Abstract
Pole development is coordinated with the Caulobacter crescentus cell cycle by two-component signaling proteins. We show that an unusual response regulator, PleD, is required for polar differentiation and is sequestered to the cell pole only when it is activated by phosphorylation. Dynamic localization of PleD to the cell pole provides a mechanism to temporally and spatially control the signaling output of PleD during development. Targeting of PleD to the cell pole is coupled to the activation of a C-terminal guanylate cyclase domain, which catalyzes the synthesis of cyclic di-guanosine monophosphate. We propose that the local action of this novel-type guanylate cyclase might constitute a general regulatory principle in bacterial growth and development.
Keywords: Caulobacter development, GGDEF domain, protein localization, response regulator
During developmental transitions, localized changes of cellular morphology are mediated by adaptation in levels and arrangement of proteins. Temporal and spatial control often relies on the timed synthesis or activation of transcriptional regulators and on the establishment of gradients through the compartmentalization of signaling complexes. Although the regulatory mechanisms of gene expression are relatively well understood, it is often not clear how morphogenetic changes are controlled and coordinated locally. In prokaryotes, the major paradigm for signal transduction is the two-component regulatory system (Parkinson and Kofoid 1992). On signal input, the first component, a sensor kinase, autophosphorylates on a histidine residue. The second component, a soluble response regulator, often functions as a transcriptional regulator. Its phosphorylation by the cognate histidine kinase on a conserved aspartate residue in the N-terminal receiver domain usually results in increased DNA binding affinity (Parkinson and Kofoid 1992). Here we present evidence that a novel-type response regulator acts at a distinct subcellular site where it contributes to local changes in cell morphology through the production of a novel signaling molecule.
The unicellular bacterium Caulobacter crescentus goes through an obligate developmental transition that allows it to switch between a sessile, adhesive, and a motile, planktonic cell during its cell cycle. As a consequence, cell poles are continuously remodeled during cell differentiation to facilitate assembly and removal of motility and surface adherence organelles at the right time and in the correct order. Asymmetry is established in the predivisional cell with a single flagellum, a chemotaxis machinery, and pili being assembled at one pole, whereas the opposite pole consists of a stalk and an adhesive organelle, the holdfast (Fig. 1). As a result, division generates two cell types with distinct properties: a surface-attached stalked cell and a motile swarmer cell. The swarmer progeny first differentiates into a stalked cell before it initiates DNA replication and cell division. During this transition the pili retract, flagella are released, and the adhesive organelles are synthesized at the same pole. Here we investigate the function and regulation of the PleD response regulator in C. crescentus polar development. Cells that lack a functional PleD protein are hypermotile, are unable to eject the flagellum, and fail to synthesize a complete stalk structure (Hecht and Newton 1995; Aldridge and Jenal 1999). In contrast, the presence of a constitutively active mutant protein PleD* results in elongated stalks and has a dominant negative effect on motility (Aldridge et al. 2003). The PleD* protein contains four point mutations (Asn/Thr 120, Ala/Thr 214, His/Pro 234, and Tyr/Asn 234) and retains its activity even when its phosphoryl acceptor site Asp 53 is modified, suggesting that this mutant form does not rely on phosphorylation input by a cognate kinase (Aldridge et al. 2003). In vivo phosphorylation experiments indicate that the polar kinases DivJ and PleC are involved in modulating the phosphorylation status of PleD. Whereas PleD∼P is reduced to about 10% in cells lacking DivJ, it is undetectable in a mutant lacking both DivJ and PleC (Aldridge et al. 2003). DivJ and PleC are asymmetrically positioned at opposite cell poles, with DivJ localizing to the stalked pole coinciding with the requirement for active PleD during cell differentiation (Fig. 1; Wheeler and Shapiro 1999; Ohta and Newton 2003). Here we provide evidence that DivJ and PleC directly interact with PleD to modulate its phosphorylation state, suggesting that together they are responsible for PleD phosphorylation in vivo. We show that the PleD regulator dynamically localizes to the differentiating stalked pole during the cell cycle as a function of its phosphorylation state. Our results indicate that only activated PleD is sequestered to the stalked pole, providing a mechanism that spatially restricts PleD activity to the emerging stalked pole, where it coordinates polar morphogenesis.
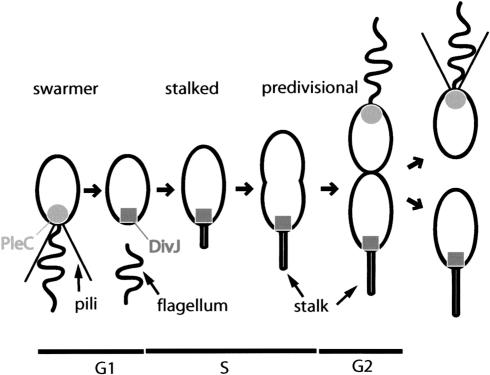
Figure 1.
Dynamic localization of the PleC and DivJ sensor protein kinases during the C. crescentus cell cycle. The positioning of PleC (circle) and DivJ (rectangle; Wheeler and Shapiro 1999) during the cell cycle are indicated. Polar organelles and cell cycle stages are specified.
PleD is a multidomain protein with two N-terminal receiver modules arranged in tandem and a C-terminal domain apparently serving as an output module (Hecht and Newton 1995). This putative output domain, termed “GGDEF” or “DUF1,” is widespread and highly conserved in many bacterial species. Postulating a local activity of PleD at one cell pole calls for a molecular mechanism that converts the phosphorylation input into a readout that affects downstream targets. We propose that the PleD readout is the production of a cyclic nucleotide, which acts as secondary messenger. In vitro experiments with C. crescentus crude extracts and with purified PleD protein show that PleD contains an intrinsic nucleotide cyclase activity, which converts two molecules of GTP into cyclic diguanylic acid (c-di-GMP). Cyclase activity correlates with PleD activation by phosphorylation and requires an intact PleD C-terminal output domain. This suggests that the GGDEF domain constitutes a novel class of guanylate cyclases, which in PleD is specifically activated in response to phosphorylation of the N-terminal receiver domain. Our findings, together with the observation that more than 900 GGDEF proteins are reported in the nonredundant SMART database (Schultz et al. 1998), implies that diguanylate cyclases are abundant in the bacterial kingdom and that the diffusible molecule c-di-GMP might be a common secondary messenger in prokaryotes.
Results
DivJ and PleC directly control PleD phosphorylation
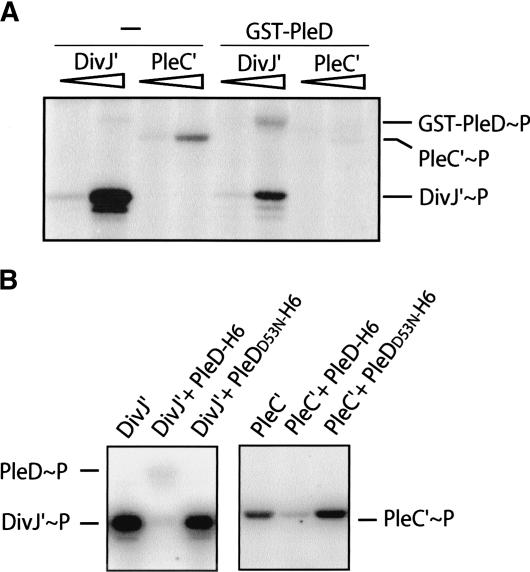
Genetic data (Sommer and Newton 1991; Aldridge et al. 2003) and in vivo phosphorylation experiments (Aldridge et al. 2003) established a role of the polar kinases DivJ and PleC in PleD control. To test whether DivJ and PleC directly modulate phosphorylation of PleD, in vitro phosphorylation assays were carried out using purified full-length PleD, fused to either a GST or a hexa-histidine tag, and purified soluble catalytic domains of DivJ (DivJ′) and PleC (PleC′). DivJ′ and PleC′ autophosphorylate in the presence of ATP and Mg2+ (Fig. 2). The addition of GST-PleD to autophosphorylated DivJ′ and PleC′ results in transfer of phosphate to PleD (Fig. 2A), whereas the purified PleD protein is not phosphorylated in the presence of ATP alone (data not shown). Autophosphorylation of purified PleC′ is relatively inefficient (Fig. 2A). Although this is in agreement with earlier findings (Hecht et al. 1995), we find that the addition of GST-PleD to the autophosphorylated soluble PleC′ kinase fragment results in a rapid loss of PleC∼P, presumably by transfer of the phosphoryl group to the response regulator (Fig. 2A). The phosphotransfer from the kinases to GST-PleD is incomplete, possibly because of interference by the N-terminal GST tag. When using a PleD-His6 fusion protein instead, efficient phosphotransfer from DivJ′ and PleC′ is observed. The addition of PleD but not PleDD53N (lacking the phosphoryl acceptor site) to autophosphorylated DivJ′ or PleC′ results in an almost complete phosphotransfer to the response regulator (Fig. 2B). Because both PleD-His6∼P and GST-PleD∼P could barely be detected, we hypothesize that under these conditions the stability of the phosphorylated form of PleD is relatively low. This suggests that both DivJ and PleC directly interact with the PleD response regulator.
