Abstract
Studies of a hybrid zone between two house mouse subspecies (Mus musculus musculus and M. m. domesticus) along with studies using laboratory crosses reveal a large role for the X chromosome and multiple autosomal regions in reproductive isolation as a consequence of disrupted epistasis in hybrids. One limitation of previous work has been that most of the identified genomic regions have been large. The goal here is to detect and characterize precise genomic regions underlying reproductive isolation. We surveyed 1401 markers evenly spaced across the genome in 679 mice collected from two different transects. Comparisons between transects provide a means for identifying common patterns that likely reflect intrinsic incompatibilities. We used a genomic cline approach to identify patterns that correspond to epistasis. From both transects, we identified contiguous regions on the X chromosome in which markers were inferred to be involved in epistatic interactions. We then searched for autosomal regions showing the same patterns and found they constitute about 5% of autosomal markers. We discovered substantial overlap between these candidate regions underlying reproductive isolation and QTL for hybrid sterility identified in laboratory crosses. Analysis of gene content in these regions suggests a key role for several mechanisms, including the regulation of transcription, sexual conflict and sexual selection operating at both the postmating prezygotic and postzygotic stages of reproductive isolation. Taken together, these results indicate that speciation in two recently diverged (c. 0.5 Ma) house mouse subspecies is complex, involving many genes dispersed throughout the genome and associated with distinct functions.
Keywords: genomics/proteomics, hybridization, mammals, speciation
Introduction
A crucial step in speciation according to the biological species concept (Dobzhansky 1937; Mayr 1942) is the formation of reproductive barriers between emerging independently evolving lineages. Postzygotic reproductive barriers such as hybrid sterility and inviability are often caused by Bateson–Dobzhansky–Muller incompatibilities (BDMIs) (Bateson 1909; Dobzhansky 1936; Muller 1940, 1942; Coyne & Orr 2004). The BDMI model is based on the accumulation of mutations in allopatric populations that cause incompatibilities between interacting genes in hybrid genomes when the populations come together. Another common phenomenon associated with postzygotic isolation is the observation that the X chromosome plays a disproportionately large role in hybrid sterility, termed the large X-effect (Charlesworth et al. 1987; Coyne & Orr 1989; Masly & Presgraves 2007). The large X-effect seems to be a consequence of a higher density of BDMIs on the X chromosome than on the autosomes, although the underlying cause of this higher density is still unclear and may be due to faster evolution of the X, sex-ratio meiotic drive or the disruption of gene regulation on the X during spermatogenesis (Presgraves 2008). BDMIs between the X chromosome and the autosomes also provide an explanation for Haldane’s rule (Muller 1940, 1942; Turelli & Orr 1995; Zeng 1996), a phenomenon whereby the heterogametic sex is the sex that suffers from reduced viability or fertility in hybrid genomes (Haldane 1922; Orr 1997). Haldane’s rule together with the large X-effect may apply more generally to taxa with differentiated sex chromosomes (Coyne & Orr 2004).
Most of the pioneering work on the genetic basis of reproductive isolation has been performed on fruit flies of the genus Drosophila (Coyne & Orr 2004). The experiments support a prominent role of the X chromosome in the formation of postzygotic reproductive barriers (Coyne & Orr 1989). However, the importance of autosomal factors in the formation of reproductive barriers has also been confirmed (True et al. 1996; Orr & Irving 2001; Tao et al. 2003; Masly & Presgraves 2007; Lu et al. 2010).
The importance of the X chromosome in the formation of a reproductive barrier has also been documented in laboratory crosses of mouse strains (Oka et al. 2004, 2007; Storchová et al. 2004; Good et al. 2008a,b, 2010). Several QTL studies identified the central portion of the X chromosome as contributing to hybrid male sterility (Oka et al. 2004, 2007; Storchová et al. 2004). In addition, Good et al. (2008b) and White et al. (2011) identified more proximal parts of the X chromosome as contributing to hybrid male sterility. Moreover, the data of Good et al. (2008b) suggest that multiple regions of the X appear to be associated with hybrid male sterility, emphasizing the importance of the X as a whole. More recently, Good et al. (2010) reported extensive overexpression of X-linked genes in the testes of sterile but not fertile hybrid mice and suggested that sterility might be due to a general disruption of X chromosome gene expression during male meiosis.
Despite the importance of the X chromosome, a significant contribution of autosomes to hybrid male sterility has also been suggested in mice. It has been known for a long time that chromosome 17 harbours gene(s) involved in hybrid male sterility (Forejt & Iványi 1974; Forejt 1996; Vyskočilová et al. 2005, 2009; White et al. 2011), and the first hybrid sterility gene (Prdm9) was positionally cloned to the proximal part of chromosome 17 (Mihola et al. 2009). However, it is clear from mapping studies that other autosomes also contribute to hybrid sterility (Oka et al. 2007; White et al. 2011). In addition, consomic strains carrying each Mus musculus musculus chromosome on a largely M. m. domesticus background showed reduced fertility for nearly every autosome (Gregorová et al. 2008). Although the male and female effects of fertility were not explored separately in this study, the consomic strains demonstrate that genes underlying hybrid sterility map to most autosomes. The genetic basis of hybrid sterility in mice is clearly complex, and laboratory crosses may be inefficient in identifying specific genes because the genomic regions identified in mapping studies tend to be quite large (Storchová et al. 2004; Good et al. 2008a; Vyskočilová et al. 2009; White et al. 2011).
From this perspective, hybrid zones, narrow regions where two genetically diversified populations meet, may be more appropriate for the study of the genetic basis of reproductive isolation (Payseur 2010). Barton & Hewitt (1985) argued that most naturally occurring hybrid zones are tension zones maintained by the dispersal of parental types and selection against hybrids as a result of incompatibilities between co-adapted parental genomes. From multilocus cline theory (Barton 1983; Barton & Gale 1993), it is known that unless the strength of selection (s) exceeds the rate of recombination (r) between loci (described by the coupling coefficient: θ = s/r), different parts of chromosomes behave partially independently enabling neutral loci to escape the influence of loci under selection, thus making it possible to distinguish regions associated with negatively selected reproductive barriers from neutral regions. In instances where hybrids represent complex mosaics of parental genomes because of multigenerational backcrossing and intercrossing, it is possible to distinguish regions under selection on a much finer scale than is possible with laboratory crosses (Rieseberg & Buerkle 2002; Buerkle & Lexer 2008; Payseur 2010; White et al. 2011; Baird & Macholán 2012).
The two house mouse subspecies (M. m. musculus and M. m. musculus) which diverged from a common ancestor approximately 500 000 BP (Boursot et al. 1996; Suzuki et al. 2004; Chevret et al. 2005; Salcedo et al. 2007; Geraldes et al. 2008, 2011; Duvaux et al. 2011) form a narrow hybrid zone that extends from Bulgaria to Denmark (Boursot et al. 1993; Macholán et al. 2003). Recently, hybrid mice have been reported also from Norway (Jones et al. 2010). The house mouse hybrid zone is a tension zone as suggested by Barton & Hewitt (1985) and confirmed by others (Payseur et al. 2004; Raufaste et al. 2005; Macholán et al. 2007). Reduced fertility of hybrid males probably plays the most important role in maintaining the tension zone as it has been demonstrated in laboratory crosses (Forejt & Iványi 1974; Forejt 1996; Storchová et al. 2004; Britton-Davidian et al. 2005; Vyskočilová et al. 2005, 2009; Good et al. 2008a,b, 2010; Mihola et al. 2009; White et al. 2011) and was recently reported directly from the centre of the hybrid zone (Turner et al. 2012). In addition, increased parasite loads in hybrids have been suggested (Sage et al. 1986; Moulia et al. 1993); however, recently, the opposite has been reported from a different part of the hybrid zone (Baird et al. 2012; Gouy de Bellocq et al. 2012). Also, hybrid female sterility has been proposed to contribute to hybrid failure (Britton-Davidian et al. 2005).
Multiple studies using cline theory (Nagylaki 1975; Barton 1979, Barton 1983, Barton & Hewitt 1985; Barton & Gale 1993; Gompert & Buerkle 2009, 2010) have demonstrated selection acting on X-linked markers in the house mouse (Tucker et al. 1992; Dod et al. 1993; Macholán et al. 2007, 2008; Teeter et al. 2008). Other studies (Payseur et al. 2004; Dufková et al. 2011; Macholán et al. 2011) have identified the central part of the X chromosome as a particularly important region harbouring loci involved in reproductive isolation, in accord with the results of laboratory crosses (Oka et al. 2004; Storchová et al. 2004). Teeter et al. (2008) found some autosomal regions to be potentially contributing to reproductive isolation, including a few that are in linkage disequilibrium with X-linked markers. However, that study used fairly sparse marker coverage, so the extent to which the X chromosome interacts epistatically with different autosomal regions is still unclear.
