Abstract
A review of data mining and analysis techniques that can be used for the mapping of knowledge domains is given. Literature mapping techniques can be based on authors, documents, journals, words, and/or indicators. Most mapping questions are related to research assessment or to the structure and dynamics of disciplines or networks. Several mapping techniques are demonstrated on a data set comprising 20 years of papers published in PNAS. Data from a variety of sources are merged to provide unique indicators of the domain bounded by PNAS. By using funding source information and citation counts, it is shown that, on an aggregate basis, papers funded jointly by the U.S. Public Health Service (which includes the National Institutes of Health) and non-U.S. government sources outperform papers funded by other sources, including by the U.S. Public Health Service alone. Grant data from the National Institute on Aging show that, on average, papers from large grants are cited more than those from small grants, with performance increasing with grant amount. A map of the highest performing papers over the 20-year period was generated by using citation analysis. Changes and trends in the subjects of highest impact within the PNAS domain are described. Interactions between topics over the most recent 5-year period are also detailed.
Scientists have always had the desire to do research of high impact. Part of this desire has been for so-called selfish reasons such as to obtain tenure, increase one's salary, or to enhance one's reputation. However, altruistic purposes also play a large role. We desire to make a difference, to advance knowledge for the benefit of our employers, our nations, or all mankind.
This raises questions that all scientists face and that collectively give rise to innovation and the advancement of science and technology: “What should I work on?” “Are my ideas any good, are they novel, or have they already been taken?” “What can I learn from others?” “How can I improve on their work?” “Who should I work with?” and “Who will fund this?”
Such questions accrue on an institutional level as well. Organizations that answer well are rewarded. Universities develop reputations that drive research agendas and secure large amounts of funding over many years. Successful companies drive markets and consumer preference, maintaining their profitability. Success often reflects an ability to stay on the leading edges of science and technology curves.
In today's world, we have unparalleled access to information, which should enable us to answer questions of a strategic nature more readily than in the past. However, with this increased information has come dilution. Fortunately, tools are now becoming available that allow us to sift, condense, and associate this information in ways that help us answer our questions.
This paper will start with a review of data mining and analysis techniques for the mapping of literatures, including their best uses and the types of questions that can be answered. Subsequent sections will use some of these techniques to provide an indicator-based characterization of the domain comprised by PNAS. Specifically, multiple data sources are combined to give a unique look at input-output (funding-impact) and import-export (diffusion between disciplines) from the perspective of this multidisciplinary, but biomedically dominated journal. A map of the highest impact research in PNAS is also introduced.
Techniques for Mapping Knowledge Domains
Mapping of scientific literature as a field has been in existence for many decades. We are indebted to Eugene Garfield, Derek de Solla Price, and others who, through their desire to understand the structure and flow of scientific advancement (1-5), started the work that has made the indexing and dissemination of bibliographic information a commodity. Electronic sources such as the Science Citation Index Expanded (SCIE), INSPEC, and Medline contain entries for millions of scientific articles, providing us with information to help answer our questions.
Historically, answers have not come without great effort. Given the lack of computing resources, early studies naturally tended to focus on small subsets and were, with some exceptions, academic in nature. With the recent availability of electronic data, exponentially increasing computing power, advanced algorithms, and visualization techniques, we are now at a point where much less effort is required to get answers. Indeed, we can almost routinely do large scale studies aimed at answering significant questions of a strategic nature (6).
Notable among recent advances is the development of the field of information visualization. The past decade has seen rapid growth in this field, and the application of many new techniques to the visualization of literature, patents, genomes (cf. ref. 7), and other information types (8, 9). However, it must be remembered that whereas visualization can be critical to understanding, it is simply a window into the rigorous, often multidimensional, analyses that have formed the basis of informatics for many years. Thus, mapping, as a term, does not merely refer to the visualization piece, but to the underlying data mining and analysis techniques as well.
Mapping knowledge domains, then, takes as its input such seemingly diverse subjects as network analysis (e.g., web, social networks, scale-free networks, and metabolic pathways), linguistics, concept or topic extraction, citation analysis, and science and technology indicators, in addition to visualization techniques. Similarly, knowledge domain can be more broadly defined than the narrow “technical field” that is commonly associated with the term. Genomes, communities, and networks are all domains with multiple attributes from which one can derive different types of knowledge. Although this paper focuses on mapping of literatures, many of the same analysis and visualization techniques have been and can be applied to other domains.
