Abstract
Mutant analysis has been tremendously successful in deciphering the genetics of plant development. However, less is known about the molecular basis of morphological variation within species, which is caused by naturally occurring alleles. In this study, we succeeded in isolating a novel regulator of root growth by exploiting natural genetic variation in the model plant Arabidopsis. Quantitative trait locus analysis of a cross between isogenized accessions revealed that a single locus is responsible for ∼80% of the variance of the observed difference in root length. This gene, named BREVIS RADIX (BRX), controls the extent of cell proliferation and elongation in the growth zone of the root tip. We isolated BRX by positional cloning. BRX is a member of a small group of highly conserved genes, the BRX gene family, which is only found in multicellular plants. Analyses of Arabidopsis single and double mutants suggest that BRX is the only gene of this family with a role in root development. The BRX protein is nuclear localized and activates transcription in a heterologous yeast system, indicating that BRX family proteins represent a novel class of transcription factors. Thus, we have identified a novel regulatory factor controlling quantitative aspects of root growth.
Keywords: Arabidopsis, BRX, root growth, root meristem, natural variation, gene family
The last several years have witnessed tremendous advances in the genetic analysis of plant development, thanks to the rigorous application of mutagenesis approaches. However, much less is known about the molecular basis for the variation observed within species. This variation is the result of natural genetic heterogeneity, which is the result of selection pressures that are created by environmental conditions. For sessile terrestrial plants, adaptation to local conditions is especially important and has been observed on a temporally and geographically very small scale (Linhart and Grant 1996). Such natural variation can be exploited to isolate novel genes or alleles involved in plant physiology and development, for instance by analysis of isogenized accessions of the model plant Arabidopsis thaliana (Alonso-Blanco and Koornneef 2000). This approach has been successful in isolating both novel genes (Johanson et al. 2000) and novel alleles of known genes (El-Din El-Assal et al. 2001; Maloof et al. 2001). A distinct advantage of exploiting natural genetic variation is its ability to detect alleles that have been subjected to selection in the wild. This approach, in essence, counter-selects against alleles that are detrimental to plant survival and can thus complement the more common mutagenesis approaches, which often target genes that are essential for the trait of interest. Here we have exploited natural variation in Arabidopsis to isolate a novel regulator of root growth.
The root system plays a pivotal role in the survival of higher plants. Roots provide the plant with physical support as well as essential nutrients and water, which they take up from the soil. Arabidopsis thaliana is a dicotyledonous plant and has a typical allorhiz root system. Initially, growth is restricted to a primary root, which is formed during embryogenesis. Later in development, the root system expands by forming lateral roots, which originate from the pericycle, an inner cell layer of the primary root. Eventually, adventitious roots might also be formed at the hypocotyls-root junction. At the cellular level, Arabidopsis roots have a simple organization, consisting of concentric layers of epidermis, cortex, and endodermis, surrounding the stele that contains the vascular tissues (Dolan et al. 1993). These tissue layers are formed through the action of a growth zone at the distal tip of the root, the apical root meristem. Within this meristem, signals emanating from a quiescent center of slowly dividing cells organize a region of stem cells, which give rise to the cell files of the tissue layers by stereotypic divisions in a reiterative fashion (van den Berg et al. 1997; Sabatini et al. 2003). The daughter cells continue to divide several times in the distal meristematic zone before entering a zone of rapid cell elongation and differentiating to maturity.
Genetic analysis has provided evidence that plant hormone signaling pathways are fundamentally important for root development. An intact auxin signaling pathway, for example, is required for proper root growth (Davies 1995; Sabatini et al. 1999), a growth-promoting effect that is mediated via signaling through another plant hormone, gibberellic acid (Fu and Harberd 2003). In addition, root patterning requires correctly localized peaks of auxin concentration gradients (Sabatini et al. 1999) as well as the action of two transcription factors, SCARECROW (SCR) and SHORT ROOT (SHR). The latter are needed for the asymmetric division of initials that give rise to the cortex and endodermis cell layers, as well as for the differentiation of these tissues (Di Laurenzio et al. 1996; Helariutta et al. 2000; Nakajima et al. 2001). Interestingly, SCR and SHR also have a fundamental role in the maintenance of the quiescent center and, thereby, the stem cell population (Sabatini et al. 2003).
The ontogenesis of the root system is highly plastic and sensitive to changes in environmental conditions. In particular, the availability of rate-limiting nutrients for plant growth, such as phosphate and nitrate, results in profound changes in root system architecture. Root systems can react to localized supplies of these nutrients by adjusting their rate and direction of growth, as well as their extent of branching and their extent of root hair formation (Zhang and Forde 1998; Malamy and Ryan 2001; Linkohr et al. 2002; Lopez-Bucio et al. 2002). These localized growth responses are mediated by pathways that appear to be coordinated with phytohormone signaling, allowing for their coordination with the cell elongation and proliferation events that underlie all growth phenomena (Lopez-Bucio et al. 2003).
Although environmental inputs have an important influence on root system architecture, it is conceivable that root growth is limited by inherent genetic boundaries. Such boundaries are, for instance, set by the cellular mechanisms controlling cell elongation and proliferation (Beemster et al. 2003). For instance, cell proliferation is a particularly important factor in the determination of root growth rate, as transgenic interference with cell cycle progression has profound effects on growth rate and sometimes also on meristem organization of the root (Doerner et al. 1996; Cockcroft et al. 2000; De Veylder et al. 2001). Furthermore, cell production is an important component of root growth rate in natural accessions of Arabidopsis (Beemster et al. 2002). To a significant degree, the effect of plant hormones on root growth also appears to be mediated by modulation of cell cycle duration (Beemster and Baskin 2000; Stals and Inze 2001; Werner et al. 2003). At the organ level, the outputs of the cellular mechanisms that control the size of the root meristem, the rate of cell proliferation, and the extent of cell elongation, are integrated to determine the overall rate of growth. However, whether or to what degree these mechanisms are acting independently from one another is not clear (Beemster et al. 2003).
The aim of this study was to isolate novel regulators of quantitative aspects of root growth that are responsible for the intraspecific variation of root system morphology in Arabidopsis. Therefore, we exploited natural genetic variation rather than mutagenesis of a particular wild-type background. This strategy also avoids the isolation of alleles that affect basic properties of the root system, such as the formation of certain tissue layers or physiological responses to nutrient availability. We were successful in isolating a novel gene that regulates the extent of cell proliferation and elongation in the root. It represents a member of a novel, plant-specific gene family and encodes a novel type of nuclear protein that appears to be involved in transcriptional regulation.
Results
Root growth parameters vary among isogenized Arabidopsis wild-type lines
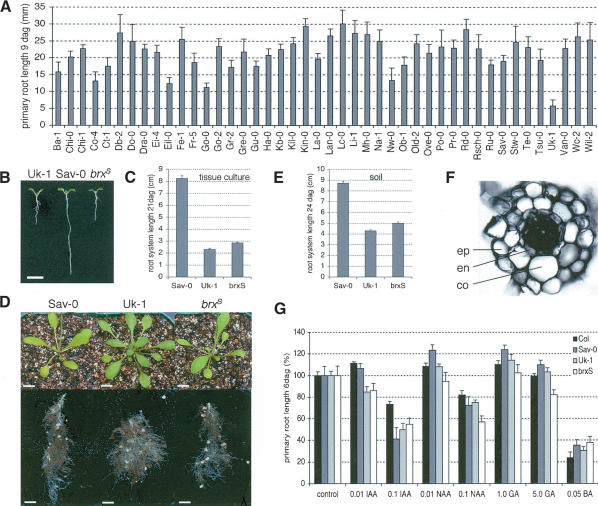
To determine natural genetic variation of root system morphology, we compared 44 arbitrarily chosen Arabidopsis accessions in tissue culture experiments. A sample of 20 seedlings of each line was grown under constant illumination on solid medium containing basic macro- and micronutrients and agar. Nine days after germination (dag), the length of the primary root, the number of lateral roots, and the number of adventitious roots were recorded. An overall two- to threefold variation in primary root length and lateral root number was observed between accessions. Adventitious roots were very rare in all accessions; however, they were observed more frequently in Umkirch-1 (Uk-1). This accession also developed a significantly shorter primary root than average (Fig. 1A) and a generally more branched root system at later stages. Because of its clearly distinct root system phenotype, we chose to analyze this line in further detail.