Here, we present a whole genome analysis of hybrid mice collected from two previously studied transects across the hybrid zone (Wang et al. 2011). One transect is located in southern Bavaria and Austria (BV), and the second transect is located 200 km to the north in Bavaria and the Czech Republic (CZ). By comparing the patterns of introgression in two transects, we identified common patterns that are likely to reflect intrinsic genetic incompatibilities. We surveyed 1401 SNP markers (Wang et al. 2011) across the genome and analysed these data using a genomic cline approach (Gompert & Buerkle 2009, 2010). This approach compares individual loci to the average pattern for all loci and assesses whether loci are neutral or under selection. Here, we focus on those loci inferred to be involved in epistasis based on their non-neutral patterns of introgression, following Gompert & Buerkle (2009). First, we compared introgression patterns between pairs of markers on the X chromosome and identified adjacent markers with similar epistatic patterns of introgression in both transects. We then compared these regions with autosomal markers to identify candidate BDMIs. We found that these regions harbour genes involved in a number of distinct processes including transcriptional regulation, sexual conflict and sexual selection.
Materials and methods
Mice
House mice were collected from the CZ and BV transects (Wang et al. 2011). Because X-linked markers are haploid in males, X-autosome comparisons are only possible using data from females. The BV transect is located 200 km south of the CZ transect. It is one-dimensional extending 176 km in a linear east–west direction from south-central Bavaria, Germany, to north-western Austria (Tucker et al. 1992; Payseur et al. 2004; Božíková et al. 2005). A total of 254 females of 432 total mice collected from 41 localities in three different years (1984, 1985 and 1992) were used. The CZ is two-dimensional and is 130 km long × 50 km wide, extending from north-eastern Bavaria, Germany, to western Bohemia, Czech Republic (Božíková et al. 2005; Macholán et al. 2007, 2008, 2011). A total of 425 females of 869 mice collected from 79 localities between 1991 and 2003, with >95.5% collected since 1997, were used.
SNP development and genotyping
A previously published data set of 1401 SNPs, diagnostic for the two subspecies and providing reliable genotyping calls (Wang et al. 2011), was used. The data matrix consisted of 1316 autosomal and 85 X-linked SNP markers. The median and mean gap sizes between two SNPs are approximately 1.86 Mb. However, several gaps are present owing either to the lack of coverage in the Mouse Diversity Array (Yang et al. 2009), from which markers were selected, or by human factors. For a detailed description of SNPs used and the matrix of genotypes, see Wang et al. (2011).
Genomic clines
To describe the behaviour of SNP markers from across the genome, we started with genomic cline analysis using the Introgress program (Gompert & Buerkle 2009, 2010). The analysis uses a multinomial regression approach to characterize the behaviour of genetic markers along a genomic admixture gradient represented by individual hybrid index scores estimated using a previously published method (Buerkle 2005). The behaviour of a marker is represented as a probability of observing a particular genotype given the hybrid index (Gompert & Buerkle 2009, 2010) and the likelihood of this model is compared to the likelihood of observing the data under a neutral model. Following Gompert & Buerkle (2009), four alternative patterns of selection are considered including overdominance (in which heterozygous genotypes are observed more than expected), underdominance (in which heterozygous genotypes are observed less than expected), directional selection (in which individual clines are shifted relative to the genome average) or epistasis (defined in this context as interactions between a single locus and the genomic background and characterized by steep homozygote clines and high and sharp heterozygote clines). Here, we focus particularly on those loci inferred to be involved in epistatic interactions. Hybrid indexes were based on 1316 autosomal loci (Wang et al. 2011). To obtain P-values estimates, the data were permuted 1000 times. The two significance levels (Pintrogress ≤ 0.01 and Pintrogress ≤ 0.001) were used to distinguish markers with patterns significantly differ from the neutral prediction.
Overall strategy
Previous work has shown that the demographic dynamics of this hybrid zone are quite different in these two transects (Wang et al. 2011), making it difficult to compare patterns directly. Because we know that the X chromosome underlies reproductive isolation between these taxa and because we were interested in identifying markers involved in BDMIs, we began by identifying regions on the X chromosome that show evidence of epistasis and where adjacent markers are not significantly different from one another, termed X-linked uniform regions (XURs; see details below). We then compared XURs between transects and identified those with similar patterns of introgression. Subsequently, these were compared to autosomal regions to identify those autosomal regions that may be important in reproductive isolation.
Comparison of introgression models between pairs of markers
The genomic cline approach was modified to compare introgression models between pairs of markers. Introgression models between pairs of X-linked markers were compared to identify adjacent markers on the X with similar patterns of introgression (described below). The mode of selection for all markers from each region was identified using results from the original genomic cline analysis.
A log-likelihood ratio of introgression models (Mm1 and Mm2) for a pair of markers (m1 and m2) was used as a measure of similarity between their models. Models Mm1 and Mm2 were inferred by the approach implemented in the original version of INTROGRESS (Gompert & Buerkle 2009, 2010), and a log-likelihood ratio of the Mm1 and Mm2 models given the marker m1 data (Dm1) was obtained as follows:
For the comparison of X-linked markers, markers m1 and m2 were assigned arbitrarily, while for the X-linked vs. autosomal markers, m2 was assigned to X-linked markers. A significance test for the two model comparison was conducted by the permutation of the m2 data. Genotypes for each individual were chosen according to assigned weights represented by conditional probabilities of genotypes given hybrid index as inferred by the approach implemented in the original version of the Introgress program (Gompert & Buerkle 2009, 2010). P-value estimates were then obtained by comparing log-likelihood ratio of two marker models (Mm1 and Mm2) with the distribution of log-likelihood ratios for Mm2 and models based on 1000 times permuted m2 data. The two significance levels (Ppairwise ≤ 0.01 and Ppairwise ≤ 0.001) were used to distinguish whether the behaviour of the two markers significantly differ from each other. All computations were conducted in the R statistical environment (R Development Core Team 2005) using a modification of the INTROGRESS package (Gompert & Buerkle 2009, 2010).
Characterizing the structure of X chromosome introgression
To identify adjacent regions exhibiting similar patterns of introgression along the X chromosome, log-likelihood ratios and P-values for pairs of markers along their physical position on the chromosome were plotted. The regions of uniform introgression, XURs, were identified using a P-value cut-off >0.01 for pairs of markers in each transect separately. This cut-off provides a conservative estimate, thus minimizing false positives when comparing X-linked markers to autosomes. The overlap between XURs in the two transects were subsequently defined as shared X-linked uniform regions (SXURs). However, only XURs comprising markers with non-neutral patterns of introgression obtained by the original version of the Introgress program (Gompert & Buerkle 2009) were used.
Distribution of autosomal markers within and between chromosomes and between transects
Autosomal markers exhibiting similar patterns to X-linked markers were plotted along their physical position. Adjacent markers were pooled into regions, and the distribution of the region sizes was tested against a distribution of region sizes based on the permutations of marker positions. The procedure was repeated 1000 times. The aim of this approach was to test whether markers putatively under selection clustered more often than expected by chance alone.
Significance of over/under enrichment was also tested using a hypergeometric distribution as suggested by Boyle et al. (2004). Bonferroni correction was used to treat for multiple comparisons. Results of these two tests were then compared between the two transects.
To reveal shared regions harbouring candidate loci associated with reproductive isolation, a comparison of identified markers between the two transects was conducted. As any overlap might have arisen by chance alone, the significance of the overlap was tested. The positions of identified regions were permuted taking into account the structure of the autosomal genome (i.e. varying size of chromosomes and regions harbouring candidate loci). This procedure was repeated 1000 times with the number of overlapping markers recorded each time. P-value estimates were obtained by comparing the observed number of markers shared between the two transects against the distribution of simulated overlaps.