Figure 2.
In vitro phosphotransfer between the protein kinases DivJ and PleC and the response regulator PleD. DivJ and PleC autophosphorylation in the presence of [γ-32P]ATP and subsequent phosphotransfer to PleD are shown. The bands corresponding to the phosphorylated proteins are marked on the side. (A) Assays contained 0.5 and 5 μg of the soluble kinase fragments and 37.5 μg GST-PleD as indicated. (B) Assays contained 12.5 μg DivJ′, 20 μg PleC′, and 50 μg PleD-H6 and PleDD53NH6, respectively, as indicated. In this experiment, DivJ′ and PleC′ were preincubated with [γ-32P]ATP for 15 min before PleD or PleDD53N were added to the reaction mix for an additional 5 min (PleC′) or 10 min (DivJ′), respectively.
The PleD response regulator dynamically localizes to the stalked pole during the cell cycle
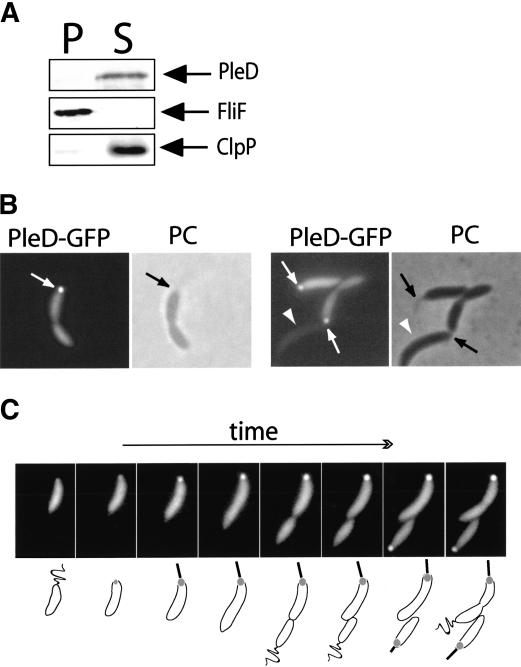
Whereas the sensor kinases DivJ and PleC are membrane-bound, the PleD response regulator is a soluble cytoplasmic protein (Fig. 3A). However, the fact that DivJ and PleC are specifically localized in the cell (Fig. 1) implies that information transfer from the sensor kinase to PleD requires the physical presence of the response regulator at the cell poles. In addition, the role of PleD in controlling assembly and function of polar organelles during development suggested that the PleD regulatory output might be restricted to the cell pole. To test the hypothesis that PleD might perform its regulatory function locally, we first analyzed the subcellular distribution of PleD during the C. crescentus cell cycle. A PleD-GFP fusion was introduced into the ΔpleD strain UJ284 on a low-copy number plasmid, and the analysis of motility and stalk formation of the resulting strain (UJ626) confirmed that the PleD-GFP fusion protein was functional (data not shown). The same fusion was also introduced into the wild-type strain CB15N (UJ627), and immunoblot analysis with anti-PleD and anti-GFP antibodies confirmed that in both strains the PleD-GFP fusion was produced at similar levels to PleD wild-type and excluded degradation of the fusion protein and the release of soluble GFP (data not shown). Analysis of strain UJ627 by fluorescence microscopy revealed that in a large fraction of stalked and predivisional cells, PleD-GFP is concentrated at the stalked pole (Fig. 3B, Table 1). From a total of 1000 cells counted, 36% had visible GFP foci at the cell pole, whereas only 4% of the cells had nonpolar foci. Localization of PleD-GFP in strains UJ626 and UJ627 was qualitatively and quantitatively indistinguishable (data not shown). Importantly, in all cases in which the identity of the cell poles could be determined unequivocally by the presence of a visible polar stalk, the GFP foci were associated with the stalked pole. This strongly implied that the PleD-GFP protein specifically localizes to the stalked pole and is absent from the flagellated swarmer pole. This, in turn, suggested dynamic behavior of the PleD protein during the C. crescentus cell cycle. To resolve the dynamic spatial distribution of PleD-GFP during the cell cycle, we performed time-lapse fluorescence microscopy with isolated swarmer cells of strain UJ627. Swarmer cells were grown directly on a microscope slide coated with a thin layer of agar, and progression though the cell cycle was visualized by phase contrast microscopy (Fig. 3C). The PleD protein is evenly distributed within C. crescentus swarmer cells, but then concentrates at the emerging stalked pole during the swarmer-to-stalked cell differentiation. With increasing time, the signal at the stalked pole increases in strength, whereas the pole opposite the stalk remains free of PleD-GFP throughout the entire cell cycle (Fig. 3C). This results in an asymmetric PleD-GFP distribution throughout most of the cell cycle and, on division, generates two different progeny cells: a swarmer cell with a uniform distribution of PleD-GFP and a stalked cell with an accumulation of PleD-GFP at the stalked pole. Only after the newborn swarmer cell has undergone the morphological transition into a stalked cell does PleD-GFP concentrate at this pole (Fig. 3C). The new poles generated by cytokinesis remain free of PleD-GFP protein.
Figure 3.
PleD is a soluble protein that localizes to the stalked pole of C. crescentus cells. (A) Immunoblot analysis of fractionated cell extracts of C. crescentus CB15N wild-type with anti-PleD, anti-FliF, and anti-ClpP antibodies. Cells of a logarithmically growing culture were lysed and soluble, and insoluble fractions were separated as indicated in Materials and Methods. Staining of the membrane-integral FliF and the soluble ClpP protein demonstrates the quality of the cell fractionation. (P) Insoluble pellet fraction; (S) soluble fraction. (B) PleD specifically localizes to the stalked pole. Phase contrast (PC) and fluorescent images of wild-type CB15N strains producing PleD-GFP from a low-copy number plasmid. The arrows indicate the polar foci of PleD-GFP in the fluorescent images and the stalk structures visible by phase contrast. (C) PleD dynamically localizes to the stalked pole during the C. crescentus cell cycle. Representative time-lapse experiment on C. crescentus wild-type cells producing PleD-GFP from a low-copy number plasmid. Fluorescent images (top) and a schematic representation of the cell cycle-dependent localization of PleD-GFP (bottom) are shown. In young swarmer cells, PleD-GFP is uniformly distributed in the cytoplasm. As the cells progress through the cell cycle and differentiate into stalked cells, PleD-GFP accumulates at the old flagellated and emerging stalked pole coinciding with flagellar ejection and stalk formation. PleD-GFP remains at the stalked pole throughout the cell cycle and is randomly dispersed in the newly formed swarmer cell. Only when the swarmer cell differentiates into a stalked cell again does PleD-GFP localize to the pole.
Table 1.
Quantitative analysis of PleD-GFP localization in Caulobacter crescentus wild-type and mutant strains
| Construct | Strain (genotype) | % cells with polar focia | % cells with non-polar foci | Total cells counted |
|---|---|---|---|---|
| PleD-GFP | CB15N (wild type) | 36 | 4 | 1000 |
| PleDD53N-GFP | CB15N (wild type) | 0 | 0 | 500 |
| PleD-GFP | UJ506 (δpleC) | 22 | 12 | 396 |
| PleD-GFP | UJ998 (divJ::ω) | 13 | 22 | 494 |
| PleD-GFP | UJ1000 (δpleC divJ::ω) | 0 | 0 | 500 |
| PleD*D53N-GFP | CB15N (wild type) | 37 | 10 | 348 |
| PleD*D53N-GFP | UJ1000 (δpleC divJ::ω) | 40 | 17 | 381 |
| PleDGG368DE-GFP | UJ284 (δpleD) | 44 | 6 | 617 |
Based on the dynamic behavior of PleD-GFP during the cell cycle (Fig. 3), about two-thirds of the cells of a mixed population are expected to show polar PleD-GFP foci. Whether this discrepancy is a biological property of the system or due to a technical property of the experiment is not clear.
