Abstract
TopBP1 (DNA topoisomerase IIβ binding protein I) contains multiple BRCT domains and is involved in replication and the DNA damage checkpoint. Through its BRCT domain, TopBP1 interacts with and represses exclusively E2F1 but not other E2F factors. This regulation of E2F1 transcriptional activity is mediated by a pRb-independent, but Brg1/Brm-dependent mechanism. TopBP1 recruits Brg1/Brm, a central component of the SWI/SNF chromatin-remodeling complex, to E2F1-responsive promoters and represses the activities of E2F1, but not E2F2 or E2F3. This regulation is crucial in the control of E2F1-dependent apoptosis during normal cell growth and DNA damage. Interestingly, TopBP1 is induced by E2F and interacts with E2F1 during G1/S transition. Thus, TopBP1 functions as a critical modulator and serves as a negative feedback regulator of E2F1 by inhibiting E2F1-dependent apoptosis during G1/S transition as well as DNA damage to promote cell survival.
Keywords: E2F1, TopBP1, apoptosis, Brg1, Brm, BRCT
E2F is a family of transcription factors that play an important role in the regulation of cell cycle progression (Dyson 1998; Nevins 1998; Trimarchi and Lees 2002). It regulates a very diverse array of genes, and is also involved in other biological processes in addition to cell cycle progression (Cam and Dynlacht 2003). Among the E2F family, E2F1 is unique in its activity to trigger apoptosis (DeGregori et al. 1997; Kowalik et al. 1998; Moroni et al. 2001; Hallstrom and Nevins 2003) and its involvement in the cellular response to DNA damage (Lin et al. 2001; Pediconi et al. 2003). E2F1 is phosphorylated by ATM (ataxia telangiectasia mutated; Lin et al. 2001) and Chk2 (Stevens et al. 2003) during DNA damage. These phosphorylation events lead to stabilization and activation of E2F1. ATM and ATR (ATM and Rad3-related) can phosphorylate E2F1, but not E2F2 and E2F3 (Lin et al. 2001). This specificity accounts for the selective induction of E2F1 among E2F family during DNA damage.
The physiologic role for E2F1-mediated apoptosis has been demonstrated in several contexts, such as thymocyte apoptosis (Field et al. 1996), negative selection of T-cells (Zhu et al. 1999), and etoposide-induced apoptosis in thymocytes (Lin et al. 2001). Furthermore, E2F1-induced apoptosis is required to avoid transformation during oncogenic stress (Pan et al. 1998; Yamasaki et al. 1998). As a result, E2F1-/- mice are prone to develop a broad spectrum of tumors (Field et al. 1996; Yamasaki et al. 1996). On the other hand, E2F1-dependent apoptosis must be tightly controlled during normal cellular proliferation because the level of E2F1 rises during G1/S transition and S phase of each cell cycle. How is the control of different E2Fs' activities orchestrated during G1/S transition, such that E2F1-mediated apoptosis is repressed, but other E2F activities are allowed to induce S-phase entry? E2F1 and E2F3 appear to be differentially regulated during the cell cycle. Whereas both E2F1 and E2F3 DNA-binding activities increase as quiescent cells are stimulated by growth factors to enter the first cell cycle, only E2F3 activity, but not E2F1 activity, reaccumulates in subsequent G1/S transitions despite reaccumulation of both E2F1 and E2F3 proteins (Leone et al. 1998). However, the nature of this E2F1-specific regulation remains unknown. Recently, it was shown that constitutively active Akt attenuated E2F1-induced apoptosis and a phosphoinositide 3-kinase inhibitor abrogated serum-mediated suppression of E2F1 apoptosis, indicating a role of the Ras-dependent Akt pathway in suppressing E2F1-induced apoptosis during normal growth (Hallstrom and Nevins 2003). Nevertheless, the mechanism by which E2F1-mediated apoptosis is blocked during normal cell growth remains to be elucidated.
To identify E2F1-specific regulators, we used an E2F1-specific fragment (N terminus) as a bait and isolated DNA topoisomerase-IIβ-binding protein I (TopBP1) in a yeast two-hybrid screen (Liu et al. 2003). TopBP1 contains eight BRCA1 C-terminal (BRCT) motifs and interacts with several other proteins, including human papilloma virus type 16 (HPV16) transcription/replication factor E2 (Boner et al. 2002), DNA polymerase ε, checkpoint protein hRad9 (Makiniemi et al. 2001), and Miz-1 (Herold et al. 2002). It appears to be involved in DNA replication because incubation of an antibody against the sixth BRCT motif of TopBP1 inhibits DNA replication in an in vitro HeLa nuclei replication assay (Makiniemi et al. 2001). TopBP1 is induced during DNA damage and is also involved in DNA damage checkpoint (Yamane et al. 2002, 2003). Upon γ-irradiation, TopBP1 colocalizes with Nbs1, BRCA1, and 53BP1 in the ionizing radiation-induced foci representing stalled replication forks (Makiniemi et al. 2001; Yamane et al. 2002). In addition to the control of DNA replication, TopBP1 is also required for cell survival. Inhibition of TopBP1 expression by antisense Morpholino oligomers induces apoptosis (Yamane et al. 2002). Thus, TopBP1 is involved in several important aspects of growth control. So far, the detailed mechanism by which TopBP1 regulates these signaling events remains poorly understood.
TopBP1 interacts with E2F1 through the sixth BRCT motif of TopBP1 and the N terminus of E2F1 (Liu et al. 2003). This interaction is induced by ATM-mediated phosphorylation of E2F1 at Ser 31 during DNA damage. The interaction between BRCT domains and phosphopeptides was later found to be a general phenomenon (Manke et al. 2003; Rodriguez et al. 2003; Yu et al. 2003). Through this interaction, the transcriptional and apoptotic activities of E2F1 are repressed, and E2F1 is recruited to DNA damage-induced foci (Liu et al. 2003). Moreover, the interaction between TopBP1 and E2F as well as the repression of E2F activity are specific to E2F1, but not seen in E2F2, E2F3, and E2F4, suggesting that TopBP1 is an E2F1-specific regulator.
In this report, we show that E2F1 is regulated by a novel Retinoblastoma protein (pRb)-independent mechanism, in which TopBP1 recruits Brg1/Brm, a central subunit of the SWI/SNF chromatin-remodeling complex, to inhibit E2F1 transcriptional activity. This regulation is specific for E2F1 and is critical for the control of E2F1-dependent apoptosis during S phase and DNA damage. TopBP1 is induced by E2F and interacts with E2F1 during G1/S transition. Thus, E2F1 and TopBP1 form a feedback regulation to prevent apoptosis during DNA replication.
Results
Regulation of E2F1 transcriptional activity by TopBP1 requires Brg1/Brm
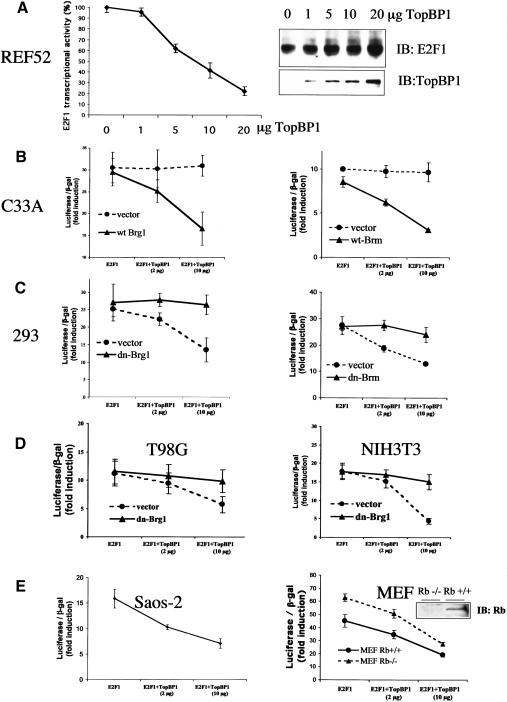
Modification of chromatin structure is a general mechanism for regulating gene transcription (Felsenfeld 1992; Kingston and Narlikar 1999). E2F activity can be inhibited by chromatin-remodeling enzymes, including the SWI/SNF complex (Trouche et al. 1997; Muchardt and Yaniv 2001; Wang et al. 2002) and histone deacetylases (HDACs; Brehm et al. 1998; Luo et al. 1998; Magnaghi-Jaulin et al. 1998). For example, pRb actively represses E2F transcriptional activity by recruiting these complexes (Harbour and Dean 2000). TopBP1 does not contain the structures that directly modify chromatin and regulate transcription. Thus, we hypothesized that TopBP1 might repress E2F1 activity by recruiting chromatin-remodeling enzymes, such as the SWI/SNF complex, which contains either Brg1 or Brm as its central subunit. To test that, we examined the effect of TopBP1 on E2F1 transcriptional activity in C33A cells, which lack Brg1 and Brm. The E2F1 activity was assessed by a p14ARF promoter-luciferase assay (Liu et al. 2003). Although TopBP1 repressed E2F1 activity in multiple cell lines including 293, 293T, MCF7, primary human fibroblasts (Liu et al. 2003), and REF52 without inhibiting E2F1 expression (Fig. 1A), it failed to suppress E2F1 activity in C33A cells (Fig. 1B, •). Strikingly, reconstitution of Brg1 or Brm in C33A cells restored TopBP1-mediated repression of E2F1 (Fig. 1B, ▴). To further test the role of Brg1/Brm in other cells, we used the dominant-negative mutants of Brg1 and Brm (mutations in ATP-binding site) to inhibit the function of endogenous Brg1 or Brm in HEK293 cells, T98G human glioblastoma cells, and NIH3T3 cells, and assessed E2F1 activity in the presence of TopBP1. Indeed, these mutants significantly inhibited the capacity of TopBP1 to repress E2F1 (Fig. 1C,D, ▴). Together, these results demonstrate that the regulation of E2F1 by TopBP1 requires Brg1/Brm. It is also apparent that this mechanism of regulation is general to both nontransformed and cancer cells.