Gene enrichment in regions exhibiting epistasis
Gene content was characterized for both X-linked and autosomal SNP markers identified as exhibiting epistasis with gene content restricted to within 1 Mb of the marker or in cases when an adjacent marker was closer than 1 Mb, to within the total distance between the two markers. Genes were downloaded from the Ensembl website (http://www.ensembl.org) using the BioMart tool (http://www.biomart.org). Only genes identified as protein-coding genes [protein-coding genes, IG_(C/D/J/V)] were included for further analysis. BioMart was also used to assign Ensembl Ids to Gene Ontology (GO) term names (Ashburner et al. 2000). GO terms with <50 genes in the whole genome (X, autosomes) were excluded from further analyses. The enrichment of particular GO terms within the identified regions was characterized. The ‘biological process’ and ‘molecular function’ domains were tested separately in each transect for autosomal regions only. The GO term enrichment was tested for X chromosomal and autosomal regions for cases in which markers overlapped between transects and then for autosomal regions for each transect separately. A hypergeometric distribution as suggested by Boyle et al. (2004) was used to identify significantly enriched GO terms. As a threshold of significance, a q-value representing a false discovery rate (FDR) provided by qvalue R package (Storey & Tibshirani 2003) was used.
Results
The pattern of introgression on the X chromosome
We characterized the patterns of introgression for 85 markers along the X chromosome in the CZ and BV transects. With the exception of a 10.5-Mb gap located between positions 23 and 33.5 Mb along the chromosome, the markers were approximately evenly spaced.
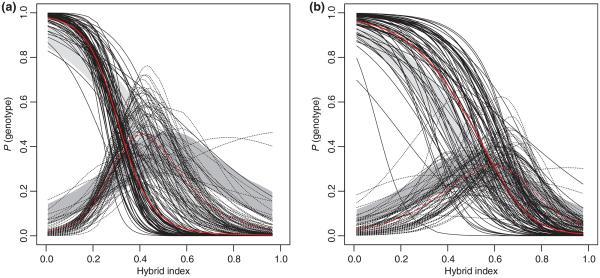
We used the Introgress program (see Material and Methods) to assess the prevailing pattern of introgression along the X chromosome in both transects and compared it to the neutral prediction based on the averaged introgression of 1316 autosomal markers (Fig. 1). Epistasis of varying strength (see Gompert & Buerkle 2009) is inferred to be a prevailing non-neutral pattern in both transects. The average genomic cline over all X-linked markers exhibits overabundance of M. m. musculus genotypes with respect to the null model prediction (autosomal average) in the BV transect, while the opposite is true for the CZ transect.
Fig. 1.
Genotype probabilities given hybrid index (genomic clines) for 85 X-linked markers as provided by the Introgress program (Gompert & Buerkle 2009) for (a) BV and (b) CZ transect (solid lines: homozygotes for M. m. domesticus alleles; dashed lines: heterozygotes). Null expectations (light grey: homozygotes for M. m. domesticus alleles; dark grey: heterozygotes) are based on the permutation of autosomal data (1316 markers). Hybrid index used from Wang et al. (2011). Red lines represent average genomic clines for X-linked markers.
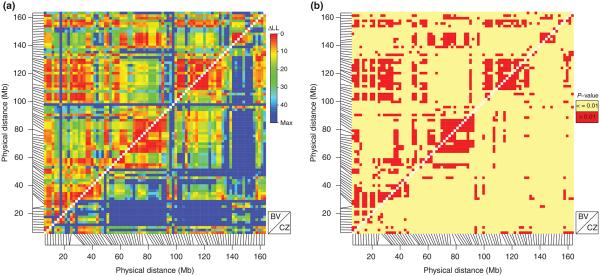
To compare the patterns of introgression along the X chromosome, the log-likelihood ratios and P-values estimates were calculated for all pairs of X-linked markers using the modified version of the Introgress program (see Material and Methods). When log-likelihood ratios were plotted against physical map position, a distinct pattern of introgression emerged (Fig. 2a). The whole X chromosome was partitioned into several large regions, interestingly, in a similar way in both transects. The only difference was found to be in the degree of independence these regions exhibited across the length of the chromosome. Chromosomal regions in the BV tran-sect introgressed in a more similar way than in the CZ transect. To find regions along the X with similar patterns of introgression, XURs were defined. They are represented by adjacent markers with a homogeneous pattern of introgression for which the log-likelihood ratio between them did not exceed the sampling error (P-value >0.01) (Figs 2b and 3). These included 19 and 16 XURs in the BV and the CZ transects, respectively (Fig. 3, Table S1a,b, Supporting information). The sizes of XURs varied from 0.7 to 17.0 Mb in the BV transect and from 1.4 to 14 Mb in the CZ transect and they included from 2 to 9 (BV) and 2 to 8 (CZ) SNP markers, respectively. Almost all of the markers from within XURs exhibited introgression patterns significantly deviated from neutral prediction as provided by the Introgress program (Gompert & Buerkle 2009). The only exceptions were XUR 2 and 3 in the CZ transect. Assuming a common reproductive basis for these patterns across hybrid populations, we then identified overlapping XURs between transects, defining them as SXURs. We identified 10 SXURs comprising 29 X-linked markers (Fig. 3, Table 1). The identified SXURs were based on the overlap between transect-specific XURs. The size of these groups ranged from 1.2 to 10.1 Mb and included from 2 to 5 markers.
Fig. 2.
Comparison of introgression models for pairs of markers plotted along their physical positions on the X chromosome (BV: above diagonal; CZ: below diagonal). (a) log-likelihood ratios (ΔLLs) for pairs of markers. The more similar behaviour of the two markers the lower ΔLL. (b) statistical significance of pairwise comparisons (red: behaviour of the two markers is not significantly different). Plotted in the R statistical environment (http://www.r-project.org/) using modified version of LDHEATMAP package (Shin et al. 2006).
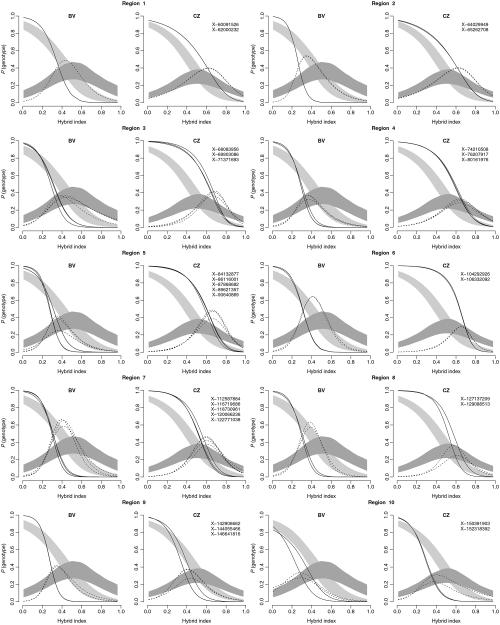
Fig. 3.

The X-linked uniform regions (XURs) were defined based on the P-value estimates for the pairwise comparisons of X-linked markers in each transect separately (black lines). Red lines signify regions containing markers not significantly different from neutral expectations based on the permutation of autosomal data (1316 markers). The overlapping regions between XURs are considered as shared X-linked uniform regions (SXURs) (blue rectangles).
Table 1.
Shared X chromosome linked uniform regions (SXURs) with candidate epistatically interacting loci as identified from both transects
| Region ID | No markers | Begin (bp) | End (bp) | Size (Mb) | Marker positions | Marker name |
|---|---|---|---|---|---|---|
| 1 | 2 | 60 091 526 | 62 000 232 | 1.909 | 28, 29 | X-60091526, X-62000232 |
| 2 | 2 | 64 029 949 | 65 262 708 | 1.233 | 30, 31 | X-64029949, X-65262708 |
| 3 | 3 | 68 083 956 | 71 371 693 | 3.288 | 33, 34, 35 | X-68083956, X-69903086, X-71371693 |
| 4 | 3 | 74 210 508 | 80 161 976 | 5.951 | 37, 38, 39 | X-74210508, X-76207917, X-80161976 |
| 5 | 5 | 84 132 877 | 90 640 869 | 6.508 | 40, 41, 42, 43, 44 | X-84132877, X-86116001, X-87868682, X-89621357, X-90640869 |
| 6 | 2 | 104 292 926 | 106 332 092 | 2.039 | 52, 53 | X-104292926, X-106332092 |
| 7 | 5 | 112 587 884 | 122 771 038 | 10.183 | 57, 58, 59, 60, 61 | X-112587884, X-116719686, X-118730961, X-120066236, X-122771038 |
| 8 | 2 | 127 137 209 | 129 088 513 | 1.951 | 64, 65 | X-127137209, X-129088513 |
| 9 | 3 | 142 906 662 | 146 641 816 | 3.735 | 74, 75, 76 | X-142906662, X-144055466, X-146641816 |
| 10 | 2 | 150 391 903 | 152 318 392 | 1.926 | 78, 79 | X-150391903, X-152318392 |
Epistasis was identified as the predominant non-neutral pattern for individual markers within the SXURs with variation in the strength of the effect between regions and transects (Figs 4 and S1a,b, Supporting information). All markers from within introgress groups in BV transect had their genomic clines shifted into M. m. domesticus range with respect to the neutral prediction. In the CZ transect, markers tended to show the opposite trend with the exception of two regions located in the distal part of the X chromosome.