Figure 1.
Natural variation in root system morphology among Arabidopsis accessions. (A) Primary root length of Arabidopsis seedlings at 9 dag, grown in 8 h dark-16 h light cycle on 0.5× MS medium. n ≥ 10. (B) Representative seedlings of the Uk-1 and Sav-0 accessions, and a seedling resulting from introgression of the Uk-1 short-root phenotype into an Sav-0 background (brxS), 9 dag grown in constant light on 0.5× MS medium containing 0.3% sucrose. Bar, 1 cm. (C) Primary root length of plants of the three genotypes grown in constant light on 0.5× MS medium containing 1.0% sucrose, 21 dag. n = 6. (D, top) Representative rosette phenotypes of the three genotypes at 24 dag, grown on soil under constant illumination. (Bottom) Root system belonging to the shoots shown in the top panel, dug out from the soil and cleaned. Bar, 1 cm. (E) Approximate primary root length of plants of the three genotypes grown on soil under constant illumination, 24 dag; n ≥ 7. (F) Transverse cryosection through the mature part of a primary root of a 7-day-old Uk-1 seedling. (ep) epidermis; (co) cortex; (en) endodermis. (G) Relative response of Col, Sav-0, Uk-1, and brxS seedlings to different exogenous plant hormone applications, 6 dag. Seedlings were grown in constant light on 0.5× MS medium containing 2.0% sucrose plus indicated hormone supplement. (IAA) indole acetic acid; (NAA) naphtalene acetic acid; (GA) gibberellic acid; (BA) benzylaminopurine. Error bars are standard error.
The short primary root of Uk-1 seedlings is largely caused by a single locus
To test whether the alleles conferring the root phenotype of Uk-1 are of a dominant or recessive nature, we crossed Uk-1 into Slavice-0 (Sav-0), an accession with an average root system as compared with other accessions in our assays. In the F2 generation of our cross we noticed that the short primary root phenotype of Uk-1 segregated as a recessive in a ratio close to 3:1. Root development is highly plastic, and although the average primary root lengths of the Uk-1 and Sav-0 lines are clearly distinct (Fig. 1A), their ranges of root length in individuals overlap. By analysis of the F3 progeny, however, it was possible to unequivocally determine the phenotype of the parental F2 plants, confirming the suspected 3:1 ratio. Thus, the short-root phenotype of Uk-1 appears to be largely caused by a single locus, which we named BREVIS RADIX (BRX), latin for “short root.”
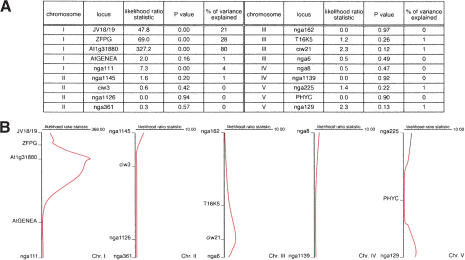
Starting from the F2 progeny of two different F1 plants, we also established a recombinant inbred line population of 206 lines by repeated selfing for six generations. The primary root length of these lines was measured, and each line was genotyped for a set of simple sequence length polymorphism markers spread over the Arabidopsis genome (Table 1). The data were then subjected to quantitative trait locus (QTL) analysis. The results indicate that a major QTL for primary root length is located on the upper arm of chromosome I and identical with BRX (see Fig. 5, below).
Table 1.
Analysis of a recombinant inbred line (RIL) population derived from a cross between the Sav-0 and Uk-1 accessions
Columns indicate the line number, average primary root length determined from a sample of 16-20 seedlings in the S6 generation, and the genotype at simple sequence length polymorphism markers distributed throughout the Arabidopsis genome. Genotypes: (A) Uk-1 allele; (B) Sav-0 allele; (H) heterozygous; (U) unknown.
Figure 5.
Quantitative trait locus (QTL) analysis of the RIL population. (A) Results from the regression analysis of the data presented in Table 1 plus the genotypes at the BRX (At1g31880) locus, with respect to primary root length. (B) Graphical presentation of the data shown in A. The different chromosomes and the relative position of the scored simple sequence length polymorphism markers are indicated, along with the likelihood statistics for the positions of QTLs.
The Uk-1 short-root phenotype does not depend on shoot-derived signals
Morphological differences between accessions were observed not only in the root system but also in the shoot system. Because it has been shown that communication between shoot and root tissues can significantly influence each other's growth rate and branching pattern (Turnbull et al. 2002; Sorefan et al. 2003), we wanted to determine whether the Uk-1 root phenotype is autonomous from shoot-derived signals. To this end, we introgressed the short primary root phenotype into a Sav-0 background, whose shoot morphology is very different from Uk-1. Sav-0 plants flower early, approximately after the sixth true leaf (under constant illumination), and form multiple shoots. By contrast, in the same conditions Uk-1 plants flower late (approximately after the 24th true leaf) and form a single shoot. From a sample of the F2 generation resulting from our Uk-1 × Sav-0 cross, we selected the seedling with the shortest primary root. This plant was then back-crossed into the parental Sav-0 line, a scheme that was in total repeated four times. From this introgression we derived plants whose genome consists of ∼97% of Sav-0 DNA and only 3% of Uk-1 DNA. In the following we refer to individuals with a short-root phenotype that have been derived from this introgression into an Sav-0 background as brxS.
The roots of brxS seedlings are as short as those of Uk-1 seedlings, both when grown in the light (Fig. 1B) or in darkness (data not shown). In the adult root system of brxS plants, the primary root is slightly longer and the root system is less branched than in Uk-1. This is true for root systems grown in tissue culture (Fig. 1C) as well as for soil-grown roots (Fig. 1D,E). In contrast to the root system, the shoot system morphology and flowering time of brxS plants resembles the Sav-0 shoot system (Fig. 1D). Moreover, grafts between Sav-0 shoots and Uk-1 roots, and vice versa, do not influence the respective root system morphologies (data not shown). Therefore, the short-root phenotype conferred by the Uk-1 allele of the BRX locus is independent from shoot-derived signals.
Physiological responses of the root system are intact in brxS plants
Because the influence of patterning genes, plant hormones, and environmental stimuli on root growth are well documented, we checked whether brxS plants are impaired in any of the corresponding pathways. Transverse sections of Uk-1 roots indicate that the cortex and endodermis cell layers are present (Fig. 1F), ruling out defects in the SCR or SHR genes. In addition, brxS seedlings respond to exogenous application of plant hormones, such as auxins, gibberellins, or cytokinins, in roughly the same proportional range as the parental Sav-0 line (e.g., Fig. 1G). Notably, the application of plant hormones was in no instance able to rescue the short-root phenotype (Fig. 1G), even when very low concentrations were applied (data not shown). Finally, we also tested the response of brxS seedlings to different nutrient conditions, as nutrient availability has been demonstrated to affect root system architecture (Lopez-Bucio et al. 2003). However, we did not observe any apparent defects in the numerous assays that we conducted, including examination of the responses to low or high nitrate or phosphate levels or to different ratios of nitrogen to carbon source. Again, brxS seedlings responded in proportional ranges similar to those of the parental Sav-0 line (data not shown). In summary, the short-root phenotype of brxS plants is not the result of a major defect in basic hormone or physiological response pathways.
