Abstract
This study presents a strategy to enhance the uptake of superparamagnetic iron oxide nanoparticle (SPIO) clusters by manipulating the cellular mechanical environment. Specifically, stem cells exposed to an orbital flow ingested almost two-fold greater amount of SPIO clusters than those cultured statically. Improvements in MR contrast were subsequently achieved for labeled cells in collagen gels and a mouse model. Overall, this strategy will serve to improve the efficiency of cell tracking and therapies.
Keywords: iron oxide nanoparticles, MR imaging, mechanotransduction, cell tracking, receptor-mediated endocytosis
1. INTRODUCTION
Stem and progenitor cells possess the potential to treat various acute and chronic diseases, and tissue defects, due to their multipotent differentiation capacity, trophic factor secretion, and immunosuppressive activities.1 In mesenchymal stem cell therapies, stem cells are isolated from a patient’s bone marrow or adipose tissue, expanded to therapeutic levels ex vivo, and then re-injected locally or systemically.2 To better understand and modulate cellular therapeutic activities, clinicians must assess the localization and bioavailability of transplanted cells in vivo using a clinical imaging modality, such as magnetic resonance imaging (MRI).
Towards this goal, extensive efforts have been made to label stem cells ex vivo with superparamagnetic iron oxide nanoparticles (SPIOs), a popular T2 contrast agent capable of highly sensitive in vivo imaging.3 One emerging, simple method to modulate SPIO size and functionality for labeling is to cluster several SPIOs together using self-assembling polymers with diverse functional groups.4, 5 In this way, a cluster is formed, containing multiple SPIOs surrounded by a polymeric coating already grafted with various biomolecules of interest. With this technique, the size of the SPIO cluster can be controlled through the concentration and chemical structure of the self-assembling molecules, in turn allowing SPIO clusters to be easily tuned for enhanced receptor-mediated endocytosis6 or maximum T2 relaxivity.7 In addition, the cluster formation process avoids the extensive conjugation and purification steps required in the direct surface modifications of SPIOs.8-10 Despite the advantages offered by this clustering technique, advanced methods are still needed to increase SPIO loading efficiency within cells, as cell proliferation and SPIO exocytosis results in a gradual reduction of the MR signal in vivo, in turn limiting the long-term effectiveness of cell tracking.11
Therefore, we sought to develop a new method to tailor the cellular uptake of SPIO clusters and improve cell tracking, apart from conventional approaches that rely on changes to SPIO size, charge, and surface chemistry12 or potentially harmful external stimuli such as electroportation.13 With this strategy, we also seek to maintain cell viability and function. According to recent cell biology studies, the extracellular mechanical environment regulates the endocytosis and exocytosis of extracellular components both in vitro and in vivo.14 For example, shear flow has been shown to affect adhesion and endocytosis of quantum dots to endothelial cells.15 Aligned with these findings, we hypothesized that cells exposed to an external flow in vitro would ingest a greater amount of SPIO clusters grafted with integrin-binding peptides.
We examined this hypothesis by co-incubating bone marrow-derived mesenchymal stem cells (BMSCs) with SPIO clusters. These SPIO clusters are coated with integrin-binding peptides containing an Arg-Gly-Asp (RGD) sequence. BMSCs were labeled with RGD-SPIO clusters on an orbital shaker rotating at controlled speeds, at which the average cluster velocity and shear stress on the cell membrane were estimated to increase. The resulting cell labeling efficiency was evaluated by measuring RGD-SPIO clusters per cell using inductively coupled plasma (ICP) spectroscopy, and independently confirmed by measuring the relaxivity of labeled BMSCs in a collagen gel. Finally, cell labeling under orbital flow was demonstrated by locally injecting BMSCs labeled with RGD-SPIO clusters into the muscle of a mouse’s hindlimb, and imaging the leg with MRI. Taken together, this study will serve to improve the effectiveness of cell tracking and, ultimately, the therapeutic activities of a wide range of cells.
2. EXPERIMENTAL SECTION
Materials.
Materials were purchased from Sigma Aldrich unless otherwise specified.
Synthesis of Oleic Acid-Coated Superparamagnetic Iron Oxide Nanoparticles (OA-SPIOs)
5 nm diameter iron oxide nanoparticles were prepared from the thermal decomposition of iron acetylacetonate.16 First, a three-neck flask was charged with 0.2 g of iron acetylacetonate, 660 μL of oleic acid, 600 μL of oleylamine, and 0.7 g of 1,2-dodecanediol. All compounds were dissolved in 6.7 mL of benzyl ether. Under nitrogen flow, the mixture was heated to 200 °C for 2 hours, and then slowly heated up to 300 °C under reflux for 1 hour with gentle stirring. To purify the SPIOs, the reaction mixture was precipitated with ethanol (Declon), and then magnetically separated (K & J Magnetics). These oleic acid-coated SPIOs (termed “OA-SPIOs”) were finally dispersed in chloroform at 10 mg/mL, and stored at −20 °C until further analysis.
Synthesis of Polysuccinimide
Polysuccinimide (PSI) was prepared from the thermal condensation of aspartic acid, as previously reported.17 In a two-neck flask, 25 g of aspartic acid was dissolved in the presence of 125 mL of mesitylene and 125 mL of sulfolane. The mixture was heated to 180 °C under nitrogen, and then 240 μL of phosphoric acid was injected to catalyze the reaction. The reaction was run overnight, after which the reaction mixture was vacuum filtered, washed thoroughly with methanol and water, and then lyophilized.
Synthesis of PHEA Substituted with Octadecyl Chains (C18) and RGD Peptides (RGD-PHEA-g-C18)
To prepare RGD-PHEA-g-C18, 0.4 g of PSI was dissolved in 10 mL of dimethylformamide (DMF). Then, 59 mg of octadecylamine was added to this mixture, and reacted overnight. Afterwards, 32 mg of GGGGRGDSP (Mimotope Peptide) was added to the mixture, and then reacted for 5 hours, followed by 1 mL of ethanolamine for 3 hours. All reaction steps were completed under nitrogen at room temperature in water-free conditions. After completion, the reaction mixture was added to a dialysis bag (MWCO 3,500; Fisher) and then dialyzed against DI water for 48 hours, while adding fresh water at least four times. NaCl was added to water for the first round of dialysis. Afterwards, the product was frozen and then lyophilized (Labconco) for 48 hours to obtain a dried product. Total yield ranged from 70 to 90%.1H-NMR (Varian VXR 500 with Unity Inova Console) was used to analyze the product dissolved in d-6 DMSO. For accurate shimming, the temperature of the NMR probe was greater than 30 °C.