Fig. 4.
Genomic clines for 29 X-linked markers from within the 10 SXURs plotted for each transect separately. Probability of genotype (M. m. domesticus homozygotes—solid line; heterozygotes—dashed line) plotted on hybrid index. Null expectations (light grey: homozygotes for M. m. domesticus alleles; dark grey: heterozygotes) are based on the permutation of autosomal data (1316 markers). Hybrid index used from Wang et al. (2011). Numbers correspond to the SXURs numbers in Fig. 3.
Analysis of epistasis for autosomes
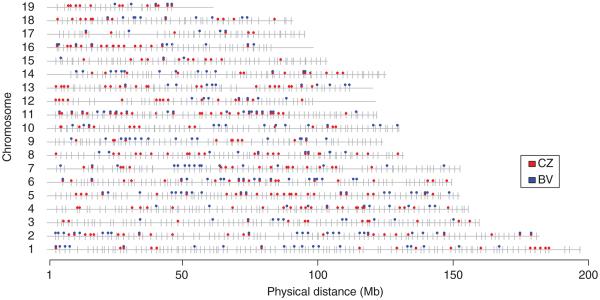
The log-likelihood ratios and P-value estimates for all X-autosomal pairs were calculated, and all pairwise comparisons having P-values above two different thresholds (P-valuepairwise > 0.01, 0.001) were considered. Subsequently, the introgression of these markers was tested against a neutral model as provided by the original version of the INTROGRESS program (Gompert & Buerkle 2009, 2010) to exclude those markers with introgression models that are not significantly different from the neutral expectation (P-valueintrogress ≤ 0.01, 0.001). Using the more conservative cut-off (P-valuepairwise > 0.01 and P-valueintrogress ≤ 0.01), we identified 204 and 237 autosomal markers showing epistasis in the BV and CZ transects, respectively (Fig. S2, Supporting information; Table 2). Using a less conservative cut-off (P-valuepairwise > 0.001 and P-valueintrogress ≤ 0.001), we identified 246 and 329 autosomal markers in BV and CZ transects, respectively (Fig. 5, Table 2). The introgression patterns of all identified markers were plotted using the original version of the INTROGRESS program (Fig. S3a,b, Supporting information).
Table 2.
Number of autosomal candidate epistatically interacting markers under two different cut-offs
| 0.01* |
0.001** |
|||
|---|---|---|---|---|
| Transect | No markers | % | No markers | % |
| BV | 204 | 15.50 | 246 | 18.69 |
| CZ | 237 | 18.01 | 329 | 25.00 |
| BV and CZ | 39 | 2.96 | 69 | 5.24 |
P-valuepairwise > 0.01 and P-valueintrogress ≤ 0.01.
P-valuepairwise > 0.001 and P-valueintrogress ≤ 0.001.
Fig. 5.
Genome location of autosomal markers. Grey ticks depict all of the 1316 autosomal markers. Blue and red circles depict candidate epistatically interacting markers in BV and CZ transect, respectively (P-valuepairwise > 0.001 and P-valueintrogress ≤ 0.001).
Comparison of introgression between the BV and CZ transects
The autosomal markers under epistasis were compared between the two transects. Using a more conservative cut-off as defined above, we identified 39 markers under putative epistasis in both transects (~3% of markers), and using the less conservative cut-off, we found 69 markers under putative epistasis (~5% of markers; Table 2). Because these markers might randomly overlap between the two transects, we simulated the data to estimate the probability of obtaining the same overlap by chance. The overlap between the two transects based on the more conservative cut-off was not significantly different from the random overlap (P-value = 0.251); however, the overlap based on the less conservative cut-off was significantly different (P-value = 0.036).
Distribution of autosomal markers under epistasis across the genome
To characterize the genome-wide architecture of reproductive isolation, the clustering of markers under putative epistasis was explored via permutations. The output of the permutation study indicated that the identified markers were grouped more often than expected by chance alone (Fig. S4a,b, Supporting information). We identified 143 (BV) and 162 (CZ) regions for the more conservative cut-off (P-valuepairwise > 0.01 and P-valueintrogress ≤ 0.01) and 163 (BV) and 203 (CZ) regions for the less conservative cut-off (P-valuepairwise > 0.001 and P-valueintrogress ≤ 0.001), respectively. The largest fraction of all regions was represented by single markers in both transects (~68%) and the fraction of regions comprising two or more markers decreased steeply with increasing number of markers per region. The largest regions identified spanned up to 11 Mb (BV) and 9 Mb (CZ), respectively. There were no differences in the distribution of sizes between transects. In our comparison between transects, we identified 59 overlapping regions exhibiting epistasis using the less conservative cut-off (P-valuepairwise > 0.001 and P-valueintrogress ≤ 0.001). Forty-nine of these are composed of a single marker.
We also conducted an analysis of marker distribution between chromosomes for each transect separately and then compared the results. When autosomes were tested for over- and under-abundance of identified markers, two chromosomes, chr. 15 in BV transect (less conservative cut-off) and chr. 8 in CZ transect (more conservative cut-off) were found to have an under-abundance and over-abundance, respectively. For overlapping markers exhibiting epistasis in the two transects, there was no significant enrichment except for chromosome 11 using the less conservative cut-off (P-valuepairwise > 0.001 and P-valueintrogress ≤ 0.001).
Gene enrichment in regions exhibiting epistasis
Gene content was characterized for X-linked and autosomal SNP markers identified as exhibiting epistasis, with attention to those regions identified in both transects (Table S3, Supporting information). Using Gene Ontology terms (Ashburner et al. 2000) under the domain ‘molecular function’, several groups were consistently enriched in all four analyses (BV and CZ including X, BV and CZ excluding X, BV and CZ, separately). The first group included terms involved in DNA binding and regulation of transcription. The group was represented by several related terms (GO:0003700, GO:0003677, GO:0003676, GO:0043565, GO:0016779). The second group included terms involved in the regulation of peptidase activity (GO:0004252, GO:0030414, GO:0016779), and the third group included chemokine and cytokine activity (GO:0008009, GO:0005125). Other GO terms were specific for the overlap between transects and included various terms involved in metal ion binding, electron carrier activity, energetic metabolism and/or a combination of these groups. Under the domain ‘biological process’, the group common to both transects included terms associated with transcription and its regulation represented by two GO terms (GO:0006355 and GO:0006351). Interestingly, in the case where we included the X chromosome in the analysis, two additional biological processes terms, skeletal muscle tissue development (GO:0007519) and DNA recombination (GO:0006310), appeared to be significant. It is also worth noting that one BV-specific group included terms involved in response to bacterium and viruses (GO:0006952, GO:0042742, GO:0009615). All the identified GO terms were found to be significant at an FDR ≤ 0.1.
Discussion
We surveyed 1401 SNPs approximately evenly spaced along the genome in two transects of the house mouse hybrid zone to identify regions of the X chromosome and autosomes that may be involved in BDMIs. Given our concerns that differences in hybrid zone movement between these transects (Wang et al. 2011) might obscure similarities between transects, we devised a method that does not rely on a direct comparison of introgression models between transects. We assessed the introgression along the X chromosome to identify regions exhibiting epistasis in both transects. Markers from these regions were subsequently used to reveal autosomal markers under putative epistasis common to both transects.
In general, genotypic patterns across a hybrid zone may be due to various forces including selection acting on intrinsic incompatibilities, selection in particular environments, drift, migration or demographic processes such as local extinction or colonization of populations across the landscape. Many of these processes may operate differently in different transects. Thus, comparisons of ‘signatures of selection’ among transects provide one means of distinguishing selection that is owing to intrinsic incompatibilities from selection that may be specific to a particular environment or false positives that may result from stochastic processes. We argue that common pattern of epistasis (as defined by Gompert & Buerkle 2009) observed in both transects is caused by selection against intrinsic incompatibilities responsible for reproductive isolation.
X chromosomal introgression
Most markers along the entire X chromosome were inferred to be involved in epistatic interactions in both transects, thus emphasizing the importance of interactions between X-linked loci and the genomic background as a cause of reproductive isolation. These patterns are consistent with the large X-effect (Charlesworth et al. 1987; Coyne & Orr 1989; Masly & Presgraves 2007) and with findings from house mice laboratory crosses (Storchová et al. 2004; Oka et al. 2004, Britton-Davidian et al. 2005, Good et al. 2008a, 2010). While these patterns of X-linked introgression are similar across the two transects, we detected a difference in the position of X-linked genomic clines with respect to the neutral prediction. This is likely due to differences in hybrid zone movement between the two transects (Wang et al. 2011), although we cannot fully discount other processes.
