Abstract
Identification of common mechanisms underlying organ development and primary tumor formation should yield new insights into tumor biology and facilitate the generation of relevant cancer models. We have developed a novel method to project the gene expression profiles of medulloblastomas (MBs)—human cerebellar tumors—onto a mouse cerebellar development sequence: postnatal days 1-60 (P1-P60). Genomically, human medulloblastomas were closest to mouse P1-P10 cerebella, and normal human cerebella were closest to mouse P30-P60 cerebella. Furthermore, metastatic MBs were highly associated with mouse P5 cerebella, suggesting that a clinically distinct subset of tumors is identifiable by molecular similarity to a precise developmental stage. Genewise, down- and up-regulated MB genes segregate to late and early stages of development, respectively. Comparable results for human lung cancer vis-a-vis the developing mouse lung suggest the generalizability of this multiscalar developmental perspective on tumor biology. Our findings indicate both a recapitulation of tissue-specific developmental programs in diverse solid tumors and the utility of tumor characterization on the developmental time axis for identifying novel aspects of clinical and biological behavior.
Keywords: cerebellar development, medulloblastoma, comparative genomics, multiscale models, metastasis, principle component analysis
In the 19th century, Lobstein and Cohnheim were among the first to theorize similarities between human embryogenesis and the biology of cancer cells (Rather 1978). The brain tumor classification system of Bailey and Cushing (1926), from which modern taxonomies derive, emphasizes the histologic resemblance to cells of the developing central nervous system (CNS; Bailey and Cushing 1926). Nevertheless, the putative relationship between underlying mechanisms in normal development and tumorigenesis remains controversial for most types of cancer, particularly the solid tumors such as medulloblastomas (MBs) and carcinomas.
Here, we focused on the relationship between genes regulated during oncogenesis in the human cerebellar tumor, MB, and the developing wild-type mouse cerebellum during postnatal days 1-60 (P1-P60). The cerebellum is the brain structure largely responsible for coordinating motor activities. Granule neurons, the most abundant cell type in the cerebellum during development, are derived from precursors of the embryonic hindbrain (Hallonet et al. 1990). In mice, the major phase of granule cell proliferation commences at birth and peaks by P8-P10 (Altman and Bayer 1987). Differentiation is complete by P60 in mice, and at ∼18 mo of age in humans. Granule neuron progenitors are thought to be the predominant dysregulated cell type from which the majority of MB cases arise (Kadin et al. 1970; Reddy and Packer 1999). MBs are the most common pediatric CNS malignancy and comprise two primary histological subtypes: desmoplastic medulloblastoma (dMB) and classic medulloblastoma (cMB). dMBs are distinguished by the presence of nodules with localized neuronal differentiation within an otherwise densely cellular sea of undifferentiated malignant cells (Kleihues and Cavenee 2000). cMBs are histologically characterized by the abundance of undifferentiated “small blue” malignant cells.
Gene expression profiling studies of human MBs have been performed (MacDonald et al. 2001; Pomeroy et al. 2002; Chopra et al. 2003; Hernan et al. 2003; Packer 2003). Pomeroy et al. (2002) found genes distinguishing human dMB from cMB, suggesting that human MBs derive from cerebellar granule cells through the activation of the sonic hedgehog (SHH) pathway. The latter studies focused on metastatic human MBs and the roles of metastasis-related genes such as PDGFRα, PDGFRβ, and ERBB2 (Chopra et al. 2003; Hernan et al. 2003; Packer 2003). Lee et al. (2003) have analyzed the expression profile of different mouse models of MB with a p53-/- mutant background. To date, a comprehensive multifactorial and multiscalar comparison of gene regulation between animal models of development and human tumorigenesis has not been performed, in part because methods for cross-species expression analyses have not been established. Existing cross-species comparative genomic studies have primarily focused on the use of DNA or protein sequence-based features to identify molecular regulatory elements, establish phylogenetic relationships, and annotate genes/proteins (Rubin et al. 2000; Hoopengardner et al. 2003; Mineta et al. 2003; Modrek and Lee 2003).
Here, we investigated human MBs against the backdrop of mouse cerebellar development on multiple scales. At a gene-by-gene or microscopic level, we find a significant and cell-type-specific segregation of down- and up-regulated genes in human MB to the late and early stages of mouse cerebellar development, respectively. At the genomic or macroscopic level, we applied a novel approach of projecting multivariate molecular features of human MBs onto the homologous genomic space of mouse cerebellar development to find a close association of human MBs with mouse cerebella stages P1-P10; and of normal human cerebella with mouse cerebella P30-P60. With respect to the mouse development trajectory, dMBs were molecularly more homogeneous than cMBs. Moreover, metastatic human MBs were, on the macroscopic scale, closest to mouse cerebella stage P5, suggesting that a clinically distinct subset of tumors is identifiable by their similarity to a precise mouse cerebellar developmental stage. Comparable results were obtained using this approach in a non-CNS environment: human squamous cell carcinoma (SCL) and normal lung on a developing mouse lung background. From a technical viewpoint, this projection method enables the multivariate analysis of human tissue against a non-human system background across different measurement platforms. Our results indicate that global expression characteristics of individual tumors with respect to a cognate developmental timeline provide a novel criterion for general segregation of tumors into meaningful biological and clinical subgroups. Moreover, these findings provide in silico evidence for the conserved, organ-specific mechanisms underlying organ development and tumorigenesis.
Results
Segregation of gene expression during mouse postnatal cerebellar development: Cerebellar Early and Late Mouse Partitions
Our development model is whole wild-type mouse cerebellum total RNA profiled at 10 postnatal days—P1, P3, P5, P7, P10, P15, P21, P30, P50, and P60—using Affymetrix Mu11K arrays, as previously described (Zhao et al. 2002). Of the 13,179 genes/ESTs present, 2552 homologous genes (Zhang et al. 2000) exist between the mouse Mu11K and human (Affymetrix HuFL) array platforms that were previously used to assay clinical samples of human MB (Pomeroy et al. 2002). To identify the predominant mouse cerebellar development profile clusters among these 2552 genes, we performed a principal component analysis (PCA) of the temporal axis of these mouse genes.
PCA is a technique commonly used to simplify large datasets. It is a transformation that reduces the dimensionality (i.e., the number of variables) in the dataset while retaining those characteristics that contribute most to its internal variance structure. PCA is particularly useful in the domain of microarray expression data, in which a lot of intracorrelations exist between a large number of variables. For example, in this experiment, expression measurements at 10 time points/variables per gene is transformed into an equivalent set of uncorrelated variables called principal components (PCs; Duda et al. 2001; Misra et al. 2002). PCs are ranked in decreasing order of how they account for the total variability in the data. Therefore, instead of using all 10 time points to characterize each gene, we may use a smaller number of variables, namely, the first few PCs, while retaining most of the intrinsic information in the original data.