The main purpose of mapping knowledge domains is to give us knowledge, or answers to our questions. Mapping is useful for the subject matter expert and nonexpert alike. For the nonexpert, mapping provides an entry point into a domain, a means of gaining knowledge on both the macro and micro levels. For the expert, mapping provides validation of perceptions and a means to quickly investigate trends and new information. Yet, even the expert can be surprised by developments on the periphery of his perception. Mapping and interactive exploration provide context for such surprises.
Commonly utilized techniques for mapping literatures are shown in Table 1 with their primary uses. Most questions of interest fall into three categories: fields and paradigms, communities or networks, and assessment of performance or opportunity. Coauthorship analysis is very similar to social network analysis. Yet, whereas social network analysis is concerned with global properties of large author databases (10), coauthorship studies aim to answer specific questions about collaboration groups (11). Author cocitation analysis is particularly suited to investigation of intellectual structure and history, and is often used with factor analysis and multidimensional scaling (12). Pathfinder network scaling is particularly effective at preparing these data for layout in a visualization program (9).
Table 1. Summary of commonly utilized literature mapping techniques and their uses.
| Questions related to
|
||||
|---|---|---|---|---|
| Unit of analysis | Fields and paradigms | Communities and networks | Research performance or competitive advantage | Commonly used algorithms |
| Authors | Social structure, intellectual structure, some dynamics | Use network characteristics as indicators | Social network packages, multidimensional scaling, factor analysis, Pathfinder networks | |
| Documents | Field structure, dynamics, paradigm development | Use field mapping with indicators | Cocitation, co-term, vector space, latent semantic analysis, principle components analysis, various clustering methods | |
| Journals | Science structure, dynamics, classification, diffusion between fields | Cocitation, intercitation | ||
| Words | Cognitive structure, dynamics | Vector space, latent semantic analysis, latent dirichlet allocation | ||
| Indicators and metrics | Comparisons of fields, institutions, countries, etc., input-output | Counts, correlations | ||
Documents are the most often used unit of analysis because they can be used to map a particular scientific or technical field and its development. Cocitation and co-word are the two most common types of document analysis, and often lead to different groupings of documents. At the finest levels, cocitation techniques cluster documents by scientific paradigm, or by the same research question and hypotheses (9), whereas co-word document clusters are more topical in nature. Alternatives to the co-word method for generating document similarities include Salton's vector space model (13) and latent semantic analysis (14, 15). Journals are used less often, and are used for larger scale studies, such as to view the relationships between different fields (16). They are also suitable for the study of diffusion between disciplines (often called import-export) by using intercitation rates (17).
Mapping of words or indexing terms as networks reveals the cognitive structure of a field (18). There is some debate as to whether co-word analyses should be used for studies of science dynamics (19). The most reliable approaches aim to combine co-word techniques with citation analyses (20). More advanced techniques using sophisticated algorithms to group and relate topics show great promise for dynamic studies (21, 22).
Similar visualization methods are applied to the mapping types mentioned above for the simple reason that authors, documents, journals, and words (or groupings of these) all work equally well as the mapping unit. Common visualizations include traditional scatterplots and link-node diagrams, such as those drawn by the PAJEK program (23). Newer, more powerful visualizations include self-organized maps (24), landscapes (25, 26), timelines and crossmaps (27), and 3D displays (9). The best of these have the capability of allowing the user to navigate the information space and get detail on demand, which facilitates analysis that helps the user to answer questions.
The power of visualization is enhanced when mapping types are combined. Combining types adds more dimensions to the information, which are more easily explored by using visualization than with traditional analysis methods. For example, Chen (9, 28) combines indicators (citation counts by year) with document cocitation analysis in a 3D display to show the growth of scientific paradigms.