Figure 1.
Regulation of E2F1 transcriptional activity by TopBP1 requires Brg1/Brm. An E2F1-expressing plasmid was cotransfected with increasing amounts of a TopBP1 plasmid in various cell lines as indicated, and the E2F1 activity was measured with a p14ARF promoter-luciferase activity assay. Luciferase activity of transfected E2F1 was determined as fold induction relative to that of empty vector control. Each sample was performed in triplicate, and the experiments were repeated multiple times with consistent results. (A) TopBP1 represses the transcriptional activity, but not the expression, of E2F1 in REF52 cells. The activity of E2F1 in the presence of TopBP1 was determined as the percentage relative to that in the absence of TopBP1. The expressional levels of E2F1 and TopBP1 shown in the right panel were determined by immunoblotting. (B) E2F1 activity was assayed in a Brg1/Brm-deficient C33A cell line with or without reconstitution of Brg1 (left) or Brm (right). (C) E2F1 activity was assayed in HEK293 cells with or without cotransfection of dominant-negative (dn) mutants of Brg1 (left) or Brm (right). (D) E2F1 activity was assayed in T98G cells (left) or NIH3T3 cells (right) with cotransfection of a dominant-negative (dn) mutant of Brg1. (E) The regulation of E2F1 by TopBP1 is independent of Rb. E2F1 activity was determined in Saos-2 cells (left) or a pair of mouse embryonic fibroblasts (MEFs) prepared from Rb+/+ or Rb-/- sibling embryos.
Regulation of E2F1 by TopBP1 is independent of pRb
pRb is the major E2F regulator and also recruits the SWI/SNF complexes to repress E2F activity (Trouche et al. 1997; Strobeck et al. 2000). To investigate whether the regulation of E2F1 by TopBP1 was mediated through the classical Rb pathway, we examined the effect of TopBP1 on E2F1 transcriptional activity in Rb+/+ and Rb-null mouse embryonic fibroblasts (MEFs) as well as Saos-2 cells in which Rb is deficient. TopBP1 effectively repressed E2F1 activity both in Rb+/+ and Rb-/- MEFs (Fig. 1E, right panel). TopBP1 also significantly inhibited E2F1 activity in Saos-2 cells (Fig. 1E, left panel). Thus, we conclude that pRb is not involved in the TopBP1-mediated regulation of E2F1.
TopBP1 facilitates the association between E2F1 and Brg1/Brm
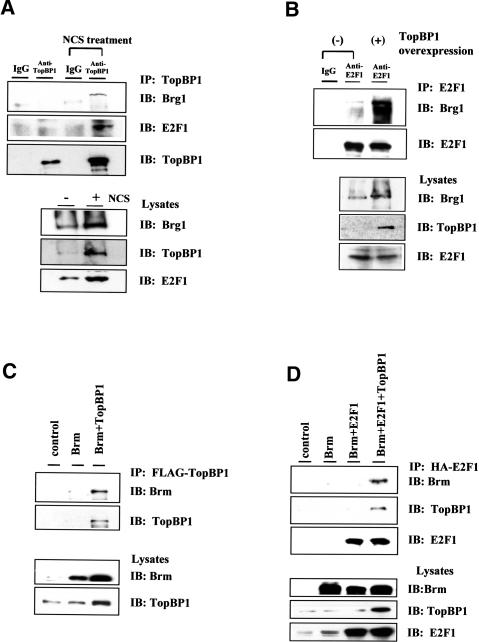
To examine whether TopBP1 could interact with Brg1/Brm and recruit the SWI/SNF complex to regulate E2F1, coimmunoprecipitation of endogenous TopBP1, Brg1, and E2F1 was performed in HEK293 cells. As shown in Figure 2A, TopBP1 interacted with both Brg1 and E2F1 in cells treated with neocarzinostatin (NCS), a radiomimetic chemical. In addition, E2F1 interacted with Brg1, which was further enhanced by overexpression of TopBP1 (Fig. 2B). We examined the interaction between Brm and TopBP1 or E2F1 as well. Similarly, transfected Brm coimmunoprecipitated with TopBP1 in HEK293 cells (Fig. 2C). In addition, Brm associated with E2F1 only in the presence of overexpressed TopBP1 (Fig. 2D). Interestingly, coexpression of TopBP1 mildly increased the expression of Brg1 or Brm (∼1.5-fold); however, the amplitude of induction in interaction between E2F1 and Brg1 (>5-fold) or Brm (>10-fold) is far greater. Taken together, the results indicate that TopBP1 interacts with Brg1/Brm, and facilitates their association with E2F1.
Figure 2.
Interaction between TopBP1, E2F1, and Brg1/Brm. (A) Interaction between endogenous TopBP1 and Brg1. HEK293 cells were untreated or treated with radio-mimetic agent neocarzinostatin (NCS) at 300 ng/mL for 3 h. Lysates were immunoprecipitated (IP) with an anti-TopBP1 antibody or a control mouse IgG antibody and immunoblotted (IB) as indicated. (Lower panel) An aliquot of the cell lysates before immunoprecipitation was analyzed by Western blot and probed with Brg1, TopBP1, and E2F1. (B) E2F1 coimmunoprecipitated with Brg1, which was further enhanced by overexpression of TopBP1. Endogenous E2F1 was immunoprecipitated with its specific antibody or a control mouse IgG antibody, and the immunoblot was probed with antibodies for Brg1 or E2F1. (C) Interaction between TopBP1 and Brm. HEK293 cells were transfected with the expression plasmids as indicated. Flag-tagged TopBP1 was then immunoprecipitated from cell lysates, and the coimmunoprecipitated Brm protein was detected by immunoblotting. (D) Interaction between E2F1 and Brm. HEK293 cells were transfected with the expression plasmids as indicated. HA-tagged E2F1 was immunoprecipitated from the cell lysates, and the coimmunoprecipitated Brm and TopBP1 were detected by immunoblotting with their specific antibodies.
Co-occupancy of TopBP1, E2F1, and Brg1/Brm on E2F1-responsive promoters
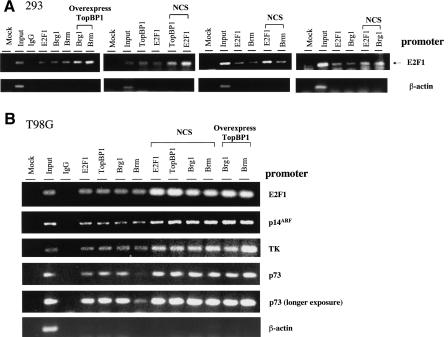
To further explore the mechanism by which TopBP1 regulates E2F1, we performed chromatin immunoprecipitation (ChIP) assay to examine the co-occupancy of TopBP1, E2F1, and Brg1/Brm on E2F1-responsive promoters. We examined the occupancy on the E2F1 promoter that is known as a direct E2F1 target (Johnson et al. 1994) in HEK293 cells. As shown in Figure 3A, E2F1 promoter sequences, but not β-actin promoter sequences, were detected in E2F1, Brg1, Brm, and TopBP1 immunoprecipitates, indicating the co-occupancy of E2F1, Brg1/Brm, and TopBP1 on the E2F1 promoter. Importantly, the occupancy of Brg1 and Brm on the E2F1 promoter was induced by overexpression of TopBP1 (Fig. 3A, left panel). Furthermore, the co-occupancy between E2F1 and TopBP1, Brm, or Brg1 was enhanced by NCS treatment (Fig. 3A, right panels). The promoter co-occupancy is not limited to the E2F1 promoter and can also be observed in T98G cells. We performed ChIP assay on several promoters of known E2F1 target genes, including E2F1, p14ARF (Bates et al. 1998), thymidine kinase (TK; Kim and Lee 1992; Tommasi and Pfeifer 1997; Ren et al. 2002), and p73 (Stiewe and Putzer 2000; Pediconi et al. 2003) in T98G cells. As shown in Figure 3B, E2F1, p14ARF, TK, and p73 promoter sequences, but not β-actin promoter sequences, were detected individually in E2F1, TopBP1, Brg1, and Brm immunoprecipitates. Thus, E2F1, TopBP1, and Brg1/Brm proteins also co-occupied on E2F1-responsive promoters in growing T98G cells. Furthermore, the co-occupancy was enhanced by NCS treatment or by overexpression of TopBP1 in T98G cells (Fig. 3B). In conjunction with the coimmunoprecipitation experiments (Fig. 2), these data strongly suggest that TopBP1 represses E2F1 activity by recruiting Brg1/Brm to interact with E2F1 on E2F1-responsive promoters. Importantly, TopBP1 also co-occupied with E2F1 in cells not treated with NCS, implying that TopBP1 regulates E2F1 even in normally growing cells.