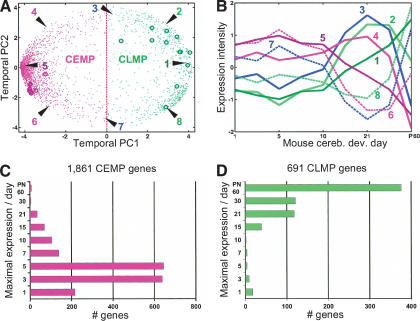
Figure 1A shows the first two temporal PC representations (capturing 70.07% of total temporal variation) for each gene—each dot signifies a gene listed in Supplementary Table 1. A general pattern revealed by this representation is that genes in the left hemisphere (Fig. 1A, magenta dots), that is, genes with a negative first PC (PC1 < 0), typically have a higher expression level between P1 and P10 and a decreased expression level during P15-P60, whereas genes in the right hemisphere (Fig. 1A, green dots), that is, mouse cerebellum temporal (PC1 > 0), generally have a higher expression level between P15 and P60 and lower expression during P1-P10. That is, representing the genes by their first two temporal PCs provides a succinct characterization of their expression profile during P1-P60 (Fig. 1A,B). We call the genes with nonpositive PC1 (left hemisphere) the Cerebellar Early Mouse Partition (CEMP), and the set of genes with positive PC1 (right hemisphere) the Cerebellar Late Mouse Partition (CLMP). The 2552 genes were further subclassified according to the days during cerebellar development that they are maximally expressed (Fig. 1C,D), justifying the early-late nomenclature.
Figure 1.
The mouse cerebellum developmental expression profile at P1-P60 of 2552 genes represented in their first two temporal principal components (PC1-2). (A) Expression profile of 2552 genes in the developing mouse cerebellum represented as dots in cerebellar temporal PC1 (53.72% temporal variance) and PC2 (16.30% temporal variance). Magenta dots indicate Cerebellar Early Mouse Partition (CEMP) genes. Green dots mark Cerebellar Late Mouse Partition (CLMP) genes. Dark-magenta and green circles indicate the 8 early-development and 11 late-development expressed genes, respectively, whose temporal expression profile was verified in situ (Table 1; Fig. 2). The units for PC1-2 are normalized (mean zero, variance one across samples/stages) Affymetrix signal intensity. (B) Representative expression profiles of genes at different sectors of the temporal PC1-2 representation (number-matched to A). Note that genes in the CLMP hemisphere (e.g., 1-3, 8) show a pattern of increasing expression levels over time. In contrast, CEMP hemisphere genes (e.g., 4-7) are expressed at high levels early on (corresponding to the peak phase of granule cell precursor proliferation) and fall off sharply at approximately P10. (C,D) Two histograms breaking down the 1861 CEMP and 691 CLMP genes into their day of maximal expression during mouse cerebellar development. There is an obvious correspondence between the large number of genes expressed at P3-P5 (C) and the density found at 9 o'clock of the principal component analysis representation in A and P60 associated genes (D) with the density at the 4-o'clock position. This temporal PC “egg-plot” (A) appears to be a useful way to display the distribution of all 2552 temporal gene expression profiles.
Overall, we observed that during mouse cerebellar development, 72.9% (1861) of the total genes are CEMP and 27.1% (691) are CLMP. As indicated by the increased density of dots (genes) at the 9- and 4-o'clock peripheries of Figure 1A, the majority of genes fall into two broadly defined patterns: genes whose expressions are high/maximal during early cerebellar development and monotonically decrease with time, or genes whose expression is absent or weak during early cerebellar development and monotonically increases to maximal levels at later stages. Indeed, only 12.5% of total genes (i.e., visually, dots situated within a circle of radius 3 centered about zero in Fig. 1A) have fluctuating expression profiles outside of these two predominant patterns.
Validation of markers identified by temporal PCA of microarray cerebellar development data
To validate the preceding temporal PCA classification of array-derived profiles with actual gene expression during mouse cerebellar development, we confirmed the expression of 19 time- and cell-type-specific markers by in situ hybridization (Fig. 2; Table 1). Expression of eight marker genes whose array profiles were classified as CEMP (Ccnd1, Ccnb1, Cdc20, Cks2, Ezh2, Nmyc1, Hmgb2, and Mcmd6; Fig. 1A, magenta circles) was found to localize to the immature proliferative external granular layer in situ at P7 with decreasing or absent expression at P15-P22 during mouse cerebellar development. Conversely, by in situ hybridization, 11 CLMP (late) marker genes (Neurod1, Neurod2, Etv1, Thra, Zfp216, Btg1, Gas7, Calb1, Cbln1, Pcp2, and Pcp4; Fig. 1A, green circles), localized to postmitotic granule cells of the internal granule layer (IGL) or the single-neuron Purkinje layer at P15-P22. These results demonstrate the robust correspondence of genes identified as CEMP/CLMP by temporal PC1-2, with actual temporal-spatial gene expression in the developing postnatal mouse cerebellum.
Figure 2.
Stage- and cell-type-specific gene expression in the developing mouse cerebellum as assessed by in situ hybridization. Representative results using several markers (Table 1) with distinct expression patterns are shown. (A,B) Expression of Hmgb2 is present at P7 but not at later stages and is restricted to granule neuron precursors of the external granular layer (EGL). (C,D) Gas7 expression is detected in both early Purkinje cells and weakly in the IGL, whereas at later stages it is strongly expressed in mature granule neurons of the IGL. (E,F) Expression of Zfp216 is restricted to the IGL and increases over time.
Table 1.
Stage- and layer-specific expression of 19 marker genes in the developing postnatal cerebellum as assessed by in situ hybridization (ISH)
| P7
|
P15, P22
|
Temporal PC1
|
Temporal PC2
|
|||
|---|---|---|---|---|---|---|
| EGL | PK/IGL | EGL | PK/IGL | |||
| Early population | ||||||
| Ccnd1, 12433 | +++ | −/− | − | −/− | −2.996 | −0.422 |
| Ccnb1, 268697 | +++ | −/− | − | −/− | −3.749 | −1.133 |
| Cdc20, 107995 | +++ | −/− | − | −/− | −3.760 | −1.415 |
| Cks2, 66197 | ++ | −/− | − | −/− | −3.714 | −1.619 |
| Ezh2, 14056 | +++ | −/− | − | −/− | −3.767 | −1.551 |
| Nmyc1, 18109 | +++ | −/− | − | −/− | −3.719 | −1.077 |
| Hmgb2, 97165 | +++ | −/− | − | −/− | −3.682 | −1.709 |
| Mcm6, 17219 | +++ | −/− | − | −/− | −3.457 | −1.521 |
| Late population | ||||||
| Neurod1, 18012 | ++ | −/++ | + | −/+++ | 2.211 | 1.819 |
| Neurod2, 18013 | ++ | −/++ | + | −/+++ | 3.422 | −1.252 |
| Etv1, 14009 | + | −/++ | − | −/+++ | 3.601 | 1.029 |
| Thra, 21833 | − | +/++ | − | +/++ | 0.703 | 1.778 |
| Zfp216, 22682 | − | +/++ | − | +/+++ | 2.918 | 1.674 |
| Btg1, 12226 | − | −/++ | − | −/+++ | 2.210 | 2.651 |
| Gas7, 14457 | − | ++/+ | − | +/+++ | 3.438 | 2.052 |
| Calb1, 12307 | − | ++/− | − | +++/− | 3.953 | 0.159 |
| Caln1, 12404 | − | ++/− | − | +++/− | 4.063 | 0.999 |
| Pcp2, 18545 | − | ++/− | − | +++/− | 3.566 | 1.049 |
| Pcp4, 18546 | − | ++/− | − | +++/− | 2.936 | 2.441 |
In situ hybridization was carried out on adjacent frozen sections of cerebellum comprising the stages: P7, P15, and P22. Following each gene name is its LocusLink identifier. The relative intensity of in situ hybridization results per gene over the time series is indicated as follows: (−) absent; (+) weak; (++) moderate; (+++) strong (see Fig. 2). Temporal principal component 1 (PC1) and PC2 coordinates of mouse cerebellar development are given where PC1 > 0 means a CEMP gene, and PC1 < 0, a CLMP gene. The markers segregated into two broad categories with strongest expression in early cerebellar populations (effectively the external granular layer) and later populations (granule neurons of the internal granular layer and Purkinje neurons), mapping to CEMP and CLMP regions, respectively, during mouse cerebellar development (Fig. 1A, dark magenta and green circles).