The identified regions common in both transects were detected in the central and distal portions of the X chromosome. This is in accord with previous findings from the hybrid zone as well as from laboratory crosses. Payseur et al. (2004), Macholán et al. (2011) and Dufková et al. (2011) identified reduced gene flow in a central portion of the X chromosome in the BV and the CZ transects, respectively. In addition, laboratory crosses between wild-derived inbred strains suggested several loci along the X chromosome are associated with various sterility phenotypes. Recently, lowered testis weight, smaller sperm counts and reduced sperm velocity have been reported from the hybrid zone (Turner et al. 2012). Storchová et al. (2004) and Vyskočilová et al. (2009) found the central portion of the X to be associated with testis weight as well as sperm count phenotypes. In this part of the X, approximately between 60 and 80 Mb, Storchová et al. (2004) obtained the strongest signal. This region corresponds to five of the SXURs (1, 2, 3, 4 and 5) identified in our study (Fig. 3). Oka et al. (2004) also implicated the central portion of the X. However, in their study, this region was associated with a sperm head morphology phenotype. Recently, Good et al. (2011) reported five candidate genes potentially involved in reproductive isolation from the central portion of the X chromosome. All are involved in male reproduction, and all have elevated rates of evolution. However, only two of them overlapped with regions we identified in the hybrid zone (SXUR 4: Pbsn and SXUR 5: Tsga8). The distal part of the X was found to be associated with either testis weight (Storchová et al. 2004) or testis weight and sperm counts (Oka et al. 2004; Vyskočilová et al. 2009). We identified two large regions comprising three (6, 7 and 8) and two (9 and 10) SXURs, respectively, which correspond with these previous findings. However, some recent studies found that the proximal part of the X is associated with testis weight and/or sperm counts, while the central and distal parts are associated with sperm head morphology (Good et al. 2008b; White et al. 2011). This discrepancy might be due to the fact that these studies crossed different inbred strains. Because M. m. musculus and M. m. domesticus have been isolated for only a short time (c. 0.5 Ma), alleles associated with reproductive isolation may not be fixed across populations. This was shown by Vyskočilová et al. (2005, 2009) who found a polymorphism in hybrid sterility in M. m. musculus populations. The fact that some wild-derived inbred strains are mosaics of M. m. musculus and M. m. domesticus genomes (Yang et al. 2011) might also contribute to the discrepancy in results between various crosses. Alternatively, all of these studies may be limited by their inability to precisely map the QTL of interest.
Autosomal introgression
We identified 69 autosomal markers that (i) showed similar patterns of introgression with the SUXR’s, (ii) were inferred to be involved in epistatic interactions and (iii) were shared between the BV and CZ transect. These regions, which constitute ~5% of the autosomal markers, are good candidates for BDMIs underlying reproductive isolation.
A total of 69 markers shared between transects is a relatively small subset of the markers identified in the two transects separately (246 in BV and 329 in CZ), highlighting the importance of comparing transects for filtering out potential ‘false positives’. However, the lack of a more extensive overlap between the two transects may be due to polymorphisms associated with hybrid sterility (Forejt & Iványi 1974; Britton-Davidian et al. 2005; Vyskočilová et al. 2005, 2009; Good et al. 2008) or stochastic processes (Gompert & Buerkle 2009, 2011; Dufková et al. 2011; Polechová & Barton 2011). In addition, the density of markers may not be fine enough to cover the entire genome. As markers in close proximity may exhibit different behaviour in a hybrid zone (e.g. Macholán et al. 2011), we might have missed some of the incompatibility loci, thus underestimating the number of regions involved in reproductive isolation.
Incompatibility loci were shown to be primarily equally distributed along the autosomal genome as there were no significant differences in the abundance of identified markers between chromosomes consistent over the two transects. The only exception was chromosome 11 where we found significant enrichment for markers shared between transects. The predominantly equal distribution of incompatibility loci is in agreement with other laboratory studies (Gregorová et al. 2008; White et al. 2011) and suggests that incompatibilities gradually accumulated as a function of time and strength of selection which is consistent with an allopatric model (Barton & Charlesworth 1984; Geraldes et al. 2008, 2011; Nosil et al. 2009; Duvaux et al. 2011).
Gene content of the regions under putative epistasis
The gene content of X-linked and autosomal regions under putative epistasis was characterized. We were particularly interested in which GO biological processes and molecular functions were associated with genes from within these regions. Among all enriched categories, one of the most important findings was the involvement of a group of genes associated with DNA binding, transcription and its regulation, thus emphasizing the role of between-loci interactions (e.g. transcription factors) as suggested by the BDMIs. A second group involved genes with peptidase inhibitory activity that was recently related to sexual conflict and sexual selection in the female reproductive tract (Dean et al. 2008, 2009, 2011; Dorus et al. 2010). Interestingly, three out of four rapidly evolving genes (Serpine2, Spink3, Spinkl) identified in the seminal vesicles and subsequently referenced to proteolytic inhibitory activity by Dean et al. (2009), fall into epistatically interacting regions. Other studies found proteolytic inhibitory activity enriched in rapidly evolving genes functioning mostly on the outer surface of sperm (Dorus et al. 2010; Dean et al. 2011). Thus, these observations are consistent with a role for sexual conflict in reproductive isolation. The next GO term, chemotaxis, was enriched in the overlap of the two transects. The ability of sperm to find their way through the female reproductive tract is key to successful reproduction (Clark et al. 2006). From this perspective, chemotaxis may play a very important role in reproduction and may be under strong selection as well. The selective force operating at this stage may be sperm competition when females undergo multiple mating (Dean et al. 2006; Dean & Nachman 2009).
Finally, genes involved in defence response to bacteria and viruses were identified in the BV transect. Clark et al. (2006) suggested a role for resistance against various pathogens and female immune system in sperm. Our findings on defence response are also in accord with Dean et al. (2008) who found the same biological processes (defence response, defence response to bacterium) enriched in mouse epididymal proteins.
Our GO analyses do not suggest the involvement of intraspecific individual recognition mechanisms, which would be in accord with previous analyses (Laukaitis et al. 1997; Talley et al. 2001; Smadja & Ganem 2002, 2005, 2008; Smadja et al. 2004; Cheetham et al. 2007; Stopková et al. 2007; Ganem et al. 2008; Thom et al. 2008; Vošlajerová Bímová et al. 2011) including MUPs (Cheetham et al. 2007; Stopková et al. 2007; Thom et al. 2008) and ABPs (Karn & Dlouhy 1991; Laukaitis et al. 1997; Talley et al. 2001). The reason for this failure may be simply that genes involved in intraspecific recognition have not been annotated or their role in reproductive isolation is not large enough to be captured by the approach used. Also, there is a possibility that these genes fall within several gaps where there is no marker coverage.
Comparing autosomal regions exhibiting epistasis to other studies
We compared autosomal regions identified in the hybrid zone to be under epistasis to regions and/or genes identified in previous studies. We focus only on the shared regions under epistasis between the two transects as they are most likely to harbour loci involved in reproductive isolation (Fig. 6, Table S2, Supporting information).
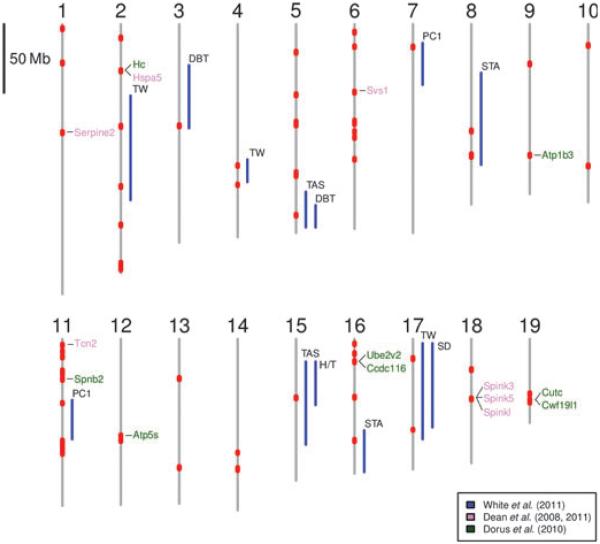
Fig. 6.