brxS seedlings have shorter and fewer root cells
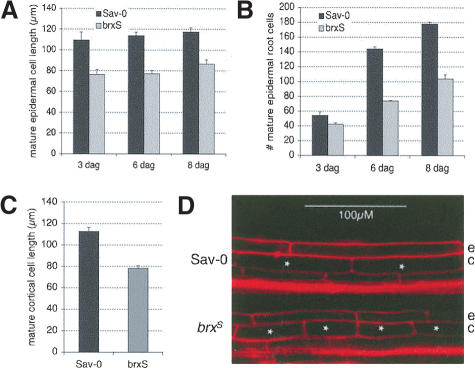
To characterize the brxS phenotype in further detail, we analyzed the primary roots of brxS seedlings at the cellular level. In principal, the brxS short-root phenotype could be caused by one of two phenomena: either shorter cells or fewer cells. To distinguish between these two possibilities, we microscopically analyzed mature epidermal cell files (i.e., the root hair-bearing region distal to the meristem). Analysis of the size and number of epidermal cells revealed that brxS roots are composed of shorter (Fig. 2A) as well as fewer (Fig. 2B) cells. These parameters remained relatively constant throughout the period of observation (3-8 dag). In Sav-0, the production rate of mature epidermal cells was 19-24 cells per day, and their average length was 110-117 μm, whereas in brxS, 11-13 cells per day with a length of 76-87 μm were produced. Because the root growth rate in both genotypes remained roughly the same up to 21 dag, it is reasonable to assume that these parameters did not change throughout development. In line with the observations in epidermal cell files, confocal microscopy revealed that the more evenly sized cortical cells are also shorter in brxS roots (Fig. 2C,D). In summary, both cell elongation and cell production rate are decreased in brxS seedlings, contributing approximately one-third and two-thirds, respectively, to the overall difference in root length as compared with Sav-0 seedlings.
Figure 2.
Mature cell size and number in the primary roots of Sav-0 and brxS seedlings. (A) Mature epidermal cell length at 3, 6, and 8 dag. For each genotype, three seedlings were measured per time point. The number of cells measured in each seedling was ≥13 at 3 dag, ≥46 at 6 dag, and ≥34 at 8 dag. (B) Number of mature epidermal cells in a cell file of the root at 3, 6, and 8 dag. For each genotype, three seedlings were counted per time point. (C) Mature cortical cell length at 6 dag. For each genotype, three seedlings were measured. The number of cells measured was ≥50. (D) Confocal microscopy images of the mature region of Sav-0 and brxS roots. Asterisks mark cortical cells. (e) epidermis; (c) cortex. Error bars are standard error.
The BRX locus affects cell proliferation in the apical root meristem
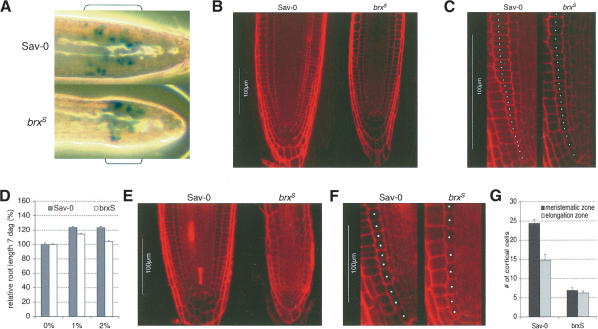
To visualize the meristematic region of the root, we crossed a transgenic reporter of cell proliferation, a fusion protein between cyclin B1;1 (CYCB1;1) and β-glucuronidase (GUS) expressed under control of the CYCB1;1 promoter (de Almeida Engler et al. 1999), into the brxS and Sav-0 lines. GUS staining of roots of these seedlings revealed that the root meristems of brxS seedlings are smaller than Sav-0 meristems (Fig. 3A). When investigated by confocal microscopy, the organization of brxS root meristems appears normal (Fig. 3B). However, compared with Sav-0 meristems, cells in the meristematic zone in brxS appear to increase in size earlier, and the number of cells undergoing division appears to be reduced (Fig. 3C). This phenotype (shown for 0.5% sucrose concentration in Fig. 3B,C) becomes more pronounced in growth-promoting conditions. In our physiological assays we noticed that the difference in root length between Sav-0 and brxS seedlings increased when root growth rate was stimulated by increasing the amount of sucrose in the medium (Fig. 3D; Benfey et al. 1993). This correlates with a further size reduction of brxS meristems at a higher growth rate (e.g., 2% sucrose; Fig. 3E). In these conditions, they are composed of fewer cells that are less organized and not as isodiametric (Fig. 3F).
Figure 3.
Root meristem morphology and size in the primary roots of Sav-0 and brxS seedlings. (A) Activity of a CYCB1;1::CYCB1;1:GUS reporter gene in the meristems of brxS and Sav-0 seedlings, detected by GUS staining. Brackets indicate the meristematic region as defined by the GUS signal. (B) Confocal images of root meristems grown on 0.5× MS medium containing 0.5% sucrose. (C) Magnification of cortical cell files (marked by white dots), starting from the initial cell, shown in B. (D) Response of root growth of Sav-0 and brxS seedlings to increasing amounts of sucrose (given in percentages) in the medium, scored 7 dag. n ≥ 8. (E) Confocal images of root meristems grown on 0.5× MS medium containing 2.0% sucrose. (F) Magnification of cortical cell files (marked by white dots), starting from the initial cell, shown in E. (G) Number of cells in cortical cell files of the root meristematic and elongation zones as defined in the text, grown on 0.5× MS medium containing 2.0% sucrose and scored 6 dag. n ≥ 10. Error bars are standard error.
To quantify our observations, we measured the size of the meristematic and elongation zones of Sav-0 and brxS seedlings that were grown on 2% sucrose at 6 dag by analyzing cell files. We took the number of cortical cells, counted from the initial cell up to the first rapidly elongating cell, as an indicator of root meristem size (Casamitjana-Martinez et al. 2003). By this measure, brxS root meristems consist of ∼25% of the number of cells in Sav-0 meristems (Fig. 3G). We also took the number of cortical cells, counted from the first rapidly elongating cell up to the first cell of mature size, as an indicator of elongation zone size. By this measure, brxS elongation zones consist of ∼40% of the number of cells in Sav-0 elongation zones (Fig. 3G). Therefore, the ratio between the number of cells in the meristematic zone and the number of cells in the elongation zone is shifted to close to 1.0 in brxS from ∼1.7 in Sav-0. Thus, the size of both the meristematic and elongation zones of the root tips of brxS seedlings are decreased, but the meristematic zone is affected more severely.
Isolation of the BRX gene by positional cloning
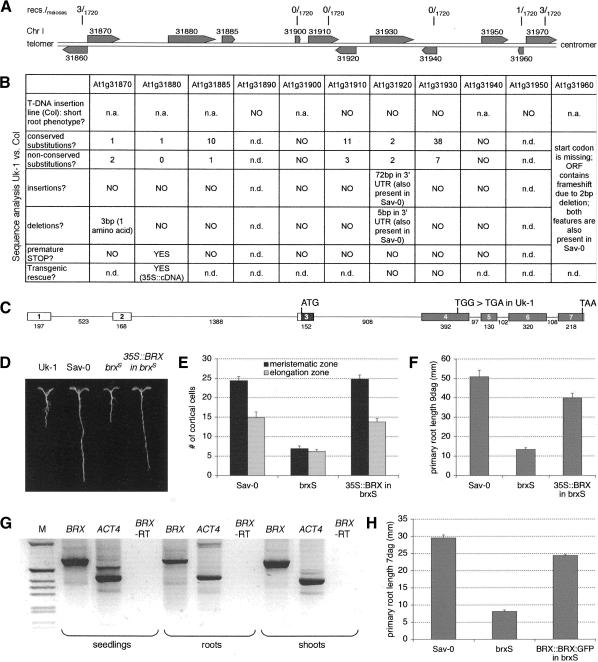
To identify the BRX gene at the molecular level, we followed a positional cloning approach. To this end, genomic DNA was isolated from 860 individuals of the F2 population from the Uk-1 × Sav-0 cross and genotyped with molecular markers that showed polymorphism between the two accessions. The root phenotype of the F2 plants was unequivocally scored by analysis of the F3 progeny. Recombination mapping placed the BRX locus on the upper arm of chromosome I. Subsequently, novel markers were generated from PCR-amplified DNA fragments arbitrarily chosen from the Arabidopsis genome sequence. This strategy allowed us to locate the BRX gene in a zero-recombination interval of ∼45 kb, flanked by proximal and distal markers indicating three and one recombination events, respectively (Fig. 4A).