Differentially down- and up-regulated genes in human MBs segregate to late and early mouse cerebellar development, respectively
The human samples in our study are previously published microarray data of four normal cerebella: nine dMBs and 22 cMBs (Pomeroy et al. 2002). With this data, we separately compared gene expression levels in dMB and cMB cases with respect to normal human cerebella. dMBs have 239 genes twofold down-regulated and 120 genes twofold up-regulated from normal human cerebella (Fig. 3A,B; Supplementary Table 2). We investigated the expression profiles of murine homologs to these human genes in the mouse cerebellar development data and found a significant segregation of dMB-down-regulated genes to the CLMP and of dMB-up-regulated genes to the CEMP (χ2; p < 0.0001) with a 7.19 odds ratio (o.r.). The o.r. is a measure of the relative likelihood of an event belonging to one of two categories. Here it is defined as the odds of a dMB up-regulated gene being CEMP divided by the odds that it is CLMP. An o.r. > 1 implies a higher likelihood for the gene to be CEMP than CLMP, and conversely, an o.r. less than 1 implies that the gene is more likely to be CLMP than CEMP. Analogous results were obtained for cMB twofold regulated genes (Fig. 3C,D; Supplementary Table 3). Together, these findings indicate that the genes down- and up-regulated in human cerebellar cancers show a significant and robust association with the temporal regulation of their murine homologs during cerebellar development.
Figure 3.
Murine homologs of genes twofold regulated in human desmoplastic medulloblastomas (dMB) and classic medulloblastomas (cMB; with respect to normal human cerebella) on the mouse cerebellum developmental background (as described, Fig. 1A). (A) Murine cerebellar development expression spread of 359 mouse genes whose human homologs are twofold regulated in human dMB. Magenta crosses mark genes twofold up-regulated in dMB; green triangles are genes twofold down-regulated in dMB. (B) Histograms of the mouse cerebellar temporal PC1 coordinates of twofold up-regulated (top magenta histogram) and down-regulated (bottom green histogram) genes in human dMB. The number of the 359 genes in each compartment is indicated. The odds ratio (o.r.) and its 95% confidence interval ([left endpoint, right endpoint]), χ2, and its associated p value are given. These data indicate a significant and robust association between genes up-regulated in the dMB with the CEMP, as well as down-regulated dMB genes with the CLMP. (C,D) Identical analysis as described in A and B, but for 263 human cMB twofold regulated on a mouse cerebellar development background. Similar conclusions can be drawn as for the dMB concerning the significant and strong correspondence of tumor up-/down-regulated genes and CEMP/CLMP genes.
Associations between tumorigenesis and development are organ specific
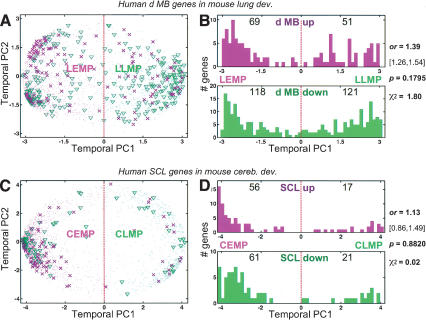
A relevant follow-up question to consider is whether this observed genetic association between cerebellar development and MB tumorigenesis is merely and entirely a consequence of non-cell-type-specific programs inherent in all dividing cells—for example, cell cycle regulation, metabolism, protein synthesis—that may be dominating the MB expression profile. To address this, we performed cross-organ analyses of development and tumorigenesis. Because the developmental origins of the lung and of lung cancer are distinct from the CNS, we repeated the foregoing experimental/analytic strategy on genes that were twofold up- and down-regulated in 21 human lung SCL samples with respect to 17 normal human lung samples (Bhattacharjee et al. 2001). The background development model here was the whole mouse lung assayed at 11 time points: embryonic days 12, 14, 16, 18 (E12, E14, E16, E18); P1, P4, P7, P10, P14, P21; and adult (>60 d; Mariani et al. 2002). A temporal PCA of the mouse lung development dataset was performed exactly as described for the cerebellum above (Fig. 4A,B). Genes in the left hemisphere (i.e., mouse lung temporal PC1 < 0; Fig. 4, magenta dots) are more highly expressed earlier rather than later in lung development. We call these the Lung Early Mouse Partition (LEMP), in keeping with our earlier nomenclature for cerebellar development. Genes in the right hemisphere (i.e., temporal PC1 > 0) are more highly expressed later in lung development; these we call the Lung Late Mouse Partition (LLMP). Of the 2552 genes, 63.1% (1610) are LEMP and 36.9% (942) are LLMP during mouse lung development. A comparison of the murine cerebellar and lung development profiles of these genes shows that they are more similar early in development than late; 53.5% are more highly expressed during both early cerebellar and lung development, whereas only 17.3% are expressed at late stages of both the developing lung and cerebellum. These percentages obviously mirror the progressively divergent genetic mechanisms underlying cerebellum and lung development. Genes twofold down- and up-regulated in human SCL exhibit a strong and significant enrichment for LLMP and LEMP genes, respectively (χ2; p < 0.0001, o.r. = 7.10; Fig. 4C,D; Supplementary Table 4), similar to results shown above for the cerebellar tumors.
Figure 4.
Mouse lung developmental expression profile at E12 to more than P60 of 2552 genes represented in their first two temporal principal components (PC1-2), and murine homologs of genes twofold regulated in human squamous cell lung carcinoma (SCL), with respect to normal human cerebella, on the mouse lung developmental background. (A) Expression profile of 2552 genes in the developing mouse lung represented as dots in the lung temporal PC1 (49.10% temporal variance) and PC2 (13.68% temporal variance). Magenta dots mark Lung Early Mouse Partition (LEMP) genes. Green dots mark Lung Late Mouse Partition (LLMP) genes. The units for PC1-2 are normalized (mean zero, variance one across samples) Affymetrix signal intensity. (B) Representative lung expression profiles of genes at different sectors of the temporal PC1-2 representation (number-matched, A). Genes from the LLMP (e.g., 1-3, 8) and LEMP (e.g., 4-7) hemispheres fluctuate over time in inverse profiles. (C) Expression profile of 155 mouse genes whose human homologs are twofold regulated in human SCL on a mouse lung developmental background (A). Magenta crosses mark genes twofold up-regulated in SCL; green triangles are genes twofold down-regulated in SCL. Interestingly, we note the focused and higher concentration of SCL up-regulated genes to the 10-o'clock sector, despite the higher density of normal developmental genes that map to the 8-o'clock sector (A). (D) Histograms of the mouse lung temporal PC1 coordinates of twofold up-regulated (top magenta histogram) and down-regulated (bottom green histogram) genes in human SCL. The number of the 155 genes in each compartment is indicated. The odds ratio (o.r.) and its 95% confidence interval ([left endpoint, right endpoint]), χ2, and its associated p value are given. These data indicate a significant and strong association between genes up-regulated in the SCL tumors with the LEMP and down-regulated tumor genes with the LLMP.