Quantification of Octadecyl Chain Engraftment to PHEA
Using 1H-NMR and an established protocol,18 the degree of octadecylamine substitution (DSC18) was quantified by the following equation:
| [1] |
Preparation of SPIO Clusters with RGD-PHEA-g-C18
To prepare RGD-SPIO clusters, 400 μL of 5 mg/mL OA-SPIOs suspended in chloroform was added to 8 mL of 5 mg/mL RGD-PHEA-g-C18 in DI water. The mixture was quickly sonicated (Fisher Scientific) in order to form a cloudy suspension. Using a rotary evaporator (Heidolph), the chloroform phase was removed from the suspension after about 20 minutes. The concentrated RGD-SPIO cluster suspension was first transferred to a centrifuge tube and then centrifuged at 4,000 rpm for 10 minutes (Eppendorf 5180R). Afterwards, the supernatant was collected. By collecting the supernatant, sediments containing non-encapsulated and unstable SPIOs were removed and then discarded. In this way, we could only retain RGD-SPIO clusters for further purification. Next, to remove free RGD-PHEA-g-C18, the supernatant was centrifuged at 15,000 rpm for 20 minutes (Eppendorf 5424), allowing RGD-SPIO clusters to be separated out. Finally, RGD-SPIO clusters were reconstituted in Dulbecco’s Modified Eagle’s Medium (DMEM, Corning Cellgro) with gentle bath sonication (Fisher), and stored at 4 C until further usage.
Analysis of Iron Content in RGD-SPIO Clusters and Cells
Iron content in RGD-SPIO clusters and in BMSCs was quantified through inductively coupled plasma spectrometry (ICP, Perkinelmer 2000 DV). All samples were dissolved in nitric acid (Macron) and then diluted to 5 mL with DI water. NIST primary standards were used for instrument calibration. Samples were fed into the instrument with a peristaltic pump, and an internal standard was used to account for variations in sample fluidity.
Measurement of RGD-SPIO Size with Dynamic Light Scattering (DLS)
After purification, RGD-SPIO clusters were redispersed at a dilute concentration, and then analyzed with dynamic light scattering (Malvern Zetasizer). At least three measurements were made, and RGD-SPIO diameter was reported as the peak of the number distribution.
Image Analysis of OA-SPIOs and RGD-SPIO Clusters
OA-SPIOs and RGD-SPIO clusters were visualized with transmission electron microscopy (TEM). The samples were air-dried on a holey carbon-coated grid and then imaged at 200kV (JEOL 2100 TEM). To determine the diameter of the OA-SPIOs, at least 50 particles were analyzed using ImageJ software.
Analysis of Magnetic Resonance (MR) Relaxivity of SPIO Clusters
MR relaxivity of RGD-SPIO clusters was measured using a spin-echo sequence (14.1 T Varian MR system). Prior to imaging, the instrument was shimmed with a single-pulse sequence until linewidth was around 600 Hz. Imaging parameters included receiver gain: 44; repetition time (trepetition): 300 milliseconds; echo time (techo): 9, 10, 11,12, 13, 14 milliseconds; data matrix: 256 × 256; field of view: 30 × 30 mm; slice thickness varied as needed. Total acquisition time was between 10 to 20 minutes, and a copper sulfate solution was used as a marker. To calculate T2 values, the mean gray value (termed “relative brightness”) from ImageJ was plotted against echo time. Using an exponential fit, the inverse of T2 was calculated. An example of this graph is presented in Figure S1. T2 relaxivity was quantified from the slope of the curve between the inverse T2 and iron concentration. For relaxivity measurements, at least five different iron concentrations were used.
Bone Marrow-Derived Mesenchymal Stem Cell (BMSC) Culture
Mouse bone marrow-derived D1 mesenchymal stem cells (ATCC) were cultured in DMEM (Corning Cellgro) supplemented with 10 % fetal bovine serum (Thermo Scientific) and 1 % penicillin/streptomycin (Gibco). Cells were incubated at 5 % CO2 and 37 °C in sterile conditions. Cell media was changed every 3 days, and cells were passaged when over 90 % confluent. No passage numbers higher than 28 were used.
Co-Incubation of Cells with RGD-SPIO Clusters
BMSCs were plated in T-25 cell culture flasks at a density of 10,000 cells/cm2, and then incubated overnight. Afterwards, fresh DMEM supplemented with 10 or 1 % FBS was added, and then the cell culture flasks were placed on an orbital shaker set to rotate at 0, 20, or 50 rpm (Heidolph Rotamax 120). The cell culture flasks were shaken for approximately 10 hours to allow cells to adapt to their new environment. In previous studies, at least 6 hours were necessary for noticeable cellular gene expression changes to take place under shear.19 After 10 hours of continuous shaking, media was removed, and fresh media containing RGD-SPIO clusters at a concentration of 0.32 mM Fe was added. Once again, FBS concentration was kept consistent at either 10 or 1 %. Then, cells were exposed to orbital flow and RGD-SPIO clusters for the next 14 hours. After incubation, cells were washed three times with PBS to remove free RGD-SPIO clusters. In parallel, cells in a stationary flask were incubated for 10 hours without RGD-SPIO clusters, and then for 14 hours with media containing RGD-SPIO clusters at a concentration of 0.32 mM Fe, again with a constant FBS concentration. Unlabeled cells incubated for 24 hours in a stationary flask at an FBS concentration of 10 % served as a control.
Phase Contrast Imaging of Cells Incubated with SPIO Clusters
Following co-incubation of BMSCs with RGD-SPIO clusters, cells were fixed with 3.7 % paraformaldehyde in phosphate buffer saline (PBS) for 30 minutes. After washing with PBS, Perls reagent (1:1 10 % potassium ferrocyanide/1 N HCl) was added for 20 minutes. In this step, poisonous hydrogen cyanide is liberated. Then, cells were washed thoroughly with PBS. Finally, cellular images were taken with a Zeiss Axiovert 200M apotome microscope at 20× and 63×.
ICP Analysis of Cellular Uptake of RGD-SPIO Clusters
After co-incubation with RGD-SPIO clusters, cells were washed, trypsinized, counted, and then centrifuged to form a pellet. The pellet was dissolved in concentrated nitric acid overnight at room temperature before analysis. ICP measurements were taken as previously described. At least three aliquots containing 100,000 cells each were used for each sample for ICP analysis.
