Abstract
HuR is a regulator of mRNA turnover or translation of inflammatory genes through binding to adenylate-uridylate–rich elements and related motifs present in the 3′ untranslated region (UTR) of mRNAs. We postulate that HuR critically regulates the epithelial response by associating with multiple ARE-bearing, functionally related inflammatory transcripts. We aimed to identify HuR targets in the human airway epithelial cell line BEAS-2B challenged with TNF-α plus IFN-γ, a strong stimulus for inflammatory epithelial responses. Ribonucleoprotein complexes from resting and cytokine-treated cells were immunoprecipitated using anti-HuR and isotype-control Ab, and eluted mRNAs were reverse-transcribed and hybridized to an inflammatory-focused gene array. The chemokines CCL2, CCL8, CXCL1, and CXCL2 ranked highest among 27 signaling and inflammatory genes significantly enriched in the HuR RNP-IP from stimulated cells over the control immunoprecipitation. Among these, 20 displayed published HuR binding motifs. Association of HuR with the four endogenous chemokine mRNAs was validated by single-gene ribonucleoprotein-immunoprecipitation and shown to be 3′ UTR-dependent by biotin pull-down assay. Cytokine treatment increased mRNA stability only for CCL2 and CCL8, and transient silencing and overexpression of HuR affected only CCL2 and CCL8 expression in primary and transformed epithelial cells. Cytokine-induced CCL2 mRNA was predominantly cytoplasmic. Conversely, CXCL1 mRNA remained mostly nuclear and unaffected, as CXCL2, by changes in HuR levels. Increase in cytoplasmic HuR and HuR target expression partially relied on the inhibition of AMP-dependent kinase, a negative regulator of HuR nucleocytoplasmic shuttling. HuR-mediated regulation in airway epithelium appears broader than previously appreciated, coordinating numerous inflammatory genes through multiple posttranscriptional mechanisms.
Introduction
Posttranscriptional gene regulation is a powerful adaptive mechanism implemented in eukaryotic cells that coordinates the processes of mRNA splicing, transport, turnover, and translation. In response to cell perturbations such as stress, proliferation, or immune activation, these cytoplasmic events act in coordination with transcriptional regulation through shared kinase pathways, and they are critical determinants of the magnitude and timing of the protein output for early-response and inflammatory genes (1–3). Genome-wide studies indicate that ∼50% of stress-induced genes are chiefly regulated posttranscriptionally, indicating the impact of this regulatory mode on gene expression (4). Posttranscriptional gene regulation relies on the dynamic formation of messenger ribonucleoprotein (mRNP) complexes, in which RNA-binding proteins associate, in the context of specific RNA secondary structures, to regulatory sequences mostly present in the untranslated regions (UTRs) of the transcripts, recently defined as untranslated sequence elements for regulations (USER) (2, 5). The adenylate-uridylate–rich elements (ARE) present within the transcript 3′ UTR represent the most conserved and well-studied group of USER shown to regulate early-response genes, and they are now considered central cis-elements of immune gene regulation, given the high number of immune genes regulated through these motifs (6, 7). In addition, the expression of chemokine transcripts, which in airway epithelium is highly selective in response to different inflammatory stimuli, displays a strong posttranscriptional regulatory component (8–10).
HuR is the sole ubiquitous member of the Hu RNA-binding protein family (11). It binds to a heterogeneous group of cis-elements, including AREs, in mRNAs encoding key regulators of proliferation and stress responsiveness, as well as transcription factors (12–14). Functionally, HuR has been found to act in vitro as a positive regulator of mRNA stability and translation (15), although findings in a transgenic mouse macrophage model show that its activity in vivo can lead to inhibition of inflammatory gene expression in this cell type (16). Among the asthma-related cytokine genes clustered on chromosome 5q, IL-3, GM-CSF, IL-4, and IL-13 have all been found to be associated with HuR (17–20), which regulates also multiple genes linked to asthma pathophysiology, such as TNF-α, IL-3, IL-6, IL-8, GM-CSF, cyclooxygenase 2, VEGF, TGF-β, inducible nitric oxide synthase, CD154 (the CD40L), and the β-adrenergic receptor (17, 18, 21–31). Our previous work revealed that treatment of human primary airway epithelial cells with TNF-α and IL-4 triggers the stabilization of the CC chemokine CCL11/eotaxin mRNA in a 3′ UTR-dependent fashion, an increase in cytoplasmic levels of HuR and the association of HuR to the endogenous CCL11 mRNA (8). These studies provide compelling support for a role of HuR as an important intracellular mediator of inflammation.
On the basis of genome-wide studies probing the relationship between RNA-binding proteins and the functional profile of their associated transcripts, it has been established that multiple, functionally related mRNAs that bear a common USER, such as the ARE, can be regulated by one or more cognate RNA-binding proteins, thus creating subsets of transcripts whose fate is determined posttranscriptionally in a coordinate, operon-like fashion (5, 32). Studies on the role of HuR in coordinating posttranscriptional regulation of angiogenic and growth-related factors in cancer provide support for such a ribonomic paradigm in human disease (23, 33). Immune responses and inflammation are also characterized by proliferative responses and by remodeling processes that include angiogenesis (34), making such a paradigm applicable to immune-mediated processes as well. Interestingly, HuR is encoded in chromosome 19 (19p13.2) (35), which has been associated with candidate genes in asthma and other atopic disorders (36, 37).
The profiles of transcripts associated with several RNA-binding proteins such as HuR, TIA, tristetraprolin (TTP), and KSRP have been characterized in diverse cellular models (14, 38–40) using a ribonomic approach, in which protein-bound transcripts are isolated by immunoprecipitation of mRNPs using Abs specific for an RNA-binding protein, and the associated mRNA is then identified by gene arrays (41). With the aim of testing the role of HuR as an intracellular amplifier of epithelial responses in the context of airway inflammation, we set to investigate the spectrum of epithelial-derived genes that associate with HuR upon acytokine challenge, that with TNF-α and IFN-γ, that polarizes epithelial gene expression toward a strong upregulation of a subset of chemokines and cytokines (42, 43). Upon hybridization of HuR-associated mRNAs obtained by immunoprecipitation of mRNPs to a focused inflammatory gene array, we identified as HuR targets, in the airway epithelial cell line BEAS-2B, a transcript pool containing a considerable cluster of chemokines and of signaling molecules. The chemokines CCL2, CCL8, CCL13, CXCL1, and CXCL2 displayed putative HuR binding sites in their 3′ UTR and showed 3′ UTR-dependent association with HuR by biotin pull-down. However, association with HuR had a different effect on the mRNA expression of these targets. Among the genes examined, only the transcripts that displayed an increase in mRNA turnover following cytokine treatment—CCL2 and CCL8—selectively showed changes in mRNA levels that were dependent on HuR levels, modified exogenously by transient overexpression and silencing.
HuR translocation from the nucleus, where it predominantly resides, to the cytoplasm is critical to its functions (44, 45), because it correlates with the increased stability of several target mRNAs (8, 46, 47). We previously established that cytokine treatment of airway epithelial cells increases cytoplasmic HuR, in parallel with stabilization of the HuR target CCL11 (8). Stress-induced HuR translocation in the cytoplasm is inhibited by the AMP-activated protein kinase (AMPK), an ubiquitous enzyme that functions as a cellular sensor of metabolic stress (48) and mediates HuR nuclear import through phosphorylation of the adaptor protein importin α (49, 50). By promoting HuR nuclear localization, the AMPK-mediated pathway impairs its cytoplasmic effects on mRNA stability and translation. Conversely, inhibition of AMPK causes a marked increase in the cytoplasmic levels of HuR, which correlates with increased HuR function (47). It is therefore conceivable that the effect of cytokines on HuR localization involves modulation of AMPK function. Our data support this hypothesis, because cytokine treatment blocked the activation of AMPK induced by the synthetic agonist 5-amino-imidazole-4-carboxamide riboside (AICAR) and conversely, activation of AMPK by AICAR inhibited cytokine-driven nucleocytoplasmic shuttling of HuR as well as expression of its chemokine targets.
Our data show that cytokine-induced activation of the airway epithelium induces the association of HuR with a subset of signaling and effector molecules, and this event contributes to the increased expression levels of validated chemokines targets. The AMPK signaling pathway, by regulating HuR function, appears to participate to the inflammatory response in the airway epithelium.
Materials and Methods
Cell culture, plasmid constructs, siRNA, and transfection protocols
The BEAS-2B cell line, derived from human tracheal epithelium transformed by an adenovirus 12-SV40 hybrid virus (51) (ATCC, Manassas, VA) was maintained in F12/DMEM (Life Technologies/Invitrogen, Frederick, MD) containing 5% heat-inactivated FCS, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 mg/ml). This medium is referred to as complete medium. Primary bronchial epithelial cells (PBECs) were isolated by pronase digestion from bronchi of cadaveric lungs as described previously (52). The purity of these preparations was confirmed by immunohistochemical staining for cytokeratin (data not shown). PBECs were cultured on collagen-coated flasks and maintained in serum-free LHC-9 medium (Biofluids, Rockville, MD). Cultures of PBECs were used during the first passage only, whereas BEAS-2B cells were used from passages 36 to 45. All cells were cultured at 37°C in humidified air containing 5% CO2.
HuR was overexpressed in PBECs and BEAS-2B cells by infection with an adenoviral vector (AdCMV-HuR) in parallel with the empty vector (Ad-EV; ViraQuest, North Liberty, IA). Cells were cultured in six-well plates in serum-free DMEM, and infections were performed at 80% cell confluency. Titration experiments showed that significant upregulation of HuR levels was achieved with 6.25 PFUs per cell. The same protocol was used for transduction of Ad (CA)AMPK, expressing a cDNA encoding residues 1 to 312 of the α1 subunit of AMPK, which encodes for a constitutively active form of the enzyme (53). For HuR silencing, PBECs and BEAS-2B cells were transiently transfected with a previously described siRNA (5′ -aaGAGUGAAGGAGUUGAAACU-3′) or a scrambled control (5′ -GCCAAUUCAUCAGCAAUGG-3′; Qiagen, Valencia, CA) (54). Cells were seeded in six-well plates at a density of ∼250,000 cells per well and transfected 24 h later, when cells reached ∼50% confluence, using the non-liposomal cationic vehicle FuGene (Roche Applied Science, Indianapolis, IN) according to the manufacturer's specifications. Briefly, 3 μl of FuGene per sample were resuspended (5 min at room temperature) in serum-free Opti-MEM (Life Technologies, Carlsbad, CA) and allowed to complex with 1 μg plasmid DNA for 15 min at room temperature. The plasmid-FuGene mixture was then overlayed on the cells in a final volume of 2 ml of complete medium. After incubation for 48 h at 37°C, cells were treated according to the experimental protocols. The efficiency of the transient transfection of cells, assessed with a CMV promoter-driven expression vector coding for green fluorescence protein (Promega, Madison, WI), was ∼22% of total cell count performed by nuclear staining with Hoechst dye. Changes in HuR levels were determined by Western blot 48 h after transfection.