Having demonstrated analogous relationships between gene regulation in cerebellar and lung tumors with their respective murine development counterparts, we went on to investigate the cell type/organ-specificity of these findings. We examined the murine homolog profiles of differentially expressed genes in MB during lung development and, conversely, the murine homolog profiles of differentially expressed genes in SCL during cerebellar development. As shown (Fig. 5), the gene profile segregations with respect to these noncognate development backgrounds are not significant (χ2; p > 0.1). The cross-organ comparison shows a much less pronounced relationship between down-regulated genes in the human tumor and the late phase of mouse development. This is reflected by the more than fivefold drop in o.r.—as compared with earlier analyses with respect to a cognate development background. In addition, with regard to the 120 dMB up-regulated genes, their marked bias toward the early phase of development is no longer distinct (Fig. 5B). Together, these cross-organ comparisons suggest that the study of tumorigenesis from a developmental perspective is only meaningful in an organ-specific context. It follows that the strong association between the expression profile of human MBs and the mouse CEMP/CLMP is based on common regulation of organ-specific rather than general genetic programs.
Figure 5.
Profile distribution of genes twofold regulated in human tumors on noncognate mouse organ developmental backgrounds: dMB on lung development, and SCL on cerebellar development. (A) The 359 genes twofold regulated in human dMB on a mouse lung developmental background (Fig. 4A). Magenta crosses mark genes twofold up-regulated in dMB; green triangles are genes twofold down-regulated in dMB. (B) Histograms of the mouse lung temporal PC1 coordinates of twofold up-regulated (top magenta histogram) and down-regulated (bottom green histogram) genes in human dMB. The number of the 359 genes in each compartment is indicated. The odds ratio (o.r.) and its 95% confidence interval ([left endpoint, right endpoint]), χ2, and its associated p value are given. Note the sharp loss of significance and o.r. in the association between LEMP/LLMP and the fact that dMB down-regulated genes are almost equally distributed across LEMP-LLMP. (C,D) Analogous to A and B, but for 155 genes twofold regulated in human SCL on a mouse cerebellum developmental background (Fig. 1A). Again, we note a sharp loss in significance and o.r. in the association between CEMP/CLMP and SCL up-/down-regulated genes: 74.4% of the 82 SCL down-regulated genes are CEMP, which is not much different from a CEMP percentage bias occurring by chance—recall that because 72.9% of all 2552 genes are CEMP, any randomly selected group of genes would typically have a 72.9% CEMP bias.
Global expression characteristics of individual human tumor samples can be associated with distinct mouse development stages
We next asked whether the individual human MBs and normal cerebella were classifiable with respect to the global (genomic) characteristics that they share with particular mouse cerebellar development stages. Rather than inspecting individual genes as we had done previously, we switched to a global or “macroscopic” view, whereby the genomic profiles of mouse samples/stages—instead of the temporal profile of single genes—are analyzed by PCA. This methodology effectively reduces the dimensionality from 2552 gene measurements per sample/stage to three, while capturing 87.88% of the total genomic variation (Duda et al. 2001; Misra et al. 2002). Human samples—comprising four normal cerebella, nine dMBs, and 22 cMBs—were then projected into the genomic developmental trajectory of the mouse cerebellum (Fig. 6). In the three mouse genomic PC representations, mouse samples form four temporally contiguous groups: P1-P3, P5-P10, P15-P30, and P50-P60. Genomic PC1 appears to correspond with developmental progression. Individual human cases were then subclassified by their relatedness to a particular developmental stage—as defined by the Euclidean distance along the first 20 genomic PCs (capturing 100% of the total genomic variation in the mouse development data) between the human and each mouse sample. Twenty-eight of thirty-one (90.3%) MBs were closest to either P5 (71.0%, 22 MBs) or P7 (19.4%, six MBs; Fig. 6B,D; Supplementary Table 5). We note that the one normal human cerebellum that was associated with P10 possessed the lowest percentage of Present calls (probably because of poorer assay quality). These results suggest that, at the level of the overall expression profile, human MBs most closely resembled the developing mouse cerebellum at stages before P10, whereas normal human cerebella most closely resembled the mouse cerebellum at P30-P60. In addition, the dMBs appear to be molecularly more homogeneous than the cMBs, a possible reflection of distinct underlying tumorigenic mutations. An identical genomic PCA was performed on the abovementioned mouse lung development data (Mariani et al. 2002) and on the human SCL and normal lung (Bhattacharjee et al. 2001) projected therein, which demonstrated analogous results in which human SCLs most resemble mouse lung at E14, whereas normal human lung resembled a later stage—E18 (data not shown). Together, these findings indicate that global gene expression within individual tumors can be associated with particular organ-specific stages of development.
Figure 6.
Genomic features of human normal cerebella and MBs projected into mouse genomic cerebellar development space. (A) Mouse and human samples in the genomic PC1 (60.15% genomic variance) and PC3 (6.97% genomic variance) of mouse cerebellar development P1-P60. Reference circles mark mouse cerebellum developmental stages. Projected human samples into this mouse genomic space are marked ×: (green) normal cerebella (n = 4); (dark magenta) nonmetastatic (M0) dMB (n = 7); (light magenta) metastatic (M+) dMB (n = 2); (blue) M0 cMB (n = 14); (light blue) M+ cMB (n = 8). Note the configurations of human samples (×) in relation to the mouse stages (○). (B) A stacked histogram of the mouse cerebellum developmental stage/day most closely associated with each human case. Note the higher concentration of M+ MBs with P5 association. Detailed quantitation/distances are shown (C,D; Supplementary Table 5). (C) The average Euclidean distance between each mouse cerebellum development stage to nonmetastatic dMB (n = 7, solid dark-magenta line), metastatic dMB (n = 2, solid light-magenta line), and normal human cerebellum (n = 4, solid green line). Color-matched dotted lines mark the one standard deviation envelope around the average distance. The first 20 genomic mouse PCs are used for calculating distances between human and mouse, which account for 100% of total mouse genomic variance. Note that on average, dMBs (M0 and M+) are closest to the early-developing mouse cerebellum around P5 than to normal human cerebella. The latter are, on average, closest to the late development stages of mouse cerebellum P30-P60 than dMBs. (D) Similar to B, but for cMBs: nonmetastatic cMB (n = 14, solid blue line), metastatic cMB (n = 8, solid light-blue line), and normal human cerebellum (n = 4, solid green line). Again, we note that on average, cMBs (especially M+ cases) are closest to the early-developing mouse cerebellum around P5 than to normal human cerebella. M0 cMBs are closest to P5-P7.