MR Imaging of Cells Labeled with RGD-SPIO Clusters
After co-incubation with RGD-SPIO clusters, cells were washed, trypsinized, and then resuspended in DMEM at 1×106 cells/mL. Cells were then embedded in bovine type I collagen gel (PureCol, Advanced Biomatrix) by mixing cells with collagen solution and then increasing pH and temperature to form a gel, as previously reported.20 Care was taken to avoid air bubbles in the viscous collagen suspension, and the gels were formed within a glass capillary tube at 37 °C. The total concentration of cells in the pre-gelled solution was approximately 300 cells/μL. After embedding, labeled BMSCs were imaged with 14.1 T Varian MR system using a spin-echo sequence. Prior to imaging, the instrument was shimmed using a single pulse sequence until linewidth was less than 600 Hz. MR imaging parameters were as follows—receiver gain: 44; trepetition: 300 milliseconds; techo: 1, 2, 4, 6, 8, 10 milliseconds; data matrix: 256 × 256; field of view: 30 × 30 mm. Total acquisition time ranged from 10 to 20 minutes, and a copper sulfate solution was used as a marker. To quantify a mean T2 value per condition, at least two 0.7 mm thick slices were selected in different locations of each gel.
Analysis of Cell Viability Following Cellular Uptake of RGD-SPIO Clusters
The viability of BMSCs incubated with RGD-SPIO clusters in static or dynamic conditions was assessed with a Trypan Blue and a (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent assay (ATCC). For Trypan Blue assays, cells were washed with PBS and then incubated with 0.4 % Trypan Blue for 5 minutes. Then, the percentage of live cells was counted using a light microscope (Leica). For the MTT assay, 10 μL of (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide was added to 100 μL of phenol red-free DMEM with cells, and incubated at 37 °C for 4 hours. Then, cells were digested in 100 μL of detergent for 2.5 hours. Absorbance of cell lysate was read at 570 nm (BioTek Synergy HT).
Immunostaining of β1Integrins
Cells were fixed with 3.7 % paraformaldehyde for 10 minutes. After washing cells with PBS, samples were incubated with 1 % bovine serum albumin, 5 % FBS, and 0.3 % Triton-X for 1 hour to block non-specific binding and permeabilize cells. Then, cells were incubated with a mouse monoclonal antibody to integrin β1 (Abcam) overnight at 4 °C. After washing, cells were incubated with goat polycloncal 6563 secondary anitbody-Cy5 (Abcam) for 1 hour at room temperature. Dilutions ratios were 1:1000 and 1:100 for the primary and secondary antibodies, respectively. Lastly, 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI, Invitrogen) was used to stain cell nuclei at a working concentration of 100 ng/mL.
Nuclei and cellular integrins were imaged using Zeiss LSM 700 confocal microscope at 40×. ImageJ software was used to quantify the integrin expression level and integrin cluster formation. Each image was separated into three channels, and the mean gray value of the green channel (termed “mean green value”) was determined over the cell area. Here, a higher mean green value corresponds to a higher relative level of integrin expression. At least 15 cells were analyzed per condition.
Intramuscular Hindlimb Injection of D1 Cells into a Mouse Model
All mice work was carried out in accordance with university and federal regulations (Institutional Animal Care and Use Committee approval number: 11089). We used three four month old Balb/c and two three month old C57Black mice for our in vivo work. Before cell injection, all mice were anesthetized with isoflurane. Then, a 50 μL BMSC suspension (2×106 cells/mL) was injected into the muscles of the right caudal thigh approximately between semimembranosus and semitendinosus muscles, using a 1.0 mL syringe with 25G1 needle. BMSCs labeled in static conditions or at 50 rpm were used in this study, and unlabeled BMSCs were used as a negative control. To help localize cells using a collagen gel, cells labeled at 50 rpm were mixed with 1 mg/mL of type I collagen (PureCol), followed by injection.
In Vivo MR-based Tracking of BMSCs
Mice were humanely sacrificed approximately 1 hour after injection. Then, both legs of each mouse were imaged with a spin-echo sequence(14.1 T Varian MR system) to locate the transplanted BMSCs. A custom-built coil was used to contain the sacrificed mice, the details of which are described elsewhere.21 Prior to imaging, the instrument was shimmed manually until linewidth was around 200 Hz. Imaging parameters include trepetition: 40 milliseconds; techo: 1000-1300 milliseconds; data matrix: 256 × 256; slice thickness: 0.5-1.0 mm; field of view: varied as needed. Coronal cuts were used to identify the hypointense regions, which were subsequently processed in ImageJ. Total acquisition time was around 20 minutes, and a copper sulfate solution was used as a marker. Afterwards, mouse bodies were disposed of according to university regulations.
Computational Simulation of Average RGD-SPIO Cluster Velocity
The particle motion in unsteady, free-surface flow inside a 25 mm2 cell culture flask was simulated using finite element method (FEM) commercial software (Comsol Multiphysics 3.3). To begin this analysis, 4 mL media rotated at three different speeds (f) of 0, 20 and 50 rpm was simulated. Then, the free surface motion in the liquid–air interface and the particle flow patterns inside the well were analyzed. The orbital shaker imparts the same two-dimensional, in-plane movement to all points on the plate. The velocity of the plate walls (U) are thus given by:
| [2] |
where Rgis the orbital radius (~ 20 mm in this study) and Ω is the angular velocity that is given by:
| [3] |
Statistical Analysis
All data were analyzed using a two-tailed t-test with equal variance (Microsoft Excel). P < 0.05 was considered the threshold for significance. All samples were done in triplicate unless otherwise noted. Data are represented as mean ± standard error of mean (SEM).