Only activated PleD localizes to the pole
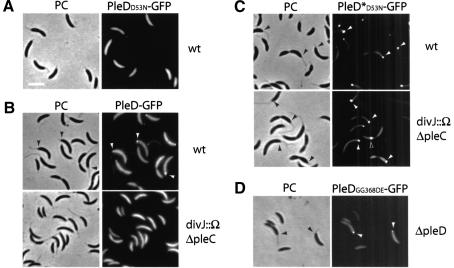
It is evident from the illustrations in Figure 3B and C that even in stalked and predivisional cells only a fraction of the PleD-GFP protein accumulates at the pole, whereas the rest seems to be evenly distributed in the cytoplasm. This is most evident from the observation that few cells, which do not seem to express the pleD-gfp copy, not only lack polar foci but also have a lower cytoplasmic fluorescence signal (Fig. 3B, short arrows). One possible explanation for this is that PleD exists in two different forms that have different targeting properties. To test whether phosphorylation of PleD is required for dynamic localization, we fused GFP to an inactive PleD mutant form that lacks the aspartic acid phosphoryl acceptor residue at position 53 (Asp 53). Immunoblot analysis confirmed that the resulting fusion protein PleDD53N-GFP is stable and produced at wild-type levels (data not shown). However, in contrast to PleD-GFP, PleDD53N-GFP is homogenously distributed in all cells and fails to accumulate at the stalked pole (Fig. 4A; Table 1), irrespective of the genetic background (data not shown). PleD-GFP also fails to localize in a mutant strain lacking both the DivJ and PleC kinases (Fig. 4B; Table 1). In this mutant PleD phosphorylation is reduced below detectable levels in vivo (Aldridge et al. 2003). A partial loss of PleD localization to the stalked pole was observed in mutants lacking either PleC or DivJ (Table 1).
Figure 4.
Dynamic localization of PleD to the stalked pole requires its activation by phosphorylation and is dependent on the polar kinases DivJ and PleC. (A) A nonphosphorylatable PleD mutant is impaired in polar localization. Phase contrast (PC) and corresponding fluorescent image of wild-type strain CB15N producing PleDD53N-GFP from a low-copy number plasmid. Bar: left panel, 2 μm. (B) DivJ and PleC are required for the polar positioning of wild-type PleD-GFP. Phase contrast (PC) and corresponding fluorescent images of wild-type and divJ pleC mutant strain producing PleD-GFP from a low-copy number plasmid. (C) DivJ and PleC are not required for localization of the constitutive mutant PleD*D53N-GFP. Phase contrast (PC) and corresponding fluorescent images of wild-type and divJ pleC mutant strain producing PleD*D53N-GFP from a low-copy number plasmid. (D) An active GGDEF output domain is not required for localization of PleD. Phase contrast (PC) and corresponding fluorescent image of a pleD mutant strain producing PleDGG368DE-GFP from a low-copy number plasmid. Filled arrows point to stalked poles in the phase contrast images (black) and to polar PleD-GFP foci in the fluorescent images (white). Open arrows point to nonpolar PleD-GFP foci.
These data suggest that phosphorylation plays a critical role in sequestering PleD to the pole. To support this and to distinguish between the possibilities that phosphorylation itself might constitute the targeting signal or, alternatively, that PleD preferentially binds to the cell pole in its active conformation, we analyzed the dynamic behavior of a constitutively active PleD mutant protein, PleD*D53N. PleD*D53N is dominant over wild-type PleD, but because it lacks the aspartic acid phosphoryl acceptor site at position 53, its activity is not dependent on phosphorylation (Aldridge et al. 2003). As shown in Figure 4C, PleD*D53N-GFP localizes almost exclusively to the cell poles in both the wild-type and the divJ pleC double mutant. The polar foci of PleD*D53N-GFP are considerably stronger than the foci observed for wild-type PleD-GFP, and the cytoplasmic signal is significantly reduced. The fact that cellular levels of the two fusion proteins are similar (data not shown) suggests that a larger fraction of the activated PleD protein is concentrated at the pole compared with the wild-type PleD-GFP fusion. Pole selectivity of PleD*D53N-GFP is similar to wild-type PleD in that the protein has a strong preference for the stalked pole and is absent from newly formed poles at division and from flagellated swarmer poles (Fig. 4C; Table 1). Pole selectivity of PleD*D53N-GFP is unaltered even in the absence of the stalked pole-specific kinase DivJ (Fig. 4C), indicating that DivJ contributes to PleD localization mainly by activating the response regulator. Together, these data are consistent with the idea that activated PleD protein is specifically targeted to the emerging stalked pole. The observation that a PleDGG368DE-GFP fusion protein, which lacks an active C-terminal output domain (see below), still localizes to the stalked pole (Fig. 4D) suggests that an activated conformation of PleD, rather than the PleD readout itself, is required for polar sequestration of the regulator.
The PleD response regulator is a di-guanylate cyclase
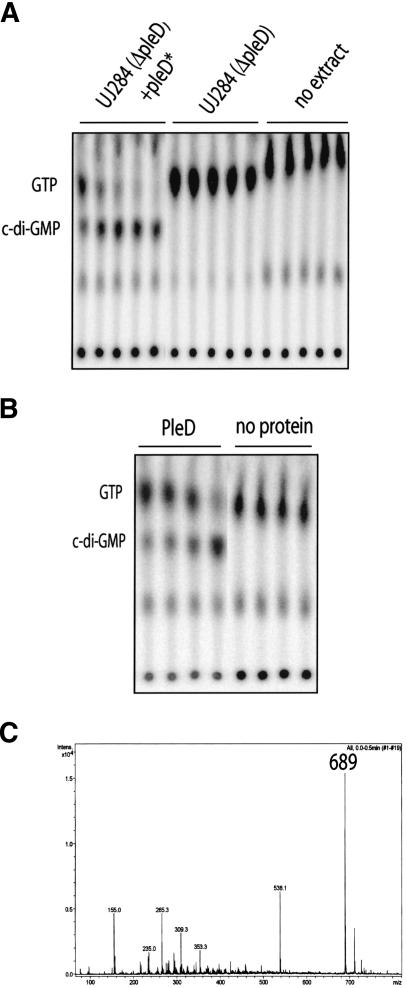
The experiments described above suggest that PleD accumulates at the old pole of the cell only in its activated state. Because genetic data indicated that PleD∼P is required for the differentiation of a flagellated into a stalked pole (Hecht and Newton 1995; Aldridge et al. 2003), PleD could act locally at this subcellular site, coordinating the developmental events involved in pole remodeling. However, what could be the output signal generated by the activated PleD response regulator, which in turn controls these downstream events? A report has established a link between a multidomain protein family containing the GGDEF domain and the metabolism of cyclic di-GMP (c-di-GMP), a compound discovered as a cofactor of cellulose synthase in Gluconacetobacter xylinum (Ross et al. 1991; Tal et al. 1998). To examine the possibility that the PleD output domain harbors di-guanylate cyclase activity, we attempted to biochemically assay its ability to convert GTP into c-di-GMP. Although extracts of C. crescentus wild-type strain CB15N and CB15N ΔpleD showed no activity (Fig. 5A), GTP was readily converted into a novel nucleotide compound when extracts of a strain containing the pleD* or pleD*D53N alleles were used (Fig. 5A; data not shown). To demonstrate that PleD was responsible for this activity, PleD with a C-terminal His-tag was overexpressed in Escherichia coli and purified to homogeneity. In the presence of purified PleD protein, GTP rapidly disappeared and was replaced by a nucleotide with a retardation factor (RF) value identical to the one observed with crude extracts (Fig. 5B).
Figure 5.