Immunoprecipitation of endogenous messenger ribonucleoprotein complexes and Western blot analysis
BEAS-2B cell monolayers cultured in T-175 flasks were stimulated at ∼70% confluence in the absence or presence of TNF-α (50 ng/ml) in combination with IFN-γ (50 ng/ml; R&D Systems, Minneapolis, MN). Cytokine stimulation did not affect cell viability, measured by trypan blue exclusion. Cells were then harvested by trypsinization, and their viability was assessed by trypan blue exclusion (consistently >95%). An equal number of cells per condition (30 × 106) was pelleted and resuspended in approximately two cell-pellet volumes of polysome lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES, pH 7.0, 0.5% Nonidet P-40 with 1 mM DTT, 100 U/ml RNaseOUT (Life Technologies), 0.2% vanadyl-ribonucleoside complex (Life Technologies), 0.2 mM PMSF, 1 mg/ml pepstatin A, 5 mg/ml bestatin, and 20 mg/ml leupeptin added fresh immediately before use. Cell lysates were then frozen at −80°C for storage. At time of use, cell lysates were thawed and centrifuged at 16,000 × g for 10 min at 4°C. The cell lysate contained ∼30 mg/ml total protein and was immunoprecipitated with anti-HuR or the irrelevant Ab, according to adaptation of an established protocol (8). Protein-A sepharose beads (Sigma-Aldrich, St. Louis, MO) were swollen 1:1 v/v in NT2 buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM MgCl2/0.05% Nonidet P40) supplemented with 5% BSA. A 100-μl aliquot of the preswollen protein-A bead slurry was used for immunoprecipitation (IP) reaction and incubated for 4 h at room temperature with excess immunoprecipitating Ab (30 μg), using either a mouse mAb specific for HuR, 3A2 (Santa Cruz Biotechnology, Santa Cruz, CA) (55) or an IgG1 isotype control Ab (B&D Life Sciences, Franklin Lanes, NJ). The Ab-coated beads were washed with ice-cold NT2 buffer and resuspended in 900 μl NT2 buffer supplemented with 100 U/ml RNaseOUT, 0.2% vanadyl-ribonucleoside complex, 1 mM DTT, and 20 mM EDTA. The beads were vortexed briefly, and 100 μl of the mRNP cell lysate was added and immediately centrifuged. A 100-μl aliquot was removed to represent total cellular RNA. The IP reactions were tumbled at room temperature for 2 h and then washed six times with ice-cold NT2 buffer. Washed beads were resuspended in 100 μl NT2 buffer supplemented with 0.1% SDS and 30 μg proteinase K, incubated for 30 min at 55°C, and cytoplasmic RNA was extracted using phenol-chlorophorm-isoamylalcohol and precipitation in ethanol.
Western blot was performed as described (8), using as primary Abs 2 μg/ml mouse monoclonal anti-HuR 3A2 (55) or 0.3 μg/ml anti–β-tubulin (both from Santa Cruz) or a 1:1000 dilution of the AMPK or pAMPK Abs (Cell Signaling, Danvers, MA) in 1× PBS containing 1% Tween for 1 h at room temperature with continuous shaking. After washes with 1× PBS containing 0.1% Tween (10 min, room temperature), membranes were incubated for 1 h at room temperature with an HRP-conjugated goat anti-mouse secondary Ab. After a final washing step (1× PBS containing 0.1% Tween, 10 min, room temperature), immunoreactive bands were visualized by ECL (Amersham Biosciences, Piscataway, NJ).
Inflammatory gene arrays
The IP RNA was reverse-transcribed according to an established labeling protocol to generate 33P-labeled probes, which were then hybridized following the manufacturer's instructions against a GEArray S series Human Autoimmune and Inflammatory Response Gene Array (SuperArray Bioscience, Frederick, MD), which contains full-length cDNAs for 364 known human genes. The results were collected by exposure to a phosphoimager screen and then scanning by a Typhoon 8600 Variable Mode Image (GE Healthcare). Analysis was performed with the P-SCAN analysis software using a custom array option available (56).
Arrays were normalized within each treatment or control set using a subset of genes that were reproducibly precipitated by IgG control only. Fold changes were independently calculated between HuR Ab and IgG matched replicates within each set and tested for significance using the paired t test. Genome ontology analysis was performed using the DAVID gene functional classification tool (National Insitute of Allergy and Infectious Diseases, National Institutes of Health 2008). Results are accessible at http://www.ncbi.nlm.nih.gov/geo/info/linking.html, NCBI GEO accession number GSE25264.
RNA isolation and analysis
Total RNA was extracted using TRIzol (Invitrogen/Life Technologies) (57). Cytoplasmic RNA was isolated and DNase-treated using the RNAeasy kit (Qiagen). Nuclear and cytoplasmic RNA were separated and isolated as described (58). RNA was reverse transcribed using the Gene Amp Kit (Perkin-Elmer, Waltham, MA) and PCR amplified using the SYBR Green Reagent Kit (Perkin-Elmer). The PCR primers (forward/reverse) for detection of CCL2, CCL8, CCL13, CXCL1, CXCL2, and GAPDH, used for normalization, are listed in Table I. The conditions for amplification by real-time PCR for all targets were as follows: 50°C for 2 min for UNG activation, 95°C for 10 min (1 cycle each, hot-start to activate the Taq polymerase), 95°C for 15 s, and 63°C for 1 min for 40 cycles. All samples were run in triplicate (SD < 0.1) for real-time fluorimetric determination in an ABI 7300 sequence detector (Applied Biosystems/LifeTechnologies) and quantified using the comparative cycle threshold (CT) method (8, 59).
Table I. Primers for real-time PCR assay (I) and for generation of biotinylated RNA (II).
| Human Gene Name | Accession No. | Forward/Reverse |
|---|---|---|
| CCL2 | NM_002982 | F-5′-CCTTCTGTGCCTGCTGCTCATAG-3′ |
| R-5′-TCTTCGGAGTTTGGGTTTGCTTGT-3′ | ||
| CCL8 | NM_005623 | F-5′-CCGAGGAGCAGAGAGGTTGAGAAC-3′ |
| R-5′-CTTGGGACATTGGATGTTGGTGATT-3′ | ||
| CCL13 | NM_005408 | F-5′-GTGCTTCTGTGCCTGCTGCTCAT-3′ |
| R-5′-ATTCTGGACCCACTTCTCCTTTGG-3′ | ||
| CXCL1 | NM_001511 | F-5′-GCAGCAGGAGCGTCCGTGGC-3′ |
| R-5′-CAGTTGGATTTGTCACTGTTCAGCAT-3′ | ||
| CXCL2 | NM_002089 | F-5′-TCACCTCAAGAACATCCAAAGTGTG-3′ |
| R-5′-CTTCAGGAACAGCCACCAATAAGC-3′ | ||
| CCL5 | NM_002985 | F-5′-CCATATTCCTCGACCAC-3′ |
| R-5′-GGGTGACAAAGACGACTGCT-3′ | ||
| GAPDH | NM_002046 | F-5′-TGCACCACCAACTGCTTAGC-3′ |
| R-5′-GGCATGGACTGTGGTCATGAG-3′ | ||
| CCL2-CR (coding region) | F-5′-GTGAATTGTAATACGACTCACTATAGGGAACCTTCTGTGCCTGCTGCTCATAG-3′ | |
| R-5′-TCTTCGGAGTTTGGGTTTGCTTGT-5′ | ||
| CCL2 3′UTR | F-5′-CCAAGCTTCTAATACGACTCACTATAGGGAGAACACTCACTCCACAACCC-3′ | |
| R-5′-TGTACAAAAATATATTTATTTGGTGTAATAGTTAC-3′ | ||
| CCL2 3′UTR-A (374-499) | F-5′-CCAAGCTTCTAATACGACTCACTATAGGGAGAACACTCACTCCACAACCC-3′ | |
| R-5′-ACTTAAGGCATAATGTTTCACATCAACAAAC-3′ | ||
| CCL2 3′UTR-B (500-624) | F-5′-CCAAGCTTCTAATACGACTCACTATAGGGAGAGCCTTAAGTAATGTTAATT-CTTATTTAAG-3′ | |
| R-5′-ACATCCCAGGGGTAGAACTGTG-3′ | ||
| CCL2 3′UTR-C (625-749) | F-5′-CCAAGCTTCTAATACGACTCACTATAGGGAGAGGGTCTTTGCAAGAATCA-3′ | |
| R-5′-TGTACAAAAATATATTTATTTGGTGTAATAGTTAC-3′ | ||
| CCL8-CR | F-5′-GTGAATTGTAATACGACTCACTATAGGGAAGGTTTCTGCAACGCTTCTGTG-3′ | |
| R-5′-TATTTGGTCCAGATGCTTCATGG-3′ | ||
| CCL8-3′UTR | F-5′-GTGAATTGTAATACGACTCACTATAGGGAGCCTTCATACATGGACTGAGAGT-3′ | |
| R-5′-CAACAGCAGTACAAAGCACACATTTG-3′ | ||
| CCL13-CR | F-5′-GTGAATTGTAATACGACTCACTATAGGGCGAGTGCTTCTGTGCCTGCTGCTCAT-3′ | |
| R-5′-ATTCTGGACCCACTTCTCCTTTGG-3′ | ||
| CCL13-3′UTR | F-5′-GTGAATTGTAATACGACTCACTATAGGGAGCTACCCGTACTGAAATCAAGC-3′ | |
| R-5′-GCAAAATGTCAGCAGTCCTACTATTG-3′ | ||
| CXCL1-CR | F-5′-GTGAATTGTAATACGACTCACTATAGGGCTCCTGCGAGTGGCACTGCTG-3′ | |
| R-5′-TCAGTTGGATTTGTCACTGTTCAG-3′ | ||
| CXCL1-3′UTR | F-5′-GTGAATTGTAATACGACTCACTATAGGGACCAGAAGGGAGGAGGAAGC-3′ | |
| R-5′-CAGAAACACTGTAAAACTACCATTAAAC-3′ | ||
| CXCL2-CR | F-5′-GTGAATTGTAATACGACTCACTATAGGGACGCTCTCCGCCGCCCCCAG-3′ | |
| R-5′-GTTGGATTTGCCATTTTTCAGCATCT-3′ | ||
| CXCL2-3′UTR | F-5′-GTGAATTGTAATACGACTCACTATAGGGACCAGAAGGAAGGAGGAAGC-3′ | |
| R-5′-CTGCTCTAACACAGAGGGAAACAC-3′ |
In vitro biotin pull-down assay
To generate the labeled RNA probes, PCR products encompassing the coding regions or 3′ UTR of the selected HuR targets were generated by RT-PCR of BEAS-2B RNA, using the primers listed in Table I and purified from agarose gels as described (12). The resulting cDNA was used as a template for synthesizing biotinylated RNA using T7 RNA polymerase and biotin-conjugated CTP (MaxiScript T7 In Vitro Transcription Kit, Ambion, Austin, TX). Biotinylated transcripts were amplified by RT-PCR using specific primers (Table I). All 5′ primers are preceded by a T7 tag, shown in Table I. Following an established protocol (14), cytoplasmic fractions of unstimulated BEAS-2B cells (40 μg) were incubated for 1 h at room temperature with 1 μg of biotinylated transcripts, and ribonucleoprotein complexes were isolated with streptavidin-conjugated Dynabeads (Invitrogen/Life Technologies). The presence of HuR in the pull-down pellet was verified by standard Western blot analysis using the mouse monoclonal anti-HuR Ab 3A2 (Santa Cruz Biotechnology) and the ECL protocol (GE Healthcare) as described (8).