Figure 3.
Co-occupancy of TopBP1, E2F1, and Brg1/Brm on E2F1-responsive promoters. (A) HEK293 cells were transfected with an empty vector or a TopBP1-expressing plasmid. Some empty-vector-transfected cells were treated with NCS (300 ng/mL) for 3 h. A chromatin immunoprecipitation (ChIP) assay was performed using antibodies against E2F1, TopBP1, Brm, or Brg1, respectively, as indicated. Mock immunoprecipitations correspond to control reactions lacking antibodies. The IgG lane represents an additional control reaction using normal mouse IgG for chromatin immunoprecipitation. The precipitated DNA was amplified with two primers derived from an E2F1 promoter or a β-actin promoter. The input represented 0.5% of total amount of chromatin added to each immunoprecipitation reaction. (B) T98G cells were transfected with an empty vector or a TopBP1-expressing plasmid. Some empty-vector-transfected cells were treated with NCS (300 ng/mL) for 3 h. ChIP was performed as above. The precipitated DNA was amplified with primer pairs derived from each promoter as indicated. A longer exposure of p73 promoter ChIP assay is shown to demonstrate the occupancy of Brm.
TopBP1 siRNA derepresses the transcriptional activity of E2F1, but not E2F2 or E2F3
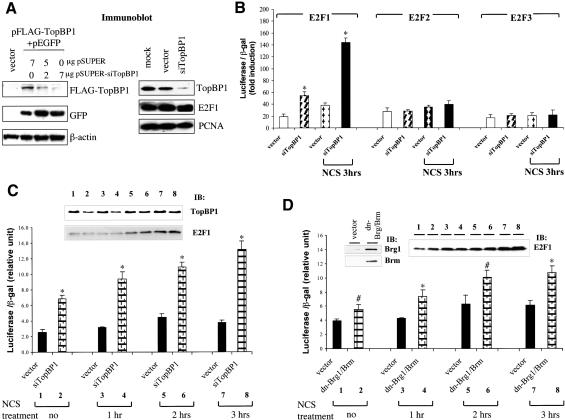
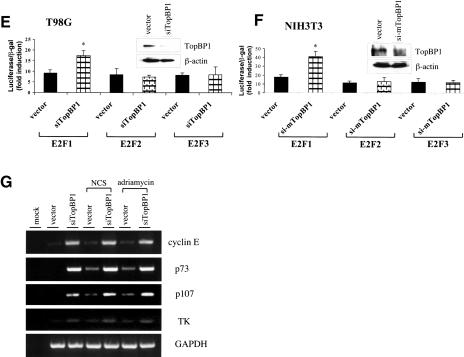
Previous studies have demonstrated that TopBP1 specifically represses E2F1 activity in an overexpression system (Liu et al. 2003). To understand the physiological function of TopBP1 in the regulation of E2F1, the expression of endogenous TopBP1 was knocked down by a TopBP1-specific short interfering RNA (siRNA) in HEK293 cells transfected with pSUPER-siTopBP1. TopBP1 siRNA significantly inhibited the expression of cotransfected Flag-TopBP1 without affecting the protein levels of coexpressed GFP (green fluorescence protein; Fig. 4A). TopBP1 siRNA also selectively inhibited the expression of endogenous TopBP1, but not E2F1, PCNA (Fig. 4A, right panel), E2F2, and E2F3 (Fig. 5A, top panel). The activities of E2F1, E2F2, and E2F3 were assayed in HEK293 cells transfected with pSUPER-siTopBP1. TopBP1 siRNA specifically increased the transcriptional activity of E2F1, but not E2F2 and E2F3 (Fig. 4B). The increased E2F1 activity observed in TopBP1 siRNA-transfected cells is not a result of increased E2F1 expression because the E2F1 protein level was not induced in siTopBP1-transfected cells (Figs. 4A,C, 5), but rather reflects derepression of E2F1 activity. The derepression of E2F1 activity by TopBP1 siRNA became more significant upon NCS treatment when E2F1 protein was induced, indicating significant regulation of E2F1 by TopBP1 during DNA damage. We further tested the effect of TopBP1 siRNA on the activity of endogenous E2F1. As shown in Figure 4C, although there was significant induction of E2F1 proteins by NCS treatment, the activity of E2F1 was only mildly elevated in the vector-transfected cells. Moreover, E2F1 activity started to decline at 3 h after NCS treatment despite continuous rise of E2F1 protein. In contrast, E2F1 activity in TopBP1 siRNA-transfected cells was derepressed and allowed to increase by NCS treatment (Fig. 4C). Interestingly, the derepression was consistently observed in cells without NCS treatment (Fig. 4B,C). This correlates with the observation of promoter co-occupancy of TopBP1 and E2F1 in normally growing cells. Thus, TopBP1 regulates E2F1 not only during DNA damage, but also in normal growth conditions. Similarly, inhibition of endogenous Brg1/Brm function by dominant-negative mutants of Brg1/Brm also derepressed the endogenous E2F1 activity (Fig. 4D), consistent with their role in the repression of E2F1 activity. We confirmed the TopBP1-mediated regulation of E2F1 in two additional cell lines, T98G and NIH3T3, in which TopBP1 knockdown also specifically derepressed the transcriptional activity of E2F1 (Fig. 4E,F). The endogenous transcripts of E2F1 target genes were also examined in TopBP1 siRNA-transfected T98G cells. TopBP1 knockdown activated the endogenous expression of E2F1 target genes, including cyclin E, p73, and p107 as well as TK in growing T98G cells and in T98G cells treated with NCS or adriamycin (Fig. 4G). These results provide strong evidence that TopBP1 regulates E2F1 transcriptional activity during normal growth as well as during DNA damage.
Figure 4.
Knockdown of TopBP1 derepresses E2F1 transcriptional activity. (A) HEK293 cells were transfected with a Flag-tagged TopBP1-expressing plasmid, increasing amounts of pSUPER-siTopBP1 and pEGFP (left panel), or with either pSUPER or pSUPER-siTopBP1 (right panel). The expression of GFP protein in each transfectant serves as a control. The expression of endogenous TopBP1, E2F1, and PCNA was analyzed by immunoblotting. (B) E2F transcriptional activities were determined by p14ARF promoter-driven luciferase assay in HEK293 cells transfected with E2F and TopBP1 siRNA expression plasmids. Cells were left untreated or treated with NCS for 3 h before analysis. (*) p < 0.001 (t test) compared with their counterpart control groups. (C) The endogenous E2F1 activity was assayed in HEK293 cells transfected with TopBP1 siRNA expression plasmids. Cells were treated with NCS for different periods of time before analysis. The expression of endogenous TopBP1 and E2F1 was analyzed by immunoblotting. (*) p < 0.001 (t test) compared with their counterpart control groups. (D) The endogenous E2F1 transcriptional activity was assayed in HEK293 cells transfected with the expression vectors of dominant-negative mutants of Brg1 and Brm. Cells were treated with NCS for different periods of time as indicated before analysis. The expression of E2F1, Brg1, and Brm in the whole-cell lysates was detected with their specific antibodies, respectively. (#) p < 0.05 (t test) compared with vector control; (*) p < 0.01 (t test) compared with vector control. (E) E2F transcriptional activity assay as described in B was performed in T98G cells in the presence of TopBP1 siRNA. The expression of endogenous TopBP1 was analyzed by immunoblotting. (*) p < 0.001 (t test) compared with their counterpart control groups. (F) E2F transcriptional activity assay as described in B was performed in NIH3T3 cells in the presence of mTopBP1 siRNA. The expression of endogenous TopBP1 was analyzed by immunoblotting. (*) p < 0.001 (t test) compared with their counterpart control groups. (G) T98G cells were transfected with an empty vector or pSUPER-siTopBP1. Two days later, cells were either left untreated or treated with NCS (300 ng/mL) or adriamycin (1 μM) for 5 h before harvesting. RNA was then extracted and RT-PCR analysis was performed using primers specific for selected E2F1 target genes or GAPDH as indicated. Mock RT-PCR represents a control reaction without addition of RNA.