Candidate autosomal regions associated with reproductive isolation in the house mouse. Shared regions between the two transects exhibiting epistasis (this study) are shown in red. The QTL regions associated with hybrid sterility phenotypes (White et al. 2011) are represented as blue lines. Phenotype abbreviations, TW, relative testis weight; SD, sperm density; PC1, sperm head morphology; STA, seminiferous tubule area; DBT, distal bent tail; TAS, total abnormal sperm; H/T, headless/tailless correspond to those in White et al. (2011). Genes identified in Dean et al. (2008, 2011) are coloured in violet and those identified by Dorus et al. (2010) are coloured in green.
Several studies focused on the association of hybrid sterility phenotypes with particular markers in the genome using QTL analysis (Mihola et al. 2009; Vyskočilová et al. 2009; White et al. 2011). White et al. (2011) recently mapped traits underlying hybrid male sterility in a large F2 mapping population. In general, the identified QTL were in wide genomic intervals. Nonetheless, the results of White et al. (2011) provide a useful comparison to the present study, because similar hybrid male sterility phenotypes are observed in the hybrid zone (Turner et al. 2012). Several markers identified in overlapping regions between the two transects were found to fall within five QTL regions associated with relative testis weight or sperm density phenotypes (2-74558017, 2-118591908, 4-103390028, 17-14320574 and 17-65932862). Other markers (3-74249497, 5-139601762, 7-16643634, 8-77982361, 8-95400455, 8-96505302, 11-46639287, 15-42990323 and 16-74152290) fall within nine QTL regions associated with sperm morphology. Although QTL regions sometimes span large portions of a chromosome making an assessment on the overlap with our data difficult, for certain cases, the overlap is very precise and/or markers identified reside very close to regions with the highest likelihood (4-103390028, 5-139601762, 7-16643634, 8-77982361, 8-95400455, 8-96505302, 16-74152290). Although an autosomal region involved in hybrid male sterility was found to be on chromosome 17 (Forejt & Iványi 1974; Forejt 1996) and, recently, a particular gene (Prdm9—Chr17:15682083-15701318) was identified (Mihola et al. 2009), we found no correspondence with our data from the hybrid zone. This is in contrast to Vyskočilová et al. (2009) and White et al. (2011) who found an association between the proximal portion of chromosome 17 and sperm counts and relative testis weight. Given the large span of this QTL in both studies, it is possible that a different region (i.e. marker 17-14320574)isinvolved.
Also, we found several genes (Atp1b3, Atp5s, Ccdc116, Cutc, Cwf19l1, Hc, Hspa5, Serpine2, Spink3, Spink5, Spinkl, Spnb2, Svs1, Tcn2, Ube2v2) in the regions of overlap whose products are known to be involved in male reproduction (Dean et al. 2009a, 2011; Dorus et al. 2010). Although the study of Harr (2006) who searched for genomic regions of high differentiation between house mouse subspecies suffered from potential methodological biases (Boursot & Belkhir 2006), two autosomal regions clearly emerged. One of them in the proximal region of chromosome 2 correlates with our results based on the two-transect comparison. However, the second of them on chromosome 10 does not appear to overlap with any of our markers under putative epistasis.
Although there is always a possibility that correspondence between our data and previous studies is by chance, in some cases, the overlap is very precise, thus reinforcing the evidence for epistasis involving these particular regions. Agreement between our data and other studies and our ability to more finely map regions exhibiting epistasis demonstrate the utility of using naturally occurring hybrid populations exhibiting multi-generational backcrossing and intercrossing to explore the genetic basis of reproductive isolation.
Indeed, our analysis of gene content in regions exhibiting epistasis implicates a key role in reproductive isolation for several mechanisms, involving the regulation of transcription, sexual conflict and sexual selection operating at both the postmating prezygotic (e.g. genes expressed on the outer surface of the sperm) and postzygotic (e.g. candidate loci associated with hybrid male sterility) stages of isolation. These findings, together with information that epistatically interacting loci are distributed throughout the genome, provide strong evidence that reproductive isolation in emerging house mouse species is polygenic, complex and likely resulted from the accumulation of incompatibilities between M. m. musculus and M. m. domesticus during the relatively short time period in which these populations diverged in allopatry.
Supplementary Material
Acknowledgements
We thank Richard Sage who collected all of the mice from the Bavaria transect and, together with the MVZ at the University of California, Berkeley, provided DNA samples and/or tissues to the Tucker laboratory. Czech-Bavarian samples were obtained through a warm collaboration of many local farmers and students and their help is appreciated. Research was supported by NSF DEB0746560 to PKT, CSF 206-08-0640 to JP, MM and PM, and VJ received support from the SVV project of Charles University in Prague.
Footnotes
This work resulted from collaboration among several research groups who use naturally occurring hybrid populations to understand the genetic underpinnings of reproductive isolation in house mice.
Data accessibility
The genotype data matrix is available from http://hdl.handle.net/2027.42/83674.
Supporting information
Additional supporting information may be found in the online version of this article.
References
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SJE, Macholán M. What can the Mus musculus musculus/M. m. domesticus hybrid zone tell us about speciation? In: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the House Mouse. Cambridge Studies in Morphology and Molecules. New Paradigms in Evolutionary Biology. Cambridge University Press; Cambridge, UK: 2012. in press. [Google Scholar]
- Baird SJE, Ribas A, Macholán M, et al. Where are the wormy mice? A re-examination of hybrid parasitism in the European house mouse hybrid zon. Evolution. 2012 doi: 10.1111/j.1558-5646.2012.01633.x. doi: 10.1111/j.1558-5646.2012.01633.x. [DOI] [PubMed] [Google Scholar]
- Barton NH. Dynamics of hybrid zones. Heredity. 1979;43:341–359. [Google Scholar]
- Barton NH. Multilocus clines. Evolution. 1983;37:454–471. doi: 10.1111/j.1558-5646.1983.tb05563.x. [DOI] [PubMed] [Google Scholar]
- Barton NH, Charlesworth B. Genetic revolutions, founder effects, and speciation. Annual Review of Ecology and Systematics. 1984;15:133–164. [Google Scholar]
- Barton NH, Gale KS. Genetic analysis of hybrid zones. In: Harrison RG, editor. Hybrid Zones and the Evolutionary Process. Oxford University Press; Oxford, UK: 1993. pp. 13–45. [Google Scholar]
- Barton NH, Hewitt GM. Analysis of hybrid zones. Annual Review of Ecology and Systematics. 1985;16:113–148. [Google Scholar]
- Bateson W. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and Modern Science. University Press; Cambridge, UK: 1909. pp. 85–101. [Google Scholar]
- Boursot P, Belkhir K. Mouse SNPs for evolutionary biology: beware of ascertainment biases. Genome Research. 2006;16:1191–1192. doi: 10.1101/gr.5541806. [DOI] [PubMed] [Google Scholar]
- Boursot P, Auffray JC, Britton-Davidian J, Bonhomme F. The evolution of house mice. Annual Review of Ecology and Systematics. 1993;24:119–152. [Google Scholar]
- Boursot P, Din W, Anand R, et al. Origin and radiation of the house mouse: mitochondrial DNA phylogeny. Journal of Evolutionary Biology. 1996;9:391–415. [Google Scholar]
- Boyle EI, Weng S, Gollub J, et al. GO::TermFinder – open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Božíková E, Munclinger P, Teeter KC, Tucker PK, Macholán M, Piálek J. Mitochondrial DNA in the hybrid zone between Mus musculus musculus and Mus musculus domesticus: a comparison of two transects. Biological Journal of the Linnean Society. 2005;84:363–378. [Google Scholar]
- Britton-Davidian J, Fel-Clair F, Lopez J, Alibert P, Boursot P. Postzygotic isolation between the two European subspecies of the house mouse: estimates from fertility patterns in wild and laboratory-bred hybrids. Biological Journal of the Linnean Society. 2005;84:379–393. [Google Scholar]
- Buerkle CA. Maximum-likelihood estimation of a hybrid index based on molecular markers. Molecular Ecology Notes. 2005;5:684–687. [Google Scholar]
- Buerkle CA, Lexer C. Admixture as the basis for genetic mapping. Trends in Ecology and Evolution. 2008;23:686–694. doi: 10.1016/j.tree.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex-chromosomes and autosomes. American Naturalist. 1987;130:113–146. [Google Scholar]