PleD is a di-guanylate cyclase. (A) Cyclic-di-GMP is produced by C. crescentus cell extracts. The soluble fraction of total cell extracts of CB15N ΔpleD (UJ284) or CB15N ΔpleD producing PleD* from a low-copy number plasmid were used to assay for di-guanylate cyclase activity. Control reactions without cell extracts are also shown. Samples were taken at 5, 10, 15, 20, and 30 min after addition of extracts and were analyzed on thin-layer chromatography plates. (B) Cyclic-di-GMP is produced by purified PleD protein. PleD-His6 (50 μg) was tested for di-guanylate cyclase activity, and the products of the enzymatic reactions were analyzed for 30, 45, 60, and 300 sec (lanes 1-4) after addition of purified PleD-His6, as indicated in A. (Lanes 5-8) Control reactions without PleD are also shown. (C) Analysis of products synthesized by PleD in vitro. The reaction products of PleD (200 μg) and GTP (1 mM) were separated by HPLC (cf. Fig. 6B). Peaks were collected and applied to mass spectrometry analysis. The fragmentation pattern shown corresponds to the main reaction product with a high-pressure liquid chromatography retention time of 6.73 min. The main peak had a molecular mass of 689 (theoretical molecular weight of c-di-GMP: 688.4). Reisolation and analysis of the substance with the molecular mass of 689 resulted in an identical fragmentation pattern.
To confirm that the novel spot indeed corresponds to the cyclic dimeric form of GMP, the reaction product of PleD and GTP was analyzed by mass spectrometry. The major peak resulting from the mass fingerprinting corresponds to a molecular mass of 689, which exactly matches the molecular weight of c-di-GMP (Fig. 5C). To gather additional evidence for the proposed enzymatic reaction, product inhibition was investigated. When chemically synthesized c-di-GMP was added to the reaction mix in a concentration range similar to the GTP substrate, strong inhibition was observed (Fig. 6A). This suggests that c-di-GMP effectively competes with GTP for the binding site. To exclude the possibility that PleD also possesses phosphodiesterase (PDE) activity and catalyzes the cleavage of c-di-GMP into two GMP monomers, c-di-GMP concentration was measured quantitatively by HPLC after incubation with purified PleD-His6 protein for several hours. Neither a decrease of the c-di-GMP concentration nor a conversion of c-di-GMP into GMP or any other degradation product was observed during a prolonged incubation period (Fig. 6B). The absence of PDE activity is not caused by the loss of PleD enzyme activity because the PleD sample used in this experiment had a high di-guanylate cyclase (DGC) activity (Fig. 6B). Together, this is consistent with the view that the PleD protein harbors a di-guanylate cyclase activity, which specifically catalyzes the conversion of GTP into the di-cyclic form of guanosine monophosphate, and that this activity constitutes the signaling output of the PleD response regulator.
Figure 6.
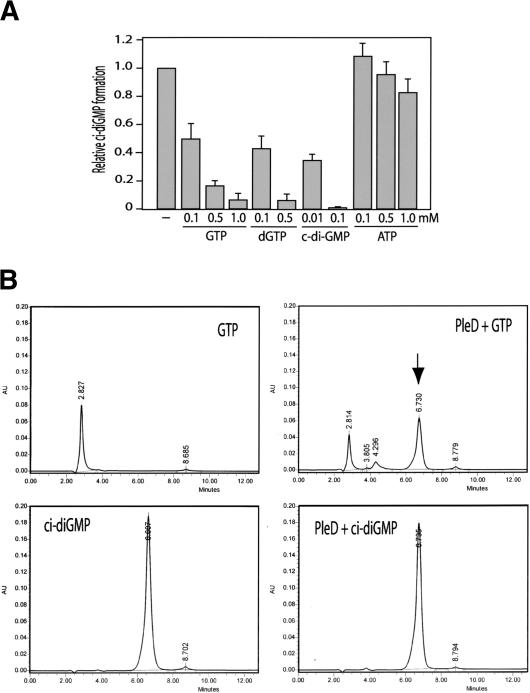
The PleD nucleotide cyclase activity is GTP-specific. (A) GTP, deoxyGTP, and c-di-GMP, but not ATP, specifically inhibit the PleD-dependent conversion of [32P]GTP into [32P]c-di-GMP. The relative c-di-GMP formation corresponds to the initial velocity determined for the enzymatic reactions. The reaction mixtures routinely contained 25 μg PleD and 100 μM GTP (see Materials and Methods) and were supplemented with nonlabeled nucleotides, as indicated below the graph. (B) PleD is a di-guanylate cyclase but lacks phosphodiesterase activity. High-pressure liquid chromatography analyses of GTP and c-di-GMP (left) and their reaction products with PleD (right) are indicated. The reaction mixtures contained 100 μg PleD and GTP and c-di-GMP at 200 μM each. Conversion of GTP into c-di-GMP by PleD is shown in the top panel by the appearance of a novel peak (arrow), which corresponds to chemically synthesized c-di-GMP (panels in second row). Incubation of PleD with chemically synthesized c-di-GMP for several hours did not lead to the cleavage or disappearance of the cyclic substance.
The PleD nucleotide cyclases activity is GTP specific
Nucleotide cyclases have been described for both adenosine and guanosine nucleotides (Domino et al. 1991; Johnson and Salomon 1991). To investigate whether the nucleotide cyclase activity of PleD is specific for GTP, we measured the synthesis of radiolabeled [32P]c-di-GMP from [α32P]GTP (0.1 mM) on addition of nonlabeled nucleotides (see Materials and Methods). As expected, an excess of unlabeled GTP efficiently inhibits the formation of [32P]-labeled c-di-GMP (Fig. 6A). Similarly, deoxyGTP was able to effectively compete with radiolabeled GTP, suggesting that GTP and deoxyGTP bind to PleD with comparable affinities (Fig. 6A). In contrast, the addition of ATP had only a marginal effect on [32P]c-di-GMP formation. Although the addition of 100 μM nonlabeled GTP reduced the formation of [32P]-labeled c-di-GMP by about 50%, the activity was unchanged in the presence of 100 and 500 μM ATP and dropped by only 10%-20% in the presence of a 10-fold higher concentration of ATP (Fig. 6A). This suggests that guanosine nucleotides bind specifically to the PleD nucleotide cyclases, whereas the affinity for ATP is significantly lower. However, HPLC analysis showed that neither ATP nor deoxyGTP were converted into their respective dimeric forms (data not shown). In summary, the nucleotide cyclase domain of PleD seems to specifically bind to guanosine nucleotides, but only GTP serves as a substrate for the formation of a dimeric product.
PleD-dependent synthesis of c-di-GMP is stimulated by phosphorylation of the receiver domain and requires an intact GGDEF output domain
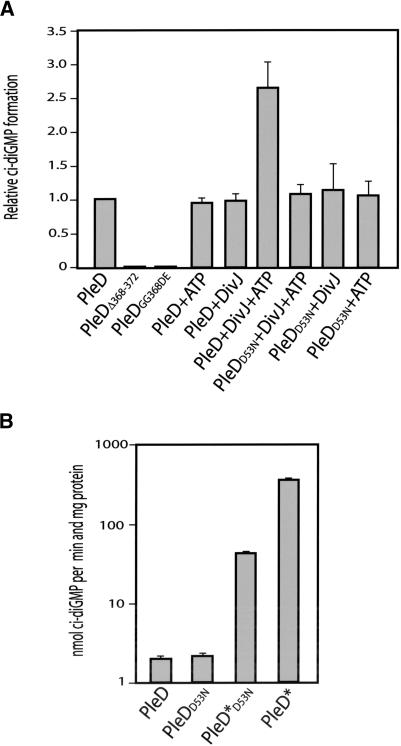
The modular architecture of the PleD response regulator suggests that the receiver domain or domains are involved in information input and that the C-terminal GGDEF domain constitutes the regulatory output of the molecule (Parkinson and Kofoid 1992). To test whether the guanylate cyclase activity is indeed localized in the GGDEF domain, we determined the activity of wild-type PleD with the activity of two mutant proteins with amino acid changes in the highly conserved GGDEF signature motif. The mutant alleles pleDΔ368-372 (lacking the entire GGDEF motif) and pleDGG368DE (two highly conserved Gly residues in GGDEF replaced by Asp and Glu) failed to complement the pleD mutant phenotype, even though their products were stably expressed in C. crescentus (Aldridge and Jenal 1999). Consistent with their functional deficiency in vivo, both mutant proteins completely lack di-guanylate cyclase activity in vitro (Fig. 7A). This is in line with the idea that the C-terminal GGDEF domain represents the output domain of PleD and is responsible for the enzymatic activity observed.
Figure 7.