Indicators have been used for as long as people have wanted to compare things. Science and technology indicators were largely developed from the 1950s through the 1970s (29) by the Organization for Economic Cooperation and Development and the National Science Foundation, and have resulted in publications such as National Science Foundation's biannual Science and Engineering Indicators (30). Although activity measures (31), and specifically economic activity measures, have been the dominant component of such reports, scientific output measures such as counts of papers, patents, and citations have also played a large role. Measures of converging partial indicators have been used with the aim of identifying areas of science and technology likely to yield the greatest benefits (32, 33). Output measures have been correlated to economic activity at a macro level to show the relative strengths of countries, states, and/or technical fields (30). Several studies have reported correlation between aggregated scientific outputs and funding (34-39), but none have reported any such correlations at the individual grant level.
Characterization of PNAS
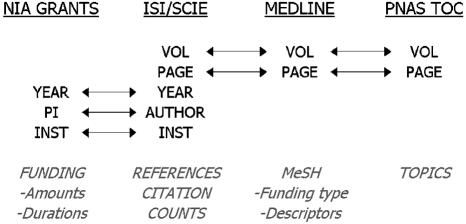
Data Sources. Data from four sources (see Fig. 1) were merged to provide the basis for a characterization of PNAS. Most studies merging databases do so to provide deeper coverage of a field (40, 41). However, this study merges multiple data sources to get more detailed information on a single journal and its impact. The base set to which other sources were merged was data from the SCIE. These data consist of 47,073 records covering the 20 years of PNAS from 1982 to 2001, including full reference lists and citation counts to each paper as of December 31, 2002. Citation counts were determined by matching of Institute for Scientific Information (ISI) reference lists (journal name variations were accounted for) with bibliographic data.† For this analysis, only the 45,326 articles, letters, notes, and reviews (commonly referred to as ALNR) were considered. The balance of the records, from editorials, corrections, book reviews, etc., contribute little or no original research, and are commonly discounted in such analyses.
Fig. 1.
Data sources, field joins (arrows), and unique properties from each source (italics).
PNAS records were also extracted from Medline, and were joined to the SCIE records primarily for use of the MeSH (medical subject heading) terms. MeSH terms are desirable for several reasons: (i) SCIE keywords are sparse, uncontrolled, and available only back to 1991; (ii) MeSH is a rich, controlled vocabulary added by human indexers; and (iii) MeSH contains specific funding-related terms. Joining MeSH terms to the ISI citation counts enables input-output studies with respect to funding type.
PNAS has a topic structure that is clearly visible in both the print and web versions of the journal Tables of Contents. First-level topics are broad: Biological Sciences, Physical Sciences, and Social Sciences. Within each of these first-level topics are secondary topics, such as Biochemistry, Biophysics, and Cell Biology within the Biological Sciences topic. First- and second-level topics for each paper were extracted from the Tables of Contents and added to the SCIE data. Joining of topics to the other data enables import-export studies as well as the correlation between impact and topic.
Finally, grant data from the National Institute on Aging (one of the institutes of the National Institutes of Health) containing principal investigator (PI) names, institutions, and funding amounts by year were joined to the other data. These data were obtained from the National Institute on Aging as part of a previous study (39). An effort was made to match grants to PNAS papers that were likely to have resulted from specific grants. For a paper to be linked to a specific grant the following conditions were required (also see Fig. 1):
PNAS author = Grant PI (last name + first initial)
and PNAS author institution = Grant PI institution
and PNAS publication year ≥ Grant initial year
and (PNAS publication year ≤ Grant initial year + 5
or PNAS publication year ≤ Grant final year + 2)
A total of 1,862 PNAS papers were found to be probable matches to specific grants. Although we cannot say with certainty that these papers are from National Institute on Aging-funded studies, they were authored by National Institute on Aging-funded PIs and were written at a time consistent with their National Institute on Aging funding. Joining of grant data to the balance of the data enables correlation of impact to funding amount, something that has to date been very difficult to quantify.