Figure 5.
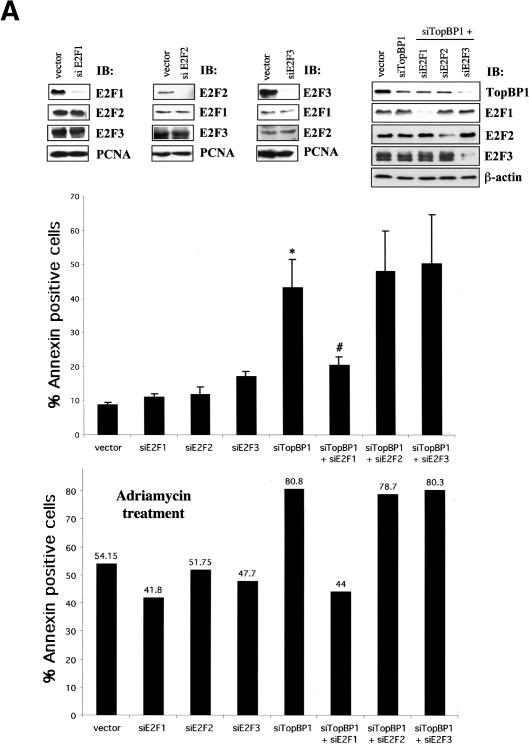
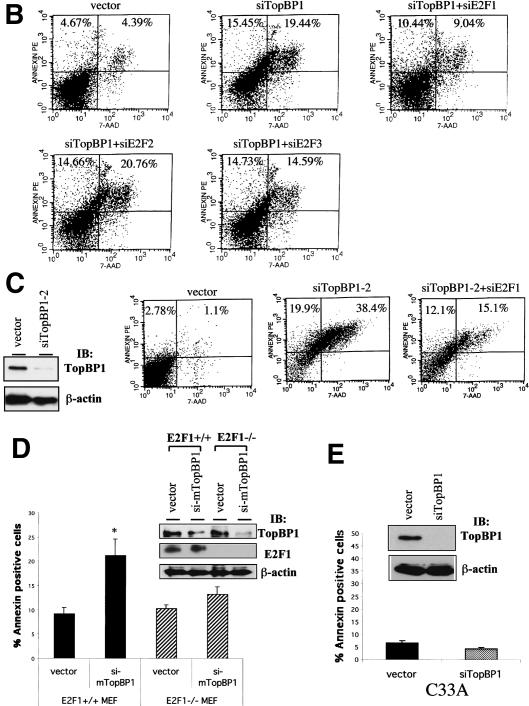
TopBP1 siRNA induces E2F1-dependent apoptosis. (A, upper panel) Immunoblotting of endogenous E2F1, E2F2, E2F3, and TopBP1 in HEK293 cells transfected with corresponding pSUPER-siRNA. The expression of PCNA and β-actin serves as a protein-loading control. (Middle panel) HEK293 cells were transfected with the plasmids expressing various siRNAs and GFP. Cells were then stained with annexin V-PE and 7-AAD. GFP-positive cells were gated and analyzed by flow cytometry. The data shown are the mean ± S.E. of three independent experiments. (Lower panel) The siRNA-transfected 293 cells were treated with 1 μM adriamycin for 6 h and analyzed as described above. (*) p < 0.05 (t test) compared with pSUPER vector group; (#) p < 0.05 (t test) compared with siTopBP1 group. (B) A representative profile of apoptosis from 293 cells expressing different siRNAs as shown above. (C) The apoptosis assay as described above was performed in HEK293 cells transfected with pSUPER-siTopBP1-2 construct. The Western blot analysis of endogenous TopBP1 is shown in the left panel. (D) Wild-type and E2F1-/- MEFs were transfected with pEGFP and the TopBP1 siRNA expression plasmids. Apoptosis was analyzed as described in A. The data shown are the mean ± S.E. of four independent experiments. (Upper panel) A representative immunoblot showing the expression of endogenous TopBP1, E2F1, and β-actin in E2F1+/+ or E2F1-/- MEFs. (*) p < 0.05(t test) compared with control group. (E) C33A cells were transfected with pEGFP and the TopBP1 siRNA expression plasmids. Apoptosis was analyzed as described in A. The data shown are the mean ± S.E. of three independent experiments. (Upper panel) An immunoblot showing the expression of endogenous TopBP1 in TopBP1-siRNA-transfected C33A cells.
To rule out the possibility that TopBP1 siRNA could induce DNA damage and, therefore, activate E2F1, the activation of several other DNA damage response proteins such as p53, Chk1, and Chk2 was also examined in TopBP1 siRNA-transfected cells. The induction or activation of p53, Chk1, and Chk2 was assayed using antibodies against p53, phospho-Ser15 p53, phospho-Ser20 p53, phospho-Ser345 Chk1, and phospho-Thr68 Chk2, respectively. Neither p53, Chk1, nor Chk2 was induced or activated in the TopBP1 siRNA-transfected cells (data not shown), indicating that no DNA damage was associated with TopBP1 siRNA transfection.
Taking these data together, we conclude that TopBP1 selectively interacts with E2F1 and represses the transcriptional activity of E2F1 under physiological conditions.
TopBP1 siRNA-induced apoptosis was rescued by E2F1 siRNA, but not by E2F2 or E2F3 siRNA
Previously it has been shown that inhibition of TopBP1 expression by TopBP1-specific antisense oligomers induces apoptosis (Yamane et al. 2002). Here, we show that TopBP1 siRNA can derepress E2F1 transcriptional activity (Fig. 4). Based on these findings, we postulated that TopBP1 might be required to inhibit E2F1-mediated apoptosis. To test this hypothesis, we further knocked down the expression of endogenous E2Fs (E2F1, E2F2, and E2F3) by individual E2F-specific siRNA and assessed their effects on TopBP1 knockdown-induced apoptosis. In this experiment, pEGFP expression vector was transiently cotransfected into HEK293 cells with pSUPER-siTopBP1 and/or each pSUPER-siE2F expressing E2F-specific siRNA. As shown in upper panel of Figure 5A, these siRNAs specifically knocked down the expression of corresponding endogenous proteins without altering the expression of other E2Fs. Apoptosis was analyzed by annexin/7-amino-actinomycin (7-AAD) staining in GFP-positive cells. Indeed, our results showed that suppression of endogenous TopBP1 expression in HEK293 cells significantly induced apoptosis; however, inhibition of endogenous E2F proteins by E2F-specific siRNA did not affect cell survival (Fig. 5A, middle panel). Remarkably, TopBP1 knockdown-induced apoptosis was greatly abrogated by E2F1 siRNA, but not E2F2 siRNA or E2F3 siRNA (Fig. 5A, middle panel), suggesting that suppression of TopBP1 expression specifically derepresses E2F1 activity and therefore induces apoptosis. Figure 5B is a representative result showing the annexin V-PE/7-AAD profiles of gated GFP-positive cells. Taken together, these results reveal a pivotal role for TopBP1 to regulate E2F1-induced apoptosis.
We also assessed the role of TopBP1 and E2F1 during DNA damage by examining adriamycin-induced apoptosis in HEK293 cells expressing TopBP1 siRNA and/or E2F siRNA. TopBP1 knockdown potentiated apoptosis during adriamycin treatment, which was again blocked by E2F1 siRNA but not E2F2 siRNA or E2F3 siRNA (Fig. 5A, lower panel). To verify the genuine effect of TopBP1 on the functional suppression of E2F1, we used another TopBP1 siRNA expression vector, pSUPER-siTopBP1-2, in similar experiments. This TopBP1 siRNA was directed against different sequences of TopBP1. It also significantly induced apoptosis. Likewise, the apoptosis was inhibited by a siRNA specific to E2F1 (Fig. 5C, right panel).
To further verify the physiological function of TopBP1 in regulating E2F1-mediated apoptosis, we assessed TopBP1 siRNA-induced apoptosis in primary mouse embryonic fibroblasts (MEFs) derived from wild-type and E2F1-null sibling embryos. Here pEGFP expression vector was transiently transfected into MEFs with either pSUPER empty vector or pSUPER-si-mTopBP1 expressing siRNA against murine TopBP1. Annexin assay was performed by flow cytometry in GFP-positive cells. Our results showed that suppression of endogenous murine TopBP1 by TopBP1 siRNA in E2F1+/+ MEFs induced apoptosis, which was significantly abrogated in E2F1-/- MEFs (Fig. 5D). It should be noted that the extent of TopBP1 siRNA-mediated knockdown of endogenous TopBP1 in MEFs would be much greater because the best transfection efficiency of MEFs we could reach was ∼50%. To test the dependency of TopBP1 siRNA-induced apoptosis on Brg1/Brm status, we performed similar experiments in C33A cells, which lack Brg1 and Brm. In contrast to HEK293 and MEFs cells, TopBP1 siRNA failed to induce apoptosis in C33A cells despite almost completely blocking the expression of TopBP1 (Fig. 5E). This result is consistent with the idea that TopBP1 requires Brg1/Brm to repress E2F1-mediated apoptosis. Taking all the data together from the different approaches shown above, we conclude that TopBP1 plays an essential role in the inhibition of E2F1-mediated apoptosis during both DNA damage response and normal cellular proliferation.
