.Y., and Jan, Y.N. 2001a. Two types of asymmetric divisions in the Drosophila sensory organ precursor cell lineage. Nat. Cell Biol. 3: 58-67. [DOI] [PubMed] [Google Scholar]
- ____. 2001b. Bazooka is required for localization of determinants and controlling proliferation in the sensory organ precursor cell lineage in Drosophila. Proc. Natl. Acad. Sci. 98: 14469-14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Bor V. and Giangrande, A. 2001. Notch signaling represses the glial fate in fly PNS. Development 128: 1381-1390. [DOI] [PubMed] [Google Scholar]