- Cheetham SA, Thom MD, Jury F, Ollier WER, Beynon RJ, Hurst JL. The genetic basis of individual-recognition signals in the mouse. Current Biology. 2007;17:1771–1777. doi: 10.1016/j.cub.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Chevret P, Veyrunes F, Britton-Davidian J. Molecular phylogeny of the genus Mus (Rodentia: Murinae) based on mitochondrial and nuclear data. Biological Journal of the Linnean Society. 2005;84:417–427. [Google Scholar]
- Clark NL, Aagaard JE, Swanson WJ. Evolution of reproductive proteins from animals and plants. Reproduction. 2006;131:11–22. doi: 10.1530/rep.1.00357. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Patterns of speciation in Drosophila. Evolution. 1989;43:362–381. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates, Inc.; Sunderland, Massachusetts: 2004. [Google Scholar]
- Dean MD, Nachman MW. Faster fertilization rate in conspecific versus heterospecific matings in house mice. Evolution. 2009;63:20–28. doi: 10.1111/j.1558-5646.2008.00499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Ardlie KG, Nachman MW. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus) Molecular Ecology. 2006;15:4141–4151. doi: 10.1111/j.1365-294X.2006.03068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Good JM, Nachman MW. Adaptive evolution of proteins secreted during sperm maturation: an analysis of the mouse epididymal transcriptome. Molecular Biology and Evolution. 2008;25:383–392. doi: 10.1093/molbev/msm265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Clark NL, Findlay GD, et al. Proteomics and comparative genomic investigations reveal heterogeneity in evolutionary rate of male reproductive proteins in mice (Mus domesticus) Molecular Biology and Evolution. 2009;26:1733–1743. doi: 10.1093/molbev/msp094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean MD, Findlay GD, Hoopmann MR, et al. Identification of ejaculated proteins in the house mouse (Mus domesticus) via isotopic labeling. BMC Genomics. 2011;12:306. doi: 10.1186/1471-2164-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Studies on hybrid sterility. 11. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics. 1936;21:113–135. doi: 10.1093/genetics/21.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the Origin of Species. Columbia University Press, New York; New York: 1937. [Google Scholar]
- Dod B, Jermiin LS, Boursot P, Chapman VH, Nielsen JT, Bonhomme F. Counterselection on sex-chromosomes in the Mus musculus European hybrid zone. Journal of Evolutionary Biology. 1993;6:529–546. [Google Scholar]
- Dorus S, Wasbrough ER, Busby J, Wilkin EC, Karr TL. Sperm proteomics reveals intensified selection on mouse sperm membrane and acrosome genes. Molecular Biology and Evolution. 2010;27:1235–1246. doi: 10.1093/molbev/msq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufková P, Macholán M, Piálek J. Inference of selection and stochastic effects in the house mouse hybrid zone. Evolution. 2011;65:993–1010. doi: 10.1111/j.1558-5646.2011.01222.x. [DOI] [PubMed] [Google Scholar]
- Duvaux L, Belkhir K, Boulesteix M, Boursot P. Isolation and gene flow: inferring the speciation history of European house mice. Molecular Ecology. 2011;20:5248–5264. doi: 10.1111/j.1365-294X.2011.05343.x. [DOI] [PubMed] [Google Scholar]
- Forejt J. Hybrid sterility in the mouse. Trends in Genetics. 1996;12:412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- Forejt J, Iványi P. Genetic studies on male-sterility of hybrids between laboratory and wild mice (Mus musculus) Genetical Research. 1974;24:189–206. doi: 10.1017/s0016672300015214. [DOI] [PubMed] [Google Scholar]
- Ganem G, Litel C, Lenormand T. Variation in mate preference across a house mouse hybrid zone. Heredity. 2008;100:594–601. doi: 10.1038/hdy.2008.20. [DOI] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Gibson B, et al. Inferring the history of speciation in house mice from autosomal, X-linked, Y-linked and mitochondrial genes. Molecular Ecology. 2008;17:5349–5363. doi: 10.1111/j.1365-294X.2008.04005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldes A, Basset P, Smith KL, Nachman MW. Higher differentiation among subspecies of the house mouse (Mus musculus) in genomic regions with low recombination. Molecular Ecology. 2011;20:4722–4736. doi: 10.1111/j.1365-294X.2011.05285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA. A powerful regression-based method for admixture mapping of isolation across the genome of hybrids. Molecular Ecology. 2009;18:1207–1224. doi: 10.1111/j.1365-294X.2009.04098.x. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA. INTROGRESS: a software package for mapping components of isolation in hybrids. Molecular Ecology Resources. 2010;10:378–384. doi: 10.1111/j.1755-0998.2009.02733.x. [DOI] [PubMed] [Google Scholar]
- Gompert Z, Buerkle CA. Bayesian estimation of genomic clines. Molecular Ecology. 2011;20:2111–2127. doi: 10.1111/j.1365-294X.2011.05074.x. [DOI] [PubMed] [Google Scholar]
- Good JM, Handel MA, Nachman MW. Asymmetry and polymorphism of hybrid male sterility during the early stages of speciation in house mice. Evolution. 2008a;62:50–65. doi: 10.1111/j.1558-5646.2007.00257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Dean MD, Nachman MW. A complex genetic basis to X-linked hybrid male sterility between two species of house mice. Genetics. 2008b;179:2213–2228. doi: 10.1534/genetics.107.085340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Giger T, Dean MD, Nachman MW. Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genetics. 2010;6:e1001148. doi: 10.1371/journal.pgen.1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM, Vanderpool D, Smith KL, Nachman MW. Extraordinary sequence divergence at Tsga8, an X-linked gene involved in mouse spermiogenesis. Molecular Biology and Evolution. 2011;28:1675–1686. doi: 10.1093/molbev/msq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy de Bellocq J, Ribas A, Baird SJE. New insights into parasitism in the house mouse hybrid zone. In: Macholán M, Baird SJE, Munclinger P, Piálek J, editors. Evolution of the House Mouse. Cambridge Studies in Morphology and Molecules. New Paradigms in Evolutionary Biology. Cambridge University Press; Cambridge, UK: 2012. In press. [Google Scholar]
- Gregorová S, Divina P, Storchová R, et al. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Research. 2008;18:509–515. doi: 10.1101/gr.7160508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics. 1922;12:101–109. [Google Scholar]
- Harr B. Genomic islands of differentiation between house mouse subspecies. Genome Research. 2006;16:730–737. doi: 10.1101/gr.5045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones EP, Van der Kooij J, Solheim R, Searle JB. Norwegian house mice (Mus musculus musculus/domesticus): distributions, routes of colonization and patterns of hybridization. Molecular Ecology. 2010;19:5252–5264. doi: 10.1111/j.1365-294X.2010.04874.x. [DOI] [PubMed] [Google Scholar]
- Karn RC, Dlouhy SR. Salivary androgen-binding protein variation in Mus and other rodents. Journal of Heredity. 1991;82:453–458. doi: 10.1093/oxfordjournals.jhered.a111127. [DOI] [PubMed] [Google Scholar]
- Laukaitis CM, Critser ES, Karn RC. Salivary androgenbinding protein (ABP) mediates sexual isolation in Mus musculus. Evolution. 1997;51:2000–2005. doi: 10.1111/j.1558-5646.1997.tb05121.x. [DOI] [PubMed] [Google Scholar]
- Lu X, Shapiro JA, Ting CT, et al. Genome-wide misexpression of X-linked versus autosomal genes associated with hybrid male sterility. Genome Research. 2010;20:1097–1102. doi: 10.1101/gr.076620.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholán M, Kryštůfek B, Vohralík V. The location of the Mus musculus/M. domesticus hybrid zone in the Balkans clues from morphology. Acta Theriologica. 2003;48:177–188. [Google Scholar]
- Macholán M, Munclinger P, Sugerková M, et al. Genetic analysis of autosomal and X-linked markers across a mouse hybrid zone. Evolution. 2007;61:746–771. doi: 10.1111/j.1558-5646.2007.00065.x. [DOI] [PubMed] [Google Scholar]
- Macholán M, Baird S, Munclinger P, Dufková P, Bímová B, Piálek J. Genetic conflict outweighs heterogametic incompatibility in the mouse hybrid zone? BMC Evolutionary Biology. 2008;8:271. doi: 10.1186/1471-2148-8-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholán M, Baird SJE, Dufková P, Munclinger P, Vošlajerová Bímová B, Piálek J. Assessing multilocus introgression patterns: a case study on the mouse X chromosome in central Europe. Evolution. 2011;65:1428–1446. doi: 10.1111/j.1558-5646.2011.01228.x. [DOI] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biology. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Systematics and the Origin of Species. Columbia University Press, New York; New York: 1942. [Google Scholar]