Figure 4.
Positional cloning of the BRX gene. (A) Schematic representation of recombination mapping of the BRX locus to an ∼45-kb interval on chromosome I of Arabidopsis. Solid bars indicate predicted genes, numbers indicate their unicode. (B) Summary of the genetic and sequence analysis of the genes in the region of interest. (n.a.) Not available; (n.d.) not determined. (C) Schematic presentation of the intron-exon structure of the BRX gene. Boxes represent exons, lines represent introns, and their sizes are given in nucleotides below. The shaded boxes indicate the open reading frame. The position of the mutation resulting in a premature stop codon in the Uk-1 accession is shown. (D) Representative Uk-1, Sav-0, and brxS seedlings and brxS seedlings carrying a 35S::BRX transgene, 9 dag grown in constant light on 0.5× MS medium containing 0.3% sucrose. (E) Number of cells in cortical cell files of the root meristematic and elongation zones as defined in the text, grown on 0.5× MS medium containing 2.0% sucrose and scored 6 dag. n ≥ 10. (F) Primary root length of seedlings grown in constant light on 0.5× MS medium containing 1.0% sucrose, 9 dag. n ≥ 15. (G) RT-PCR of BRX and the control gene actin4 (ACT4) from RNA isolated from different sources. Control reactions for BRX in which the reverse transcriptase was lacking (BRX-RT) are shown as well. (M) DNA size marker. (H) Primary root length of seedlings grown in constant light on 0.5× MS medium containing 1.0% sucrose, 7 dag. n ≥ 15. Error bars are standard error.
Crosses of Uk-1 with the Arabidopsis reference accession Columbia (Col) result in segregation of a recessive short-root phenotype as well. Thus, we tested five of the 10 candidate genes in the 45-kb interval by analyzing respective T-DNA insertion mutants in Col background that were available (Alonso et al. 2003). A short-root phenotype was not observed in any of these mutants (Fig. 4B). We also analyzed 8 of the 10 BRX candidate genes by comparing the sequence of the Uk-1 alleles with the corresponding Col alleles. We found no Uk-1 alleles with obvious implications for gene functionality (Fig. 4B), with the exception of the gene represented by unicode At1g31880. This gene contains a base pair change in the fourth exon, which results in a premature stop codon in the open reading frame and, therefore, a truncated protein missing approximately two-thirds of the C terminus (Fig. 4C). This stop codon is not present in the respective alleles of other accessions with long primary roots (determined for accessions Sav-0, Wassilewskaja, Landsberg erecta, Freiburg-1, Eilenburg-0, Loch Ness-0, Chisdra-0, Goettingen-0, and Kindalville-0). Moreover, the stop codon is also missing from the sequence of the accessions Uk-2, Uk-3, and Uk-4, whose BRX alleles are nearly identical to the Col allele apart from very few silent polymorphisms or one conserved substitution. These three accessions have long primary roots and were collected in the immediate vicinity of Uk-1 (The Arabidopsis Information Resourse, TAIR, http://www.arabidopsis.org). Introduction of a transgenic construct expressing the open reading frame of At1g31880 under control of the 35S cauliflower mosaic virus gene promoter (35S) into brxS seedlings largely rescues the short-root phenotype (Fig. 4D) and restores the meristem size to Sav-0 dimensions (Fig. 4E). Finally, this is also true for a transgene expressing a BRX open reading frame in its native start codon context (i.e., including the untranslated exons and introns up to the ATG; Fig. 4C) under control of a 1.9-kb fragment of the BRX promoter (data not shown). Thus, the combined evidence demonstrates that At1g31880 and BRX are identical.
BRX is expressed in the root at very low levels
From the brxS phenotype it can be expected that BRX is expressed in the root. To determine whether this is the case, we analyzed whole seedlings, shoots, and roots by RT-PCR. In these experiments, BRX expression can be detected in all three samples (Fig. 4G). To visualize BRX expression at spatiotemporal resolution, we also constructed transgenic plants expressing the green fluorescent protein (GFP) or a fusion protein of BRX and GFP under control of the BRX promoter (constructs BRX::GFP and BRX::BRX:GFP, respectively). Importantly, the BRX::BRX:GFP transgene rescues the brxS root phenotype, demonstrating expression and functionality of the BRX:GFP fusion protein (Fig. 4H). However, in (confocal) fluorescence microscopy, neither the BRX:GFP fusion protein nor native GFP could be detected. In line with these observations, Western analysis of the transgenic lines using an anti-GFP antibody yields a very faint signal, and only does so if an excess amount of protein extract is loaded, whereas GFP produced in a 35S::GFP line is readily detectable in very little extract (data not shown). Therefore, in summary our results indicate that BRX is expressed in the shoot and root, albeit at very low levels.
BRX explains most of the variance in primary root length between Uk-1 and Sav-0
We observed rescue of the short-root phenotype of brxS seedlings in several transgenic lines derived from independent primary transformants. However, we noticed that rescue was not complete in any of these lines (e.g., Fig. 4F,H). This finding is consistent with the idea that BRX represents the major QTL for primary root length predicted on chromosome I from regression analysis of our recombinant inbred line population. The creation of a BseGI restriction enzyme polymorphism by the base pair change in the Uk-1 allele of BRX allowed us to directly score the BRX genotype in the recombinant inbred lines and to include this information in the regression analysis. The results indicate that the BRX locus explains ∼80% of the observed variance in primary root length in the population (Fig. 5).
BRX is a member of a novel, plant-specific gene family
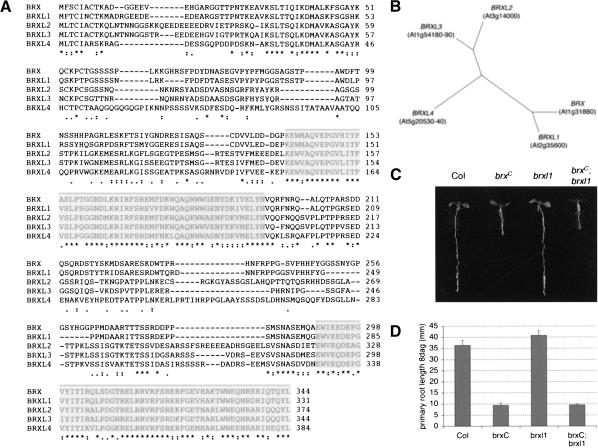
At the time of its identification, the BRX gene was not correctly annotated in public databases, with most of the open reading frame predicted to be fused with the neighboring gene and consequently considered a novel type of aquaporin (Johanson et al. 2001). However, the annotation of a related gene, which we named BRX-like 1 (BRXL1; unicode At2g35600), enabled us to determine the correct intron-exon structure of BRX by comparison, including two noncoding exons representing 5′ untranslated regions (Fig. 4C). Based on the gene structure of BRX and BRXL1, we were able to identify and annotate three more genes of this type in the Arabidopsis genome, BRXL2, BRXL3, and BRXL4 (unicode or fusion of parts of unicodes At3g14000, At1g54180-At1g54190, and At5g20530-At5g20540, respectively). Subsequently, full-length cDNA clones became available for four out of the five genes and confirmed the predicted gene models. The BRX family genes and the proteins they encode are highly conserved (Fig. 6A) and are found in all higher plants for which data are available, but are absent from unicellular organisms or animals. Therefore, this gene family appears to be specific to multicellular plants.