Interaction between TopBP1 and E2F1 during G1/S transition
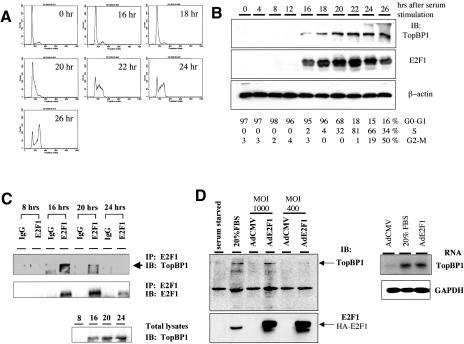
The ChIP assay (Fig. 3) showed that the co-occupancy of TopBP1 and E2F1 on the E2F1 promoter occurred during normal cellular proliferation. In addition, TopBP1 siRNA derepressed E2F1 activity, thereby enhancing E2F1-dependent apoptosis in the absence of DNA damage (Fig. 5). These results raised the hypothesis that TopBP1 might regulate E2F1 function during cell growth through the interaction with E2F1 at a specific phase of cell cycle. To answer that, we synchronized primary human foreskin fibroblasts (HFF) by serum starvation in medium containing 0.1% fetal bovine serum for 48 h, which was confirmed by propidium iodide staining of DNA (Fig. 6A). The entry of the cell cycle was then induced with 20% fetal bovine serum, and cells were harvested at different time points to analyze the cell cycle profile and the expressional levels of TopBP1 and E2F1. As shown in Figure 6B, the expression of TopBP1 was dramatically induced at G1/S transition and S phase, which is consistent with previous observation (Makiniemi et al. 2001). Interestingly, the expressional pattern of TopBP1 during the cell cycle correlates very well with that of E2F1 (Fig. 6B). We further performed coimmunoprecipitation of TopBP1 with E2F1 from cell lysates prepared at different time points. As shown in Figure 6C, TopBP1 was coimmunoprecipitated with E2F1 in cell lysates prepared at 16 h, when cells entered G1/S transition. Although the expression of both E2F1 and TopBP1 continued to increase at 20 h, when cells were in S phase, the interaction decreased significantly. This interaction was not detected in cell lysates prepared at 24 h, when cells started to enter G2/M, although both TopBP1 and E2F1 were still expressed. Taken together, our results indicate that TopBP1 associates with E2F1 during G1/S transition of cell cycle, thereby regulating E2F1 function during normal cell cycle progression. More importantly, TopBP1 inhibits E2F1 apoptosis before the initiation of DNA replication.
Figure 6.
Interaction between TopBP1 and E2F1 during G1/S transition. (A) Primary human foreskin fibroblasts (HFF) were brought to quiescence by serum starvation for 48 h and then stimulated by 20% FBS for indicated periods of time. The cells were fixed and analyzed with propidium iodide staining by flow cytometry. (B) Cell lysates from synchronized HFF were subjected to Western blot analysis, and the immunoblot was probed with antibodies specific to TopBP1, E2F1, and β-actin, respectively. (Bottom panel) The fractions of each phase of cell cycle quantitated by FACS analysis. (C) The endogenous E2F1 in the whole-cell lysates of synchronized HFF was immunoprecipitated with an anti-E2F1 antibody (KH95) or control mouse IgG and resolved by SDS-PAGE; the immunoblotting was performed as indicated. (Lower panel) An aliquot of lysates was subjected to Western blot analysis. The immunoblot was probed with a TopBP1 antibody. (D) TopBP1 is an E2F target. HFF cells were serum-starved for 48 h and then infected with an adenovirus expressing HA-E2F1 (AdE2F1) or that harboring a CMV empty vector (AdCMV) at a multiplicity of infection of 400 and 1000. Some AdCMV-infected cells were stimulated with 20% serum, and the others remained in 0.1% serum. The cells were harvested at 21 h after infection. (Left panel) The expression of TopBP1 and E2F1 in the whole-cell lysates was detected with their specific antibodies. (Right panel) Poly(A) RNA was prepared from infected HFF, and Northern blot analysis was carried out with a TopBP1 cDNA probe or GAPDH probe.
TopBP1 is an E2F target
Many proteins induced in S phase are known to be E2F target genes. Because the induction of TopBP1 expression coincided with the surge of E2F1 levels during G1/S transition (Fig. 6B), we speculated that the expression of TopBP1 might be induced by E2F. Interestingly, we found that the human TopBP1 gene derived from the Celera human genome database contains potential E2F-binding elements (consensus sequence TTTc/gGCGCc/g) arranged in an overlapping fashion (TTTCGCGCCAAC; overlapping E2F-binding sites are indicated by underlines and italics, respectively) at its 5′-flanking region. This E2F site is followed by two GC boxes. The organization is very similar to the promoters of several E2F target genes. To test whether E2F could transactivate TopBP1, E2F1 was overexpressed in serum-starved HFF cells by infection with E2F1 adenovirus (AdE2F1). The expressional levels of TopBP1 protein and RNA were analyzed by immunoblotting and Northern blotting, respectively. Our results showed that overexpression of E2F1, indeed, induced the expression of TopBP1 protein (Fig. 6D, left panel) and RNA (Fig. 6D, right panel) in the absence of serum. These results strongly suggest that TopBP1 is an E2F target during G1/S transition and S phase.
Discussion
Many lines of evidence have indicated that E2F activity is crucial for the G1/S transition and DNA replication, whereas the pRb is the primary negative regulator of E2F. More recently, several new functions of E2F have been discovered, particularly in DNA damage and repair checkpoint. The distinct roles for each E2F family member are also appreciated. Specifically, the role of E2F1 protein as a signal for apoptosis and the inducer of p73 (Irwin et al. 2000; Lissy et al. 2000; Pediconi et al. 2003), p53 (Kowalik et al. 1998), caspase (Nahle et al. 2002), and Apaf1 (Moroni et al. 2001; Furukawa et al. 2002) has defined a pathway that links Rb/E2F cell cycle control with apoptosis. Therefore, new important questions need to be answered regarding the differential regulation in controlling different E2F activities, particularly E2F1-mediated apoptosis. The current work is aimed at addressing this issue. We have identified a novel regulatory mechanism specifically controlling E2F1-dependent apoptosis during normal growth and DNA damage. Through the interaction between a BRCT motif of TopBP1 and the N terminus of E2F1, TopBP1 selectively regulates E2F1 activity by recruiting Brg1/Brm to E2F1-responsive promoters and suppresses E2F1-mediated apoptosis.
pRb-independent regulation of E2F1 function by TopBP1
E2F family members, E2F1-E2F5, share the C-terminal pocket-protein binding domain that binds to the pRb family. Thus, a regulation specific to E2F1 might involve an interaction that is unique to E2F1. Although E2Fs contain several fairly conserved domains, the sequences in the N terminus of E2F are quite diverse. In addition, the N terminus of E2F1 is uniquely phosphorylated by ATM/ATR (Lin et al. 2001). Therefore, the interaction mediated by this domain is likely specific to E2F1.
Using Rb-deficient cells (Saos-2 and Rb-/- MEFs), we demonstrate that TopBP1-mediated regulation of E2F1 does not require pRb. It is worth noting that the inhibition of E2F1 activity by TopBP1 is rather specific and is not due to a general transcriptional repression. TopBP1 does not inhibit the activity of E2F2, E2F3, E2F4, or several E2F1 mutants that lose the TopBP1-binding capacity (Liu et al. 2003). Moreover, expression of TopBP1 in serum-starved REF52 by AdTopBP1 infection inhibits E2F1-stimulated, but not serum-stimulated BrdU incorporation (Liu et al. 2003). Thus, TopBP1 is not a general cell cycle regulator like the pRb family, but rather an E2F1-specific modulator. Several features distinguishing this newly identified regulation from classical Rb/E2F regulation are: (1) TopBP1 binds specifically to E2F1 and only controls E2F1; pRb binds and controls E2F1, E2F2, E2F3, and E2F4. (2) TopBP1 binds to the N terminus of E2F1 through a BRCT domain; pRb binds to the C terminus of E2F through the pocket domain. (3) E2F1 binding to TopBP1 is induced by ATM-dependent phosphorylation of E2F1; E2F binding to pRb is inhibited by phosphorylation of pRb. (4) TopBP1 recruits E2F1 to nuclear foci during DNA damage (Liu et al. 2003).