Immunofluorescence
BEAS-2B cells and PBECs were seeded at 40–50% confluency on sterile coverslips and incubated in complete medium overnight. Following treatment, cells were fixed for 15 min in PBS containing 4% paraformaldehyde for a period of 15 min, followed by permeabilization in PBS containing 0.4% TritonX-100 for an additional 15 min. After incubation in blocking buffer (PBS containing 2% BSA and 0.1% Tween 20) for 16 h, coverslips were incubated in a 1:500 dilution of mouse anti-HuR Ab (Santa Cruz Biotechnology) in blocking buffer for 1 h. After washes in blocking buffer, samples were incubated with a mixture of horse anti-mouse Texas Red (1:200; Jackson Immunoresearch Laboratories, West Grove, PA) and Hoechst 33342 (1:5,000; Molecular Probes/Life Technologies) or DAPI nuclear staining (1 μg/ml, Roche Applied Science, Indianapolis, IN) for 1 h. After washes with blocking buffer, coverslips were mounted in Vectashield (Vector Laboratories, Burlingame, CA). A Zeiss Axiovert 200 M microscope (×63 lens) was used for visualization using separate channels for the analysis of phase-contrast images, red fluorescence, and blue fluorescence. Images were then processed with the Zeiss AxioVision program, version 3.0.
Statistical analysis
Data were analyzed using repeated measures ANOVA test with post hoc analysis or paired t test when appropriate.
Results
Identification of transcripts associated with HuR in BEAS-2B cells after cytokine challenge
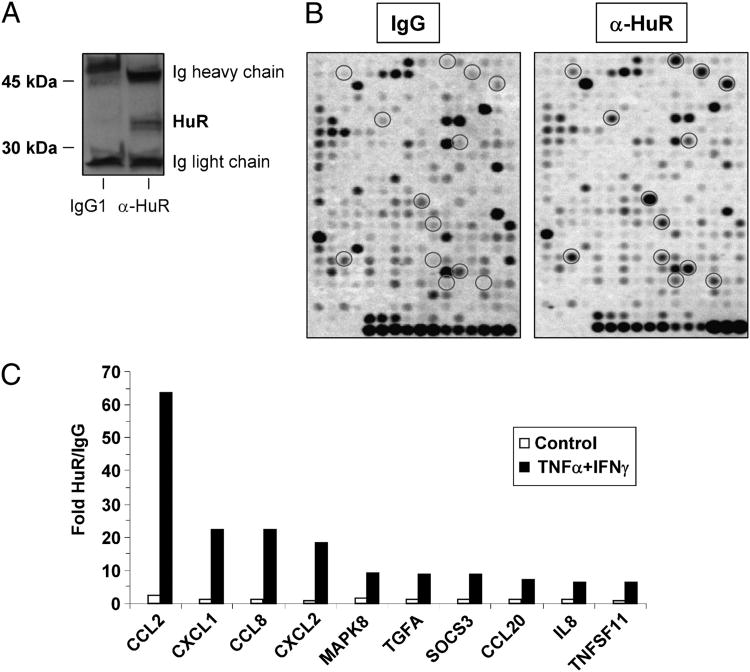
To identify the mRNAs associated with HuR, we used a ribonucleoprotein-IP protocol coupled to an array-based hybridization assay. The study was performed with the human tracheal epithelial cell line BEAS-2B, which closely recapitulates several features of the nontransformed PBEC, including HuR levels, cytoplasmic translocation upon cytokine treatment, and response to cytokine challenge (8). After stimulation of BEAS-2B cells for either 3 or 18 h with a combination of TNF-α with IFN-γ (50 ng/ml each stimulus; n = 3), cytoplasmic lysates were prepared using conditions that would retain the integrity of ribonucleoprotein complexes. The lysates were subjected to IP using the monoclonal anti-HuR Ab or an isotype-matched control. Labeled DNA probes, obtained by reverse transcription from the mRNA pools eluted from the IP, were hybridized to an array platform displaying 364 genes related to inflammation and immunity. Specificity of the IPs was confirmed by Western blot analysis of HuR in both IP lysates (Fig. 1A). Association with HuR was considered for genes in which the signal detected, for each condition, in HuR IPs was statistically significant over the background signal from IgG1 IPs (Fig. 1B, representative hybridization). Transcript association with HuR occurred in a time-dependent fashion, being detected almost exclusively in cells stimulated for 18 h. In this condition, we identified 27 transcripts that were significantly enriched in the transcript pool immunoprecipitated with anti-HuR over the mRNA levels found in the control Ab-immunoprecipitates (p < 0.05; Table II; Fig. 1C shows the ten highest ranking targets in HuR IPs). Several known HuR targets were detected in this pool, such as IL-8 and IL-6 (25), thus validating the array detection. An additional pool of 95 genes showed consistent, above-threshold enrichment that did not reach statistical significance because of variability within the experiments (Supplemental Table I). Only two genes, CCL2 and CXCL1, showed a small but significant enrichment after 3 h of cytokine treatment, whereas transcript enrichment in the HuR IP was neither consistent nor significant at any time in unstimulated cells (Supplemental Table I).
Figure 1.
Identification of inflammatory transcripts as targets of HuR. Cell lysates from resting and cytokinetreated BEAS-2B were subjected to IP assay with either anti-HuR Ab or IgG1 control. A, Western blot showing specificity of HuR IP. B, Representative hybridization profile from RNA pools obtained by anti-HuR (right panel) and IgG1 IPs using a focused array with 364 genes related to inflammation and immunity. Targets marked by circles indicate genes for which fold increase of the signal detected, for each condition, in HuR IPs over the background signal from IgG1 IPs was statistically significant (see Table II and Supplemental Table I). C, Fold enrichment for the ten highest ranking targets in HuR IPs in the indicated conditions (p < 0.05; see Table II).
Table II. Gene array-based analysis of HuR-associated transcripts following ribonucleoprotein-IP in BEAS-2B cells.
| Mean Enrichment (HuR/IgG) | |||||
|---|---|---|---|---|---|
| Symbol | Gene Name | Description | GenBank Accession No. | Control | TNF-α + IFN-γ |
| CCL2 | MCP1/SCYA2 | Chemokine (C-C motif) ligand 2 | NM_002982 | 2.20 | 63.73 |
| CXCL1 | GROα/MGSA | Chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, α) | NM_001511 | 1.24 | 22.47 |
| CCL8 | MCP-2 | Chemokine (C-C motif) ligand 8 | NM_005623 | 1.32 | 22.21 |
| CXCL2 | MIP-2α/GROβ | Chemokine (C-X-C motif) ligand 2 | NM_002089 | 0.91 | 18.12 |
| MAPK8 | JNK1 | MAPK 8 | NM_002750 | 1.68 | 9.14 |
| TGFA | TGF-α | TGF, α | NM_003236 | 1.07 | 8.86 |
| SOCS3 | SSI-3 | Suppressor of cytokine signaling 3 | NM_003955 | 1.01 | 8.70 |
| CCL20 | MIP-3α/SCYA20 | Chemokine (C-C motif) ligand 20 | NM_004591 | 1.11 | 7.03 |
| IL8 | IL-8 | IL 8 | NM_000584 | 1.19 | 6.52 |
| TNFSF11 | TRANCE | TNF (ligand) superfamily, member 11 | NM_003701 | 0.83 | 6.32 |
| TRPV6 | ECAC2/Cat1 | Transient receptor potential cation channel, subfamily V, member 6 | NM_018646 | 0.82 | 5.50 |
| VAV1 | Vav1 | Vav 1 oncogene | NM_005428 | 0.78 | 5.28 |
| TNFRSF1B | TNFR2/p75 | TNF receptor superfamily, member 1B | NM_001066 | 1.16 | 5.26 |
| TYK2 | Tyk2 | Tyrosine kinase 2 | NM_003331 | 0.92 | 5.22 |
| CSNK2A1 | CK-II α | Casein kinase 2, α 1 polypeptide | NM_001895 | 1.26 | 5.14 |
| ITGB7 | Integrin b7 | Integrin, β 7 | NM_000889 | 1.00 | 4.97 |
| TNFRSF7 | CD27 | TNF receptor superfamily, member 7 | NM_001242 | 0.92 | 4.78 |
| TGFBR1 | ALK-5 | TGF, β receptor I (activin A receptor type II-like kinase, 53kDa) | NM_004612 | 1.20 | 4.58 |
| PPP3R1 | CALNB1 | Protein phosphatase 3, regulatory subunit B, 19kDa, α isoform (calcineurin B, type I) | NM_000945 | 1.46 | 4.50 |
| TNFRSF21 | TNFRSF21 | TNF receptor superfamily, member 21 | NM_014452 | 0.99 | 4.48 |
| RNF110 | ZNF144 | Ring finger protein 110 | NM_007144 | 0.77 | 4.30 |
| IL6 | IL-6 | IL 6 (IFN, β 2) | NM_000600 | 1.02 | 4.17 |
| MAP3K2 | MEKK2 | MAPK kinase kinase 2 | NM_006609 | 0.98 | 4.05 |
| TGFBR2 | TGFbR2 | TGF, β receptor II (70/80kDa) | NM_003242 | 1.05 | 4.00 |
| TRAF1 | TRAF1 | TNF receptor-associated factor 1 | NM_005658 | 0.91 | 3.87 |
| CSNK2B | CK-II β | Casein kinase 2, β polypeptide | NM_001320 | 0.93 | 3.53 |
| SMAD2 | MADH2 | SMAD, mothers against DPP homolog 2 (Drosophila) | NM_005901 | 1.03 | 2.45 |
Paired T ns All p < 0.05
The newly identified HuR targets are coding for important signal transduction proteins, transcription factors, cytokine and chemokine receptors, as well as for several chemokines (Supplemental Table II). A cluster of chemokines ranks the highest in the target list and includes CC chemokine members of the MCP subfamily, such as CCL2/MCP-1, CCL8/MCP-2, CCL20, and members of the GRO (melanoma growth-stimulating activity) subfamily of CXC chemokines, CXCL1/GRO-α and CXCL2/GRO-β. Additional chemokines were detected in the HuR IP, although at lower statistical power (p = 0.05–0.1), such as CCL13/MCP-4, CCL7/MCP-3, CXCL3/GRO-γ as well as other known HuR targets such as IL-8, IL-6, Jun D, VEGF, and TNF-α (Table II; Supplemental Table I). We then investigated the presence of putative HuR consensus sites, previously identified computationally by global analysis of HuR-associated transcripts (14), in the newly identified HuR targets. Genes represented on the Superarray and those listed in the published HuR motif database (14) were mapped to identical GenBank accession numbers, and 125 of the 364 genes (34%) represented on the array were present in the HuR motif database (Supplemental Table III). Among the 27 genes that had a statistically significant enrichment over the IgG IP (p < 0.05), 20 displayed HuR motifs (p = 1.4 × 10−5, Fisher's exact test). Moreover, 64 of the 95 genes with p = 0.05–0.1 also contained HuR sites (p = 1 × 10−14, Fisher's exact test; Supplemental Table IV). Thus we were able to identify the large majority of HuR-motif bearing genes in our IP experiments.