Figure 6.
The BRX family of genes. (A) Sequence alignment of the predicted sequences of BRX family of proteins. Asterisks indicate identity, two dots indicate conserved substitutions, and one dot indicates substitutions with similar basic characteristics. A highly conserved domain, occurring twice in each protein, is highlighted. (B) Unrooted phylogenetic tree based on the amino acid sequences shown in A. (C) Analysis of brxC;brxl1 double mutants. Representative seedlings of the indicated genotypes, grown in constant light on 0.5× MS medium containing 1.0% sucrose, are shown at 8 dag. (D) Primary root length of seedlings grown in constant light on 0.5× MS medium containing 1.0% sucrose, scored 8 dag. n ≥ 9. Error bars are standard error.
To test whether other BRX-like genes act partially redundant with BRX in root growth, we obtained presumed null mutants for BRXL1, BRXL2, and BRXL3 from the SALK T-DNA insertion mutagenesis project (Alonso et al. 2003). Insertions in the BRXL4 gene could not be confirmed. Interestingly, none of these mutants display a brx root phenotype. However, partial and asymmetric redundancy has been observed in other cases and might only become apparent in a brx mutant background. Thus, we created double mutants between the Uk-1 brx allele, twice introgressed into a Col background (we refer to these plants as brxC), and the other brxl mutants. In our analysis, we focused on the brxC;brxl1 double mutant, because of the high similarity of BRXL1 to BRX both in gene structure (only these two BRX-like genes possess the untranslated exons) and amino acid sequence (Fig. 6A,B). In this double mutant, we did not observe any abnormalities in the root system that would indicate an enhancement of the brxC phenotype (Fig. 6C,D). Similar results were obtained for the brxC;brxl2 and brxC;brxl3 double mutants (data not shown). Therefore, BRX likely is the only gene in this family with a role in root development.
The BRX protein is nuclear localized and can activate transcription in yeast
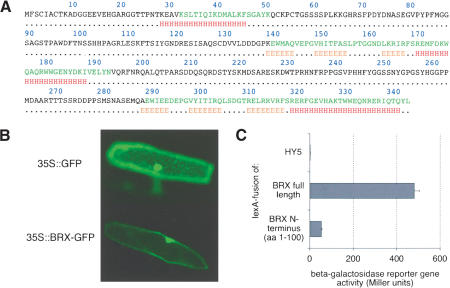
The BRX protein does not contain any previously characterized motifs that would indicate its biochemical function. However, sequence alignment of the BRX family proteins reveals that all five of them contain three highly conserved domains (Fig. 6A). One domain is located at the N terminus, between amino acids 28 and 45 of BRX, whereas two more domains that are highly similar to each other are located between amino acids 169 and 182 and between 320 and 334, respectively. Interestingly, in secondary structure predictions, these three domains all have a high probability of forming α-helical secondary structures (Fig. 7A).
Figure 7.
Analysis of the BRX protein. (A) Secondary structure prediction for the BRX protein. The domains highly conserved between BRX-like proteins are indicated in green. (H) Regions with a high probability of forming α-helical structures are indicated; (E) regions with high probability of forming extended β sheets. (B) Nuclear localization of BRX. Fluorescent microscopy of transiently transformed epidermal onion cells expressing a BRX-GFP fusion protein or GFP alone. (C) Reporter gene activity in yeast expressing the indicated lexA fusion proteins. n = 8. Error bars are standard error.
α-Helices are characteristic for transcription factor proteins and are often found in DNA binding and protein interaction domains (Luscombe et al. 2000). Transcription factors are nuclear proteins, and therefore we tested whether BRX accumulates in the nucleus. To this end, a fusion between GFP and BRX was transiently expressed in epidermal onion cells and its subcellular localization was monitored by fluorescence microscopy. In this assay, the GFP:BRX fusion protein is found primarily in the nucleus (Fig. 7B), unlike GFP by itself, indicating that BRX is actively transported into the nucleus.
We also tested whether the BRX protein can activate transcription in a heterologous yeast system. To this end, we cloned the BRX open reading frame into a yeast expression vector, in frame with the lexA DNA binding domain of Escherichia coli. Expression of this fusion protein in the presence of a β-galactosidase reporter gene controlled by lexA promoter binding sites results in strong reporter activity (Fig. 7C). This is not the case if a control fusion protein between the Arabidopsis transcription factor HY5, which lacks transactivation potential (Ang et al. 1998), and lexA is expressed instead. The transactivation potential is largely reduced in a truncated BRX protein comprising the 100 N-terminal amino acids. Thus, the data indicate that BRX contains a transcription activation domain.
Discussion
Natural genetic variation in root system morphology of Arabidopsis
The goal of our study was to isolate novel regulators of root growth that are responsible for the intraspecific variation in root system morphology. Such genes should not be essential for root development per se, based on the assumption that alleles that are selected in the wild are not detrimental to basic plant development and that evolution preferentially acts on genes controlling nonessential aspects of growth. Because of the well-developed genetic resources and the ease of manipulation, we chose to analyze natural genetic variation in isogenized accessions of the model plant Arabidopsis.
Mutagenesis approaches in Arabidopsis have been tremendously successful in isolating genes involved in different aspects of root development, such as pattern formation, growth rate, or cell shape (e.g., Benfey et al. 1993; Hauser et al. 1995). Although the analysis of these genes has greatly enhanced our knowledge of root development, less is known about the factors that specifically control quantitative aspects of root biology, such as the rate of growth. Although it is clear that control of cell proliferation has an important role in root growth (Beemster et al. 2003), to our knowledge, loss-of-function mutants that are specific to cell proliferation in the root meristem have not been isolated to date. Rather, experimental evidence for pathways controlling root growth has been gathered from transgenic gain-of-function approaches, which usually involve the ectopic or overexpression of candidate genes. By these means, for example, the control of cell cycle progression (Doerner et al. 1996; Cockcroft et al. 2000; De Veylder et al. 2001) and CLAVATA-type pathways (Casamitjana-Martinez et al. 2003; Hobe et al. 2003) have been implicated in the control of root growth or meristem size.
Notably, it has been observed that there is detectable variation in root growth between Arabidopsis accessions and that this is, to a significant degree, the result of differences in mature cell size or the rate of cell proliferation (Beemster et al. 2002), supporting the notion that genetic analysis of natural variation can identify factors controlling these processes. Consistent with this previous report, we observed an average two- to threefold variation in root growth parameters of Arabidopsis accessions. The reduction of growth in the Uk-1 line as compared to average was, however, remarkable. The occurrence of this phenotype in the wild might be related to the fact that the Uk-1 accession has been reportedly collected from a river embankment (TAIR, http://www.arabidopsis.org). Thus, water availability might not be as limiting a growth factor in the natural environment of this line, and this might have permitted the evolution of a shorter root as compared with accessions that grow in more arid environments.
The characterization of QTLs by genetic mapping is a well-established procedure; however, isolation of a gene corresponding to a QTL of interest is still an arduous task. Our success in isolating the BRX gene was greatly aided by two factors. First, the effect of the Uk-1 allele of BRX on root growth is a strong one and is, therefore, easily detectable. Second, the unmatched genomic resources for Arabidopsis enable fine mapping within a reasonable time frame (Borevitz and Nordborg 2003). Nevertheless, it can be expected that increased availability of molecular markers and automatization of mapping procedures will soon enable the routine isolation of small effect QTLs in Arabidopsis (Borevitz et al. 2003; Schmid et al. 2003; Torjek et al. 2003).