Activation of the PleD di-guanylate cyclase by phosphorylation and the requirement of an intact GGDEF output domain. (A) Influence of phosphorylation and mutations in the GGDEF domain on the PleD in vitro di-guanylate cyclase activity. The enzymatic reactions contained 100 μM GTP and were carried out with 25 μg of the following purified proteins: PleD, PleDD53N, PleDΔ368-372, and PleDGG368DE. DivJ (12.5 μg) and ATP (200 μM) were added where indicated. The relative c-di-GMP formation corresponds to the initial velocity measured for the enzymatic reactions. (B) Constitutive active forms of PleD mimic the activated state of the di-guanylate cyclase. The enzymatic reactions contained 200 μM GTP and were carried out with 12.5 μg of the following purified proteins: PleD, PleDD53N, PleD*D53N, and PleD*. The relative c-di-GMP formation corresponds to the initial velocity measured for the enzymatic reactions.
Because no phospho-donors for PleD were present in the assays, our data indicate that activation of PleD by phosphorylation is not strictly required for the in vitro synthesis of c-di-GMP. This is supported by the fact that the activity of the mutant protein PleDD53N, lacking the phosphoryl acceptor side Asp 53, is comparable to wild-type PleD (Fig. 7A,B). However, when purified DivJ kinase was added to the reaction mix, the di-guanylate cyclase activity of wild-type PleD was significantly stimulated (Fig. 7A). This stimulation was not only dependent on the presence of ATP but also required residue Asp 53, as a DivJ-dependent increase of enzyme activity was not observed for the PleDD53N mutant protein (Fig. 7A). Consistent with an increase of PleD enzyme activity on activation of the molecule by phosphorylation, we found that the constitutively active mutant proteins PleD*D53N and PleD* had a considerably higher specific activity than unphosphorylated wild-type PleD (Fig. 7B). This dramatic increase of the in vitro di-guanylate cyclase activity suggests that the dominant phenotypic effects of the pleD* and pleD*D53N alleles is caused by an uncontrolled overproduction of c-di-GMP (Aldridge et al. 2003). The observation that PleD in the presence of DivJ and ATP is by far less active than purified PleD* may be the result of the modest efficiency of PleD phosphorylation in vitro (Fig. 2A). Although the molecular mechanism of PleD activation remains to be elucidated, these data clearly indicate that phosphorylation of the first receiver domain leads to an increased activity of the C-terminal guanylate cyclase domain.
Discussion
Localization of signaling molecules is a conserved mechanism for the establishment of cell polarity in both prokaryotes and eukaryotes (Shapiro et al. 2002; Nelson 2003). In C. crescentus, cell polarity and the developmental program are controlled by sensor histidine kinases, which are asymmetrically positioned at the ends of the cell (Shapiro et al. 2002). Here we present data indicating that two of these polar kinases, DivJ and PleC, control the activity and the dynamic localization of the soluble response regulator PleD during the cell cycle. The activated form of PleD, PleD∼P, possesses catalytic guanylate cyclase activity and is specifically sequestered to one pole of the cell, arguing that spatially confined synthesis of a diffusible secondary messenger might contribute to the temporal and spatial control of pole development in this organism.
Time-lapse experiments with a PleD-GFP fusion revealed a highly dynamic behavior and precise localization mechanism for the PleD response regulator during the C. crescentus cell cycle. PleD-GFP is randomly dispersed in the cytoplasm of swarmer cells but then localizes to the emerging stalked pole during cell differentiation. This dynamic positioning coincides with the localization of DivJ to the same pole (Wheeler and Shapiro 1999) and presumably precedes flagellar release and stalk formation, both of which are dependent on activated PleD (Hecht and Newton 1995; Aldridge and Jenal 1999). We have presented several lines of evidence indicating that activation of PleD by phosphorylation is critical for polar targeting and that only the activated form of the response regulator accumulates at the pole. Localization experiments with PleD*D53N-GFP suggest that it is not phosphorylation itself but, rather, an activated conformation of the protein that provides the information for polar localization. Two different mechanisms can be envisioned to explain the coupling between activity and polar localization of PleD. PleD could auto-catalytically control its own subcellular positioning, for instance, by altering the nature of the cell pole. Alternatively, pole recognition might be restricted to the activated form of PleD. The observation that the inactive PleDD53N-GFP fusion protein does not accumulate at the stalked pole in the presence of a chromosomal pleD wild-type gene, and the finding that GFP fused to PleDGG368DE, which is unable to generate c-diGMP, still sequesters to the pole in a pleD mutant, favors the second mechanism. Preliminary evidence suggests that purified PleD is able to form dimers, raising the possibility that activation and targeting might be a consequence of PleD dimerization. The observed coupling between PleD activity and polar localization is reminiscent of the mechanism observed for the single-domain response regulator DivK (Jacobs et al. 2001; Lam et al. 2003). However, although DivK∼P localizes to both the swarmer and the stalked pole, PleD∼P shows no detectable affinity for the flagellated pole. The affinity of DivK for the cell poles is also mediated through the DivJ and PleC kinases, but in contrast to PleD, which requires both kinases to be sequestered to the stalked pole, DivK targeting to the poles is mediated by DivJ, whereas PleC controls its release from the swarmer pole late in the cell cycle (Jacobs et al. 2001). Neither PleD-GFP nor the constitutive active form, PleD*D53N-GFP, localize to the pole in swarmer cells, irrespective of the presence or absence of PleC. This argues for a marker at the stalked pole that appears or is unmasked during the swarmer-to-stalked cell transition and is recognized by activated PleD. Such a marker has already been postulated for the localization of DivJ (Wheeler and Shapiro 1999), but its molecular identity remains unknown.
Targeting of active PleD to the differentiating stalked pole might serve to position the output domain of the response regulator in close proximity to the machinery that is responsible for the morphogenetic changes during cell differentiation. PleD is a di-guanylate cyclase, which on activation by phosphorylation, synthesizes c-di-GMP. This low-molecular weight molecule was originally identified as a positive allosteric effector of cellulose synthase in G. xylinum and Agrobacterium tumefaciens (Ross et al. 1987; Amikam and Benziman 1989). The conversion of glucose moieties into cellulose polymers is energetically costly for the cell, and it has been postulated that the committing step is tightly regulated by c-di-GMP to adjust the polymerization process to the cell's metabolism (Ross et al. 1991). The intracellular concentration of c-di-GMP in G. xylinum seems to be controlled by the opposing activities of DGCs and c-di-GMP-specific PDEs (Ross et al. 1987). Tal and coworkers were able to identify three operons in G. xylinum involved in cellular turnover of c-di-GMP (Tal et al. 1998; Chang et al. 2001). Each operon contains a pair of paralogous genes termed pde and dgc, which code for multidomain proteins with an N-terminal PAS/PAC domain (Ponting and Aravind 1997), a central GGDEF (DUF1) domain, and a C-terminal EAL (DUF2) domain (Galperin et al. 2001). Although genetic data suggested that a PleD homolog in Rhizobium had DGC activity (Ausmees et al. 2001), the following evidence presented in this work strongly suggests that the GGDEF domain of PleD possesses DGC, but no PDE activity: First, in vitro synthesis of c-di-GMP with C. crescentus cell extracts is dependent on the presence of a constitutively active form of PleD. Second, purified PleD protein is able to efficiently convert GTP into a nucleotide species with a molecular mass corresponding exactly to that expected for c-di-GMP. Third, the DGC activity of purified PleD is dependent on an intact GGDEF domain and the PleD cyclase activity is specific for GTP. Fourth, purified PleD possesses no detectable PDE activity (in contrast, PDE activity can readily be detected in C. crescentus whole-cell extracts; data not shown). Fifth, the in vitro DGC activity of PleD is stimulated several fold by phosphorylation through its cognate kinase DivJ. Sixth, the specific DGC activity of a phosphorylation-independent form of PleD was up to two orders of magnitude higher than that of wild-type PleD. This dramatic increase of c-di-GMP synthesis is consistent with the dominant phenotype of the pleD* allele with respect both to motility and to stalk formation (Aldridge et al. 2003). Taken together, these results support the view that the GGDEF domain represents a novel signaling domain with a bona fide DGC activity. This is in line with a recent structure prediction (Pei and Grishin 2001), which shows an excellent correspondence between GGDEF and the catalytic domain of adenylate cyclases.