Recently, pRb was reported to contain a second E2F-binding domain in the C terminus (Dick and Dyson 2003). Unlike the previously identified E2F-binding site that interacts with E2F1-E2F4, this second domain is specific for E2F1. A mutant pRb, which is defective in general E2F binding but retains the E2F1-specific binding domain, can inhibit E2F1-induced apoptosis. Interestingly, in contrast to the DNA damage-induced interaction between TopBP1 and E2F1, binding of E2F1 to the E2F1-specific domain of pRb is greatly diminished following etoposide treatment. This further manifests distinct features between TopBP1/E2F1 and Rb/E2F1 control. The potential interaction between these two modes of control for E2F1 is very intriguing. We speculate that TopBP1 binds to E2F1 and prevents it from further binding to pRb during DNA damage.
The mechanism by which TopBP1 regulates E2F1 transcriptional activity involves chromatin remodeling
SWI/SNF was originally identified in yeast as a set of positive regulators of the HO gene (mating type switch, SWI) and the SUC2 gene (sucrose nonfermenting, SNF; for review, see Winston and Carlson 1992). The SWI/SNF complex consists of 10-12 proteins that form a 2-MD complex. This complex alters nucleosome structure by disrupting histone-DNA interactions in an ATP-dependent manner. The mammalian SWI/SNF complex contains either Brg1 or Brm as its central subunit. Here, we demonstrate that the mechanism by which TopBP1 represses E2F1 is through recruiting Brg1/Brm. We provide evidence to support this assertion: (1) TopBP1-mediated repression of E2F1 is defective in a Brg1/Brm-deficient cell line, C33A. The repression is restored by reconstitution with Brg1/Brm. (2) Dominant-negative mutants of Brg1 or Brm inhibit TopBP1 to repress E2F1. (3) TopBP1 interacts with Brg1/Brm and facilitates the interaction between E2F1 and Brg1/Brm. (4) TopBP1 recruits Brg1/Brm to E2F1-responsive promoters. (5) Dominant-negative mutants of Brg1/Brm derepress E2F1 activity during DNA damage. (6) Whereas TopBP1 siRNA induces E2F1-dependent apoptosis in HEK293 cells and wild-type MEFs, it fails to induce apoptosis in Brg1/Brm-deficient C33A cells.
Another E2F regulator that depends on Brg1/Brm is prohibitin, a potential tumor suppressor in breast cancer (Wang et al. 2002). However, prohibitin interacts with the conserved marked-box region of E2F1-E2F5 (Wang et al. 1999b) as well as all members of the pRb family, and inhibits the transcriptional activities of all E2F1-E2F5 (Wang et al. 1999a). Moreover, prohibitin-mediated transcriptional repression requires pRb (Wang et al. 2002). Although Brg1/Brm appears to be a general executor that represses E2F activity, TopBP1 recruits Brg1/Brm to regulate exclusively E2F1 because of the selective interaction between TopBP1 and E2F1, contrary to the general repression of E2F by the pRb family or prohibitin.
A critical role for TopBP1 in the control of E2F1-induced apoptosis during normal cell growth and DNA damage
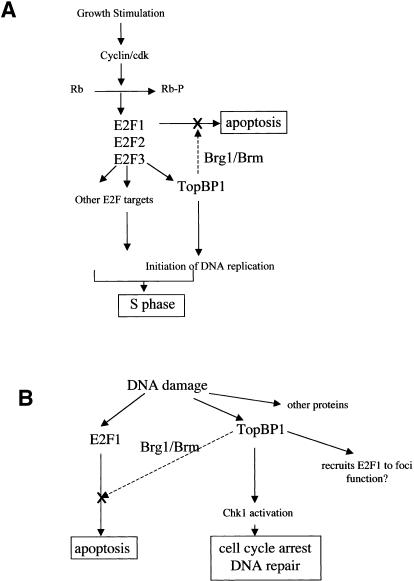
TopBP1 is induced upon DNA damage and is involved in DNA damage checkpoint, but also appears to play a very important role in the initiation of DNA replication. TopBP1 is required for DNA replication in an in vitro HeLa nuclei replication system (Makiniemi et al. 2001). The Xenopus homolog of TopBP1, Xmus101 or Xenopus Cut5, is required for the loading of Cdc45 and DNA polymerases α and ε onto replication origins and is essential for DNA replication in Xenopus egg extracts (Van Hatten et al. 2002; Hashimoto and Takisawa 2003). Thus, TopBP1 appears to be directly involved in controlling replication initiation. In this regard, its role in repressing E2F1-mediated apoptosis at the G1/S transition would be crucial to ensure suppression of apoptosis before initiation of DNA replication. Indeed, the regulation between TopBP1 and E2F1 forms a feedback control during G1/S transition. Like other proteins that are directly involved in DNA replication, TopBP1 is induced by E2F during G1/S transition. Therefore, TopBP1 acts as a critical coordinator to enforce the execution of S phase. On one hand, it promotes replication firing; on the other hand, it inhibits apoptosis (Fig. 7A). The selectivity of TopBP1 toward E2F1, but not E2F2 or E2F3, would allow E2F2 and E2F3 to function (and induce S-phase entry) while E2F1 apoptosis is inhibited.
Figure 7.
A model for the role of TopBP1 in cell survival during G1/S transition and DNA damage. (A) TopBP1 is induced during G1/S transition and participates in the initiation of DNA replication. It is also required to inhibit E2F1 activity and prevent E2F1-mediated apoptosis during S-phase entry. (B) TopBP1 recruits checkpoint activation machinery to activate Chk1 during DNA damage. It also inhibits E2F1-mediated apoptosis to allow completion of DNA repair.
Both TopBP1 and BRCA1 control the G2-M cell cycle checkpoint and are required for activation of Chk1 in response to DNA damage (Yamane et al. 2003). The Xenopus homolog of TopBP1 is also required for genotoxin-induced Chk1 activation and is essential for the binding of ATR, Rad1, and DNA polymerase α to damaged chromatin (Parrilla-Castellar and Karnitz 2003). The loading of these proteins to the stalled replication forks, in turn, mediates activation of Chk1 and cell cycle arrest. Our studies uncover an additional role of TopBP1 for cell survival during DNA damage. Thus, TopBP1 functions as a central modulator during DNA damage to ensure the execution of damage repair. On one hand, it recruits key elements of the checkpoint signaling machinery for cell cycle arrest and damage repair; on the other hand, it inhibits E2F1-mediated apoptosis to allow completion of repair (Fig. 7B). Interestingly, TopBP1 also recruits E2F1 to the BRCA1-containing nuclear foci during DNA damage (Liu et al. 2003). The function of E2F1 in these foci remains to be investigated.
The present study defines a novel mechanism that controls E2F1-mediated apoptosis by TopBP1 during G1/S transition of the cell cycle, which may bring new insights for cancer therapy. Because TopBP1 siRNA-induced apoptosis is due to derepression of E2F1 activity, cells harboring higher E2F1 levels would be more susceptible to inhibitors of TopBP1. Most cancer cells contain excessive E2F1 activities because of deregulation in the Rb pathway. Thus, TopBP1 is a potential therapeutic target to harness E2F1 for cancer treatment.
Materials and methods
Cell culture and transfection
C33A, HEK293, T98G, Saos-2, NIH3T3, MEFs, REF52, and HFF (human foreskin fibroblast) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Primary MEFs were isolated from 13.5-d-old embryos by standard methods. A standard calcium phosphate method was used for transfection of C33A cells. HEK293 cells were transfected with calcium phosphate method or the Gene Pulser Xcell electroporation system (Bio-Rad) according to the manufacturer's instructions for 293 cells. NIH3T3 cells were transfected with the Gene Pulser Xcell electroporation system. Saos-2, T98G, MEFs (Rb+/+ and Rb-/-), and REF52 cells were transfected using Lipofectamine 2000 (Invitrogen).
Luciferase assay
The expression constructs (2 μg for pcDNA3-HA-E2F1, pcDNA3-HA-E2F2, or pcDNA3-HA-E2F3; 2 or 10 μg for pcDNA3-TopBP1; 10 μg for Brg1, Brm, and their dominant-negative mutants expression plasmids; 10 μg for pSUPER constructs expressing siRNA of TopBP1, E2F1, E2F2, and E2F3), the promoter plasmids (1 μg for p14 ARF-Luc), and 1 μg of β-galactosidase plasmid were transfected into HEK293 cells. Cells were harvested 2 d later. An aliquot of cells was lysed in SDS lysis buffer for Western blot analysis in some experiments; the other cell extracts were lysed in reporter lysis buffer (Promega) for luciferase activity and β-galactosidase activity as described (Liu et al. 2003). The luciferase activity was normalized against the β-galactosidase activity. All transient expressions in this assay were carried out in triplicate.