Specificity of the brx phenotype
By introgression of the Uk-1 allele of BRX into the Sav-0 background, we have demonstrated that the brxS phenotype does not depend on shoot-derived signals. Moreover, in our phenotypic analysis we could not detect any abnormalities in the shoot system of brxS plants. Thus BRX activity is specifically needed in the root. Although many genes influencing root growth have been isolated by mutagenesis approaches, such specificity is still rare. Notably, the majority of root growth mutants isolated to date are involved in hormone signaling pathways. In general, they also display conspicuous defects outside the root system. For instance, root growth is affected in the gai and rga mutants, which disrupt gibberellic acid signaling (Fu and Harberd 2003). However, these genes also have a central role in the growth of stems. The issue is further complicated in mutants affecting the auxin signaling pathway, which include several gain-of-function mutants that might occasionally represent neomorphic phenotypes (Leyser 2002). An auxin signaling gene that appears to be required only in the root is SHY2 (Tian and Reed 1999). Although shy2 gain-of-function mutants have shoot and root phenotypes, corresponding loss-of-function mutants only display a root growth phenotype. In addition, shy2 loss-of-function results in reduced root growth only in light-grown conditions, and this reduction can be rescued by exogenous application of auxin (Tian and Reed 1999). By contrast, the phenotype of brxS seedlings is not conditional and cannot be rescued by plant hormone application. It also has to be stressed that under all growth conditions tested, brxS roots always grow at a rate that is two- to threefold lower than in roots of seedlings carrying the functional Sav-0 allele. Further, the reduction in meristem size in brxS can be observed early in development, does not change as the roots become older, and does not result in growth arrest. This differs significantly from other studies (Casamitjana-Martinez et al. 2003; Hobe et al. 2003), where the root meristem has normal size in early stages and becomes consumed over time, eventually resulting in the shutdown of growth. In summary, compared to other root growth mutants the phenotype of brxS seedlings is unique in many aspects, and BRX appears to be a very basic factor, required for an optimal rate of root growth in any condition.
The brx phenotype: cell proliferation versus cell elongation
The slow primary root growth of brxS seedlings is the result of a reduction in mature cell size as well as cell proliferation. The reduced cell proliferation quantitatively contributes more to the brxS phenotype than to the reduced cell size. It has to be noted, however, that in our introgression we always selected the seedlings with the shortest primary root, thereby likely introducing all the genetic factors affecting root growth in Uk-1 into brxS seedlings. Because transgenic expression of BRX in brxS seedlings restores mature cortical cell size to wild-type dimensions but does not rescue total root length to 100%, we must assume that indeed additional, smaller effect QTLs have been introgressed and would have to be complemented to fully restore the cell proliferation rate to Sav-0 levels.
The different contributions of cell proliferation and cell elongation to overall root growth have been difficult to dissect. To date, it is not clear whether these processes are controlled independently (Beemster and Baskin 1998; De Veylder et al. 2001). This issue is also complicated by the fact that cells still divide, although at a much lower frequency, in the elongation zone (Beemster et al. 2003). It is, however, conceivable that a reduction of cell proliferation in the meristematic region results in a decreased supply of cells to the elongation zone, thus decreasing its size. It also has been suggested that it is the time a cell spends as part of the elongation zone rather than elongation zone size per se that determines final mature cell length (Beemster and Baskin 1998, 2000). Because decreased cell proliferation in the meristem would also result in slower displacement of cells from the elongation zone, the time they spend elongating consequently might not change dramatically, even if the elongation zone is physically smaller. This explanation accounts for the observation that interference with cell proliferation in the root meristem, resulting in reduced size of the meristematic region, always results in a reduction of elongation zone size, whereas mature cell size is usually not affected to the same degree (Beemster and Baskin 2000; De Veylder et al. 2001; Casamitjana-Martinez et al. 2003; Werner et al. 2003).
Genetically, we cannot separate the roles of BRX in cell proliferation and elongation. However, several arguments support the notion that the reduced mature cell size might be a secondary consequence of reduced cell proliferation. In brxS seedlings, the growth zone of the root is reduced in size. This phenotype is enhanced if cell proliferation is stimulated by increased sucrose concentration of the medium. Compared with Sav-0, the cell number in the meristematic zone is affected to a greater extent in brxS seedlings than the cell number in the elongation zone. Interestingly, this phenotype shows significant similarity to root tips of seedlings in which cell proliferation has been slowed down; for instance, by cytokinin treatment (Beemster and Baskin 2000) or by overexpression of inhibitors of cell cycle progression (De Veylder et al. 2001). Finally, previous analyses suggest that the rate of root growth is primarily controlled at the step of cell proliferation (Beemster and Baskin 1998; Beemster et al. 2002, 2003). Thus, the primary cause of the brxS phenotype might be the reduction of cell proliferation in the root meristem.
Implications from the low expression level of BRX
Our expression analyses determined that BRX is expressed in the root as well as the shoot of young seedlings. Thus, BRX might also have a yet-unknown function in the shoot, which could be masked by redundantly acting BRX-like genes in brxS plants.
We could not detect GFP fluorescence in our reporter lines in situ. In this context, it is important to note that we demonstrate that the BRX::BRX:GFP transgene can substitute for native BRX. The transgenic proteins, that is, BRX:GFP or native GFP, are also barely detectable in Western blots, supporting our conclusion that the BRX expression level is very low. This result is corroborated independently by the very rare occurrence of BRX cDNAs in public databases (two hits at time of publication) and BRX signatures in MPSS experiments (http://mpss.udel.edu/at/java.html). Finally, based on the low expression level of BRX and the transgenic rescue of brxS seedlings with a 35S::BRX construct, it can be concluded that overexpression of BRX does not stimulate root growth beyond the rate observed in Sav-0, therefore indicating that BRX is one of several factors that determine the rate of root growth.
The BRX gene family of Arabidopsis: a novel class of transcription factors?
The BRX family proteins are remarkably well conserved in Arabidopsis (64%-93% similarity at amino acid level), indicating that most of their structure is important for their function. However, with the possible exception of BRXL4, for which we could not confirm a T-DNA insertion mutant, only BRX appears to have a role in root growth, as demonstrated by the analysis of the single and double mutants with brxl1, brxl2, and brxl3. This could indicate that, despite the similarity between these genes, there are functional differences in the activity of the encoded proteins, or that these genes act only partially redundantly because of differential expression patterns. Which of these possibilities is the case will be the subject of future investigations. Orthologous BRX-like genes can be found in all other multicellular plant species for which data are available, but not in unicellular organisms or animals. Notably, the corresponding proteins are very well conserved within and between species (C.S. Hardtke, K.F.X. Mayer, G.C. Brigg, N. Strack, and T. Hindemitt, unpubl.). Therefore, we propose that BRX is part of an important gene family with conserved functions in general plant development.
In the absence of previously defined functional domains, it is difficult to assign a biochemical activity to the BRX protein. However, the high level of conservation of distinct domains between BRX family proteins indicates that these regions might be especially important for their activity. These domains are predicted to form α-helical structures, which are often found in protein-protein interaction or DNA binding domains (Luscombe et al. 2000). Our findings that BRX can localize to the nucleus and can activate transcription in a heterologous system support the notion that BRX family proteins represent a novel class of transcription factors. Thus, although their exact biochemical activity remains elusive for now, BRX family proteins are novel nuclear localized regulatory factors of plant development.