3. RESULTS AND DISCUSSION
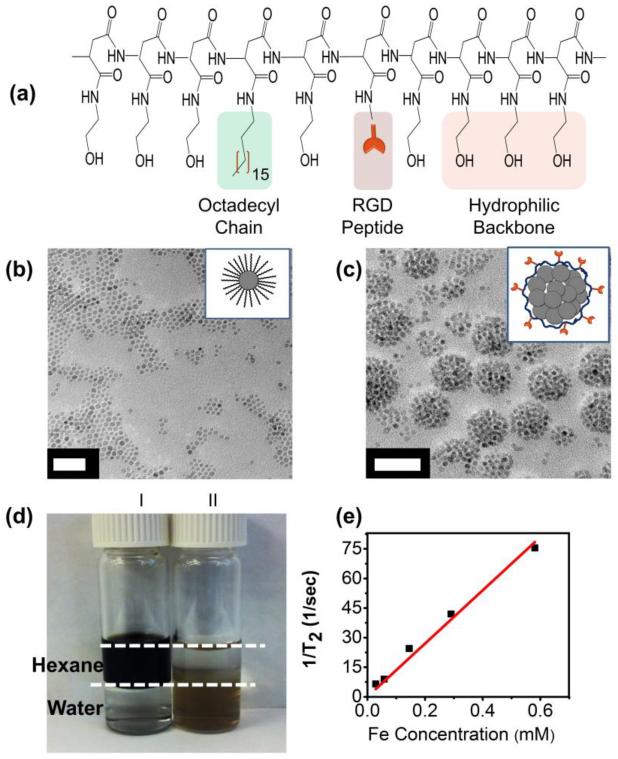
We first synthesized poly(2-hydroxyethyl aspartamide) (PHEA) grafted with octadecyl chains and RGD peptides (termed RGD-PHEA-g-C18) for use in SPIO clustering. The octadecyl chains allow PHEA to associate with a hydrophobic moiety.5 Separately, the RGD peptides of PHEA bind with cellular integrin domains, including β1.22 The sequential addition of octadecylamine, GGGGRGDSP peptide, and ethanolamine to polysuccinimide (PSI) resulted in PHEA substituted with octadecyl chains, GGGGRGDSP, and hydroxyl groups, as confirmed through 1H-NMR (Figure 1a, S2 -S4).18 The degree of substitution (DSC18) of octadecyl chain was approximately 6.7 %, as calculated with 1H-NMR.Next, the resulting RGD-PHEA-g-C18 was mixed with OA-SPIOs prepared from the thermal decomposition of iron acetylacetonate (Figure 1b).16 Adding an aqueous solution of RGD-PHEA-g-C18 to OA-SPIOs suspended in chloroform resulted in RGD-SPIO clusters. After removing the chloroform, the resulting clusters were readily suspended in DI water without noticeable aggregation (Figure 1c & 1d). Here, cluster formation is driven by the intercalation of the oleic acid ligand from OA-SPIOs and the octadecyl chains of PHEA.23 The mean diameter of cluster was approximately 43 nm, as reported by DLS. (Figure S5). A polydispersity index of less than 0.2 is reported, demonstrating a good degree of cluster size control. In addition, the RGD-SPIO clusters were less than 50 nm, which was suggested as a size range to facilitate the receptor-mediated endocytosis.6 The T2 relaxivity of RGD-SPIO clusters was around 135 mM−1s−1(Figure 1e), comparable to that of FDA-approved contrast agents such as Feridex.24 The RGD-SPIO clusters suspended in PBS remained stable at 4 °C for over three weeks.
Figure 1.
Characterization of RGD-PHEA-g-C18 and RGD-SPIO cluster. (a) Structure of RGD-PHEA-g-C18. (b) TEM image of oleic acid-coated SPIOs. (c) TEM image of RGD-SPIO clusters. Both scale bars represent 50 nm. (d) OA-coated SPIOs (I) are suspended in hexane, while RGD-SPIO clusters (II) are dispersed in water. Dotted white lines help indicate the interface between hexane and water. (e) A plot of 1/T2 vs. iron concentration used to determine T2 relaxivity of RGD-SPIO clusters.
Separately, computational simulations and mathematical calculations were conducted to estimate the average velocity of RGD-SPIO clusters and the shear stress on the cell membrane. According to a COMSOL simulation, the average velocity of RGD-SPIO clusters in the static condition was around 0.2 mm/s (Figure S6a-I & S6b). The cluster velocity was minimally changed by increasing the orbital speed to 20 rpm (Figure S6a-II & S6b). Further increasing the orbital speed to 50 rpm resulted in a significant increase of cluster velocity to 1.5 mm/s, an order of magnitude difference from the speed in the static condition (Figure S6a-III & S6b). In parallel, shear stress on the cell membrane was approximated by:
| (4) |
where R is the orbital radius of the shaker (20 mm), ρ is the density of the culture medium (0.9973 g/mL), μ is the dynamic viscosity of the medium (0.0102 Pa-s), and Ω is the angular velocity (rad/s).25
According to this calculation, increasing the orbital speed from 0 to 20 and 50 rpm increased shear stress from 0 to 0.6 dyne/cm2 and 2.4 dyne/cm2, respectively (Figure S7). Based on previous studies, shear stress levels as low as 0.2 dyne/cm2 have been shown to influence cellular gene incorporation, suggesting that a minimum shear level is needed to stimulate cells.26, 27 Alternatively, increasing the orbital speed past a certain limit, such as 100 rpm, was shown to cause extensive media drying due to the increasingly violent motion of the media.28
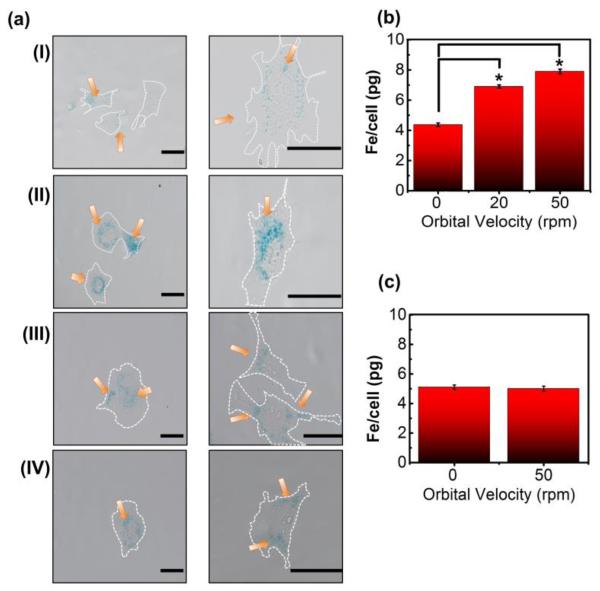
To assess whether or not shear stress and cluster velocity influenced RGD-SPIO cluster uptake, we labeled BMSCs under orbital speeds ranging from 0 to 20 and 50 rpm. As hypothesized, BMSCs labeled at orbital velocity of 50 rpm took up more RGD-SPIO clusters than those labeled under static conditions, as evidenced by Prussian blue staining (Figure 2a-I & 2a-II). An ICP analysis further confirmed a significant increase of the iron content per cell (Figure 2b). BMSCs labeled at 20 and 50 rpm took up 1.6- and 1.8-fold greater amount of RGD-SPIO clusters than those labeled under static condition, respectively. At 50 rpm, the iron content per cell was approximately 7 pg Fe, which was over four times larger than the widely-accepted benchmark for cell tracking in vivo (i.e., 1.5 pg/cell).29 The iron mass per cell was linearly related to the calculated shear stress on the cell membrane, but not to the computationally-estimated average velocity of RGD-SPIO clusters (Figure S9). Note that the cellular uptake of RGD-SPIO clusters was significantly increased even at an orbital velocity of 20 rpm, at which the cluster velocity was not significantly increased. Despite such high iron loading per cell, over 85 % of cells remained viable across all orbital speeds, as determined through a Trypan Blue assay (Figure S8). This viability level was comparable to that of cells cultured in static conditions without RGD-SPIO clusters, suggesting that neither shaking nor SPIO loading affects viability.