HuR association with chemokine transcripts involves discrete regions of the 3′ UTR
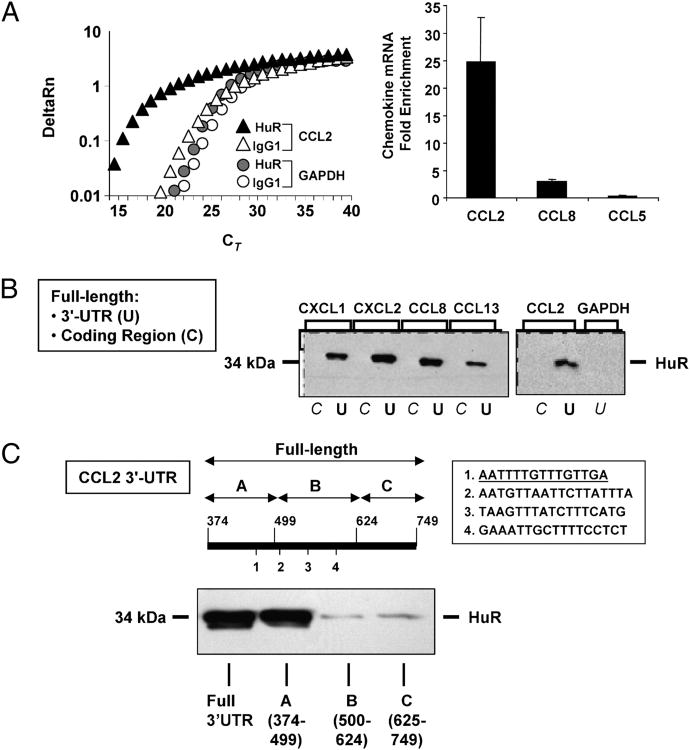
We focused further study on the highest-enriched chemokine cluster in light of their relevance in the regulation of inflammatory cell trafficking, a key epithelial-driven process in airway inflammation. We validated, by single-gene ribonucleoprotein-IP, the presence of selected chemokine transcripts in the IP mRNA pools obtained in BEAS-2B cells stimulated with or without TNF-α plus IFN-γ. For CCL2 mRNA, we confirmed a consistent enrichment in the samples obtained by IP with anti-HuR over the background mRNA levels in the control Ab-IP (n = 3). The mean difference of 4.5 ± 0.5 CT between the HuR and the mock IP after normalization to GAPDH indicated a 2(−4.5) = 22.1-fold enrichment of the CCL2 mRNA in the HuR IP (Fig. 2A). Cytokine-induced CCL8 mRNA was enriched in the HuR IP as well (n = 2; mean fold enrichment, 3.0). As negative control, GAPDH (Fig. 2A) and CCL5 (not shown), which were not significantly associated to HuR according to the array analysis, showed no difference between the two IP conditions, indicating a lack of specific enrichment.
Figure 2.
Association of HuR with chemokines mRNA through the 3′ UTR. A, Representative real-time PCR amplification plot of fluorescence intensity over background (Δ Rn) against PCR cycle (cycle threshold, CT) of CCL2 and GAPDH mRNA in BEAS-2B cells stimulated with TNF-α plus IFN-γ and subjected to IP with the anti-HuR Ab (filled symbols) or with the isotype-matched Ab (open symbols). The bar graph on the right indicates the mean enrichment in the IP with anti-HuR over the control Ab for the listed genes (n = 3 for CCL2; n = 2 for CCL8 and CCL5). B, Western blot analysis showing HuR detection following biotin pull-down of BEAS-2B cell lysates with the biotinylated transcripts spanning the either the coding regions (C) or the 3′ UTR (U) of the indicated chemokines and GAPDH (representative of n = 3). C, Biotin pull-down assay using either the full length 3′ UTR (nucleotides 374–749 of the CCL2 mRNA) or segments of the CCL2 3′ UTR (A–C) containing different putative HuR sites (indicated as 1 to 4 in the figure, sequences listed in the box). Underlined is sequence 1, which is included in the biotinylated probe A that retained HuR binding.
Although HuR is known as an ARE-binding protein, its association with the IL-4 mRNA has been reported to occur within the coding region despite the presence of ARE in this transcript (19). We therefore aimed at defining the site of interaction of HuR with the target chemokine mRNAs by biotin pull-down (Fig. 2B). Cytoplasmic protein extracts from BEAS-2B cells were incubated with in vitro-transcribed, biotinylated transcripts spanning the coding region or the full-length 3′ UTR of CXCL1, CXCL2, CCL8, and CCL2, or the full-length 3′ UTR of GAPDH as a negative control. We included CCL13 in the analysis, because it showed a robust enrichment and a trend toward statistical significance (20.7-fold over IgG; p = 0.09; Supplemental Table I). The presence of HuR in the pulled-down pellets was then detected by Western blot. HuR was undetectable in the IP samples pulled down with the probe spanning the chemokines' coding regions, as well as with the GAPDH probe. In contrast, HuR was strongly detected in association with the labeled transcripts spanning the chemokine 3′ UTR regions. These transcripts display multiple putative HuR consensus sites in their 3′ UTRs (14) (Table III). We selected the CCL2 3′ UTR to better define the role of these discrete regions in mediating HuR binding (Fig. 2C). Biotin pull-down assay was performed using biotinylated transcripts spanning the CCL2 3′ UTR in three segments: probe A, including the first 125 bp, which displayed the first binding site; probe B, which included three closely spaced sites; and probe C, which contained none. Detection of HuR after pull-down with probe A was the only one comparable to that obtained with the probe spanning the full 3′ UTR. The first region of the CCL2 3′ UTR, encompassed by probe A, contains an ARE consisting of a hexamer with flanking U, which is known to bind TTP (60), which may be the functional region to displacement by HuR (61). This sequence is: 5′ -CCCTGTTTTATTTTATTATAATGAATTTTGTTTGTTGATGTGAAACATTATG-3′.
Table III. Putative HuR binding sites in chemokine transcripts associated with HuR.
| Gene | Position | Sequence |
|---|---|---|
| CCL2 | 457 | 5′-AATTTTGTTTGTTGAT-3′ |
| 493 | 5′-AATGTTAATTCTTATTT-3′ | |
| 526 | 5′-TAAGTTTATCTTTCATG-3′ | |
| 575 | 5′-GAAATTGCTTTTCCTCT-3′ | |
| CCL8 | 831 | 5′-GTGACATTATTTTATTAT-3′ |
| 869 | 5′-TTTAAATAATTTAAA-3′ | |
| 1109 | 5′-TTGGTATTCTTTGGCAA-3′ | |
| CCL13 | 524 | 5′-GTAATATTGGCTATTAT-3′ |
| 654 | 5′-TGGCAGTGGGTTTGTAT-3′ | |
| CXCL1 | 695 | 5′-TTCATATTTAATTTGAA-3′ |
| 849 | 5′-TGCACATCTGTTTTGTA-3′ | |
| 887 | 5′-TTGTTATTTATTGAAAT-3′ | |
| 974 | 5′-GGACATTTTATGTCTTT-3′ | |
| CXCL2 | 589–605 | 5′-TGCGCCTAATGTGTTTG-3′ |
| 693–709 | 5′-AAATAAGGTTATGATTG-3′ | |
| 885–901 | 5′-AGGAATCCAAGAAAATG-3′ | |
| 1008–1024 | 5′-AACATTTCTCATGTTG-3′ |
HuR targets display different mRNA turnover rates in response to cytokines
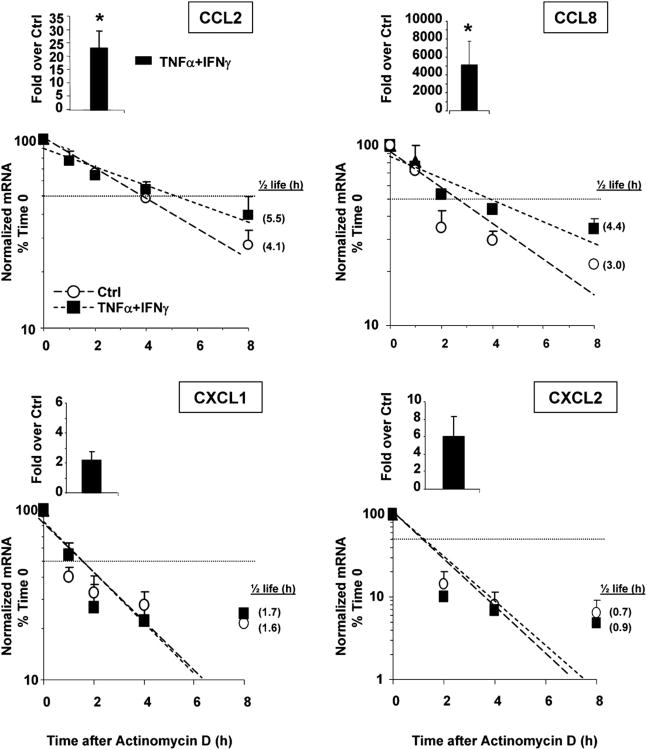
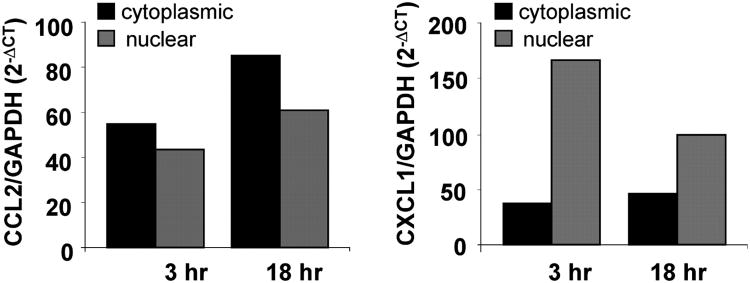
We next assessed whether changes in mRNA stability would occur under conditions in which association of HuR with chemokine mRNAs was detected. BEAS-2B cells were left untreated or stimulated with or without TNF-α plus IFN-γ (50 ng/ml each) for 18 h. Cells were then either harvested (at time 0) or cultured in the presence of the transcriptional inhibitor actinomycin D (3 μg/ml) for an additional 1, 3, or 5 h. Total RNA was isolated to assess chemokine levels by real-time RT-PCR. As expected, steady-state mRNA levels of all chemokines, calculated as fold induction over unstimulated cells at time 0, increased after challenge, although with different amplitudes. The chemokines of the MCP family (CCL2 and CCL8) showed a stronger upregulation than the members of the GRO family (CXCL1 and CXCL2), with the strongest response being elicited for CCL8 (Fig. 3, bargraphs). Treatment with actinomycin D (Fig. 3, line plots) revealed that challenge with the cytokine combination for 18 h induced a small but consistent increase in CCL2 and CCL8 mRNA stability, as demonstrated by an increase in the mRNA half life, which represents the time (expressed in hours) at which mRNA expression is 50% of the initial level [Fig. 3; t1/2 = Ln(0.5)/slope]. For CCL2, t1/2 increased from baseline by 63% in TNF-α plus IFN-γ–stimulated cells. For CCL8, cytokine treatment increased mRNA half life by 49%. In unstimulated cells, baseline levels of CCL8 were very low, and decay curves were obtainable in only two experiments. In contrast, CXCL1 and CXCL2 mRNA decay was faster than that of the CC chemokines, but its rate was unaffected by the cytokine challenge. Given the different effect of cytokines on the stability of HuR targets, we investigated potential differences between nuclear and cytoplasmic mRNA levels using CCL2 and CXCL1 as models, as HuR can be specifically acting on mRNA transport (58). We found that after cytokine stimulation for 3 and 18 h, CXCL1 mRNA is mostly represented in the nuclear fraction, whereas a higher percentage CCL2 mRNA resides in the cytoplasm (Fig. 4).