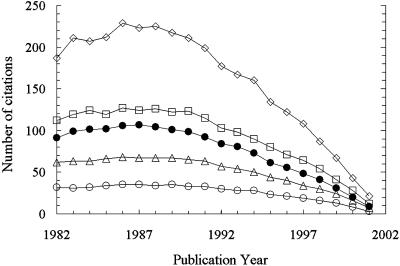
In this study, impact is equated with a ranking measure derived from citation counts. Papers were ranked by citation count for each publication year. Absolute rankings were then converted to percentile rankings. Percentile rankings are used for two reasons. First, it provides normalization across time such that papers from different years can be directly compared. This result is particularly important for recent papers, because they have typically not had enough time after publication to accumulate large numbers of citations. Second, given the skewed nature of citation count distributions, it keeps a few highly cited papers from dominating citation statistics. For example, mean citation counts for the PNAS papers range between the 64th and 70th percentile from 1982 to 1999. Related data are shown in Fig. 2.
Fig. 2.
Mean number of citations (•) to PNAS ALNR are compared with several different percentiles: 90th (⋄), 75th (□), 50th or median (▵), and 25th (○). Citation counts are as of December 31, 2002.
Whereas there are certainly factors other than citation measures in what constitutes a full definition of impact, and while the validity of using citation measures has been debated (cf. refs. 42 and 43), they are widely used (44), and will be the basis for impact in this study.
Impact and Funding. Medline MeSH terms contain three main funding source designators: Support, U.S. Gov't, P.H.S., Support, U.S. Gov't, Non-P.H.S., and Support, Non-U.S. Gov't. The first two designators refer to publications funded by the U.S. Public Health System (P.H.S.) and all other U.S. government agencies (OG), respectively. In a practical sense, P.H.S. refers to the National Institutes of Health. Support, Non-U.S. Gov't (nG) could refer to either U.S. nongovernmental sources (e.g., industry, nonprofit) or to foreign sources, but has not been segmented further. Papers with no funding source designators are tagged as Unknown. Very few papers in this category exclude a funding acknowledgment inadvertently (45). Thus, Unknown can be considered as a distinct category.
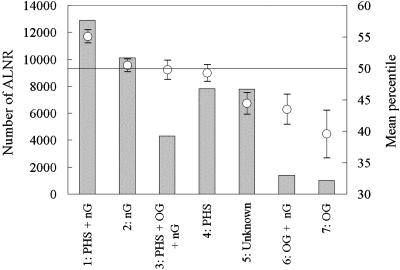
Given that each paper is tagged with anywhere from none to all three of the funding source designators, eight unique funding categories can be constructed. Two of the smaller categories, PHS+OG and PHS+OG+nG, have been combined to make a category of sufficient size for statistical purposes. Thus, seven funding categories are shown in Fig. 3 along with their numbers of papers (ALNR) and mean percentiles. The highest ranked category, with a mean percentile >55, is papers jointly funded by the U.S. Public Health System and non-U.S. government sources. By contrast, papers funded solely by the U.S. Public Health System have a mean percentile of 49.2. Yet, this is still higher than the mean percentile of 44.4 associated with papers of Unknown funding source, indicating that PHS funding has a positive impact with respect to a lack of U.S. Public Health System funding. The differences between impacts of these three categories are statistically significant at the P < 0.001 level by using a Scheffé test (ref. 46 and Table 2).
Fig. 3.
Numbers of papers (ALNR) and impact (mean citation percentile) for seven funding categories. Categories are shown in order of decreasing mean percentile. Bars indicate the number of papers (Left); circles and standard error bars indicate impact (Right). PHS, U.S. Public Health System; OG, other U.S. government; nG, non-U.S. government (includes foreign).
Table 2.
Scheffé test results for comparisons between percentile means of different funding categories (from Fig. 3)
| Category | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|
| 1 | P < .001 | P < .001 | P < .001 | P < .001 | P < .001 | P < .001 |
| 2 | NS | NS | P < .001 | P < .001 | P < .001 | |
| 3 | NS | P < .001 | P < .001 | P < .001 | ||
| 4 | P < .001 | P < .001 | P < .001 | |||
| 5 | NS | P < .001 | ||||
| 6 | P = .085 |
NS, no significant difference between means.
Other studies have shown that the mean impact of a group of papers increases with the number of authors, presumably due to multidisciplinarity (36). In general, the number of authors increases with the increasing percentile in Fig. 3. However, there are local differences that cannot be explained by number of authors. For example, for categories 1 and 2 (4.82 and 5.04 authors, respectively), and categories 4 and 5 (3.99 and 4.11 authors, respectively), the mean number of authors is anti-correlated with mean percentile.