Figure 2.
Analysis of the effect of orbital velocity on cellular uptake of RGD-SPIO clusters at varied FBS concentrations (ΦFBS). (a) Phase contrast images of BMSCs labeled at various conditions. Prussian blue stain indicates the presence of iron. (a-I) BMSCs labeled with RGD-SPIO clusters under static conditions (ΦFBS = 10 %), (a-II) BMSCs labeled with RGD-SPIO clusters at 50 rpm (ΦFBS = 10 %), (a-III) BMSCs labeled with RGD-SPIO clusters under static conditions (ΦFBS = 1 %), (a-IV) BMSCs labeled with RGD-SPIO clusters at 50 rpm (ΦFBS = 1 %). Scale bars represent 40 μm. The images on the 2nd column in (a) are magnified images of cells from the same condition in the 1st column. Orange arrows indicate RGD-SPIO clusters stained by Prussian blue, and white dotted lines indicate cellular periphery. (b) ICP analysis to quantify the dependence of the iron content per cell on orbital velocity at ΦFBS of 10 %. Differences of values between conditions were statistically significant (*p < 0.05). (c) ICP analysis to show the independence of the iron content per cell on orbital velocity at ΦFBS of 1 %.
These interesting effects of orbital flow on cell labeling efficiency became insignificant when the concentration of FBS in the cell culture media (ΦFBS) was decreased from 10 to 1 %. As characterized with both Prussian blue staining and ICP iron content per cell was independent of orbital velocity (Figure 2a-III, 2a-IV, & 2c). Accordingly, the iron content per cell was independent of changes in cluster velocity and shear stress at low ΦFBS. In addition, for cells exposed to orbital flow, the iron content per cell was decreased by reducing ΦFBS from 10 to 1 %. In contrast, labeling efficiency in static conditions was independent of ΦFBS.
To further address the underlying mechanism by which the orbital velocity and the concentration of FBS (ΦFBS) affect the cellular uptake of RGD-SPIO clusters, we first examined whether one or both of these experimental variables modulated the frequency of cellular division. According to previous gene transfection studies, more frequent cell division is correlated with improved uptake of plasmid DNA complexes.30 According to an MTT assay, decreasing ΦFBS to 1 % significantly limited cell metabolic activity and proliferation, as assessed with a decrease in the amount of MTT reagent reduced by metabolically active cells over time (Figure S10). However, at a given ΦFBS, the degree of increase in cell proliferation was independent of the orbital velocity. Therefore, the iron content per cell was not dependent on cell proliferation.
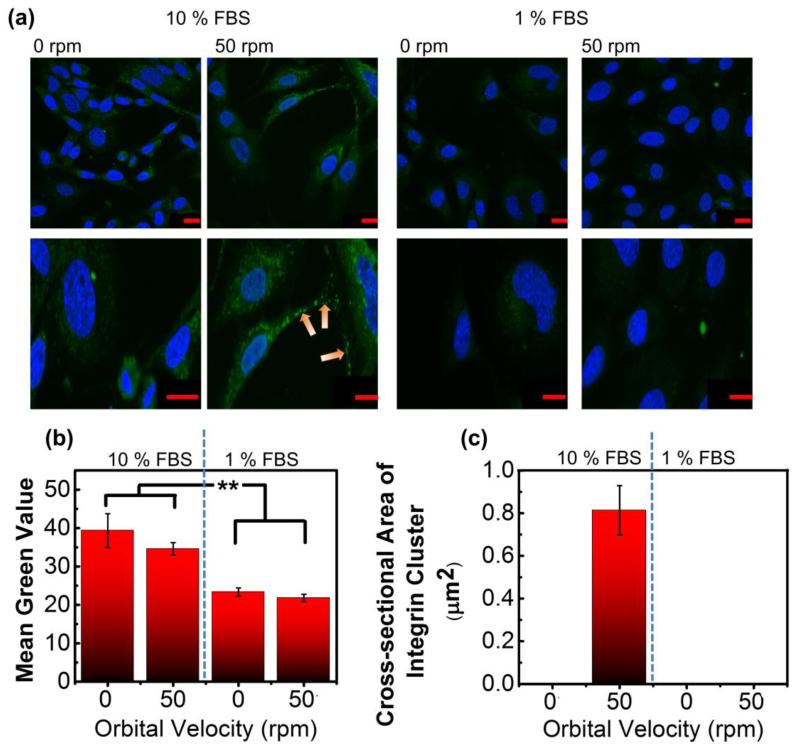
In parallel, we examined the effects of orbital velocity and ΦFBS on β1 integrin cluster formation, as integrin clustering is reported to be a key step in the cellular incorporation of RGD-coated gene complexes.31 Total cellular β1 integrin expression level was significantly reduced with decreasing ΦFBS in the cell culture media (Figure 3a & 3b). There was not a noticeable difference of the total β1 integrin expression between cells cultured in the static and shaking conditions at a given ΦFBS (Figure 3a & 3b). In contrast, at ΦFBS of 10 %, cells cultured under orbital flow displayed a large number of integrin clusters, marked by large fluorescent islands with a cross-sectional area of approximately 0.8 μm2 (Figure 3a & 3c). Almost no integrin clusters were observed within cells cultured in a static condition (Figure 3a & 3c). In addition, no significant integrin cluster formation was visualized with cells cultured at ΦFBS of 1 %. Overall, the iron content per cell, modulated by the orbital flow velocity and ΦFBS, could be related to the degree of intracellular integrin cluster formation, but not to the total integrin expression level.
Figure 3.