Figure 3.
Stimulus-dependent increase in mRNA stability in a subset of HuR targets. Chemokine mRNA expression in BEAS-2B cells harvested 18 h after the indicated treatment (50 ng/ml each stimulus, time 0) and following actinomycin D treatment (3 μg/ml) for the indicated times (n = 3–4). mRNA was detected by real-time RT-PCR. Results, normalized to GAPDH mRNA levels, are expressed as fold over unstimulated cells (ctrl) (as 2–ΔΔCt; see Materials and Methods). Levels shown in bar graphs represent the mean ± SEM of results at time 0, showing the induced steady-state mRNA levels. In the line plots, mRNA levels are expressed as mean ± SEM of the percent of maximum (mRNA at time 0). Shown in parenthesis, adjacent to each symbol, is the mRNA half-life, calculated for each condition as the time (in hours) required for the transcript to decrease to 50% (dotted horizontal line) of its initial abundance [t1/2 = Ln(0.5)/slope]. *p < 0.05 versus unstimulated cells.
Figure 4.
Differential distribution of CCL2 and CXCL1 mRNA in nuclear and cytoplasmic compartments in cytokine-treated BEAS-2B cells. Levels of nuclear and cytoplasmic CCL2 and CXCL1 mRNA detected by real-time PCR in BEAS-2B cells treated with TNF-α plus IFN-γ (50 ng/ml each) for 3 and 18 h (n = 2).
Changes in HuR levels affects CCL2 and CCL8 mRNA expression
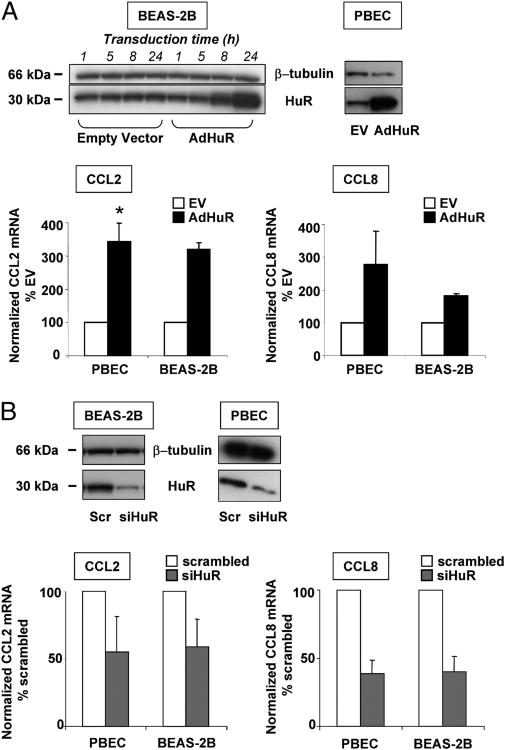
To assess the functional role for HuR association in chemokine expression, we transiently transfected epithelial cells with either an adenoviral HuR expression vector or an HuR-specific siRNA, in parallel with the appropriate controls, to achieve overexpression and silencing of HuR, respectively. In both series of experiments, consistent changes in HuR levels were achieved in PBECs and BEAS-2B cells, as verified by Western blot (Fig. 5A, 5B). Transfected cells were stimulated with or without TNF-α plus IFN-γ and harvested for chemokine mRNA analysis by real-time PCR. Expression of GAPDH mRNA, used to normalize the data, was not altered by changes in HuR levels (data not shown). In both cell types, cytokine-induced CCL2 mRNA increased >3-fold in cells overexpressing HuR (percentage of mock-transfected cells: PBECs, 342.7 ± 55.3, p < 0.05; BEAS-2B, 319 ± 26). Levels of CCL8 mRNA were also consistently increased by HuR upregulation, although with greater variability between experiments in PBECs (percentage of mock-transfected cells: PBECs, 280 ± 101.3; BEAS-2B, 182 ± 5; n = 4 and 3, respectively, for both genes; Fig. 5A).
Figure 5.
Changes in HuR levels affect expression of CCL2 and CCL8 mRNA. A, Upper left panel: Western blot analysis of HuR or β-tubulin levels (lower and upper blot, respectively) showing a time-dependent increase in HuR levels following transduction of BEAS-2B with 6.25 PFUs per cell of an adenoviral HuR expression vector (AdHuR) or the control empty vector (EV). Similarly increased levels of HuR are shown in PBECs after 24 h of transduction. Representative of three independent transductions for BEAS-2B, four for PBECs. Bar graphs below: mean ± SEM of CCL2 and CCL8 mRNA levels induced by TNF-α plus IFN-γ (50 ng/ml each) in the indicated transduced cell types and detected by real-time PCR. B, Upper panel: Western blot showing HuR and β-tubulin levels in BEAS-2B and PBECs after transient transfection with an siRNA for HuR (siHuR) or with a scrambled (Scr) siRNA as control. Bar graphs below: CCL2 and CCL8 mRNA levels detected by real-time PCR in transfected PBECs (n = 3 independent transfections) and BEAS-2B cells (n = 3–4) treated as described for the experiments in A.
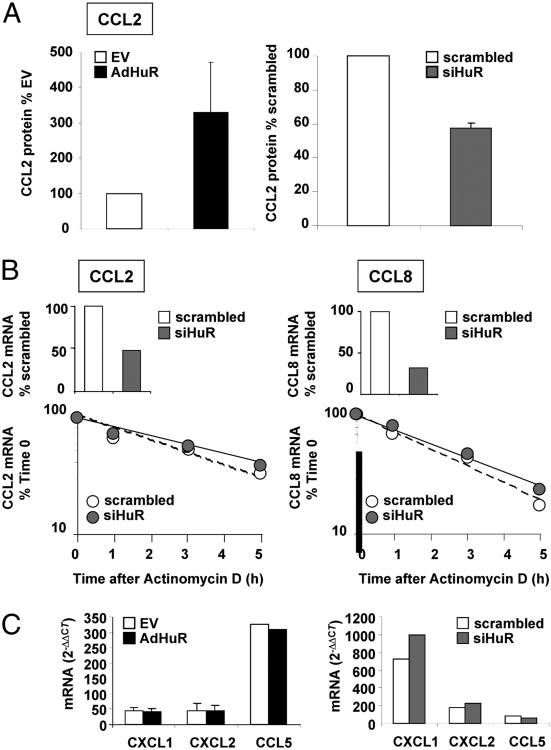
In cells in which HuR expression was silenced (Fig. 5B), levels of CCL2 mRNA were also consistently decreased by an average of 40–45%, compared with the levels achieved after cytokine challenge in cells transfected with the control scrambled siRNA (percentage of scrambled-transfected cells: PBECs, 55 ± 26.1; BEAS-2B, 59.2 ± 20.2). For CCL8, the decrease in mRNA expression was less variable among experiments and, as for CCL2, almost identical in the two cell types (percentage of scrambled-transfected cells: PBECs, 38.6 ± 10.3; BEAS-2B, 40.1 ± 11.2). Changes in HuR levels also affected the increase in cytokine-induced CCL2 protein, measured in BEAS-2B supernatants by ELISA, in a degree similar to mRNA levels (Fig. 6A). HuR overexpression induced a 3.3-fold increase in protein levels (n = 4; percentage of mock-transfected cells, 330.2 ± 141), and HuR silencing lowered CCL2 levels by 42% (n = 3; percentage of scrambled-transfected cells, 57.4 ± 2.7). The effect of HuR silencing was detectable only at an earlier time point (8 h after challenge), because at 24 h CCL2 protein levels were maximal in the scrambled-treated cells. However, CCL2 and CCL8 mRNA turnover, assessed using actinomycin D chase was not affected in cytokine-treated BEAS-2B cells after HuR silencing (Fig. 6B). Moreover, similar to results in untransfected cells (Fig. 3) changes in HuR levels did not affect the mRNA levels of CXCL1 and CXCL2, similarly to those of CCL5, which is not an HuR target (Fig. 6C.)
Figure 6.
Differential effect of changes in HuR levels on expression and decay of HuR-associated chemokines. A, CCL2 protein levels induced by TNF-α plus IFN-γ (50 ng/ml each) in BEAS-2B cells with HuR overexpressed or silenced, respectively, as described in Fig. 5 and expressed as percent over mock or scrambled-transfected cells. HuR levels were detected by a specific ELISA. B, Cytokine-induced CCL2 and CCL8 steady-state mRNA levels (bar graphs) and mRNA decay after actinomycin D (line graphs) assessed by real-time PCR in the same group of experiments (n = 2) after HuR silencing, showing a lack of change in mRNA turnover rate between cells treated with scrambled or HuR siRNA, despite a sensible change in total mRNA levels. C, Cytokine-induced CXCL1, CXCL2, and CCL5 steady-state mRNA levels assessed by real-time PCR (as fold over unstimulated cells) in BEAS-2B with HuR overexpressed or silenced, respectively, as described in Fig. 5.
Cytokine treatment inhibits activation of AMPK, a negative regulator of HuR nucleocytoplasmic shuttling
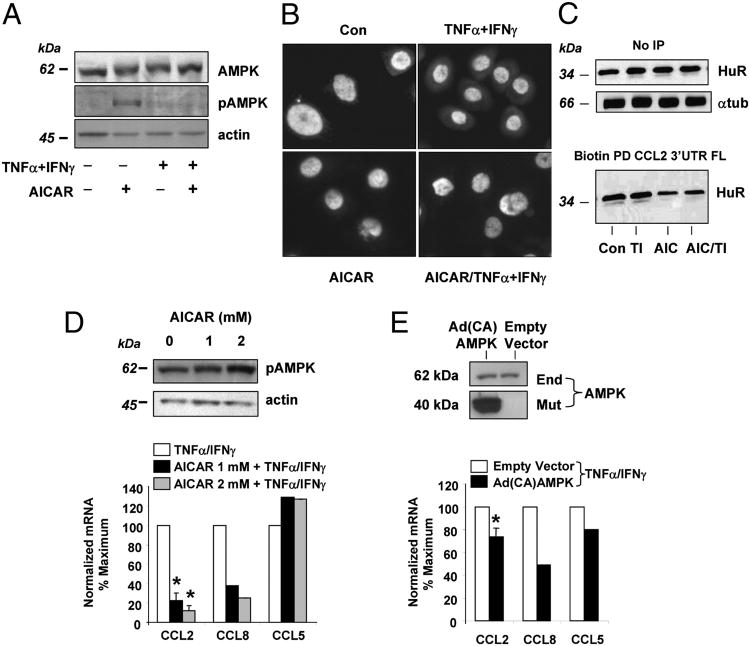
To investigate the role of AMPK in our experimental system, BEAS-2B cells were challenged with TNF-α plus IFN-γ for 18 h and subsequently treated with a potent and specific activator of AMPK, the 5-amino-imidazole-4-carboxamide riboside (AICAR). This adenosine analog mimics the physiologic activator of this pathway, AMP activating both an upstream AMPKK as well as AMPK (62). Fig. 7A shows that cytokine treatment potently inhibited AMPK activation induced by AICAR (1 mM, 4 h), as demonstrated using a specific Ab recognizing the phosphorylated form of Thr172 within the α subunit (63), without changing the total levels of the enzyme. We then investigated by immunofluorescence the effect of AMPK activation on cytokine-modulated cytoplasmic levels of HuR in PBECs. Cells were treated in the absence or presence of AICAR (1 mM, 4 h) prior to cytokine stimulation (Fig. 7B). As expected, HuR appeared mostly nuclear in unstimulated and AICAR-treated cells, and cytokine treatment strongly increased HuR cytoplasmic levels. Cell pretreatment with AICAR inhibited cytokine-induced cytoplasmic translocation of HuR. Along these lines, AICAR pretreatment did not change total HuR levels, but decreased association of HuR with biotinylated full-length CCL2 mRNA (Fig. 6C) in both control and cytokine-treated cells (Fig. 7C). As a downstream effect, activation of AMPK leads to decreased levels of HuR targets (12); therefore, we examined by real-time PCR the expression of CCL2 and CCL8, as well as of the non-HuR target CCL5 mRNA, in PBECs treated with or without AICAR (1–2 mM) prior to cytokine challenge (Fig. 7D). AICAR treatment, which caused a concentration-dependent increase in AMPK phosphorylation and inhibited the expression of CCL2 and CCL8, but not CCL5, in PBECs. GAPDH mRNA, another non-HuR target, was not changed as well (data not shown). A partial inhibition of CCL2 and CCL8 mRNA was also detected in PBECs transduced with an adenoviral vector expressing a constitutively active form of the the α1 subunit of AMPK (53) (Fig. 7E).