Genetic regulation in metastatic human MB is most tightly coupled to developing mouse cerebellum at P5
Human MBs are heterogeneous with respect to both their histology and their aggressiveness, as indicated by the presence of metastases. The human MB samples in our dataset had been clinically annotated using the World Health Organization metastatic stage classification label (Pomeroy et al. 2002). Because of the small sample size of MBs with metastatic stages M1 to M4 (10/31), we pooled these into one class, denoted M+ to distinguish them from the nonmetastatic cases (21/31), which were denoted M0. Because we have already observed an association between human cerebellar tumor samples and early stages of mouse cerebellar development before P10 (Fig. 6B,C; Supplementary Table 5), we next considered whether a developmental association could be established with tumor clinical behavior. Among cMBs, 50.0% (7/14) of P5-associated cases are M+, whereas only 12.5% (1/8) of non-P5-associated cMBs are M+, with an o.r. of 7.0. Conversely, tumors most highly associated with >5 postnatal days of development were more likely to have a less aggressive character. Consistent with this, the genomic profile of tumors from the Ptch+/- mouse model (Goodrich et al. 1997; Kim et al. 2003) of nonmetastatic MB most closely associates with P7 (Fig. 7; Supplementary Tables 5, 6). These data suggest that the developmental perspective on tumorigenesis can provide insights into the clinical behavior of solid tumors.
Figure 7.
Gene regulation and genomic features of MBs and normal cerebellar tissue from Ptch+/- mutant mice on the background of mouse cerebellum development. (A) Murine cerebellar development expression profile of 373 mouse genes that are twofold regulated in MB with respect to normal cerebellar tissue in Ptch+/- mouse mutants. Magenta crosses mark genes twofold up-regulated in dMB; green triangles are genes twofold down-regulated in dMB. (B) Histograms of the mouse cerebellar temporal PC1 coordinates of twofold up-regulated (top magenta histogram) and down-regulated (bottom green histogram) genes in mouse MB (Ptch+/-). The number of the 373 genes in each compartment is indicated. The odds ratio (o.r.) and its 95% confidence interval ([left endpoint, right endpoint]), χ2, and its associated p value are given. These data indicate a significant and strong association between genes up-regulated in the Ptch+/- mouse MB with the CEMP, as well as down-regulated MB genes with the CLMP. (C) Genomic profile of Ptch+/- mouse MB and normal cerebellar tissue in the genomic PC1 (60.15% genomic variance) and PC3 (6.97% genomic variance) of mouse cerebellar development P1-P60. Reference circles mark the mouse cerebellar development stages. Projected Ptch+/- mouse samples into this mouse cerebellar development space are marked ×: (green) normal cerebella (n = 4); (dark magenta) MB (n = 9). Note the configurations of Ptch+/- mouse samples (×) in relation to the mouse development stages (○). (D) A stacked histogram of the mouse cerebellar development stage/day most closely associated with each Ptch+/- mouse case. The Euclidean distance from each Ptch+/- mouse sample to each development stage is calculated along the first 20 genomic mouse cerebellar development PCs as in Figure 6C and D (Supplementary Table 5). Note the higher frequency of Ptch+/- MBs with P7 association.
Discussion
We report that the genomic study of solid tumors from a developmental perspective allows the identification of both specific genes and global regulatory patterns shared with organogenesis. Although our findings indicate that common programs underlie development and tumori-genesis, this is not to say that tumors are identical to, or possess all the programs of, their developing counterpart. Clearly, the impact of tumor suppressor loci mutations constitutes a profound difference from the normal developing organ. In fact, our data may well indicate that such mutations are necessary for the permissive state that allows recapitulation of developmentally associated programs of gene regulation. Because SHH signaling is required for proliferation of cerebellar granule cells during development, one particularly striking illustration of this concept has been the finding of MBs in patients with mutations of the hedgehog-pathway repressor molecules Patched/PTCH and SUFU (Rubin and Rowitch 2002). Activating mutations of the SHH pathway are associated with dMB but not cMB. Nevertheless, we have observed a significant correspondence in the mouse cerebellar development profile with genes regulated in both tumor subtypes. This suggests that primary tumors recapitulate programs of their developing counterpart, irrespective of whether the inciting tumorigenic mutation involves a developmentally relevant pathway per se. The database of genes expressed during mouse organogenesis provides a framework for interpreting genetic regulation in human cancers.
Genetic developmental profile provides insight into the biological behavior of cancer
At the macroscopic/genomic scale, we find that a subset of MBs resemble cells of the early-developing murine cerebellum. Taking into account all 2552 genes in the human samples with murine homologs across the two Affymetrix microarray platforms, we find that the global (2552 genes) features of human tumors put them closest to the early-developing cognate organ, whereas normal human tissue are closest to the late or mature stages of the developing organ. Thirty of 31 (96.8%) of the human MBs were segregated from normal human cerebella by their genomic similarity to mouse cerebellar developmental P1-P10 (especially P5-P7) versus the P30-P60 association of normal human cerebella. In particular, we found a higher likelihood for metastasis (o.r. = 7.0) in P5-associated cMBs compared with non-P5-associated cMBs. Genetic regulation within metastatic human MBs has been previously investigated (MacDonald et al. 2001, 2003; Chopra et al. 2003; Gilbertson and Clifford 2003; Hernan et al. 2003). Indeed, 37 of the 59 genes that MacDonald et al. (2001; Gilbertson and Clifford 2003) found to be up-regulated in metastatic human MBs versus nonmetastatic cases are present in our mouse cerebellar development data with suitable reproducibility (Supplementary Table 5). Of these, 75.7% (28/37) are CEMP, and the largest fraction (27.0%, 10/37) are maximally expressed at P5: PDGFRα, TM4SF1, CTSC, NME1, ADAM17, NR4A3, RFC4, DDR2, EMP1, and RPA3. The second-largest group (16.2%, 6/37) comprise genes maximal at P7: PDGFRβ, IGFBP2, IGFBP7, MSX1, POLD2, and IQGAP1. Interestingly, in mice, the highest levels of granule neuron precursor proliferation and migration characterize P5-P7 of cerebellar development—both Shh and the proliferating granule cell marker, Atoh1/Math1, are CEMP and are maximally expressed at P5 in our data. We therefore speculate that the P5-P7 cerebellum constitutes an appropriate focus for identification of mechanisms common to development and the metastatic subset of MBs in children, which carry the worst prognosis.