Fig. 3 shows only mean percentiles for the entire 20-year period of study. Mean percentiles by year are relatively stable for the larger funding categories. Smaller categories showed much more scatter by year.
Does Grant Size Matter? As previously mentioned, the correlation between impact and the amount of funding has historically been difficult to quantify. This correlation is largely due to the difficulty of accurately linking funding information with the publications resulting from those funds. Agencies and institutions, although they track many things, are uniformly poor at keeping track of input-output linkages.
A total of 1,862 PNAS papers were identified as likely having resulted from National Institute on Aging funding. We assume this to be a small fraction of the total number of National Institute on Aging-funded papers, although the exact fraction is not known. Yet, the number deduced here is consistent with the relative sizes of the National Institute on Aging and the National Institutes of Health.‡ Many of these papers can be matched to multiple grants, and conversely, many of the grants seem to have given rise to multiple papers. For these data, we have identified 3,059 grant-paper pairs. This finding corresponds well to what we know to be true in research; in many cases, institutions receive multiple grants in complementary areas, and certainly the work from a single grant can spawn more than one publication. Multiple linkages between papers and grants indicate a concentration of activity at an institution. The more money received by a particular PI from a focused organization such as the National Institute on Aging, and the more that PI publishes, the more likely it is that the funds and publications are truly linked.
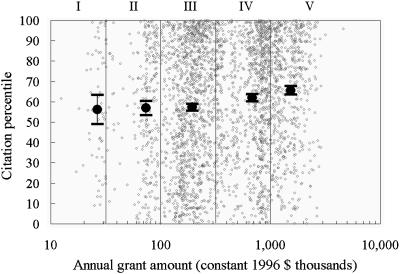
Fig. 4 shows the correlation between citation percentile and average annual grant amount for the 3,059 grant-paper pairs. Dollar amounts were normalized by GDP deflators to remove inflation biases (30). Annual grant amounts were averaged over the publication year of paper and the three previous years. Five different grant amount ranges were identified: <$31,600, $31,600 to $100,000, $100,000 to $316,000, $316,000 to $1,000,000, and >$1,000,000. Mean citation percentiles and grant amounts were calculated for the grant-paper pairs in each of the five grant ranges. The mean citation percentiles remain constant at 56-57 through the first three ranges (up to $316k), then increase to 62 and 65.6 for ranges IV and V.
Fig. 4.
Correlation between impact (citation percentile) and grant amount. Individual grant-paper pairs (small circles) and mean percentiles with standard errors (large circles) are shown for the five grant size regions that are numbered I-V.
The number of authors was also considered here as a potentially confounding variable. Cumulative probability density functions of numbers of authors per paper are nearly identical for funding ranges III-V. Thus, number of authors has little impact on the mean percentiles in these funding ranges.
Several observations can be drawn from these data. First, papers from large grants tend to outperform (in terms of mean citation percentiles) those from smaller grants, with the average performance increasing with increasing grant amount above $300,000. Second, even for small grants, papers funded by the National Institute on Aging tend to outperform the average PNAS paper; mean percentiles for each grant amount group are well over 50. Third, a high level of funding does not guarantee publication of a high impact paper. Fig. 4 shows many highly funded papers with a low citation percentile. However, the fraction of papers in the lowest quartile for ranges II-V decreases with range (0.199, 0.195, 0.130, and 0.095, respectively), which is consistent with the general increase in mean percentile. Fourth, the variance in individual paper impact appears to be very orthogonal to impact. However, this is to be expected in a single journal study of a high impact journal. If lower impact journals were included in the study, the percentile ranking for most PNAS papers would be shifted much higher.
These observations are specific to National Institute on Aging funding and PNAS papers, and cannot be directly applied to other funding sources or journals. Neither can we claim any direct cause and effect between funding and impact in the results shown here. However, this work shows a similar qualitative correlation between government funding and impact to what has been observed before. Early work by Narin and coworkers (34, 35) showed a positive correlation between National Institutes of Health funding amounts and biomedical publication counts, but did not address impact or quality. Lewison and Dawson (36) used the U.K. Research Outputs Database to show that the mean impact for groups of papers in gastroenterology increased with increases in the number of authors and the number of funding sources. They also found that papers acknowledging funding sources had significantly higher impact than those without such acknowledgments (37). Butler (38) found that whereas acknowledgment data on the whole accurately reflected the total research output of a funding body, there was no ability to track research back to the grant level.