Figure 7.
A, Cytokine treatment inhibits activation of AMPK. Western blot analysis of whole PBEC extracts treated with (+) or without (−) TNF-α plus IFN-γ (50 ng/ml each) for 18 h prior to challenge with AICAR (1 mM) or medium for 4 h. Upper panel, Total AMPK; middle panel, phosphorylated AMPK; lower panel, equal loading shown by actin. Representative of n = 2. B, Activation of AMPK inhibits the cytokine-induced cytoplasmic localization of HuR. PBECs were treated in the presence or absence of AICAR (1 mM) for 4 h prior to challenge without (Con) or with TNF-α plus IFN-γ (50 ng/ml each) for 18 h. Immunofluorescence staining with anti-HuR is superimposed with Hoechst nuclear staining, confirming HuR cytoplasmic staining in cytokine-treated cells and its absence in cells pretreated with AICAR. C, Activation of AMPK inhibits the association of HuR with CCL2 mRNA. Upper blot, Western blot analysis of HuR and tubulin (as loading control) without pull-down in cells stimulated as in B; lower blot, detection of HuR by Western blot after biotin pull-down (PD) using a biotinylated full-length (FL) 3′ UTR of CCL2 mRNA. TI, TNF-α plus IFN-γ. D and E, Activation of AMPK inhibits the expression of HuR targets. D, Upper blot: concentration-dependent phosphorylation of AMPK in PBECs induced by AICAR. Lower blot, Actin, as loading control. The bar graphs (left) show mean ± SEM of CCL2, CCL8 (HuR targets), and CCL5 (non-HuR target) mRNA in the conditions indicated, assessed by real-time PCR in PBECs (n = 3 for CCL2; n = 2 for CCL8 and CCL5). *p < 0.05 versus unstimulated cells. E, Upper blots: expression of endogenous AMPK (End) and of the mutant active form of the 1 subunit of AMPK (Mut) by Western blot, in PBECs transduced with 6.25 PFUs per cell of Ad (CA)AMPK or its control empty vector (lower panel). Bar graphs (right) show mean ± SEM of chemokine mRNA in transduced PBECs (n = 3 for CCL2; p < 0.05 versus empty vector; n = 2 for CCL8 and CCL5). CA, constitutively active
Discussion
In chronic inflammatory airway diseases such as asthma and chronic obstructive pulmonary disease (COPD), the activation of airway epithelium leads to specific changes in signaling and gene expression, which support the host mucosal innate and adaptive responses through the production of cytokines, chemokines, lipid-derived mediators, and many more classes of mediators (64). The epithelial inflammatory response is therefore considered a key pathophysiologic factor and a prime therapeutic target (65).
The RNA-binding protein HuR is a critical regulator of the mRNA stabilization or translation of an ever-growing number of immune-related genes (9). Our previous work showed that in airway epithelial cells, inflammatory cytokines can induce an increase in cytoplasmic HuR, in parallel to stabilization of the mRNA of the CC chemokine CCL11 (8). Immunomodulatory cytokines derived from Th1 or Th2 cells, such as IFN-γ or IL-4, respectively, and macrophage-derived proinflammatory cytokines such as TNF-α exert a strong synergistic effect on epithelial gene expression and are potent inducers of rather specific profiles of epithelial chemokine expression, which are controlled at transcriptional and posttranscriptional levels (42, 66, 67). On these bases, we sought to test the hypothesis that in airway epithelial cells, upon specific inflammatory stimulation, HuR associates with multiple, functionally related inflammatory transcripts bearing HuR motifs, and we set to investigate the spectrum of genes that associate with HuR upon cytokine challenge and the functional outcome of this event.
Genome ontologic analysis of the genes identified as HuR targets indicated two major classes of genes involved in the inflammatory process: chemokines and serine-threonine protein kinases. The CC chemokines CCL2 and CCL8 belong to the MCP family, which bind to the CCR2 receptor and recruit circulating memory T cells and monocytes, supplying the tissues with macrophages and dendritic cell precursors. They also function as chemoattractants for NK cells, participate to CD4+ T cell differentiation, and are potent activators of basophil and mast cell mediator release; in vivo, CCR2-mediated responses are central in many inflammatory diseases of the lung (68). The CXC chemokines CXCL1 and CXCL2 are potent neutrophil chemoattractants, found in the early phase of experimental and clinical asthma and in the inflamed airway mucosa in COPD (69, 70), and they are critical mediators of IL-17–mediated responses in the lung (71, 72). Importantly, TLR3-mediated challenge of airway epithelium elicited a strong chemokine response including CCL2 and CXCL1, underscoring their shared role in the response to microbial stimuli (73–76) and in virus-induced disease exacerbations (77). In line with this biologic profile, both type 1 and 2 IFNs are potent inducers of members of the MCP and the GRO chemokine families (78). These biologic characteristics are shared by other chemokines—CCL13, CCL20, and IL-8—found to be associated with HuR in this or earlier studies. Posttranscriptional regulatory events have been identified for several chemokines, and the molecules mediating these effects are being increasingly recognized (9, 10). In particular, CCL2 and CXCL1 have been found to be targets of TTP, an ARE-binding protein mediating increased mRNA turnover, as well as of miRNAs (124a and 155, respectively) (79–82). As described in other models (15), it can be hypothesized that in the condition of TNF-α and IFN-γ overexpression in airway epithelium, such as after viral infection, activation of HuR may counteract TTP- or microRNA-mediated destabilization, or both, to coordinate the timing and amplitude of the expression of inflammatory gene required for the host immune response. This hypothesis is supported by our data, identifying an UAUUUUAU hexamer, which binds TTP (60), in the CCL2 3′ UTR segment preferentially associated with HuR.
Among protein kinases significantly enriched in the HuR-associated transcript pool, the stress-activated protein kinase 1 (SAPK1, or JNK1) and its upstream activating kinase, MEKK2, are key signaling molecules that regulate the inflammatory response acting at transcriptional and posttranscriptional levels (83). These kinases regulate the expression of CCL2, IL-8, and CXCL1 (84–86) and are activated, among other stimuli, by the combination of TNF-α and IFN-γ and by other IFN species (87, 88). The two main classes of HuR targets found in this study are therefore strongly related from the functional standpoint, and it is conceivable that in inflammatory settings, HuR may control the expression level of its chemokine targets by sustaining appropriate levels of supporting signaling molecules.
The mechanism of HuR-mediated effects on chemokine levels appears complex. The time-dependent association of HuR with epithelial transcript, with only two genes enriched after 3 h of cytokine challenge indicates that activating stimuli likely present in the inflamed airway mucosa (TNF-α and IFN-γ in our model) appear to drive HuR-mediated posttranscriptional functions in a dynamic fashion, likely in integration with timing and degree of transcriptional activation.
Validated association of HuR with transcrips did not always correlate with positive changes in mRNA turnover. The half-life of CCL2 and CCL8 mRNA, but not that of CXCL1 and CXCL2, increased consistently after cytokine challenge, albeit by a limited degree, and because small variations in mRNA decay can convey substantial changes in the total RNA levels (89), this change could contribute to selective chemokine overexpression during inflammation. The mRNA decay rates that we observed only at a specific time after challenge are likely part of changes occurring throughout the time that cells are exposed to cytokines, and they ultimately represent the net result of broader stimulus-mediated remodeling of the RNA-binding protein complexes, which may influence mRNA turnover as well as mRNA transport and translation. Stimulus-induced changes in mRNA stability were in fact paralleled, for CCL2, by a larger cytoplasmic mRNA pool compared with the nuclear component. In contrast, CXCL1 mRNA remained mostly nuclear after cytokine stimulation. It could be hypothesized that for some targets, such as CXCL1 and CXCL2, additional signaling may be necessary to fully activate the 3′ UTR-dependent HuR function, whereas the latter could be more critical in sustaining the strong stimulus-dependent increase of other targets, such as CCL2 and CCL8, by increasing mRNA stability and export from the nucleus, possibly through transcript-specific additional regulatory signals. A more significant role for HuR in regulating CCL2 and CCL8 expression is further supported by the consistent changes in their mRNA levels following overexpression or silencing of HuR. We could not, however, document appreciable changes in CCL2 or CCL8 mRNA stability in cells where HuR was silenced. Although we cannot exclude that the residual HuR present in the siRNA-transfected cells, or the binding of compensatory protein factors, may have a role in influencing these results, our data suggest that HuR is necessary but not sufficient to determine the turnover rates of these chemokines upon signaling from TNF-α plus IFN-γ, similarly to what was reported for IL-2 expression (90). In agreement with this hypothesis, recent data show that stabilization of CCL2 mRNA by TNF-α plus IL-4 in BEAS-2B cells was not present in a reporter bearing the CCL2 3′ UTR (10). Our data also suggest that HuR might also be specifically required for the nuclear export of at least some of its epithelial targets, similar to what was reported for CD83 (58). We are currently investigating whether HuR regulates chemokine and kinase expression also at the level of translation, as was well established for other targets (15, 16, 58). Along these lines, translation has been shown to be globally upregulated in cytokine-stimulated BEAS-2B cells, and also to contribute to CCL2 expression (10). HuR may promote translation of CCL2 and other targets also indirectly by relieving microRNA-mediated translational repression (91).