At the gene-by-gene scale, we find human MB up-regulated genes to largely and significantly belong to the early mouse cerebellar developmental program, whereas MB down-regulated genes belong to a later, complementary program. It follows that similar genetic regulation underlies the biology of CNS precursors and tumorigenic cells in these cases. Indeed, recent work supports the notion that several tumorigenic pathways may yield a common MB phenotype, with gene expression characteristics similar to the early-developing cerebellum. Lee et al. (2003) found a common set of molecular markers for mouse MBs generated on a p53-/- mutant background combined with deficiencies of a variety of other genes including Ptch, Lig4, and cyclin-dependent kinase inhibitors. Interestingly, despite genetic differences, the various mouse MBs were quite homogeneous in terms of overall gene expression: 21 genes were up-regulated and nine were down-regulated consistently with respect to the mouse P5 cerebellum. Of these, 15 of 21 of the up-regulated and 7 of 9 of the down-regulated genes were present in our mouse cerebellar development dataset with suitable reproducibility (Supplementary Table 5). We find that 86.7% (13/15) of the mouse MB-associated up-regulated genes are CEMP, and 46.7% (7/15) are maximal at P5. Only 28.6% (2/7) of the mouse MB down-regulated genes are CLMP. This latter finding does not contradict an anticipated CLMP-enrichment because their choice of a baseline tissue was the mouse P5 cerebellum, an early stage of active proliferation that expresses very few, if any, cerebellar late development genes. In keeping with our findings in human tumors, the observations of Lee et al. (2003), therefore, suggest that the association with the early-developmental profile will apply to mouse models of MB as well. Indeed, the genomic profile of tumors from the Ptch+/- mouse model (Goodrich et al. 1997; Kim et al. 2003) of nonmetastatic MB most closely associates with P7 (Fig. 7; Supplementary Tables 5, 6).
Relevance of the developmental-genetic association to the design and characterization of preclinical models of tumorigenesis
Tumorigenic progenitors of the CNS have been proposed to arise from multipotent stem cells (Reynolds and Weiss 1996; Holland 2001; Singh et al. 2003) and somatic cells that have de-differentiated (Bachoo et al. 2002; Uhrbom et al. 2002; Katsetos et al. 2003). Our analyses demonstrate a conservation of developmental mechanisms in MB tumorigenesis, consistent with a stem-progenitor cell origin of such tumors. A related question is whether certain types of tumor cells are “stalled” progenitors. For instance, in acute myelogenous leukemia, evidence exists for the disruption of normal hematopoietic differentiation programs (Reya et al. 2001; Tenen 2003). However, the case for solid tumors is less clear. The general approach of investigating a pathologic system from the vantage point of a developmental sequence will test ideas such as the stem cell origin of tumors and can be extended to general cases of tumorigenesis and disease and injury, as demonstrated by our human SCL analysis. Acquisition of genomic information on humans and other vertebrate species provides new opportunities for answering biologic questions about one species by investigating an “equivalent” system that, on practical or ethical grounds, is more amenable to experimentation. Our analytic-projective technique enables the analysis of human tissue against a nonhuman system background across different measurement platforms. As a corollary, our findings indicate the importance of analyzing human primary tumors with a cognate development model: The robust association between development and tumorigenesis was lost when cerebellar tumors were analyzed against the developing lung and, vice versa, lung cancer against cerebellar development. Thus, nonhuman development models—carefully chosen and validated by global similarities to human tumors—may constitute a relevant system for preclinical testing of candidate antineoplastic therapeutics.
Although our work illustrates how mouse developmental models can be exploited to learn more about human tumors, it remains to be seen how generally applicable this approach will be to a large range of different tumors. It may be that the strong correlations we observed reflect the relative homogeneity of the developing cerebellum, in which a significant number of granule neuron precursors show coordinate patterns of gene expression at specific developmental stages. However, this might not be the case for many other tissues that are composed of a large variety of different cell types, obscuring the specific profile of the cell type that resembles the tumor. Further investigation is needed to determine whether this issue will ultimately limit the applicability of our approach. However, such potential problems could be overcome by improved linear amplification methods combined with microdissection of mouse tissues (Vincent et al. 2002; Zirlinger and Anderson 2003).
Possible diagnostic and prognostic relevance of genetic developmental associations in human cancers
Given the complex nature of solid cancers, which includes inter- and intratumor cellular-molecular heterogeneity, multivariate methods have proven critical in the accurate characterization of case-specific tumor phenotype (Golub 2003). Our results suggest that cross-species analysis of a cognate model developmental system might generally provide novel insight into the biological and clinical behavior of solid tumors in several ways. First, one might achieve an objective and robust molecular definition of an undifferentiated versus a mature state, using multivariate methods that may be more reliable than current diagnostic practices, which are primarily based on histopathology and a small number of immunohistochemical markers. We suggest that a comprehensive organ-specific database of gene expression, such as the one described here for the cerebellum, might have further application in the clinical domain. In a recent study, using the extensively studied Ptch+/- mouse model of MB, Kim et al. (2003) observed expression of known markers both for immature cells (e.g., Atoh1/Math1) and for differentiated neurons (e.g., Neuna60/NeuN), indicating that reliance on a small number of markers and morphological criteria alone is not adequate for documenting the degree of tumor differentiation (Kim et al. 2003). Our present data, using global measures of gene expression in a large number of human MBs, indicate that it is possible to associate the expression profile with an individual tumor case by its similarity to a particular phase-day of development. As such, it may provide a means for describing level of differentiation within a tumor via comprehensive measures of gene expression.
Second, the strong association between metastatic human MBs and mouse cerebellum P5 suggests that, distinct from diagnostic parameters regarding the degree of differentiation with a tumor, it may be possible to make prognostic prediction on the basis of global similarities to a particular developmental stage. As noted, P5 of cerebellar development is characterized both by high levels of granule cell proliferation and by migration, two features present in aggressive tumors. A clearer understanding of the operative mechanisms for particular phases of brain development will undoubtedly provide further insights into human CNS tumor biology, prognosis, and possible therapeutic interventions.
Materials and methods
Assessment of gene expression during wild-type mouse cerebellar development and in cerebellar tumors
Pooled whole cerebella were profiled using Affymetrix Mu11K (MAS 5.0) arrays at 10 separate time points: P1, P3, P5, P7, P10, P15, P21, P30, P50, and P60. Each time point except P50 was assayed in duplicate with split aliquots (Zhao et al. 2002). A Pearson correlation exceeding 0.50 between duplicate measurements P1-P60 (without P50) was found for 5826 unique LocusLink identified genes. In situ hybridization has been described (Zhao et al. 2002). Genotyping and propagation of tumors in Ptch+/- mice was carried out as described in Kim et al. (2003). Tumor and grossly normal-appearing cerebellar tissue were dissected in several matched samples for RNA profiling using Affymetrix Mu11K arrays as described above.
Human MB, normal human cerebella, human squamous cell lung carcinoma, normal human lung samples, and wild-type mouse lung development data
The microarray data, materials, tumor, and patient clinical phenotypes have been described—dataset_B (Pomeroy et al. 2002); LUNG_scans_SQ, LUNG_scans_NORM (Bhattacharjee et al. 2001)—and are available at http://www-genome.wi.mit.edu/cancer. The mouse lung development data have been described and are available at http://lungtranscriptome.bwh.harvard.edu (Mariani et al. 2002). Murine cerebellar development series and Ptch+/- MB data are available at http://www.chip.org/resources/data/mouse_cerebellum.