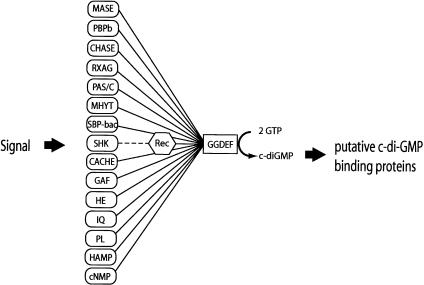
The GGDEF proteins constitute one of the largest known families of orthologs with undefined function and three-dimensional structure (Schultz et al. 1998; Tatusov et al. 2001). Whereas proteins containing a GGDEF domain are found in most bacterial species for which the genome sequence is available, they are absent in archea and eukaryotes. The analysis of the domain architecture of GGDEF proteins listed in the nonredundant protein databases reveals an intriguing pattern. The GGDEF domain seems to be highly “promiscuous,” as it is found associated as a module with a multitude of different domains. Intriguingly, all of these domains are known or proposed to be involved in signal sensing in the periplasm, the membrane, or the cytoplasm (Fig. 8). Although the nature of the signals is unclear in most cases, it has been well defined for others, like the PAS domain or hemerythrin (Gong et al. 1998; Terwilliger 1998). We propose that the GGDEF domains represent the output of a complex bacterial signal transduction network, which converts signals from different cellular compartments into the production of a secondary messenger, c-di-GMP (Fig. 8). The only two domains, which are often found associated with GGDEF and do not seem to be involved in signal sensing, are specialized metal-dependent phosphohydrolases (HD-GYP) and EAL domains. One could speculate that proteins containing both a GGDEF and an HD-GYP or EAL domain might have opposing cyclase and hydrolase activities, which contribute to the cellular level of c-di-GMP (Chang et al. 2001). The presence of a large number of potential DGCs in single bacterial species (e.g., 39 in Vibrio cholerae) raises the question of how the output specificity of parallel signaling pathways might be achieved. Our finding that C. crescentus polarized cells spatially restrict the distribution of an active DGC to the site of morphogenetic changes could offer an explanation for this dilemma. Physical proximity between c-di-GMP synthesis and action could very well be of general regulatory significance. This is in agreement with the observation that in G. xylinum, most of the c-di-GMP present in the cell seems to exist in a protein-associated rather than in a freely diffusible form (Ross et al. 1991; Weinhouse et al. 1997). An example for the compartmentalized production of a secondary messenger has been presented recently by Kriebel and collaborators, who showed that in Dictyostelium discoideum, asymmetric cellular distribution of adenylate cyclase is essential for cells to stream, possibly by contributing to the local secretion of the chemoattractant cAMP (Kriebel et al. 2003). Similarly, the observation that type VIII adenylate cyclase is enriched at cell-cell borders of endothelial cells could explain how localized changes in calcium-dependent cAMP concentrations regulate intercellular gap formation (Cioffi et al. 2002).
Figure 8.
The GGDEF domain is coupled in a modular fashion with different sensory input or information transfer domains. The domain composition of GGDEF proteins listed in the SMART protein database (Schultz et al. 1998) is shown schematically. Recognized or putative signal sensing domains are indicated in rounded rectangles, the signal transfer domain (Rec) is indicated as a polygon, and the GGDEF domain is indicated as a rectangle. Connecting bars indicate the association of domains found in a single protein. The broken line symbolizes information transfer between sensor histidine kinases (e.g., DivJ) and the receiver domain of their cognate response regulator (e.g., PleD). (Rec) Receiver domain of response regulators; (MASE) membrane-associated sensor (MASE1 and MASE2; Nikolskaya et al. 2003); (PBPb) high-affinity periplasmic solute-binding protein of ABC-type amino acid transport system; (CHASE) cyclases/histidine kinases associated sensory exracellular (Anantharaman and Aravind 2001; Mougel and Zhulin 2001; Zhulin et al. 2003); (RXAG) permease component of ribose, xylose, arabinose, galactoside ABC transporter; (PAS/C:PAS) Drosophila period clock, aryl hydrocarbon receptor, and single-minded proteins (putative signaling domain; Ponting and Aravind 1997); (PAC) PAS C-terminal motif (Ponting and Aravind 1997); (MHYT) integral membrane sensor domain (Galperin et al. 1999); (SBP-bac) bacterial extracellular solute binding protein; (SHK) Sensor histidine kinase; (CACHE) signaling domain common to Ca2+ channels and chemotaxis receptors (Anantharaman and Aravind 2000); (GAF) cGMP-specific and -stimulated phosphodiesterases/adenylate cyclases (Anabaena)/FhlA (E. coli; Galperin et al. 2001); (HE) Hemerythrin, oxygen-binding protein (Stenkamp et al. 1978); (IQ) sequence motifs for calmodulin recognition (Rhoads and Friedberg 1997); (PL) phospholamban, small protein that regulates the affinity of the cardiac sarcoplasmic reticulum Ca2+-ATPase for calcium (Smith et al. 2001); (HAMP) Histidine kinases, adenylyl cyclases, methyl-accepting proteins, phosphatases (Aravind and Ponting 1999); (cNMP) cyclic nucleotide-monophosphate-binding domain.
The finding that bacterial cells produce c-di-GMP as a regulatory compound highlights an added layer of complexity in bacterial signaling networks. What could be the cellular functions controlled by these regulatory mechanisms? Bacterial genetics has so far provided only a limited number of functional analyses of GGDEF proteins, but the results have revealed a recurring theme. In all cases, GGDEF proteins seem to be involved in the regulation of cell adhesion or cell surface colonization (Ausmees et al. 1999; Romling et al. 2000; Gronewold and Kaiser 2001; Boles and McCarter 2002; D'Argenio et al. 2002; Spiers et al. 2002; Bomchil et al. 2003). An interesting but so far poorly understood example is the HmsT protein, which allows the colonization and blockage of the flea foregut by Yersinia pestis and, as a result, the effective transmission of the plague bacillus to the mammal (Jones et al. 1999). A specific role for these novel regulatory components in adhesive behavior of bacterial cells could also help to explain why decades of (planktonic) bacterial genetics did not lead to the identification of the GGDEF network, which had to await the arrival of large-scale microbial genome analysis, bacterial cell biology, and an intensified scientific interest in microbial surface colonization.
Materials and methods
Strains, plasmids, and media
Bacterial strains and plasmids used in this study are shown in Table 2. C. crescentus strains were grown in complex peptone-yeast extract or in minimal glucose media (Ely 1991). Cultures of C. crescentus were synchronized by density gradient centrifugation as described previously (Jenal and Shapiro 1996). For conjugal transfer into C. crescentus, E. coli strain S17-1 was used as donor strain. E. coli strains were grown in Luria Broth (LB) media supplemented with antibiotics for selection, where necessary. The exact procedure of strain and plasmid construction (Table 2) is available on request.
Table 2.