Importantly, little is known of the effect of inflammatory stimuli on HuR phosphorylation, methylation, or both, which critically regulate the subcellular localization, binding pattern, and overall function of HuR (63, 92–94). Because AMPK regulates HuR nuclear import (47, 50), we hypothesize that this pathway could be influenced by inflammatory cytokines, which trigger an increase in cytoplasmic HuR. Along these lines, activation of AMPK by AICAR reduced the severity of experimental autoimmune encephalomyelitis in mice (95), and treatment with AICAR inhibited inducible NO synthase synthesis (96). We found that cell challenge with TNF-α and IFN-γ prevents AICAR-mediated activation of AMPK and, conversely, that this pathway may have a role in controlling the levels of CCL2—and potentially other chemokines—also by diminishing HuR:transcript association. Although it cannot be excluded that the observed effect may be, at least in part, due to a nonspecific effect of AICAR on other AMP-dependent enzymes (97) or on other AMPK-dependent pathways (98), our results are in agreement with the established role of AMPK as a regulator of HuR function (47). The overall relevance of AMPK contribution will ultimately need to be verified in the context of all the pathways controlling HuR nucleocytoplasmic shuttling and function, such as p38 MAPK/MK2, PKCα, PKCδ, Chk2, and CDK1 (99).
In conclusion, the ability of inflammatory cytokines to induce selective HuR binding and activation could lead to the coordinate regulation of multiple, functionally related chemokines and signaling pathways in airway epithelium. It is likely that as a global controller of epithelial inflammatory responses, HuR regulates the expression of CCL2, CCL8, and other epithelial genes through combinatorial, and possibly in some cases independent, posttranscriptional mechanisms. Unmasking the molecular mechanisms of HuR function may likely uncover novel therapeutic strategies for blockade of proinflammatory pathways pathogenetic for asthma, COPD, and other lung inflammatory diseases.
Supplementary Material
Acknowledgments
We thank Dr. Alan Berger at the Lowe Family Genomics Core at the Johns Hopkins University School of Medicine, Bayview Campus for kind assistance in statistical analysis of the array data.
This work was supported by National Institutes of Health Grant R01 AI060990-01A1 and by the American Academy of Allergy, Asthma and Immunology/Aventis Women Physician in Allergy Junior Faculty Development Award (to C.S.). M.G. is supported by the National Institute on Aging–Intramural Research Program, National Institutes of Health.
Abbreviations used in this article
- AICAR5
amino-imidazole-4-carboxamide riboside
- AMPKAMP
activated protein kinase
- ARE
adenylate-uridylate–rich elements
- COPD
chronic obstructive pulmonary disease
- IP
immunoprecipitationm
- RNP
messenger ribonucleoprotein
- PBEC
primary bronchial epithelial cell
- TTP
tristetraprolin
- USER
untranslated sequence elements for regulations
- UTR
untranslated region
Footnotes
Disclosures: The authors have no financial conflicts of interest.
The online version of this article contains supplemental material.
References
- 1.Kracht M, Saklatvala J. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine. 2002;20:91–106. doi: 10.1006/cyto.2002.0895. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 2.Wilusz CJ, Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 3.Anderson P, Phillips K, Stoecklin G, Kedersha N. Post-transcriptional regulation of proinflammatory proteins. J Leukoc Biol. 2004;76:42–47. doi: 10.1189/jlb.1103536. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 4.Fan J, Yang X, Wang W, Wood WH, III, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci USA. 2002;99:10611–10616. doi: 10.1073/pnas.162212399. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 6.Keene JD. Ribonucleoprotein infrastructure regulating the flow of genetic information between the genome and the proteome. Proc Natl Acad Sci USA. 2001;98:7018–7024. doi: 10.1073/pnas.111145598. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bakheet T, Frevel M, Williams BR, Greer W, Khabar KS. ARED: human AU-rich element-containing mRNA database reveals an unexpectedly diverse functional repertoire of encoded proteins. Nucleic Acids Res. 2001;29:246–254. doi: 10.1093/nar/29.1.246. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atasoy U, Curry SL, López de Silanes I, Shyu AB, Casolaro V, Gorospe M, Stellato C. Regulation of eotaxin gene expression by TNF-alpha and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J Immunol. 2003;171:4369–4378. doi: 10.4049/jimmunol.171.8.4369. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 9.Fan J, Heller NM, Gorospe M, Atasoy U, Stellato C. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur Respir J. 2005;26:933–947. doi: 10.1183/09031936.05.00120204. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 10.Anderson P. Intrinsic mRNA stability helps compose the inflammatory symphony. Nat Immunol. 2009;10:233–234. doi: 10.1038/ni0309-233. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 11.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci USA. 2004;101:2987–2992. doi: 10.1073/pnas.0306453101. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, Kontoyiannis DL. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell. 2005;19:777–789. doi: 10.1016/j.molcel.2005.08.007. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 17.Ford LP, Watson J, Keene JD, Wilusz J. ELAV proteins stabilize deadenylated intermediates in a novel in vitro mRNA deadenylation/degradation system. Genes Dev. 1999;13:188–201. doi: 10.1101/gad.13.2.188. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–8151. doi: 10.1074/jbc.271.14.8144. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 19.Yarovinsky TO, Butler NS, Monick MM, Hunninghake GW. Early exposure to IL-4 stabilizes IL-4 mRNA in CD4+ T cells via RNA-binding protein HuR. J Immunol. 2006;177:4426–4435. doi: 10.4049/jimmunol.177.7.4426. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 20.Casolaro V, Fang X, Tancowny B, Fan J, Wu F, Srikantan S, Asaki SY, De Fanis U, Huang SK, Gorospe M, Atasoy UX, Stellato C. Posttranscriptional regulation of IL-13 in T cells: role of the RNA-binding protein HuR. J Allergy Clin Immunol. 2008;121:853–859. e854. doi: 10.1016/j.jaci.2007.12.1166. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean JL, Wait R, Mahtani KR, Sully G, Clark AR, Saklatvala J. The 3′ untranslated region of tumor necrosis factor alpha mRNA is a target of the mRNA-stabilizing factor HuR. Mol Cell Biol. 2001;21:721–730. doi: 10.1128/MCB.21.3.721-730.2001. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ming XF, Stoecklin G, Lu M, Looser R, Moroni C. Parallel and independent regulation of interleukin-3 mRNA turnover by phosphatidylinositol 3-kinase and p38 mitogen-activated protein kinase. Mol Cell Biol. 2001;21:5778–5789. doi: 10.1128/MCB.21.17.5778-5789.2001. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nabors LB, Suswam E, Huang Y, Yang X, Johnson MJ, King PH. Tumor necrosis factor alpha induces angiogenic factor up-regulation in malignant glioma cells: a role for RNA stabilization and HuR. Cancer Res. 2003;63:4181–4187. Abstract/FREE Full Text. [PubMed] [Google Scholar]
- 24.Dixon DA, Tolley ND, King PH, Nabors LB, McIntyre TM, Zimmerman GA, Prescott SM. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J Clin Invest. 2001;108:1657–1665. doi: 10.1172/JCI12973. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3′ untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res. 2001;61:2154–2161. Abstract/FREE Full Text. [PubMed] [Google Scholar]
- 26.Goldberg-Cohen I, Furneauxb H, Levy AP. A 40-bp RNA element that mediates stabilization of vascular endothelial growth factor mRNA by HuR. J Biol Chem. 2002;277:13635–13640. doi: 10.1074/jbc.M108703200. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Pascual F, Hausding M, Ihrig-Biedert I, Furneaux H, Levy AP, Förstermann U, Kleinert H. Complex contribution of the 3′-untranslated region to the expressional regulation of the human inducible nitric-oxide synthase gene. Involvement of the RNA-binding protein HuR. J Biol Chem. 2000;275:26040–26049. doi: 10.1074/jbc.M910460199. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 28.Levy NS, Chung S, Furneaux H, Levy AP. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 29.Sakai K, Kitagawa Y, Saiki M, Saiki S, Hirose G. Binding of the ELAV-like protein in murine autoimmune T-cells to the nonameric AU-rich element in the 3′ untranslated region of CD154 mRNA. Mol Immunol. 2003;39:879–883. doi: 10.1016/s0161-5890(03)00007-5. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 30.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog. 2000;28:76–83. CrossRefMedline. [PubMed] [Google Scholar]
- 31.Blaxall BC, Pellett AC, Wu SC, Pende A, Port JD. Purification and characterization of beta-adrenergic receptor mRNA-binding proteins. J Biol Chem. 2000;275:4290–4297. doi: 10.1074/jbc.275.6.4290. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 32.Keene JD, Tenenbaum SA. Eukaryotic mRNPs may represent posttranscriptional operons. Mol Cell. 2002;9:1161–1167. doi: 10.1016/s1097-2765(02)00559-2. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 33.López de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 34.Vignola AM, Mirabella F, Costanzo G, Di Giorgi R, Gjomarkaj M, Bellia V, Bonsignore G. Airway remodeling in asthma. Chest. 2003;123(3 Suppl):417S–422S. doi: 10.1378/chest.123.3_suppl.417s. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 35.Ma WJ, Furneaux H. Localization of the human HuR gene to chromosome 19p13.2. Hum Genet. 1997;99:32–33. doi: 10.1007/s004390050305. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 36.Hamshere M, Cross S, Daniels M, Lennon G, Brook JD. A transcript map of a 10-Mb region of chromosome 19: a source of genes for human disorders, including candidates for genes involved in asthma, heart defects, and eye development. Genomics. 2000;63:425–429. doi: 10.1006/geno.1999.6075. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 37.Laitinen T, Ollikainen V, Lázaro C, Kauppi P, de Cid R, Antó JM, Estivill X, Lokki H, Mannila H, Laitinen LA, Kere J. Association study of the chromosomal region containing the FCER2 gene suggests it has a regulatory role in atopic disorders. Am J Respir Crit Care Med. 2000;161:700–706. doi: 10.1164/ajrccm.161.3.9810056. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 38.López de Silanes I, Galbán S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoecklin G, Tenenbaum SA, Mayo T, Chittur SV, George AD, Baroni TE, Blackshear PJ, Anderson P. Genome-wide analysis identifies interleukin-10 mRNA as target of tristetraprolin. J Biol Chem. 2008;283:11689–11699. doi: 10.1074/jbc.M709657200. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winzen R, Thakur BK, Dittrich-Breiholz O, Shah M, Redich N, Dhamija S, Kracht M, Holtmann H. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol Cell Biol. 2007;27:8388–8400. doi: 10.1128/MCB.01493-07. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 42.Stellato C, Matsukura S, Fal A, White J, Beck LA, Proud D, Schleimer RP. Differential regulation of epithelial-derived C-C chemokine expression by IL-4 and the glucocorticoid budesonide. J Immunol. 1999;163:5624–5632. Abstract/FREE Full Text. [PubMed] [Google Scholar]