Immunoprecipitation and Western blot analysis
The transfected cells were harvested 48 h later in TNN buffer, and immunoprecipitation was carried out as described (Liu et al. 2003). The specific signals were detected with appropriate antibodies. The antibodies specific to E2F1 (C20 and KH95), E2F2 (C20), E2F3 (C18 and N20), Brg1 (G7), Brm (E1), Rb (C15), and PCNA-(PC10) were purchased from Santa Cruz, Inc. The E2F1 (KH20) and β-actin antibodies were purchased from Sigma. The anti-GFP antibody was purchased from Clontech. The monoclonal and polyclonal TopBP1 antibodies were purchased from BD Transduction Laboratories and Bethyl Laboratories, respectively.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed following the protocol as described previously (Takahashi et al. 2000). Briefly, 293 cells or T98G cells grown in 15-cm2 dishes were untreated or treated with NCS for 3 h, and then cross-linked with formaldehyde. Cells were collected, and chromatin was extracted and sonicated; 0.5% of supernatants was used as input control PCR; the other chromatin was precleaned with protein G plus/protein A-agarose beads, and then immunoprecipitated with 4 μg of each antibody (E2F1, KH95; TopBP1, BD Transduction Laboratories; Brg1, G7; Brm, E1). The antibody-bound complexes were recovered on protein A/G beads. Immunoprecipitates were washed under stringent conditions, and cross-links of chromatin were reversed by incubating samples overnight at 65°C. The resulting DNA was purified and analyzed by PCR. The following pairs of primers were used for each indicated promoter as follows: E2F1, 5′-AGGAACCGCCGCCGTTGTTCCCGT-3′ and 5′-CTGCCTGCAAAGTCCCGGCCACTT-3′, size 124 bp; p14ARF, 5′-CCAGGCGTCCGGCCCCTGGGCCGT-3′ and 5′-CACGCGGGAAGGGCTGCCGGAGGC-3′, size 184 bp; TK, 5′-TCCCGGATTCCTCCCACGAG-3′ and 5′-TGCGCCTCCGGGAAGTTCAC-3′, size 200 bp; p73, 5′-CTCTGCCGAAGATCGCGGTCGG-3′ and 5′-GGCCGCGTCCAAGTCGGGGTCC-3′, size 170 bp; and β-actin, 5′-ACGCCAAAACTCTCCCTCCTCCTC-3′ and 5′-CATAAAAGGCAACTTTCGGAACGGC-3′, size 166 bp.
RT-PCR
T98G cells were transfected with pSUPER or pSUPER-siTopBP1. Cells were treated with NCS or adriamycin for 5 h, and RNA was extracted using TRIzol reagent. RT-PCR was performed using the following primer pairs: cyclin E, 5′-CTCCAGGAAGAGGAAGGCAA-3′ and 5′-GTAAAAGGTCTCCCTGTGAAG-3′, size 421 bp; p73, 5′-TTTAACAGGATTGGGGTGTC-3′ and 5′-CGTGAACTCCTCCTTGATGG-3′, size 405 bp; p107, 5′-TGGTGTCGCAAATGATGCTGG-3′ and 5′-AGGAGCTGATCCAAATGCCTG-3′, size 362 bp; TK, 5′-ATGAGCTGCATTAACCTGCCCACT-3′ and 5′-ATGTGTGCAGAAGCTGCTGC-3′, size 204 bp; and GAPDH, 5′-TGAAGGTCGGAGTCAACGGATTTGGT-3′ and 5′-AAATGAGCCCCAGCCTTCTCCATG-3′, size 325 bp. We ensured linear amplification in all cases.
Plasmid construction
The pSUPER vector and its derivatives expressing siRNA of human TopBP1, mouse TopBP1, human E2F1, E2F2, and E2F3 were constructed according to instructions (Brummelkamp et al. 2002). The 19-nt target sequences for siTopBP1 are 5′-TGATGGGCGGACGAGTATA-3′; for siTopBP1-2, 5′-CCTGAAGAAACCTATTTTG-3′; for si-mTopBP1 targeting murine TopBP1, 5′-TGATGGGTGGACGAGTATA-3′; for siE2F1, 5′-TATCTGTACTACGCAGCTG-3′; for siE2F2, 5′-GACTCGGTATGACACTTCG-3′; and for siE2F3, 5′-CGTCCAATGGATGGGCTGC-3′.
Flow cytometry
HEK293 or primary MEFs (E2F1+/+ and E2F1-/-, passage 3) were transfected with 2 μg of pcDNA3-GFP, and 20 μg of pSUPER vector, or siRNA constructs by either a calcium phosphate method or electroporation system. Cells were untreated or treated with adriamycin (1 μM) for 6 h before harvest. To assay apoptosis, cells were stained with annexin V-PE (Pharmigen) and 7-amino-actinomycin (7-AAD; Pharmingen). At least 5000 GFP-positive cells were gated for each sample, and the annexin/7-AAD profile of these cells was analyzed by flow cytometry. Annexin+, 7-AAD-, and annexin+, 7-AAD+ cells were scored as apoptotic. The DNA content profiles of the synchronized HFF were analyzed by propidium iodide staining followed by flow cytometry as described (Liu et al. 2003).
Adenovirus infection
HFF cells were starved in 0.1% FBS for 2 d and then infected with AdCMV or AdE2F1, which expresses HA-tagged E2F1 (Liu et al. 2003). Cells were harvested at 21 h after infection for either Western or Northern blot analysis. Northern blotting was carried out as described (Lin et al. 2001).
Acknowledgments
We thank Dr. Weidong Wang for providing Brg1-expressing plasmids, Dr. Christian Muchardt for Brm-expressing plasmids, and Dr. William A. May for pSUPER plasmid. We thank Marion Spell in the UAB Flow Cytometry Core facility for the FACS analyses. The work was supported by grants from General Motors Cancer Research Scholar Award (to W.C.L.), NIH/NCI K12 CA 7693705 (W.C.L.), UAB Avon/Breast Cancer SPORE and Howard Hughes Medical Institute Career Development Award (to W.C.L. and F.T.L.), and grant CA100848 from the National Institutes of Health (to F.-T.L.). All authors disclose that they have no financial interests that will pose a conflict of interest regarding the submitted article.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1180204.