Homology mapping
Curated and calculated functional orthologs (Zhang et al. 2000) between mouse and human genes were derived from HomoloGene (http://www.ncbi.nlm.nih.gov/HomoloGene, Sept 15, 2003, data freeze). Between the Affymetrix Mu11K and HuFL arrays, 3097 unique human-mouse homolog pairs were found. Uniqueness was achieved by selecting probes with the largest number of Present calls. When Present calls are tied, we select the probe with the maximal coefficient of variation across experimental conditions. We used 2552 of the 3097 genes that had a reproducible profile (Pearson correlation exceeding 0.50) between the two-replicate set of mouse cerebellar development assays P1-P60 (without P50).
Human tumor fold analysis
For the fold analysis of human normal and tumor samples, each sample is normalized by linear regression (Wu 2001) to a reference set consisting of the average of a normal cerebellum and a dMB sample that, respectively, have the maximal average Pearson correlation against the four normal cerebella samples and the 7 dMB (or 20 cMB) samples. We used a geometric fold method modified from Zhao et al. (2002) to assess the genes that are significantly changed in expression levels in tumors with respect to normal tissue/controls. Without loss of generality, suppose that c < a1 ≤ a2 ≤...≤ aN are the measured levels of a particular RNA in N tumors, and c < b1 ≤ b2 ≤...≤ bM are the same RNA's measurements in M reference/normal tissue for some positive real constant c. In our fold analyses, we threshold all RNA measurements at c = 50. We define the average geometric fold change of this RNA in the tumors with respect to normal tissue as AvgLF = ∑j = 1N log (aj)/N - ∑k = 1M log (bk)/M; and the maximal intragroup variation as Noise = max(NoiseTumor, NoiseNormal), where NoiseTumor = ∑j = [M/2] +1M log (aj)/[M/2] - ∑j = 1[M/2] log (aj)/[M/2], and NoiseNormal = ∑k = [N/2] +1N log (bk)/[N/2] - ∑k = 1[N/2] log (bk)/[N/2]. The notation [K] denotes the greatest integer less than or equal to the real number K; for example, [5.9] = 5 = [5.1], [10/2] = 5 = [11/2]. A gene was determined to be twofold regulated in dMBs with respect to normal cerebella if Abs (AvgLF) > Noise, Abs (AvgLF) > log(2) with at least one non-Absent call in all samples involved. We used the natural logarithm, in which log(2) = 0.6931.
Mouse cerebellum temporal PCA
Each mouse sample was linear regression-normalized (Wu 2001) to the P1 sample that had the highest average correlation against all samples P3-P60. Before the temporal PCA of mouse data, each of the 2552 genes was individually normalized to mean zero and variance one across P1-P60 (Duda et al. 2001; Misra et al. 2002). The percentage temporal variances captured by each of the first five temporal PCs (of 18 nonzero components) were 53.72%, 16.30%, 6.48%, 4.22%, and 2.92%, totaling 83.64%. PCA was performed using Matlab. The mouse lung development data of the same 2552 genes was similarly analyzed.
Mouse cerebellum genomic PC analysis and the projection of human samples onto murine development genomic space
Before the genomic PCA of mouse data, each individual mouse sample—a vector of 2552 genes—was first normalized to mean zero and variance one across all its 2552 measurements (Duda et al. 2001; Misra et al. 2002). The percentage genomic variance captured by each of the first five genomic PCs were 60.15%, 20.76%, 6.97%, 2.86%, and 1.89%, totaling 92.63%—with the first 20 genomic PCs covering 100% of total genomic variance. Each genomic PC is a 2552-vector, so that from the genomic PCA of the mouse cerebellar development data, we obtain a 2552 × 2552 matrix, denoted Φ, whose columns are composed of nonzero mouse genomic PCs. Φ was used to project human samples into the mouse cerebellar development genomic space. Each human sample is a 2552-vector with vector slots ordered genewise homologous to the mouse sample vectors; that is, if the 1685th slot of the human vector is the measured expression for BACH (brain acyl-CoA hydrolase), then the 1685th mouse vector slot is the measurement for its murine homolog, Bach. Call this human 2552-vector/sample, x. We rewrite x with respect to the mouse genomic PCs via the transformation y = ΦΤx; that is, y is how the human sample appears relative to the mouse cerebellar development framework/basis. Figure 6A (×s) shows the first and third components of all human samples, ys. This procedure is essentially a change of coordinates from the standard bases in R2552 into a new set of bases elements derived from the genomic mouse PCA. The distance between human-mouse samples is calculated along the first 20 mouse genomic PCs—constituting 100% of genomic variance in the mouse cerebellar development data.
Acknowledgments
We thank Emanuela Gussoni, Charles D. Stiles, David Pellman, Joseph Majzoub, Natasha Y. Frank, and Ashish Nimgaonkar for their critical review of the manuscript, Rosalind W. Picard for useful ideas, and Dong-in Yuk and Sovann Kaing for expert technical assistance. A.T.K. is grateful to the Dana-Mahoney Center for Neuro-Oncology for a postdoctoral fellowship and NIH grant NS40828-01A1 for support. Q.Z. acknowledges the American Brain Tumor Association's David Coren Fellowship for support. A.J.B. is supported by the Lawson Wilkins Pediatric Endocrinology Society and NIDDK grants R01 DK00837 and K12 DK063696. These studies were funded by grants from the National Institutes of Health (R01 NS35701 to S.L.P.; R21 NS41764-01 and RO1 NS4051 to D.H.R.; NS40828-01A1 and HL066582-01 to I.S.K) and the James S. McDonnell Foundation (D.H.R.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1182504.
Corresponding authors.
References
- Altman J. and Bayer, S.A. 1987. Development of the precerebellar nuclei in the rat: III. The posterior precerebellar extramural migratory stream and the lateral reticular and external cuneate nuclei. J. Comp. Neurol. 257: 513-528. [DOI] [PubMed] [Google Scholar]
- Bachoo R.M., Maher, E.A., Ligon, K.L., Sharpless, N.E., Chan, S.S., You, M.J., Tang, Y., DeFrances, J., Stover, E., Weiss-leder, R., et al. 2002. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell 1: 269-277. [DOI] [PubMed] [Google Scholar]
- Bailey P. and Cushing, H. 1926. A classification of the tumors of the glioma group on histogenetic basis with a correlated study of prognosis. J. D. Lippincott, Philadelphia, PA.
- Bhattacharjee A., Richards, W.G., Staunton, J., Li, C., Monti, S., Vasa, P., Ladd, C., Beheshti, J., Bueno, R., Gillette, M., et al. 2001. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc. Natl. Acad. Sci. 98: 13790-13795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A., Brown, K.M., Rood, B.R., Packer, R.J., and MacDonald, T.J. 2003. The use of gene expression analysis to gain insights into signaling mechanisms of metastatic medulloblastoma. Pediatr. Neurosurg. 39: 68-74. [DOI] [PubMed] [Google Scholar]
- Duda R.O., Hart, P.E., and Stork, D.G. 2001. Pattern classification. Wiley-Interscience, New York.