Strains and plasmids
| Strain | Relevant genotype or description | Reference or source |
|---|---|---|
| Caulobacter crescentus | ||
| CB15N | Synchronizable variant strain of CB15 | Evinger and Agabian 1977 |
| UJ284 | CB15N ΔpleD | Aldridge et al. 2003 |
| UJ417 | CB15N and plasmid pPA41 | Aldridge et al. 2003 |
| UJ506 | CB15N ΔpleC | Aldridge et al. 2003 |
| UJ626 | UJ284 and plasmid pPA53-4 | This study |
| UJ627 | CB15N and plasmid pPA53-4 | This study |
| UJ998 | CB15N divJ::Ω | Aldridge et al. 2003 |
| UJ1000 | CB15N ΔpleC::Ω | Aldridge et al. 2003 |
| UJ1168 | CB15N and plasmid pPA114-47 | Aldridge et al. 2003 |
| UJ1169 | UJ284 and plasmid pPA114-47 | Aldridge et al. 2003 |
| UJ1420 | UJ1000 and plasmid pPA53-4 | This study |
| UJ1466 | UJ998 and plasmid pPA53-4 | This study |
| UJ1875 | UJ506 and plasmid pPA534 | This study |
| UJ1909 | CB15N and plasmid pSW6 | This study |
| UJ1910 | UJ284 and plasmid pSW7 | This study |
| UJ2222 | UJ506 and plasmid pSW7 | This study |
| UJ2223 | UJ998 and plasmid pSW7 | This study |
| UJ2224 | UJ1000 and plasmid pSW7 | This study |
| UJ2262 | UJ284 and plasmid pSW8 | This study |
| Escherichia coli | ||
| DH10B | F−mcrA Δ(mrr− hsd RMS−mcrBC) ϕ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara, leu)7697 araD139 galU galK nupG rpsL thi pro hsdR− hsd+ recA RP4-2-Tc::Mu-Tn7 | Simon et al. 1983 |
| BL21 (DE3) | E. coliB F−dcm ompT hsdS(rB−mB−) | Stratagene |
| pLysS | gal λ(DE3) [pLysS CAMr] | |
| BL21-CodonPlus | E. coli B F−ompT hsdS(rB− mB−) dcm+ | Stratagene |
| (DE3)-RIL | Tetrgal 1 (DE3) endA Hte [argU ileY leuW Camr] |
| Plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| pMR20 | TetR low copy number vector | Roberts et al. 1996 |
| pBAD | AmpR expression plasmid | Invitrogen |
| pGEX4T3 | AmpR vector for creation of GFP-fusion proteins | Amersham Biotech |
| pEGFP-N1 | AmpR expression plasmid | Clontech |
| pET11 | AmpR expression plasmid | Stratagene |
| pPA53-4 | pMR20; pleD-GFP under the control of divK promoter | This study |
| pSW6 | pMR20; pleDD53N-GFP under the control of divK promoter | This study |
| pSW7 | pMR20; pleD*D53N-GFP under the control of divK promoter | This study |
| pSW8 | pMR20; pleDGG368DE-GFP under the control of divK promoter | This study |
| pPA69 | pGEX4T3; pleD, N-terminal GST-tag | This study |
| pRP49 | pBAD; pleC, N-terminal His6 tag | This study |
| pRP63 | pBAD; divJ, N-terminal His6 tag | This study |
| pCC2 | pET11; pleD, C-terminal His6 tag | This study |
| pRP87 | pET11; pleDD53N, C-terminal His6 tag | This study |
| pRP88 | pET11; pleDΔ368-372, C-terminal His6 tag | This study |
| pRP89 | pET11; pleD*, C-terminal His6 tag | This study |
| pRP90 | pET11; pleD*D53N, C-terminal His6 tag | This study |
| pRP91 | pET11; pleDGG368DE, C-terminal His6 tag | This study |
Purification of PleD, PleC′, and Div′
E. coli cells carrying the respective expression plasmid were grown in LB medium with ampicillin (100 μg/mL), and expression was induced by adding either arabinose (final concentration of 0.2%) or IPTG (final concentration of 0.4 mM). After harvesting by centrifugation, the cells were resuspended in TN-buffer (50 mM Tris-HCl at pH 8.0, 500 mM NaCl, 1 mM β-mercaptoethanol), lysed by passage through a French press cell, and the suspension was clarified by centrifugation. The supernatant was loaded onto Ni-NTA affinity resin (Qiagen), washed with TN-buffer, and eluted with an imidazol-gradient. All PleD fusion proteins and the DivJ′ fragment remained soluble and were purified in native form on Ni-NTA affinity resin (Qiagen) or Glutathione-agarose (Clontech), whereas the PleC′ fragment was solubilized from inclusion bodies in guanidine hydrochloride and renatured after purification as described previously (Hecht et al. 1995). Protein preparations were examined for purity by SDS-PAGE. Fractions containing pure protein were pooled and dialyzed. C. crescentus cell extracts were prepared after harvesting cells by centrifugation and resuspension in TN-buffer. Cells were lysed by passage through a French press cell, and the extract was clarified by centrifugation. Soluble and insoluble protein fractions were separated by a high-spin centrifugation step (100,000 × g, 1 h; Jenal et al. 1994).
Enzymatic assays
Di-guanylate cyclase assays were adapted from procedures described previously (Ross et al. 1987). The reaction mixtures with purified PleD protein or Caulobacter cell extracts contained 75 mM Tris-HCl at pH 7.8, 250 mM NaCl, 25m M KCl, 10 mM MgCl2 in 50 μL volume and were started by the addition of a mixture of 0.1 mM GTP [α-32P]GTP (Amersham Biosciences; 0.01 μCi/μL). To calculate the initial velocity of product formation, aliquots were withdrawn at regular time intervals and the reaction was stopped with an equal volume of 50 mM EDTA. Reaction products (2.5 μL) were separated on polyethyleneimine-cellulose plates (Macherey-Nagel) in 1.5 M KH2PO4 (pH 3.65). Plates were exposed to a phosphor-imager screen, and the intensity of the various radioactive species was calculated by quantifying the intensities of the relevant spots using the imageQuant software (Molecular Dynamics). Measurements were always restricted to the linear range of product formation. Reaction mixtures for HPLC analyses were incubated 90 min at 25°C and terminated by heating to 95°C.
In vitro phosphorylation assays were adapted from a method described previously (Hecht et al. 1995). The proteins were incubated at 25°C for 20 min in phosphorylation buffer (50 mM Tris-HCl at pH 7.8, 25 mM NaCl, 25 mM KCl, 5 mM MgCl2) containing 5 μCi [γ-32P]ATP (Amersham Biosciences). The reactions were stopped by adding one-third volume SDS-PAGE sample buffer (250 mM Tris-HCl at pH 6.8, 40% glycerol, 8% SDS, 2.4 M β-mercaptoethanol, 0.06% bromophenol blue, 40 mM EDTA), and 32P-labeled proteins were separated by electrophoresis on 10% SDS-PAGE gels followed by autoradiography.
Synthesis and analysis of c-di-GMP
c-di-GMP was chemically synthesized as described by Ross and coworkers (Ross et al. 1990) and was purified by semipreparative reverse phase high-pressure liquid chromatography. Merck Lichrospher RP18e was used at 37°C with a 0.01 M triethylammonium carbonate buffer pH 7 containing 7.5% of methanol as mobile phase. A flow rate of 5 mL/min was used on a Hewlett Packard 1050 series system with ultrviolet detection at 252 nm. Synthetic c-di-GMP was used in comparative HPLC runs to characterize the enzymatic assay products. The conditions described above were used with a flow rate of 1 mL/min on a Waters Alliance 2690 separative module connected to a Waters 2487 ultroviolet detector. The retention time of c-di-GMP ranges between 6 and 7 min.
Both synthetic and enzymatic c-di-GMP were also analyzed by mass spectrometry. ESI mass spectrometry was conducted on a Brucker Daltonics Esquire 3000 plus instrument.
Microscopy and photography
For fluorescence imaging, C. crescentus strains were grown in peptone-yeast extract media and placed on a microscope slide that was layered with a pad of peptone-yeast extract containing 1% agarose. The slide was placed on a microscope stage at room temperature (∼22°C). Samples were observed on an Olympus AX70 microscope through a phase contrast 100× objective with a Hamamatsu C4742-95 digital camera. Images were taken and processed with Improvision Openlab and with Adobe Photoshop software.
Acknowledgments
We thank Dr. Phillip Aldridge for construction of plasmid pPA53-4 and Dr. Haim Weinhouse for technical assistance. The work was supported by Swiss National Science Foundation fellowships 31-59050.99 to U.J.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.289504.