Conclusion
The development of plant organs is intrinsically linked to the localized control of cell proliferation (Beemster et al. 2003). Although considerable progress has been made in the characterization of the components of the cell cycle machinery and their differential activity throughout development, several studies clearly suggest that higher level controls modulating cell proliferation in a tissue-specific manner must exist. Accelerating or slowing down the cell cycle results in enhanced or reduced overall growth, respectively (Doerner et al. 1996; Cockcroft et al. 2000; De Veylder et al. 2001). However, in both cases the relative shape and size of plant organs is largely maintained, indicating that additional factors regulate the relative levels of cell proliferation in a highly localized fashion. The BRX gene represents such a regulatory factor with respect to root growth.
Materials and methods
Plant material and tissue culture
Seeds of Arabidopsis accessions and T-DNA insertion mutants were obtained from the Arabidopsis Biological Resources Center. Unless otherwise stated, seedlings were grown at 22°C under constant illumination on culture medium containing 0.5× MS salts, 0.5 g/L MES, and 0.9% agar (pH adjusted to 5.8-6.0 with 1 M KOH), plus the indicated amount of sucrose and any hormone supplements (Sigma-Aldrich). The light intensity was ∼140 μM m-2 sec-1. Grafting experiments were performed as described (Turnbull et al. 2002).
Root length measurements
To determine root lengths, seedlings were grown on vertically oriented plates, which were either scanned on a flatbed scanner or photographed with a digital camera to produce image files suitable for quantitative analysis using the NIH Image software (v 1.63).
GUS staining
To visualize GUS reporter activity, seedlings were incubated in 90% acetone for 1 h at room temperature and then washed once in GUS staining buffer (100 mM phosphate buffer at pH 7.0, 1 mM K-ferricyanide, 1 mM K-ferrocyanide, 0.1% Triton X-100) for 15 min. The wash was replaced by GUS staining buffer that contained 1 mg/mL of X-Gluc and the samples were incubated in darkness at 37°C. The reaction was stopped by replacing the staining solution with 20% EtOH.
Molecular markers and mapping procedures
For mapping purposes, PCR-based molecular markers detecting polymorphisms between Uk-1 and Sav-0 genomic DNA were generated by sequence or restriction analysis of described markers or arbitrarily chosen genomic fragments. Simple sequence length polymorphism markers were scored on 4% gels using high resolution agarose (Amresco).
For fine mapping of the BRX locus, genomic DNA was prepared from 860 F2 plants using the DNeasy™ Plant Genomic DNA Isolation Kit (Qiagen) and genotyped with molecular markers. The BRX genotype was deduced from phenotypic analysis of a sample of ∼20 seedlings of the F3 progeny. Recombination mapping followed standard procedures.
To generate marker data for the QTL analysis, genomic DNA was isolated from plants of the S6 generation and genotyped. Phenotypic measurements were taken from a sample of 16 seedlings of the same generation.
Transgenic analysis
The coding regions of the BRX open reading frame were amplified from genomic DNA by PCR, using Pfu polymerase (Fermentas). These fragments were then connected by subsequent directed ligation reactions and reamplifications to produce the full-length open reading frame. The open reading frame was then cloned into the binary vector pTCSH1 (Hardtke et al. 2000) and verified by sequencing to serve as a basis for further manipulations, such as replacement of the promoter driving expression. The GFP version used in our constructs is mGFP5. The transgenic constructs were transformed into brxS plants via the floral dip method, and transgenic lines were selected by screening the seed progeny for glufosinate ammonium resistance (15 mg/L, BASTA, Sigma-Aldrich) on medium containing 0.3% sucrose.
RT-PCR
Total RNA was prepared from Col seedlings with the RNeasy™ kit (Qiagen) according to the manufacturer's instructions, and RT-PCR reactions were performed according to standard procedures using Superscript II reverse transcriptase (Invitrogen). PCR reactions were performed with 5 μL RT reaction as a template. Oligonucleotides for the detection of BRX and actin4 were chosen to amplify fragments spanning an intron-exon border, to permit detection of genomic contamination.
QTL analysis
For QTL analysis, a recombinant inbred line population was established starting from the F2 progeny of two F1 plants resulting from a Uk-1 × Sav-0 cross. From the F2 plants, 206 lines were established by selfing and single-seed descent for six generations. The genotypes for simple sequence-length polymorphism markers were then determined for plants of the S6 generation, and root length was measured by analysis of 16 seedlings of the same generation. Genotype data and average root lengths were entered into a matrix and free marker regression analysis for selfed recombinant inbred lines was performed using the MapManager QTX for Macintosh software, version 0.27 (http://www.mapmanager.org/mmQTX.html).
Transient transformation of onion epidermis cells
Constructs for transient expression were generated by replacing the GUS gene in vector pTCSH1 with the open reading frame of the green fluorescent protein (mGFP5). For expression of a GFP-BRX fusion protein, the BRX open reading frame was inserted in frame at the C terminus of the GFP. Transient transformation of onion epidermis cells was performed using a PDS1000 helium particle gun (Bio-Rad). After 24 h incubation in darkness the cells were examined by fluorescence microscopy.
Transactivation assay
To test the transactivation potential of BRX, the full-length or part of the open reading frame was inserted into the vector pEG202 (Clontech), resulting in an in-frame fusion to the lexA DNA binding domain of E. coli. The HY5 control construct has been described (Hardtke et al. 2000). Plasmids were then introduced into the Saccharomyces cerevisiae strain EGY48 (Clontech), together with the reporter construct pSH18-34 (Clontech), which carries the β-galactosidase reporter gene under control of lexA binding sites. Transformants were grown in liquid culture overnight, diluted in the morning, and incubated for 6 more hours before β-galactosidase activity was measured by standard assay.
Bioinformatic analyses
The BRX-like genes were identified by homology searches of the Arabidopsis genome sequence with the BLAST search tools (http://www.ncbi.nlm.nih.gov/BLAST). The intron-exon structure of BRX-like genes, sequence alignments, phylogenetic trees and secondary structure predictions of BRX-like proteins were generated by using the analysis tools provided by the European Institute for Bioinformatics (http://www.ebi.ac.uk/Tools).
Acknowledgments
We would like to thank G. De Martino and Drs. T. Western and F. Schoeck for helpful comments on our manuscript, the Arabidopsis Biological Resources Center for seed stocks, and Drs. C. Luschnig and J. Celenza for the CYCB1;1::CYCB1;1:GUS reporter line. C.S.H. conceived this study, wrote the manuscript and contributed data for Figures 1A-E and 3A. C.F.M. contributed data for Figures 1F-G, 2, 3B-G, 4A-F and I, 5, and 7A-B, and Table 1, and established the introgression and recombinant inbred lines. G.C.B. contributed data for Figures 4C and G, 6, and 7C and analyzed the brxl single and double mutants. We thank K. Medlej and J. Ortiz for technical assistance in generating data for Table 1 and Figure 5. This work was funded by a National Sciences and Engineering Research Council (NSERC) Discovery Grant and an NSERC Strategic Projects Grant to C.S.H. C.S.H. is a Strategic Professor of the Fonds Québécois de la Recherche sur la Nature et les Technologies.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1187704.