- Gilbertson R.J. and Clifford, S.C. 2003. PDGFRB is overexpressed in metastatic medulloblastoma. Nat. Genet. 35: 197-198. [DOI] [PubMed] [Google Scholar]
- Golub T.R. 2003. Mining the genome for combination therapies. Nat. Med. 9: 510-511. [DOI] [PubMed] [Google Scholar]
- Goodrich L.V., Milenkovic, L., Higgins, K.M., and Scott, M.P. 1997. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277: 1109-1113. [DOI] [PubMed] [Google Scholar]
- Hallonet M.E., Teillet, M.A., and Le Douarin, N.M. 1990. A new approach to the development of the cerebellum provided by the quail-chick marker system. Development 108: 19-31. [DOI] [PubMed] [Google Scholar]
- Hernan R., Fasheh, R., Calabrese, C., Frank, A.J., Maclean, K.H., Allard, D., Barraclough, R., and Gilbertson, R.J. 2003. ERBB2 up-regulates S100A4 and several other prometastatic genes in medulloblastoma. Cancer Res. 63: 140-148. [PubMed] [Google Scholar]
- Holland E.C. 2001. Progenitor cells and glioma formation. Curr. Opin. Neurol. 14: 683-688. [DOI] [PubMed] [Google Scholar]
- Hoopengardner B., Bhalla, T., Staber, C., and Reenan, R. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832-836. [DOI] [PubMed] [Google Scholar]
- Kadin M.E., Rubinstein, L.J., and Nelson, J.S. 1970. Neonatal cerebellar medulloblastoma originating from the fetal external granular layer. J. Neuropathol. Exp. Neurol. 29: 583-600. [DOI] [PubMed] [Google Scholar]
- Katsetos C.D., Herman, M.M., and Mork, S.J. 2003. Class III beta-tubulin in human development and cancer. Cell Motil. Cytoskeleton 55: 77-96. [DOI] [PubMed] [Google Scholar]
- Kim J.Y., Nelson, A.L., Algon, S.A., Graves, O., Sturla, L.M., Goumnerova, L.C., Rowitch, D.H., Segal, R.A., and Pomeroy, S.L. 2003. Medulloblastoma tumorigenesis diverges from cerebellar granule cell differentiation in patched heterozygous mice. Dev. Biol. 263: 50-66. [DOI] [PubMed] [Google Scholar]
- Kleihues P. and Cavenee, W.K. 2000. Pathology and genetics of tumors of the nervous system, pp. 129-137. International Agency for Research on Cancer Press, Lyon, France.
- Lee Y., Miller, H.L., Jensen, P., Hernan, R., Connelly, M., Wetmore, C., Zindy, F., Roussel, M.F., Curran, T., Gilbertson, R.J., et al. 2003. A molecular fingerprint for medulloblastoma. Cancer Res. 63: 5428-5437. [PubMed] [Google Scholar]
- MacDonald T.J., Brown, K.M., LaFleur, B., Peterson, K., Lawlor, C., Chen, Y., Packer, R.J., Cogen, P., and Stephan, D.A. 2001. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat. Genet. 29: 143-152. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Corrigendum: Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat. Genet. 35: 287. [DOI] [PubMed] [Google Scholar]
- Mariani T.J., Reed, J.J., and Shapiro, S.D. 2002. Expression profiling of the developing mouse lung: insights into the establishment of the extracellular matrix. Am. J. Respir. Cell Mol. Biol. 26: 541-548. [DOI] [PubMed] [Google Scholar]
- Mineta K., Nakazawa, M., Cebria, F., Ikeo, K., Agata, K., and Gojobori, T. 2003. Origin and evolutionary process of the CNS elucidated by comparative genomics analysis of planarian ESTs. Proc. Natl. Acad. Sci. 100: 7666-7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J., Schmitt, W., Hwang, D., Hsiao, L.L., Gullans, S., and Stephanopoulos, G. 2002. Interactive exploration of micro-array gene expression patterns in a reduced dimensional space. Genome Res. 12: 1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B. and Lee, C.J. 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34: 177-180. [DOI] [PubMed] [Google Scholar]
- Packer R.J. 2003. Gene expression profiling to analyze embryonal tumors of the central nervous system. Curr. Neurol. Neurosci. Rep. 3: 117-119.12583839 [Google Scholar]
- Pomeroy S.L., Tamayo, P., Gaasenbeek, M., Sturla, L.M., Angelo, M., McLaughlin, M.E., Kim, J.Y., Goumnerova, L.C., Black, P.M., Lau, C., et al. 2002. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature 415: 436-442. [DOI] [PubMed] [Google Scholar]
- Rather J. 1978. The genesis of cancer: A study in the history of ideas. Johns Hopkins University Press, Baltimore, MD.
- Reddy A.T. and Packer, R.J. 1999. Medulloblastoma. Curr. Opin. Neurol. 12: 681-685. [DOI] [PubMed] [Google Scholar]
- Reya T., Morrison, S.J., Clarke, M.F., and Weissman, I.L. 2001. Stem cells, cancer, and cancer stem cells. Nature 414: 105-111. [DOI] [PubMed] [Google Scholar]
- Reynolds B.A. and Weiss, S. 1996. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 175: 1-13. [DOI] [PubMed] [Google Scholar]
- Rubin J.B. and Rowitch, D.H. 2002. Medulloblastoma: A problem of developmental biology. Cancer Cell 2: 7-8. [DOI] [PubMed] [Google Scholar]
- Rubin G.M., Yandell, M.D., Wortman, J.R., Gabor Miklos, G.L., Nelson, C.R., Hariharan, I.K., Fortini, M.E., Li, P.W., Apweiler, R., Fleischmann, W., et al. 2000. Comparative genomics of the eukaryotes. Science 287: 2204-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Clarke, I.D., Terasaki, M., Bonn, V.E., Hawkins, C., Squire, J., and Dirks, P.B. 2003. Identification of a cancer stem cell in human brain tumors. Cancer Res. 63: 5821-5828. [PubMed] [Google Scholar]
- Tenen D.G. 2003. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 3: 89-101. [DOI] [PubMed] [Google Scholar]
- Uhrbom L., Dai, C., Celestino, J.C., Rosenblum, M.K., Fuller, G.N., and Holland, E.C. 2002. Ink4a-Arf loss cooperates with KRas activation in astrocytes and neural progenitors to generate glioblastomas of various morphologies depending on activated Akt. Cancer Res. 62: 5551-5558. [PubMed] [Google Scholar]
- Vincent V.A., DeVoss, J.J., Ryan, H.S., and Murphy Jr., G.M. 2002. Analysis of neuronal gene expression with laser capture microdissection. J. Neurosci. Res. 69: 578-586. [DOI] [PubMed] [Google Scholar]
- Wu T.D. 2001. Analysing gene expression data from DNA microarrays to identify candidate genes. J. Pathol. 195: 53-65. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203-214. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Kho, A.T., Kenney, A.M., Yuk Di, D.I., Kohane, I., and Rowitch, D.H. 2002. Identification of genes expressed with temporal-spatial restriction to developing cerebellar neuron precursors by a functional genomic approach. Proc. Natl. Acad. Sci. 99: 5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirlinger M. and Anderson, D. 2003. Molecular dissection of the amygdala and its relevance to autism. Genes Brain Behav. 2: 282-294. [DOI] [PubMed] [Google Scholar]