Analysis of the effects of orbital velocity on cellular integrin expression and integrin cluster formation at varied FBS concentrations (ΦFBS). (a) Confocal images of β1 integrins (green color) and nuclei (blue color). The orange arrows mark β1 integrin clusters, and red scale bars correspond to 20 μm. (b) Quantification of the number of green-colored pixels (“Mean Green Value”), indicative of total β1 integrin expression. The difference of values between ΦFBS of 10 % and 1 % was statistically significant. ** corresponds to p < 0.01. (c) Analysis of the cross-sectional area of β1 integrin clusters
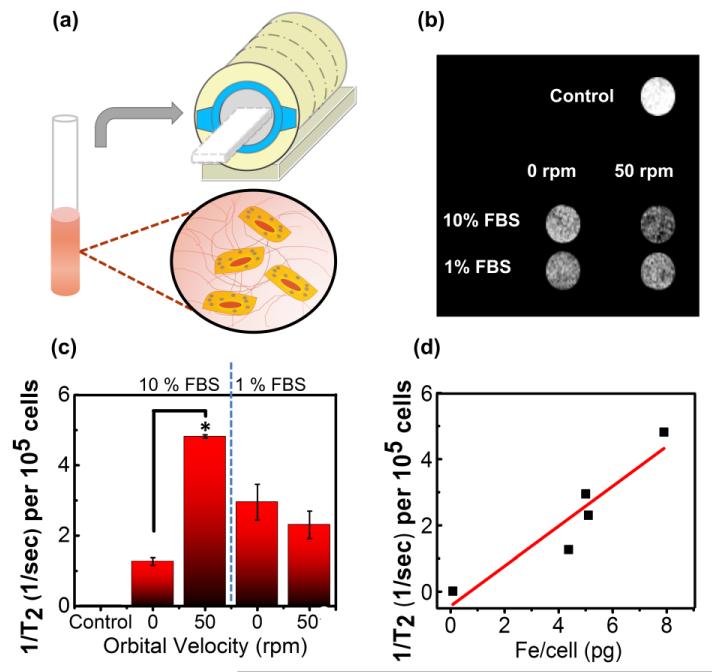
Finally, we evaluated whether this labeling protocol would generate different levels of MR contrast both in vitro and in vivo. According to MR images of BMSCs loaded in a tissue-like 3D collagen gel, the largest degree of negative contrast, or hypointensity, was observed in gels containing cells labeled with RGD-SPIO clusters at 50 rpm and ΦFBS of 10 % (Figure 4a & 4b). Similarly, the inverse of T2 (1/T2) of each gel, calculated with an array of echo times, was highest for BMSCs labeled at 50 rpm and ΦFBS of 10 % (Figure 4c). A larger 1/T2 value represents a greater amount of RGD-SPIO clusters per cell, as confirmed with the highly linear relationship between 1/T2 and iron content per cell (Figure 4d).
Figure 4.
In vitro analysis of T2 for SPIO-labeled BMSCs in a collagen gel. (a) Schematic describing in vitro MR imaging of BMSCs encapsulated in a collagen gel formed inside a glass tube. (b) Slices of collagen gels with labeled BMSCs imaged with a spin-echo sequence. (c) Effects of orbital velocity on the inverse T2, at varied FBS concentrations in the cell culture media. Difference of values between BMSCs incubated with RGD-SPIO clusters in static condition and those exposed to orbital flow at velocity of 50 rpm is statically significant (*p <0.05). (d) The linear dependency of the inverse T2 per 105 cells on the iron content per cell measured with ICP (R2> 0.8).
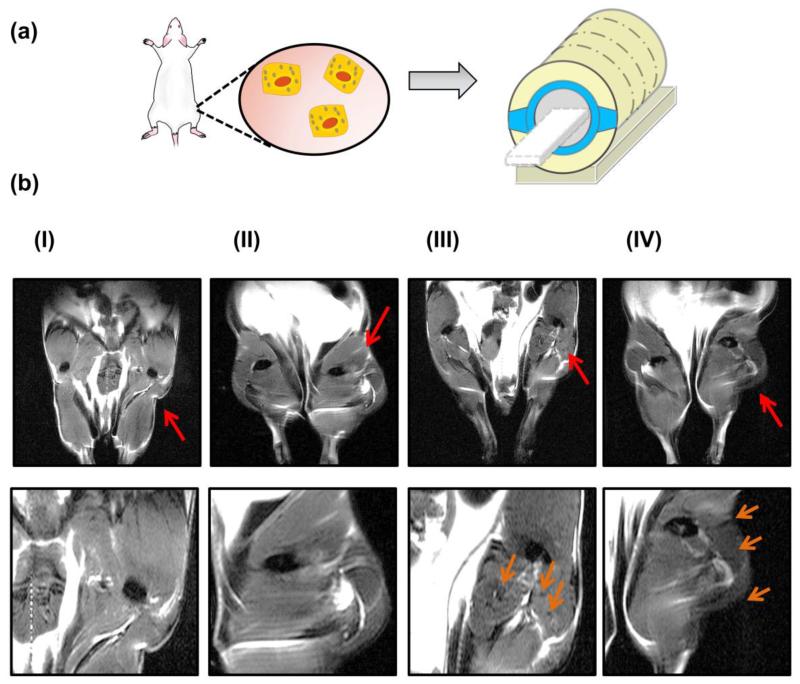
Lastly, we injected BMSCs labeled under static and shaking conditions into the right hindlimb of a mouse (Figure 5a). Injecting stem cells into this tissue is often used to treat ischemia.32 The concentration of FBS in the cell culture media was kept constant at 10 %, and BMSCs were only labeled at static conditions and at 50 rpm. Unlabeled BMSCs were used as a control. A hindlimb injected with BMSCs labeled under orbital flow displayed a greater degree of hypointensity than a hindlimb injected with cells labeled in the static condition (Figure 5b-II & 5b-III). This hypointense area was more localized by injecting a collagen gel loaded with BMSCs labeled at 50 rpm (Figure 5b-IV). No noticeable hypointense region was observed in hindlimbs injected with unlabeled MSCs (Figure 5b-I).
Figure 5.
In vivo MR images of BMSCs injected into the muscle of a mouse’s hindlimb. (a) Schematic describing hindlimb injection with labeled BMSCs. (b) MR images of both hindlimbs. The BMSCs were injected into the right hindlimb, as indicted with the red arrows. (b-I) Hindlimb transplanted with unlabeled BMSCs, (b-II) hindlimb transplanted with BMSCs labeled under static conditions, (b-III) hindlimb transplanted with BMSCs labeled at 50 rpm, (b-IV) hindlimb implanted with a BMSCs labeled at 50 rpm and then encapsulated in collagen gel in situ. In (b), images of the 2nd row are magnified view of the right hindlimb. Orange arrows in 2nd row images point the hypointense area due to the presence of transplanted cells.