This work goes further than any previous studies by correlating impact with funding level. However, it is also clear that the data are not yet sufficient to produce any definitive conclusions. Government agencies will need to create a clean and maintainable database linking grants, supported publications, patents, and policy changes to enable such analyses (39, 44). Accurate data would enable causal mechanisms to be addressed, given the temporal nature of the grant-research-publication relationship, and would also allow the overall impact (over all publications) of individual grants to be calculated. Such data have the potential to change the way research is funded.
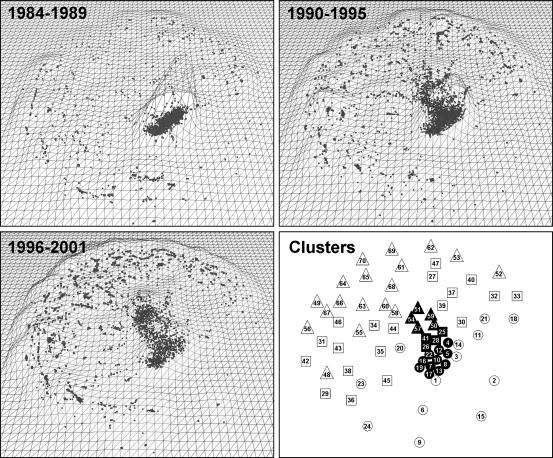
Map of High-Impact Research. To round out this characterization of research published in PNAS, a map was generated to provide information about the subjects of highest impact and related trends. Mapping of all 45,326 ALNR based on their 1.52 million references exceeded the resources available on a common desktop PC. However, a map based on the top quartile of papers from each year, those with a citation percentile of 75 or greater (see Fig. 2), could be easily generated using those same resources. This approach has the added benefit of focusing only on those topics of highest impact over the years. The resulting map contained 11,565 ALNR. Steps used in creating the map were as follows: (i) Paper-to-paper similarities were calculated using bibliographic coupling (47) and direct citations by application of the formula of Small (48), which includes normalization. Cocitation and longitudinal coupling were not considered. 1,744,258 pairs of papers (or 2.61% of the possible pairs) were linked through bibliographic coupling (i.e., having at least one common reference). In addition, the 11,565 ALNR had 411,780 references, of which 24,346 were to other papers within the set. Such direct citations were given a weight of 5. Groups of papers that cite similar sets of references are thus positioned together using this method. (ii) Paper positions were calculated from the similarities using VXORD, a force-directed placement ordination routine (49). Ordination does not assign a cluster number to each paper, but rather calculates positions for each paper on an x,y plane. (iii) Papers were assigned to clusters by using the k-means routine in MATLAB based on their x,y locations from step 2. The number of clusters was arbitrarily set at 70, and whereas 70 is not necessarily an optimum number, it is sufficient to show a distribution of topics and trends. Relative cluster positions are shown in Fig. 5. (iv) vxinsight (50) was used to interactively navigate and query the PNAS high-impact map. Fig. 5 shows landscapes for three different time periods. When used interactively, tools like vxinsight can show the growth and decay of research fronts in a visual way. (v) Diagnostic MeSH terms, i.e., those that differentiate one cluster from another, but that are not necessarily the most common terms, were generated for each cluster, and are given in Table 3. Dominant PNAS topics (from the 1997-2001 Tables of Contents) were also found for each cluster (see Table 3).
Fig. 5.
Three time periods in the PNAS high-impact map show the progression from the basic gene and protein work and techniques that dominated the 1980s to more diverse applications in the 1990s. Maps were generated by using vxinsight. Dots indicate individual papers. Wireframe mountains show the density of papers in clusters. Cluster positions are shown in Right Lower for comparison with the map panes. Clusters are numbered from oldest to youngest. Shapes indicate the first third (circles), second third (squares), and last third (triangles) in the timewise progression. Dark shapes indicate the core clusters.