References
- Alonso J.M., Stepanova, A.N., Leisse, T.J., Kim, C.J., Chen, H., Shinn, P., Stevenson, D.K., Zimmerman, J., Barajas, P., Cheuk, R., et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653-657. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C. and Koornneef, M. 2000. Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 5: 22-29. [DOI] [PubMed] [Google Scholar]
- Ang L.H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.W. 1998. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1: 213-222. [DOI] [PubMed] [Google Scholar]
- Beemster G.T. and Baskin, T.I. 1998. Analysis of cell division and elongation underlying the developmental acceleration of root growth in Arabidopsis thaliana. Plant Physiol. 116: 1515-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2000. Stunted plant 1 mediates effects of cytokinin, but not of auxin, on cell division and expansion in the root of Arabidopsis. Plant Physiol. 124: 1718-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G.T., De Vusser, K., De Tavernier, E., De Bock, K., and Inze, D. 2002. Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol. 129: 854-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beemster G.T., Fiorani, F., and Inze, D. 2003. Cell cycle: The key to plant growth control? Trends Plant Sci. 8: 154-158. [DOI] [PubMed] [Google Scholar]
- Benfey P.N., Linstead, P.J., Roberts, K., Schiefelbein, J.W., Hauser, M.T., and Aeschbacher, R.A. 1993. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 119: 57-70. [DOI] [PubMed] [Google Scholar]
- Borevitz J.O. and Nordborg, M. 2003. The impact of genomics on the study of natural variation in Arabidopsis. Plant Physiol. 132: 718-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Liang, D., Plouffe, D., Chang, H.S., Zhu, T., Weigel, D., Berry, C.C., Winzeler, E., and Chory, J. 2003. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 13: 513-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casamitjana-Martinez E., Hofhuis, H.F., Xu, J., Liu, C.M., Heidstra, R., and Scheres, B. 2003. Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13: 1435-1441. [DOI] [PubMed] [Google Scholar]
- Cockcroft C.E., den Boer, B.G., Healy, J.M., and Murray, J.A. 2000. Cyclin D control of growth rate in plants. Nature 405: 575-579. [DOI] [PubMed] [Google Scholar]
- Davies P.J.E. 1995. Plant hormones: Physiology, biochemistry and molecular biology. Kluwer Academic Publishers, Dordrect, the Netherlands.
- de Almeida Engler J., De Vleesschauwer, V., Burssens, S., Celenza Jr., J.L., Inze, D., Van Montagu, M., Engler, G., and Gheysen, G. 1999. Molecular markers and cell cycle inhibitors show the importance of cell cycle progression in nematode-induced galls and syncytia. Plant Cell 11: 793-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L., Beemster, G.T., Beeckman, T., and Inze, D. 2001. CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J. 25: 617-626. [DOI] [PubMed] [Google Scholar]
- Di Laurenzio L., Wysocka-Diller, J., Malamy, J.E., Pysh, L., Helariutta, Y., Freshour, G., Hahn, M.G., Feldmann, K.A., and Benfey, P.N. 1996. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86: 423-433. [DOI] [PubMed] [Google Scholar]
- Doerner P., Jorgensen, J.E., You, R., Steppuhn, J., and Lamb, C. 1996. Control of root growth and development by cyclin expression. Nature 380: 520-523. [DOI] [PubMed] [Google Scholar]
- Dolan L., Janmaat, K., Willemsen, V., Linstead, P., Poethig, S., Roberts, K., and Scheres, B. 1993. Cellular organisation of the Arabidopsis thaliana root. Development 119: 71-84. [DOI] [PubMed] [Google Scholar]
- El-Din El-Assal S., Alonso-Blanco, C., Peeters, A.J., Raz, V., and Koornneef, M. 2001. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 29: 435-440. [DOI] [PubMed] [Google Scholar]
- Fu X. and Harberd, N.P. 2003. Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740-743. [DOI] [PubMed] [Google Scholar]
- Hardtke C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.W. 2000. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 19: 4997-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser M.T., Morikami, A., and Benfey, P.N. 1995. Conditional root expansion mutants of Arabidopsis. Development 121: 1237-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M.T., and Benfey, P.N. 2000. The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555-567. [DOI] [PubMed] [Google Scholar]
- Hobe M., Muller, R., Grunewald, M., Brand, U., and Simon, R. 2003. Loss of CLE40, a protein functionally equivalent to the stem cell restricting signal CLV3, enhances root waving in Arabidopsis. Dev. Genes Evol.. 213: 371-381. [DOI] [PubMed] [Google Scholar]
- Johanson U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344-347. [DOI] [PubMed] [Google Scholar]
- Johanson U., Karlsson, M., Johansson, I., Gustavsson, S., Sjovall, S., Fraysse, L., Weig, A.R., and Kjellbom, P. 2001. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126: 1358-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyser H.M. 2002. Molecular genetics of auxin signaling. Ann. Rev. Plant Biol. 53: 377-398. [DOI] [PubMed] [Google Scholar]
- Linhart Y.B. and Grant, M.C. 1996. Evolutionary significance of local genetic differentiation in plants. Ann. Rev. Ecol. Systematics 27: 237-277. [Google Scholar]
- Linkohr B.I., Williamson, L.C., Fitter, A.H., and Leyser, H.M. 2002. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 29: 751-760. [DOI] [PubMed] [Google Scholar]
- Lopez-Bucio J., Hernandez-Abreu, E., Sanchez-Calderon, L., Nieto-Jacobo, M.F., Simpson, J., and Herrera-Estrella, L. 2002. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 129: 244-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bucio J., Cruz-Ramirez, A., and Herrera-Estrella, L. 2003. The role of nutrient availability in regulating root architecture. Curr. Opin. Plant Biol. 6: 280-287. [DOI] [PubMed] [Google Scholar]
- Luscombe N.M., Austin, S.E., Berman, H.M., and Thornton, J.M. 2000. An overview of the structures of protein-DNA complexes. Genome Biol. 1: REVIEWS001. [DOI] [PMC free article] [PubMed]
- Malamy J.E. and Ryan, K.S. 2001. Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 127: 899-909. [PMC free article] [PubMed] [Google Scholar]
- Maloof J.N., Borevitz, J.O., Dabi, T., Lutes, J., Nehring, R.B., Redfern, J.L., Trainer, G.T., Wilson, J.M., Asami, T., Berry, C.C., et al. 2001. Natural variation in light sensitivity of Arabidopsis. Nat. Genet. 29: 441-446. [DOI] [PubMed] [Google Scholar]
- Nakajima K., Sena, G., Nawy, T., and Benfey, P.N. 2001. Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307-311. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Beis, D., Wolkenfelt, H., Murfett, J., Guilfoyle, T., Malamy, J., Benfey, P., Leyser, O., Bechtold, N., Weisbeek, P., et al. 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99: 463-472. [DOI] [PubMed] [Google Scholar]
- Sabatini S., Heidstra, R., Wildwater, M., and Scheres, B. 2003. SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes & Dev. 17: 354-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid K.J., Sorensen, T.R., Stracke, R., Torjek, O., Altmann, T., Mitchell-Olds, T., and Weisshaar, B. 2003. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 13: 1250-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorefan K., Booker, J., Haurogne, K., Goussot, M., Bainbridge, K., Foo, E., Chatfield, S., Ward, S., Beveridge, C., Rameau, C., et al. 2003. MAX4 and RMS1 are orthologous dioxygenase-like genes that regulate shoot branching in Arabidopsis and pea. Genes & Dev. 17: 1469-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stals H. and Inze, D. 2001. When plant cells decide to divide. Trends Plant Sci. 6: 359-364. [DOI] [PubMed] [Google Scholar]
- Tian Q. and Reed, J.W. 1999. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126: 711-721. [DOI] [PubMed] [Google Scholar]
- Torjek O., Berger, D., Meyer, R.C., Mussig, C., Schmid, K.J., Rosleff Sorensen, T., Weisshaar, B., Mitchell-Olds, T., and Altmann, T. 2003. Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. Plant J. 36: 122-140. [DOI] [PubMed] [Google Scholar]
- Turnbull C.G., Booker, J.P., and Leyser, H.M. 2002. Micrografting techniques for testing long-distance signalling in Arabidopsis. Plant J. 32: 255-262. [DOI] [PubMed] [Google Scholar]
- van den Berg C., Willemsen, V., Hendriks, G., Weisbeek, P., and Scheres, B. 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390: 287-289. [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka, V., Laucou, V., Smets, R., Van Onckelen, H., and Schmulling, T. 2003. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell. 15: 2532-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. and Forde, B.G. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279: 407-409. [DOI] [PubMed] [Google Scholar]