Taken together, we have successfully demonstrated that the extracellular mechanical environment plays an important role in stem cell labeling with MR contrast agents. The use of mechanical stimuli to control cell labeling is unique, as previous attempts to improve SPIO uptake in stem cells were based mostly on the surface modification of SPIOs. In early attempts, researchers added a cell-penetrating peptide33 (such as HIV-tat) or a transfection agent34 (such as Lipofectamine or poly-l-lysine) to the SPIO surface to improve SPIO loading per cell. More recently, researchers explored a variety of polymer coatings on the SPIO surface in order to enhance cellular uptake. For example, a layer-by-layer electrostatic assembly technique was used to introduce polyethyleneimine, chitosan, dextran, and other polymers to the SPIO surface.35 Another alternative technique coated the SPIO surface with a layer of silica, allowing a variety of functional groups to be conjugated on the silica-SPIO surface.36 However, concerns have been raised over the cytotoxicity and overall usefulness of these methods. For example, HIV-tat coated SPIOs located specifically to the nucleus, which might interfere with cellular function and differentiation.33 In addition, the introduction of new coatings on the SPIO surface requires a laborious chemical synthesis and subsequent purification. Our labeling technique, on the other hand, requires a one-step method to prepare the RGD-SPIO clusters, relies on non-chemical methods to improve SPIO uptake, and maintains cell viability in a variety of cellular environments.
When compared to other methods to externally stimulate SPIO uptake, our labeling technique again has noticeable advantages. Previously reported external techniques to enhance cell labeling included electroporation13 or the use of a piezoactuator.37 However, these methods have been shown to compromise cell viability,38 and can be technically challenging.39 In contrast, our cell labeling procedure only requires an orbital shaker, which is an inexpensive, easy-to-use piece of equipment readily available in research labs and operation rooms. One shaker can contain several flasks, allowing for millions of cells to be labeled simultaneously. In addition, orbital shaking has no significant effects on cell viability. Improvements to this protocol can be made to enable highly effective stem cell labeling. For example, we suggest that cells labeled by advanced SPIO systems with high T2 relaxivity (e.g., 700 mM−1s−1) under orbital shaking would further improve stem cell tracking quality.
To better understand this protocol, we explored the changes in cell-nanoparticle interaction and cell phenotype that occurred under shaking. Initially, we hypothesized that the cellular uptake of RGD-SPIO clusters was dependent on both average particle velocity and shear stress on the cell membrane. However, a considerable increase in iron content per cell was discovered for BMSC labeling at 20 rpm, where only the shear stress was significantly increased. This trend suggested that mechanotransduction played a more significant role in regulating the cellular uptake of nanoparticle clusters than changes in average cluster velocity. The continued increase of the iron content per cell from 20 to 50 rpm likely resulted from a larger magnitude of mechanical stimulation. Further mechanistic studies on cellular division, integrin expression, and integrin cluster formation demonstrated that the mechanism by which the shear stress increased cellular SPIO cluster uptake was likely related to the upregulation of integrin clusters, a known intermediary step in integrin-mediated endocytosis. Additionally, we demonstrated that serum was an important element to switch on cellular mechanosensitivity, as confirmed with the minimal dependency of RGD-SPIO cluster uptake on orbital flow in low serum conditions. However, further work will be necessary to fully understand the specific endocytotic pathway.
4. CONCLUSION
In conclusion, this study offers the first step in a new, non-chemical method for simple but elaborate stem cell labeling based on manipulating the extracellular mechanical environment. We believe that this flow-modulated cell labeling procedure will greatly benefit MR-based cell tracking, as this method avoids the laborious chemical modifications and carefully refined protocols found in previous studies. Previous studies demonstrated that external shear flow influences cellular uptake of gene complexes and nanoparticle (e.g., quantum dot and silica nanoparticles) in microfluidic systems.15, 40 However, this is the first study to demonstrate that shear conditions readily modulated by a conventional orbital shaker can stimulate efficient mesenchymal stem cell labeling with SPIOs. Unlike complex microfluidic systems, an orbital shaker is readily available in clinical settings for cell labeling. Therefore, we believe that this protocol would be useful to labeling a wide array of stem cells and islets41 with SPIOs for diagnosing and treating various diseases and tissue defects. We suggest that the effects of external flow on cell labeling would be further amplified by tailoring other external extracellular environmental factors such as cell adhesion ligands, cell adherent matrix stiffness, and growth factors in media.42 Finally, we envision that this method would be readily utilized for improving the cellular uptake of exogenous genes (e.g., siRNA and DNA) as well as other nanoparticles used for imaging and treatments.43
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Institute of Health (1R01 HL109192 to H.J.K. and R25 CA154015A to) N.C.). We would like to thank Dr. Boris Odintsov for help with MR imaging, Dr. Mayandi Sivaguru for help with confocal and cell imaging, Dr. Wacek Swiech for help with TEM images, and Rudiger Laufhutte for ICP measurements.
Footnotes
Author Contributions
Notes
The authors declare no competing financial interests
Supporting Information. Additional figures can be found in the supporting information section. This information is available free of charge via the Internet at http://pubs.acs.org/.
REFERENCES