References
- Bates S., Phillips, A.C., Clark, P.A., Stott, F., Peters, G., Ludwig, R.L., and Vousden, K.H. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395: 124-125. [DOI] [PubMed] [Google Scholar]
- Boner W., Taylor, E.R., Tsirimonaki, E., Yamane, K., Campo, M.S., and Morgan, I.M. 2002. A functional interaction between the HPV16 transcription/replication factor E2 and the DNA damage response protein TopBP1. J. Biol. Chem. 4: 4. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska, E.A., McCance, D.J., Reid, J.L., Bannister, A.J., and Kouzarides, T. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391: 597-601. [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards, R., and Agami, R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550-553. [DOI] [PubMed] [Google Scholar]
- Cam H. and Dynlacht, B.D. 2003. Emerging roles for E2F: Beyond the G1/S transition and DNA replication. Cancer Cell 3: 311-316. [DOI] [PubMed] [Google Scholar]
- DeGregori J., Leone, G., Miron, A., Jakoi, L., and Nevins, J.R. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. 94: 7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick F.A. and Dyson, N. 2003. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol. Cell 12: 639-649. [DOI] [PubMed] [Google Scholar]
- Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes & Dev. 12: 2245-2262. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G. 1992. Chromatin as an essential part of the transcriptional mechanism. Nature 355: 219-224. [DOI] [PubMed] [Google Scholar]
- Field S.J., Tsai, F.Y., Kuo, F., Zubiaga, A.M., Kaelin Jr., W.G., Livingston, D.M., Orkin, S.H., and Greenberg, M.E. 1996. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85: 549-561. [DOI] [PubMed] [Google Scholar]
- Furukawa Y., Nishimura, N., Satoh, M., Endo, H., Iwase, S., Yamada, H., Matsuda, M., Kano, Y., and Nakamura, M. 2002. Apaf-1 is a mediator of E2F-1-induced apoptosis. J. Biol. Chem. 277: 39760-39768. [DOI] [PubMed] [Google Scholar]
- Hallstrom T.C. and Nevins, J.R. 2003. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc. Natl. Acad. Sci. 100: 10848-10853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour J.W. and Dean, D.C. 2000. The Rb/E2F pathway: Expanding roles and emerging paradigms. Genes & Dev. 14: 2393-2409. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y. and Takisawa, H. 2003. Xenopus Cut5 is essential for a CDK-dependent process in the initiation of DNA replication. EMBO J. 22: 2526-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S., Wanzel, M., Beuger, V., Frohme, C., Beul, D., Hillukkala, T., Syvaoja, J., Saluz, H.P., Haenel, F., and Eilers, M. 2002. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol. Cell 10: 509-521. [DOI] [PubMed] [Google Scholar]
- Irwin M., Marin, M.C., Phillips, A.C., Seelan, R.S., Smith, D.I., Liu, W., Flores, E.R., Tsai, K.Y., Jacks, T., Vousden, K.H., et al. 2000. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature 407: 645-648. [DOI] [PubMed] [Google Scholar]
- Johnson D.G., Ohtani, K., and Nevins, J.R. 1994. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes & Dev. 8: 1514-1525. [DOI] [PubMed] [Google Scholar]
- Kim Y.K. and Lee, A.S. 1992. Identification of a protein-binding site in the promoter of the human thymidine kinase gene required for the G1-S-regulated transcription. J. Biol. Chem. 267: 2723-2727. [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar, G.J. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes & Dev. 13: 2339-2352. [DOI] [PubMed] [Google Scholar]
- Kowalik T.F., DeGregori, J., Leone, G., Jakoi, L., and Nevins, J.R. 1998. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 9: 113-118. [PubMed] [Google Scholar]
- Leone G., DeGregori, J., Yan, Z., Jakoi, L., Ishida, S., Williams, R.S., and Nevins, J.R. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes & Dev. 12: 2120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W.C., Lin, F.T., and Nevins, J.R. 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes & Dev. 15: 1833-1844. [PMC free article] [PubMed] [Google Scholar]
- Lissy N.A., Davis, P.K., Irwin, M., Kaelin, W.G., and Dowdy, S.F. 2000. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature 407: 642-645. [DOI] [PubMed] [Google Scholar]
- Liu K., Lin, F.T., Ruppert, J.M., and Lin, W.C. 2003. Regulation of E2F1 by BRCT-domain containing protein TopBP1. Mol. Cell. Biol. 23: 3287-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R.X., Postigo, A.A., and Dean, D.C. 1998. Rb interacts with histone deacetylase to repress transcription. Cell 92: 463-473. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman, R., Naguibneva, I., Robin, P., Lorain, S., Le Villain, J.P., Troalen, F., Trouche, D., and Harel-Bellan, A. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391: 601-605. [DOI] [PubMed] [Google Scholar]
- Makiniemi M., Hillukkala, T., Tuusa, J., Reini, K., Vaara, M., Huang, D., Pospiech, H., Majuri, I., Westerling, T., Makela, T.P., et al. 2001. BRCT domain-containing protein TopBP1 functions in DNA replication and damage response. J. Biol. Chem. 276: 30399-30406. [DOI] [PubMed] [Google Scholar]
- Manke I.A., Lowery, D.M., Nguyen, A., and Yaffe, M.B. 2003. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science 302: 636-639. [DOI] [PubMed] [Google Scholar]
- Moroni M.C., Hickman, E.S., Denchi, E.L., Caprara, G., Colli, E., Cecconi, F., Muller, H., and Helin, K. 2001. Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3: 552-558. [DOI] [PubMed] [Google Scholar]
- Muchardt C. and Yaniv, M. 2001. When the SWI/SNF complex remodels... the cell cycle. Oncogene 20: 3067-3075. [DOI] [PubMed] [Google Scholar]
- Nahle Z., Polakoff, J., Davuluri, R.V., McCurrach, M.E., Jacobson, M.D., Narita, M., Zhang, M.Q., Lazebnik, Y., Bar-Sagi, D., and Lowe, S.W. 2002. Direct coupling of the cell cycle and cell death machinery by E2F. Nat. Cell Biol. 4: 859-864. [DOI] [PubMed] [Google Scholar]
- Nevins J.R. 1998. Toward an understanding of the functional complexity of the E2F and retinoblastoma families. Cell Growth Differ. 9: 585-593. [PubMed] [Google Scholar]
- Pan H., Yin, C., Dyson, N.J., Harlow, E., Yamasaki, L., and Van Dyke, T. 1998. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol. Cell 2: 283-292. [DOI] [PubMed] [Google Scholar]
- Parrilla-Castellar E.R. and Karnitz, L.M. 2003. Cut5 is required for the binding of Atr and DNA polymerase α to genotoxin-damaged chromatin. J. Biol. Chem. 278: 45507-45511. [DOI] [PubMed] [Google Scholar]
- Pediconi N., Ianari, A., Costanzo, A., Belloni, L., Gallo, R., Cimino, L., Porcellini, A., Screpanti, I., Balsano, C., Alesse, E., et al. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 5: 552-558. [DOI] [PubMed] [Google Scholar]
- Ren B., Cam, H., Takahashi, Y., Volkert, T., Terragni, J., Young, R.A., and Dynlacht, B.D. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes & Dev. 16: 245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez M., Yu, X., Chen, J., and Songyang, Z. 2003. Phosphopeptide binding specificities of BRCA1 COOH-terminal (BRCT) domains. J. Biol. Chem. 278: 52914-52918. [DOI] [PubMed] [Google Scholar]
- Stevens C., Smith, L., and La Thangue, N.B. 2003. Chk2 activates E2F-1 in response to DNA damage. Nat. Cell Biol. 5: 401-409. [DOI] [PubMed] [Google Scholar]
- Stiewe T. and Putzer, B.M. 2000. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 26: 464-469. [DOI] [PubMed] [Google Scholar]
- Strobeck M.W., Knudsen, K.E., Fribourg, A.F., DeCristofaro, M.F., Weissman, B.E., Imbalzano, A.N., and Knudsen, E.S. 2000. BRG-1 is required for RB-mediated cell cycle arrest. Proc. Natl. Acad. Sci. 97: 7748-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Rayman, J.B., and Dynlacht, B.D. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: Distinct E2F proteins mediate activation and repression. Genes & Dev. 14: 804-816. [PMC free article] [PubMed] [Google Scholar]
- Tommasi S. and Pfeifer, G.P. 1997. Constitutive protection of E2F recognition sequences in the human thymidine kinase promoter during cell cycle progression. J. Biol. Chem. 272: 30483-30490. [DOI] [PubMed] [Google Scholar]
- Trimarchi J.M. and Lees, J.A. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3: 11-20. [DOI] [PubMed] [Google Scholar]
- Trouche D., Le Chalony, C., Muchardt, C., Yaniv, M., and Kouzarides, T. 1997. RB and hbrm cooperate to repress the activation functions of E2F1. Proc. Natl. Acad. Sci. 94: 11268-11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hatten R.A., Tutter, A.V., Holway, A.H., Khederian, A.M., Walter, J.C., and Michael, W.M. 2002. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J. Cell Biol. 159: 541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Nath, N., Adlam, M., and Chellappan, S. 1999a. Prohibitin, a potential tumor suppressor, interacts with RB and regulates E2F function. Oncogene 18: 3501-3510. [DOI] [PubMed] [Google Scholar]
- Wang S., Nath, N., Fusaro, G., and Chellappan, S. 1999b. Rb and prohibitin target distinct regions of E2F1 for repression and respond to different upstream signals. Mol. Cell Biol. 19: 7447-7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhang, B., and Faller, D.V. 2002. Prohibitin requires Brg-1 and Brm for the repression of E2F and cell growth. EMBO J. 21: 3019-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F. and Carlson, M. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8: 387-391. [DOI] [PubMed] [Google Scholar]
- Yamane K., Wu, X., and Chen, J. 2002. A DNA damage-regulated BRCT-containing protein, TopBP1, is required for cell survival. Mol. Cell. Biol. 22: 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Chen, J., and Kinsella, T.J. 2003. Both DNA topoisomerase II-binding protein 1 and BRCA1 regulate the G2-M cell cycle checkpoint. Cancer Res. 63: 3049-3053. [PubMed] [Google Scholar]
- Yamasaki L., Jacks, T., Bronson, R., Goillot, E., Harlow, E., and Dyson, N.J. 1996. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85: 537-548. [DOI] [PubMed] [Google Scholar]
- Yamasaki L., Bronson, R., Williams, B.O., Dyson, N.J., Harlow, E., and Jacks, T. 1998. Loss of E2F-1 reduces tumorigenesis and extends the lifespan of Rb1+/- mice. Nat. Genet. 18: 360-364. [DOI] [PubMed] [Google Scholar]
- Yu X., Chini, C.C., He, M., Mer, G., and Chen, J. 2003. The BRCT domain is a phospho-protein binding domain. Science 302: 639-642. [DOI] [PubMed] [Google Scholar]
- Zhu J.W., DeRyckere, D., Li, F.X., Wan, Y.Y., and DeGregori, J. 1999. A role for E2F1 in the induction of ARF, p53, and apoptosis during thymic negative selection. Cell Growth Differ. 10: 829-838. [PubMed] [Google Scholar]