- 43.Schleimer RP, Kato A, Kern R, Kuperman D, Avila PC. Epithelium: at the interface of innate and adaptive immune responses. J Allergy Clin Immunol. 2007;120:1279–1284. doi: 10.1016/j.jaci.2007.08.046. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 46.Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang W, Fan J, Yang X, Fürer-Galban S, Lopez de Silanes I, von Kobbe C, Guo J, Georas SN, Foufelle F, Hardie DG, et al. AMP-activated kinase regulates cytoplasmic. HuR Mol Cell Biol. 2002;22:3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardie DG, Scott JW, Pan DA, Hudson ER. Management of cellular energy by the AMP-activated protein kinase system. FEBS Lett. 2003;546:113–120. doi: 10.1016/s0014-5793(03)00560-x. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 49.Wang W, Yang X, Kawai T, López de Silanes I, Mazan-Mamczarz K, Chen P, Chook YM, Quensel C, Köhler M, Gorospe M. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1: involvement in the nuclear import of RNA-binding protein HuR. J Biol Chem. 2004;279:48376–48388. doi: 10.1074/jbc.M409014200. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 50.Zou T, Liu L, Rao JN, Marasa BS, Chen J, Xiao L, Zhou H, Gorospe M, Wang JY. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1. Biochem J. 2008;409:389–398. doi: 10.1042/BJ20070860. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddel RR, Salghetti SE, Willey JC, Ohnuki Y, Ke Y, Gerwin BI, Lechner JF, Harris CC. Development of tumorigenicity in simian virus 40-immortalized human bronchial epithelial cell lines. Cancer Res. 1993;53:985–991. Abstract/FREE Full Text. [PubMed] [Google Scholar]
- 52.Churchill L, Friedman B, Schleimer RP, Proud D. Production of granulocyte-macrophage colony-stimulating factor by cultured human tracheal epithelial cells. Immunology. 1992;75:189–195. Medline. [PMC free article] [PubMed] [Google Scholar]
- 53.Woods A, Azzout-Marniche D, Foretz M, Stein SC, Lemarchand P, Ferré P, Foufelle F, Carling D. Characterization of the role of AMP-activated protein kinase in the regulation of glucose-activated gene expression using constitutively active and dominant negative forms of the kinase. Mol Cell Biol. 2000;20:6704–6711. doi: 10.1128/mcb.20.18.6704-6711.2000. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang JG, Collinge M, Ramgolam V, Ayalon O, Fan XC, Pardi R, Bender JR. LFA-1-dependent HuR nuclear export and cytokine mRNA stabilization in T cell activation. J Immunol. 2006;176:2105–2113. doi: 10.4049/jimmunol.176.4.2105. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 55.Gallouzi IE, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, Steitz JA. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu K, Li Y, Prabhu V, Young L, Becker KG, Munson PJ, Weng Np. Augmentation in expression of activation-induced genes differentiates memory from naive CD4+ T cells and is a molecular mechanism for enhanced cellular response of memory CD4+ T cells. J Immunol. 2001;166:7335–7344. doi: 10.4049/jimmunol.166.12.7335. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 57.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. Medline. [DOI] [PubMed] [Google Scholar]
- 58.Prechtel AT, Chemnitz J, Schirmer S, Ehlers C, Langbein-Detsch I, Stülke J, Dabauvalle MC, Kehlenbach RH, Hauber J. Expression of CD83 is regulated by HuR via a novel cis-active coding region RNA element. J Biol Chem. 2006;281:10912–10925. doi: 10.1074/jbc.M510306200. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 59.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 60.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 61.Al-Ahmadi W, Al-Ghamdi M, Al-Haj L, Al-Saif M, Khabar KS. Alternative polyadenylation variants of the RNA binding protein, HuR: abundance, role of AU-rich elements and auto-Regulation. Nucleic Acids Res. 2009;37:3612–3624. doi: 10.1093/nar/gkp223. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carling D. The AMP-activated protein kinase cascade—a unifying system for energy control. Trends Biochem Sci. 2004;29:18–24. doi: 10.1016/j.tibs.2003.11.005. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J Biol Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 64.Holgate ST. Epithelium dysfunction in asthma. J Allergy Clin Immunol. 2007;120:1233–1244. doi: 10.1016/j.jaci.2007.10.025. quiz 1245–1236. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 65.Stellato C. Glucocorticoid actions on airway epithelial responses in immunity: functional outcomes and molecular targets. J Allergy Clin Immunol. 2007;120:1247–1263. 264–1245. doi: 10.1016/j.jaci.2007.10.041. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 66.Matsukura S, Stellato C, Plitt JR, Bickel C, Miura K, Georas SN, Casolaro V, Schleimer RP. Activation of eotaxin gene transcription by NF-kappa B and STAT6 in human airway epithelial cells. J Immunol. 1999;163:6876–6883. Abstract/FREE Full Text. [PubMed] [Google Scholar]
- 67.Stellato C, Beck LA. Expression of eosinophil-specific chemokines by human epithelial cells. Chem Immunol. 2000;76:156–176. doi: 10.1159/000058785. Medline. [DOI] [PubMed] [Google Scholar]
- 68.Rose CE, Jr, Sung SS, Fu SM. Significant involvement of CCL2 (MCP-1) in inflammatory disorders of the lung. Microcirculation. 2003;10:273–288. doi: 10.1038/sj.mn.7800193. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 69.Anderson P. Pin1: a proline isomerase that makes you wheeze? Nat Immunol. 2005;6:1211–1212. doi: 10.1038/ni1205-1211. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 70.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 71.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 72.Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–969. doi: 10.1042/bst0300963. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 73.Matsukura S, Kokubu F, Kurokawa M, Kawaguchi M, Ieki K, Kuga H, Odaka M, Suzuki S, Watanabe S, Takeuchi H, et al. Synthetic double-stranded RNA induces multiple genes related to inflammation through Toll-like receptor 3 depending on NF-kappaB and/or IRF-3 in airway epithelial cells. Clin Exp Allergy. 2006;36:1049–1062. doi: 10.1111/j.1365-2222.2006.02530.x. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 74.Ritter M, Mennerich D, Weith A, Seither P. Characterization of Toll-like receptors in primary lung epithelial cells: strong impact of the TLR3 ligand poly(I:C) on the regulation of Toll-like receptors, adaptor proteins and inflammatory response. J Inflamm (Lond) 2005;2:16. doi: 10.1186/1476-9255-2-16. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Osterholzer JJ, Curtis JL, Polak T, Ames T, Chen GH, McDonald R, Huffnagle GB, Toews GB. CCR2 mediates conventional dendritic cell recruitment and the formation of bronchovascular mononuclear cell infiltrates in the lungs of mice infected with Cryptococcus neoformans. J Immunol. 2008;181:610–620. doi: 10.4049/jimmunol.181.1.610. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Serbina NV, Jia T, Hohl TM, Pamer EG. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol. 2008;26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat Rev Immunol. 2010;10:24–35. doi: 10.1038/nri2685. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 78.Hokeness KL, Kuziel WA, Biron CA, Salazar-Mather TP. Monocyte chemoattractant protein-1 and CCR2 interactions are required for IFN-alpha/beta-induced inflammatory responses and antiviral defense in liver. J Immunol. 2005;174:1549–1556. doi: 10.4049/jimmunol.174.3.1549. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 79.Liang J, Lei T, Song Y, Yanes N, Qi Y, Fu M. RNA-destabilizing factor tristetraprolin negatively regulates NF-kappaB signaling. J Biol Chem. 2009;284:29383–29390. doi: 10.1074/jbc.M109.024745. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishmael FT, Fang X, Galdiero MR, Atasoy U, Rigby WF, Gorospe M, Cheadle C, Stellato C. Role of the RNA-binding protein tristetraprolin in glucocorticoid-mediated gene regulation. J Immunol. 2008;180:8342–8353. doi: 10.4049/jimmunol.180.12.8342. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kedersha N, Chen S, Gilks N, Li W, Miller IJ, Stahl J, Anderson P. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 2002;13:195–210. doi: 10.1091/mbc.01-05-0221. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karin M, Gallagher E. From JNK to pay dirt: jun kinases, their biochemistry, physiology and clinical importance. IUBMB Life. 2005;57:283–295. doi: 10.1080/15216540500097111. Medline. [DOI] [PubMed] [Google Scholar]
- 84.Sukhumavasi W, Egan CE, Denkers EY. Mouse neutrophils require JNK2 MAPK for Toxoplasma gondii-induced IL-12p40 and CCL2/MCP-1 release. J Immunol. 2007;179:3570–3577. doi: 10.4049/jimmunol.179.6.3570. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Luo W, Reiser G. Proteinase-activated receptor-1 and -2 induce the release of chemokine GRO/CINC-1 from rat astrocytes via differential activation of JNK isoforms, evoking multiple protective pathways in brain. Biochem J. 2007;401:65–78. doi: 10.1042/BJ20060732. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wolter S, Doerrie A, Weber A, Schneider H, Hoffmann E, von der Ohe J, Bakiri L, Wagner EF, Resch K, Kracht M. c-Jun controls histone modifications, NF-kappaB recruitment, and RNA polymerase II function to activate the ccl2 gene. Mol Cell Biol. 2008;28:4407–4423. doi: 10.1128/MCB.00535-07. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim WH, Lee JW, Gao B, Jung MH. Synergistic activation of JNK/SAPK induced by TNF-alpha and IFN-gamma: apoptosis of pancreatic beta-cells via the p53 and ROS pathway. Cell Signal. 2005;17:1516–1532. doi: 10.1016/j.cellsig.2005.03.020. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 88.Yang CH, Murti A, Valentine WJ, Du Z, Pfeffer LM. Interferon alpha activates NF-kappaB in JAK1-deficient cells through a TYK2-dependent pathway. J Biol Chem. 2005;280:25849–25853. doi: 10.1074/jbc.M413721200. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Seko Y, Azmi H, Fariss R, Ragheb JA. Selective cytoplasmic translocation of HuR and site-specific binding to the interleukin-2 mRNA are not sufficient for CD28-mediated stabilization of the mRNA. J Biol Chem. 2004;279:33359–33367. doi: 10.1074/jbc.M312306200. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 91.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
- 92.Abdelmohsen K, Pullmann R, Jr, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25:543–557. doi: 10.1016/j.molcel.2007.01.011. CrossRefMedline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doller A, Huwiler A, Müller R, Radeke HH, Pfeilschifter J, Eberhardt W. Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2. Mol Biol Cell. 2007;18:2137–2148. doi: 10.1091/mbc.E06-09-0850. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin FY, Chen YH, Lin YW, Tsai JS, Chen JW, Wang HJ, Chen YL, Li CY, Lin SJ. The role of human antigen R, an RNA-binding protein, in mediating the stabilization of toll-like receptor 4 mRNA induced by endotoxin: a novel mechanism involved in vascular inflammation. Arterioscler Thromb Vasc Biol. 2006;26:2622–2629. doi: 10.1161/01.ATV.0000246779.78003.cf. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 95.Nath N, Giri S, Prasad R, Salem ML, Singh AK, Singh I. 5-aminoimidazole-4-carboxamide ribonucleoside: a novel immunomodulator with therapeutic efficacy in experimental autoimmune encephalomyelitis. J Immunol. 2005;175:566–574. doi: 10.4049/jimmunol.175.1.566. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 96.Pilon G, Dallaire P, Marette A. Inhibition of inducible nitric-oxide synthase by activators of AMP-activated protein kinase: a new mechanism of action of insulin-sensitizing drugs. J Biol Chem. 2004;279:20767–20774. doi: 10.1074/jbc.M401390200. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 97.Towler MC, Hardie DG. AMP-activated protein kinase in metabolic control and insulin signaling. Circ Res. 2007;100:328–341. doi: 10.1161/01.RES.0000256090.42690.05. Abstract/FREE Full Text. [DOI] [PubMed] [Google Scholar]
- 98.Sag D, Carling D, Stout RD, Suttles J. Adenosine 5′ -monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J Immunol. 2008;181:8633–8641. doi: 10.4049/jimmunol.181.12.8633. Abstract/FREE Full Text. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doller A, Pfeilschifter J, Eberhardt W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell Signal. 2008;20:2165–2173. doi: 10.1016/j.cellsig.2008.05.007. CrossRefMedline. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.