- 1.Uccelli A, Moretta L, Pistoia V. Nat. Rev. Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 2.Mooney DJ, Vandenburgh H. Cell Stem Cell. 2008;2:205–213. doi: 10.1016/j.stem.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Lee N, Choi Y, Lee Y, Park M, Moon WK, Choi SH, et al. Nano Lett. 2012;12:3127–3131. doi: 10.1021/nl3010308. [DOI] [PubMed] [Google Scholar]
- 4.Nasongkla N, Bey E, Ren J, Ai H, Khemtong C, Guthi JS, et al. Nano Lett. 2006;6:2427–2430. doi: 10.1021/nl061412u. [DOI] [PubMed] [Google Scholar]
- 5.Lee HJ, Jang K-S, Jang S, Kim JW, Yang H-M, Jeong YY, et al. Chem. Commun. (Cambridge, U.K.) 2010;46:3559–3561. doi: 10.1039/b925106e. [DOI] [PubMed] [Google Scholar]
- 6.Gao H, Shi W, Freund LB. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9469–9474. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pöselt E, Kloust H, Tromsdorf U, Janschel M, Hahn C, Maßlo C, et al. ACS Nano. 2012;6:1619–1624. doi: 10.1021/nn204591r. [DOI] [PubMed] [Google Scholar]
- 8.De Palma R, Peeters S, Van Bael MJ, Van den Rul H, Bonroy K, Laureyn W, et al. Chem. Mater. 2007;19:1821–1831. [Google Scholar]
- 9.Chen W, Cao Y, Liu M, Zhao Q, Huang J, Zhang H, et al. Biomaterials. 2012;33:7895–7902. doi: 10.1016/j.biomaterials.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Amstad E, Gillich T, Bilecka I, Textor M, Reimhult E. Nano Lett. 2009;9:4042–4048. doi: 10.1021/nl902212q. [DOI] [PubMed] [Google Scholar]
- 11.Hoehn M, Küstermann E, Blunk J, Wiedermann D, Trapp T, Wecker S, et al. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16267–16272. doi: 10.1073/pnas.242435499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thorek DLJ, Tsourkas A. Biomaterials. 2008;29:3583–3590. doi: 10.1016/j.biomaterials.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walczak P, Ruiz-Cabello J, Kedziorek DA, Gilad AA, Lin S, Barnett B, et al. Nanomedicine (New York, NY, U.S.) 2006;2:89–94. doi: 10.1016/j.nano.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Gerard A. Am. J. Physiol. 2002;282:F179–F190. [Google Scholar]
- 15.Samuel SP, Jain N, O’Dowd F, Paul T, Kashanin D, Gerard VA, et al. Int. J. Nanomed. 2012;7:2943–2956. doi: 10.2147/IJN.S30624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun S, Zeng H, Robinson DB, Raoux S, Rice PM, Wang SX, et al. J. Am. Chem. Soc. 2003;126:273–279. doi: 10.1021/ja0380852. [DOI] [PubMed] [Google Scholar]
- 17.Tomida M, Nakato T, Matsunami S, Kakuchi T. Polymer. 1997;38:4733–4736. [Google Scholar]
- 18.Lai M-H, Jeong JH, DeVolder RJ, Brockman C, Schroeder C, Kong H. Adv. Funct. Mater. 2012;22:3239–3246. doi: 10.1002/adfm.201102664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urbich C, Walter DH, Zeiher AM, Dimmeler S. Circ. Res. 2000;87:683–689. doi: 10.1161/01.res.87.8.683. [DOI] [PubMed] [Google Scholar]
- 20.Liang Y, Jeong J, DeVolder RJ, Cha C, Wang F, Tong YW, et al. Biomaterials. 2011;32:9308–9315. doi: 10.1016/j.biomaterials.2011.08.045. [DOI] [PubMed] [Google Scholar]
- 21.Odintsov B, Chun JL, Mulligan JA, Berry SE. Magn. Reson. Med. 2011;66:1704–1714. doi: 10.1002/mrm.22942. [DOI] [PubMed] [Google Scholar]
- 22.Seo J-H, Kakinoki S, Inoue Y, Yamaoka T, Ishihara K, Yui N. J. Am. Chem. Soc. 2013;135:5513–5516. doi: 10.1021/ja400817q. [DOI] [PubMed] [Google Scholar]
- 23.Yang H-M, Lee HJ, Jang K-S, Park CW, Yang HW, Heo WD, et al. J. Mater. Chem. 2009;19:4566–4574. [Google Scholar]
- 24.Yang X, Grailer JJ, Rowland IJ, Javadi A, Hurley SA, Steeber DA, et al. Biomaterials. 2010;31:9065–9073. doi: 10.1016/j.biomaterials.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty A, Chakraborty S, Jala VR, Haribabu B, Sharp MK, Berson RE. Biotechnol. Bioeng. 2012;109:695–707. doi: 10.1002/bit.24352. [DOI] [PubMed] [Google Scholar]
- 26.Farokhzad OC, Khademhosseini A, Jon S, Hermmann A, Cheng J, Chin C, et al. Anal. Chem. (Washington, DC, U.S.) 2005;77:5453–5459. doi: 10.1021/ac050312q. [DOI] [PubMed] [Google Scholar]
- 27.van der Meer AD, Kamphuis MMJ, Poot AA, Feijen J, Vermes I. J. Controlled Release. 2008;132:e42–e44. [Google Scholar]
- 28.Salek MM, Sattari P, Martinuzzi R. Ann. Biomed. Eng. 2012;40:707–728. doi: 10.1007/s10439-011-0444-9. [DOI] [PubMed] [Google Scholar]
- 29.Bos C, Delmas Y, Desmoulière A, Solanilla A, Hauger O, Grosset C, et al. Radiology (Oak Brook, IL, U.S.) 2004;233:781–789. doi: 10.1148/radiol.2333031714. [DOI] [PubMed] [Google Scholar]
- 30.Riddle KW, Kong H-J, Leach JK, Fischbach C, Cheung C, Anseth KS, et al. Mol. Ther. 2007;15:361–368. doi: 10.1038/sj.mt.6300017. [DOI] [PubMed] [Google Scholar]
- 31.Kunath K, Merdan T, Hegener O, Haberlein H, Kissel T. J. Gene Med. 2003;5:588–599. doi: 10.1002/jgm.382. [DOI] [PubMed] [Google Scholar]
- 32.Ishikane S, Ohnishi S, Yamahara K, Sada M, Harada K, Mishima K, et al. Stem Cells (Durham, NC, U.S.) 2008;26:2625–2633. doi: 10.1634/stemcells.2008-0236. [DOI] [PubMed] [Google Scholar]
- 33.Josephson L, Tung C-H, Moore A, Weissleder R. Bioconjugate Chem. 1999;10:186–191. doi: 10.1021/bc980125h. [DOI] [PubMed] [Google Scholar]
- 34.Frank JA, Miller BR, Arbab AS, Zywicke HA, Jordan EK, Lewis BK, et al. Radiology (Oak Brook, IL, U.S.) 2003;228:480–487. doi: 10.1148/radiol.2281020638. [DOI] [PubMed] [Google Scholar]
- 35.Schwarz S, Wong JE, Bornemann J, Hodenius M, Himmelreich U, Richtering W, et al. Nanomedicine (New York, NY, U.S.) 2012;8:682–691. doi: 10.1016/j.nano.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 36.Lu C-W, Hung Y, Hsiao J-K, Yao M, Chung T-H, Lin Y-S, et al. Nano Lett. 2006;7:149–154. doi: 10.1021/nl0624263. [DOI] [PubMed] [Google Scholar]
- 37.Vaidyanathan R, Curtis A, Mullin M. J. Nanopart. Res. 2011;13:5301–5309. [Google Scholar]
- 38.Andreason GL, Evans GA. Anal. Biochem. 1989;180:269–275. doi: 10.1016/0003-2697(89)90429-6. [DOI] [PubMed] [Google Scholar]
- 39.Canatella PJ, Karr JF, Petros JA, Prausnitz MR. Biophys. J. 2001;80:755–764. doi: 10.1016/S0006-3495(01)76055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J, Hwang I, Britain D, Chung TD, Sun Y, Kim D-H. Lab Chip. 2011;11:3941–3948. doi: 10.1039/c1lc20766k. [DOI] [PubMed] [Google Scholar]
- 41.Lee N, Kim H, Choi SH, Park M, Kim D, Kim H-C, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:2662–2667. doi: 10.1073/pnas.1016409108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Nat. Mater. 2005;4:460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- 43.Josephson L, Kircher MF, Mahmood U, Tang Y, Weissleder R. Bioconjugate Chem. 2002;13:554–560. doi: 10.1021/bc015555d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.