References
- Aldridge P. and Jenal, U. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32: 379-391. [DOI] [PubMed] [Google Scholar]
- Aldridge P., Paul, R., Goymer, P., Rainey, P., and Jenal, U. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47: 1695-1708. [DOI] [PubMed] [Google Scholar]
- Amikam D. and Benziman, M. 1989. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 171: 6649-6655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V. and Aravind, L. 2000. Cache—a signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25: 535-537. [DOI] [PubMed] [Google Scholar]
- ____. 2001. The CHASE domain: A predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem. Sci. 26: 579-582. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Ponting, C.P. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176: 111-116. [DOI] [PubMed] [Google Scholar]
- Ausmees N., Jonsson, H., Hoglund, S., Ljunggren, H., and Lindberg, M. 1999. Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii. Microbiology 145: 1253-1262. [DOI] [PubMed] [Google Scholar]
- Ausmees N., Mayer, R., Weinhouse, H., Volman, G., Amikam, D., Benziman, M., and Lindberg, M. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol Lett. 204: 163-167. [DOI] [PubMed] [Google Scholar]
- Boles B.R. and McCarter, L.L. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184: 5946-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomchil N., Watnick, P., and Kolter, R. 2003. Identification and characterization of a Vibrio cholerae gene, mbaA, involved in maintenance of biofilm architecture. J. Bacteriol. 185: 1384-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A.L., Tuckerman, J.R., Gonzalez, G., Mayer, R., Weinhouse, H., Volman, G., Amikam, D., Benziman, M., and Gilles-Gonzalez, M.A. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40: 3420-3426. [DOI] [PubMed] [Google Scholar]
- Cioffi D.L., Moore, T.M., Schaack, J., Creighton, J.R., Cooper, D.M., and Stevens, T. 2002. Dominant regulation of inter-endothelial cell gap formation by calcium-inhibited type 6 adenylyl cyclase. J. Cell Biol. 157: 1267-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Argenio D.A., Calfee, M.W., Rainey, P.B., and Pesci, E.C. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184: 6481-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domino S.E., Tubb, D.J., and Garbers, D.L. 1991. Assay of guanylyl cyclase activity. In Methods in enzymology (eds. R.A. Johnson and J.D. Corbin), pp. 345-355. Academic Press, San Diego, CA. [DOI] [PubMed]
- Ely B. 1991. Genetics of Caulobacter crescentus. Meth. Enzymol. 204: 372-384. [DOI] [PubMed] [Google Scholar]
- Evinger M. and Agabian, N. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132: 294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M.Y., Natale, D.A., Aravind, L., and Koonin, E.V. 1999. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1: 303-305. [PMC free article] [PubMed] [Google Scholar]
- Galperin M.Y., Nikolskaya, A.N., and Koonin, E.V. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203: 11-21. [DOI] [PubMed] [Google Scholar]
- Gong W., Hao, B., Mansy, S.S., Gonzalez, G., Gilles-Gonzalez, M.A., and Chan, M.K. 1998. Structure of a biological oxygen sensor: A new mechanism for heme-driven signal transduction. Proc. Natl. Acad. Sci. 95: 15177-15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronewold T.M. and Kaiser, D. 2001. The act operon controls the level and time of C-signal production for Myxococcus xanthus development. Mol. Microbiol. 40: 744-756. [DOI] [PubMed] [Google Scholar]
- Hecht G.B. and Newton, A. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177: 6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht G.B., Lane, T., Ohta, N., Sommer, J.M., and Newton, A. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14: 3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C., Hung, D., and Shapiro, L. 2001. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. 98: 4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U. and Shapiro, L. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 15: 2393-2406. [PMC free article] [PubMed] [Google Scholar]
- Jenal U., White, J., and Shapiro, L. 1994. Caulobacter flagellar function, but not assembly, requires FliL, a non-polarly localized membrane protein present in all cell types. J. Mol. Biol. 243: 227-244. [DOI] [PubMed] [Google Scholar]
- Johnson R.A. and Salomon, Y. 1991. Assay of adenylyl cyclase catalytic activity. In Methods in enzymology (eds. R.A. Johnson and J.D. Corbin), pp. 3-21. Academic Press, San Diego, CA. [DOI] [PubMed]
- Jones H.A., Lillard Jr., J.W., and Perry, R.D. 1999. HmsT, a protein essential for expression of the haemin storage (Hms+) phenotype of Yersinia pestis. Microbiology 145: 2117-2128. [DOI] [PubMed] [Google Scholar]
- Kriebel P.W., Barr, V.A., and Parent, C.A. 2003. Adenylyl cyclase localization regulates streaming during chemotaxis. Cell 112: 549-560. [DOI] [PubMed] [Google Scholar]
- Lam H., Matroule, J.Y., and Jacobs-Wagner, C. 2003. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev. Cell. 5: 149-159. [DOI] [PubMed] [Google Scholar]
- Mougel C. and Zhulin, I.B. 2001. CHASE: An extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem. Sci. 26: 582-584. [DOI] [PubMed] [Google Scholar]
- Nelson W.J. 2003. Adaptation of core mechanisms to generate cell polarity. Nature 422: 766-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolskaya A.N., Mulkidjanian, A.Y., Beech, I.B., and Galperin, M.Y. 2003. MASE1 and MASE2: Two novel integral membrane sensory domains. J. Mol. Microbiol. Biotechnol. 5: 11-16. [DOI] [PubMed] [Google Scholar]
- Ohta N. and Newton, A. 2003. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J. Bacteriol. 185: 4424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J.S. and Kofoid, E.C. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26: 71-112. [DOI] [PubMed] [Google Scholar]
- Pei J. and Grishin, N.V. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42: 210-216. [DOI] [PubMed] [Google Scholar]
- Ponting C.P. and Aravind, L. 1997. PAS: A multifunctional domain family comes to light. Curr. Biol. 7: 674-677. [DOI] [PubMed] [Google Scholar]
- Rhoads A.R. and Friedberg, F. 1997. Sequence motifs for calmodulin recognition. FASEB J. 11: 331-340. [DOI] [PubMed] [Google Scholar]
- Roberts R.C., Toochinda, C., Avedissian, M., Baldini, R.L., Gomes, S.L., and Shapiro, L. 1996. Identification of a Caulobacter crescentus operon encoding hrcA, involved in negatively regulating heat-inducible transcription, and the chaperone gene grpE. J. Bacteriol. 178: 1829-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romling U., Rohde, M., Olsen, A., Normark, S., and Reinkoster, J. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36: 10-23. [DOI] [PubMed] [Google Scholar]
- Ross P., Weinhouse, H., Aloni, Y., Michaeli, D., Weinberger-Ohana, P., Mayer, R., Braun, S., de Wroom, E., van der Marel, G.A., van Boom, J.H., et al. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325: 279-281. [DOI] [PubMed] [Google Scholar]
- Ross P., Mayer, R., Weinhouse, H., Amikam, D., Huggirat, Y., Benziman, M., de Vroom, E., Fidder, A., de Paus, P., Sliedregt, L.A., et al. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265: 18933-18943. [PubMed] [Google Scholar]
- Ross P., Mayer, R., and Benziman, M. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55: 35-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz, F., Bork, P., and Ponting, C.P. 1998. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. 95: 5857-5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L., McAdams, H.H., and Losick, R. 2002. Generating and exploiting polarity in bacteria. Science 298: 1942-1946. [DOI] [PubMed] [Google Scholar]
- Simon R., Prieffer, U., and Puhler, A. 1983. A broad host range mobilization system for in vivo genetic engineering: Transposon mutagenesis in gram negative bacteria. Biotechnology 1: 784-790. [Google Scholar]
- Smith S.O., Kawakami, T., Liu, W., Ziliox, M., and Aimoto, S. 2001. Helical structure of phospholamban in membrane bilayers. J. Mol. Biol. 313: 1139-1148. [DOI] [PubMed] [Google Scholar]
- Sommer J.M. and Newton, A. 1991. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics 129: 623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers A.J., Kahn, S.G., Bohannon, J., Travisano, M., and Rainey, P.B. 2002. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161: 33-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenkamp R.E., Sieker, L.C., Jensen, L.H., and McQueen Jr., J.E. 1978. Structure of methemerythrin at 2.8-Angstrom resolution: Computer graphics fit of an averaged electron density map. Biochemistry 17: 2499-2504. [DOI] [PubMed] [Google Scholar]
- Tal R., Wong, H.C., Calhoon, R., Gelfand, D., Fear, A.L., Volman, G., Mayer, R., Ross, P., Amikam, D., Weinhouse, H., et al.. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: Genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180: 4416-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R.L., Natale, D.A., Garkavtsev, I.V., Tatusova, T.A., Shankavaram, U.T., Rao, B.S., Kiryutin, B., Galperin, M.Y., Fedorova, N.D., and Koonin, E.V. 2001. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29: 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger N.B. 1998. Functional adaptations of oxygen-transport proteins. J. Exp. Biol. 201: 1085-1098. [DOI] [PubMed] [Google Scholar]
- Weinhouse H., Sapir, S., Amikam, D., Shilo, Y., Volman, G., Ohana, P., and Benziman, M. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416: 207-211. [DOI] [PubMed] [Google Scholar]
- Wheeler R. and Shapiro, L. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4: 683-694. [DOI] [PubMed] [Google Scholar]
- Zhulin I.B., Nikolskaya, A.N., and Galperin, M.Y. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185: 285-294. [DOI] [PMC free article] [PubMed] [Google Scholar]