- Mihola O, Trachtulec Z, Vlček C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–375. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Moulia C, Lebrun N, Dallas J, Orth A, Renaud F. Experimental-evidence of genetic determinism in high susceptibility to intestinal pinworm infection in mice – a hybrid zone model. Parasitology. 1993;106:387–393. doi: 10.1017/s0031182000067135. [DOI] [PubMed] [Google Scholar]
- Muller HJ. Bearing of the Drosophila work on systematics. In: Huxley JS, editor. The New Systematics. Clarendon Press, Oxford; UK: 1940. pp. 185–268. [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution, and temperature. Biological Symposium. 1942;6:71–125. [Google Scholar]
- Nagylaki T. Conditions for existence of clines. Genetics. 1975;80:595–615. doi: 10.1093/genetics/80.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Molecular Ecology. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Oka A, Mita A, Sakurai-Yamatani N, et al. Hybrid breakdown caused by substitution of the X chromosome between two mouse subspecies. Genetics. 2004;166:913–924. doi: 10.1093/genetics/166.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka A, Aoto T, Totsuka Y, et al. Disruption of genetic interaction between two autosomal regions and the X chromosome causes reproductive isolation between mouse strains derived from different subspecies. Genetics. 2007;175:185–197. doi: 10.1534/genetics.106.062976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. Haldane’s rule. Annual Review of Ecology and Systematics. 1997;28:195–218. doi: 10.1146/annurev.ecolsys.28.1.85. [DOI] [PubMed] [Google Scholar]
- Orr HA, Irving S. Complex epistasis and the genetic basis of hybrid sterility in the Drosophila pseudoobscura Bogota-USA hybridization. Genetics. 2001;158:1089–1100. doi: 10.1093/genetics/158.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Molecular Ecology Resources. 2010;10:806–820. doi: 10.1111/j.1755-0998.2010.02883.x. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Krenz JG, Nachman MW. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- Polechová J, Barton NH. Genetic drift widens the expected cline but narrows the expected cline width. Genetics. 2011;189:227–235. doi: 10.1534/genetics.111.129817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends in Genetics. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team . R: A Language and Environment for Statistical Computing, Reference Index Version 2.13.1. R Foundation for Statistical Computing, Vienna; Austria: 2005. ISBN 3-900051-07-0. http://www.R-project.org. [Google Scholar]
- Raufaste N, Orth A, Belkhir K, et al. Inferences of selection and migration in the Danish house mouse hybrid zone. Biological Journal of the Linnean Society. 2005;84:593–616. [Google Scholar]
- Rieseberg LH, Buerkle CA. Genetic mapping in hybrid zones. American Naturalist. 2002;159:S36–S50. doi: 10.1086/338371. [DOI] [PubMed] [Google Scholar]
- Sage RD, Heyneman D, Lim KC, Wilson AC. Wormy mice in a hybrid zone. Nature. 1986;324:60–63. doi: 10.1038/324060a0. [DOI] [PubMed] [Google Scholar]
- Salcedo T, Geraldes A, Nachman MW. Nucleotide variation in wild and inbred mice. Genetics. 2007;177:2277–2291. doi: 10.1534/genetics.107.079988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Blay S, McNeney B, Graham J. LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. Journal of Statistical Software. 2006;16 http://www.jstatsoft.org/v16/c03/paper. [Google Scholar]
- Smadja C, Ganem G. Subspecies recognition in the house mouse: a study of two populations from the border of a hybrid zone. Behavioral Ecology. 2002;13:312–320. [Google Scholar]
- Smadja C, Ganem G. Asymmetrical reproductive character displacement in the house mouse. Journal of Evolutionary Biology. 2005;18:1485–1493. doi: 10.1111/j.1420-9101.2005.00944.x. [DOI] [PubMed] [Google Scholar]
- Smadja C, Ganem G. Divergence of odorant signals within and between the two European subspecies of the house mouse. Behavioral Ecology. 2008;19:223–230. [Google Scholar]
- Smadja C, Catalan J, Ganem G. Strong premating divergence in a unimodal hybrid zone between two subspecies in the house mouse. Journal of Evolutionary Biology. 2004;17:165–176. doi: 10.1046/j.1420-9101.2003.00647.x. [DOI] [PubMed] [Google Scholar]
- Stopková R, Stopka R, Janotová K, Jedelský P. Speciesspecific expression of major urinary proteins in the house mice (Mus musculus musculus and Mus musculus domesticus) Journal of Chemical Ecology. 2007;33:861–869. doi: 10.1007/s10886-007-9262-9. [DOI] [PubMed] [Google Scholar]
- Storchová R, Gregorová S, Buckiová D, Kyselová V, Divina P, Forejt J. Genetic analysis of X-linked hybrid sterility in the house mouse. Mammalian Genome. 2004;15:515–524. doi: 10.1007/s00335-004-2386-0. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. PNAS. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Shimada T, Terashima M, Tsuchiya K, Aplin K. Temporal, spatial, and ecological modes of evolution of Eurasian Mus based on mitochondrial and nuclear gene sequences. Molecular Phylogenetics and Evolution. 2004;33:626–646. doi: 10.1016/j.ympev.2004.08.003. [DOI] [PubMed] [Google Scholar]
- Talley HM, Laukaitis CM, Karn RC. Female preference for male saliva: implications for sexual isolation of Mus musculus subspecies. Evolution. 2001;55:631–634. doi: 10.1554/0014-3820(2001)055[0631:fpfmsi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Tao Y, Chen S, Hartl DL, Laurie CC. Genetic dissection of hybrid incompatibilities between Drosophila simulans and Drosophila mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics. 2003;164:1383–1397. doi: 10.1093/genetics/164.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeter KC, Payseur BA, Harris LW, et al. Genome-wide patterns of gene flow across a house mouse hybrid zone. Genome Research. 2008;18:67–76. doi: 10.1101/gr.6757907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thom MD, Stockley P, Jury F, Ollier WER, Beynon RJ, Hurst JL. The direct assessment of genetic heterozygosity through scent in the mouse. Current Biology. 2008;18:619–623. doi: 10.1016/j.cub.2008.03.056. [DOI] [PubMed] [Google Scholar]
- True JR, Weir BS, Laurie CC. A genome-wide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics. 1996;142:819–837. doi: 10.1093/genetics/142.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK, Sage RD, Warner J, Wilson AC, Eicher EM. Abrupt cline for sex-chromosomes in a hybrid zone between two species of mice. Evolution. 1992;46:1146–1163. doi: 10.1111/j.1558-5646.1992.tb00625.x. [DOI] [PubMed] [Google Scholar]
- Turelli M, Orr HA. The dominance theory of Haldane’s rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LM, Schwahn DJ, Harr B. Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution. 2012;66:443–458. doi: 10.1111/j.1558-5646.2011.01445.x. in press. [DOI] [PubMed] [Google Scholar]
- Vošlajerová Bímová B, Macholán M, Baird SJE, et al. Reinforcement selection acting on the European house mouse hybrid zone. Molecular Ecology. 2011;20:2403–2424. doi: 10.1111/j.1365-294X.2011.05106.x. [DOI] [PubMed] [Google Scholar]
- Vyskočilová M, Trachtulec Z, Forejt J, Piálek J. Does geography matter in hybrid sterility in house mice? Biological Journal of the Linnean Society. 2005;84:663–674. [Google Scholar]
- Vyskočilová M, Prazanova G, Piálek J. Polymorphism in hybrid male sterility in wild-derived Mus musculus musculus strains on proximal chromosome 17. Mammalian Genome. 2009;20:83–91. doi: 10.1007/s00335-008-9164-3. [DOI] [PubMed] [Google Scholar]
- Wang L, Luzynski K, Pool JE, et al. Measures of linkage disequilibrium among neighbouring SNPs indicate asymmetries across the house mouse hybrid zone. Molecular Ecology. 2011;20:2985–3000. doi: 10.1111/j.1365-294X.2011.05148.x. [DOI] [PubMed] [Google Scholar]
- White MA, Steffy B, Wiltshire T, Payseur BA. Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics. 2011;189:289–304. doi: 10.1534/genetics.111.129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ding Y, Hutchins LN, et al. A customized and versatile high-density genotyping array for the mouse. Nature Methods. 2009;6:663–666. doi: 10.1038/nmeth.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HN, Wang JR, Didion JP, et al. Subspecific origin and haplotype diversity in the laboratory mouse. Nature Genetics. 2011;43:648–655. doi: 10.1038/ng.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LW. Resurrecting Muller’s theory of Haldane’s rule. Genetics. 1996;143:603–607. doi: 10.1093/genetics/143.1.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.