Table 3. Diagnostic terms and dominant topics for the 50 largest (of 70) clusters from the PNAS high-impact map.
| Cluster | Mean Year | No. of papers | MeSH term 1 | MeSH term 2 | Dominant PNAS topic 1997-2001, % |
|---|---|---|---|---|---|
| 3 | 1987.40 | 242 | *Oncogenes | DNA restriction enzymes | Biochemistry (30.8) |
| 4 | 1987.79 | 483 | *Genes, structural | DNA restriction enzymes | |
| 5 | 1987.82 | 524 | Cloning, molecular | Nucleic acid hybridization | Genetics (37.5) |
| 6 | 1988.17 | 281 | Oxidation-reduction | Lipoproteins, LDL/*metabolism | Medical Sciences (36.4) |
| 7 | 1988.46 | 339 | Electrophoresis, polyacrylamide gel | Alzheimer's disease/*pathology | Biochemistry (33.3) |
| 8 | 1988.80 | 194 | Mutation | Collagen/metabolism | Medical Sciences (33.3) |
| 9 | 1988.93 | 94 | Buthionine sulfoximine | Bacteriorhodopsins/genetics/*metabolism | Cell Biology (33.3) |
| 10 | 1988.96 | 348 | Nucleic acid hybridization | Escherichia coli genetics | Biochemistry (26.7) |
| 12 | 1989.25 | 492 | Cloning, molecular | Sequence homology, nucleic acid | Microbiology (31.3) |
| 13 | 1989.29 | 254 | Transforming growth factors | Biochemistry (20.7) | |
| 14 | 1989.37 | 162 | DNA restriction enzymes | H-2 Antigens/*genetics | Medical Sciences (50.0) |
| 16 | 1990.00 | 313 | Chromatography, affinity | Tumor necrosis factor | Biochemistry (25.6) |
| 17 | 1990.74 | 93 | Sarcoma viruses, avian | Biochemistry (34.8) | |
| 18 | 1990.93 | 127 | Neutralization tests | HIV-1/*immunology | Genetics (72.7) |
| 19 | 1991.03 | 171 | ADP-ribosylation factors | Hemochromatosis/genetics/* metabolism | Medical Sciences (36.4) |
| 22 | 1991.26 | 208 | *DNA replication | Biochemistry (34.4) | |
| 23 | 1991.41 | 144 | P-glycoprotein | Drug resistance/*genetics | Cell Biology (21.4) |
| 24 | 1991.45 | 130 | Autoradiography | Receptors, opiod/*metabolism | Biochemistry (32.4) |
| 25 | 1991.99 | 193 | Chromosome mapping | Genetics (16.7) | |
| 26 | 1992.20 | 172 | Receptors, fibroblast growth factor | Receptors, calcitriol | Biochemistry (33.3) |
| 28 | 1992.44 | 272 | Gene expression | Gene library | Biochemistry (34.5) |
| 29 | 1993.35 | 203 | Electric conductivity | Synapses/*physiology | Neurobiology (62.5) |
| 31 | 1993.77 | 117 | *Nucleic acid conformation | Biochemistry (25.5) | |
| 32 | 1993.87 | 157 | HIV-I reverse transcriptase | *Reverse transcriptase Inhibitors | Biochemistry (39.5) |
| 36 | 1994.58 | 304 | Alzheimer's disease/*metabolism | Amyloid β protein/*metabolism | Neurobiology (35.7) |
| 38 | 1994.78 | 200 | Phosphotyrosine | Protein-tyrosine kinase/*metabolism | Medical Sciences (22.2) |
| 40 | 1995.05 | 137 | Phylogeny | Bone marrow cells | Evolution (23.5) |
| 41 | 1995.10 | 229 | Comparative study | Sequence homology, amino acid | Medical Sciences (18.0) |
| 42 | 1995.10 | 90 | Magnetic resonance imaging | Photic stimulation | Neurobiology (44.9) |
| 43 | 1995.12 | 263 | Nitric oxide/ * metabolism | ω-N-Methylarginine | Medical Sciences (38.7) |
| 46 | 1995.32 | 155 | Brain-derived neurotrophic factor | Nerve tissue proteins/*pharmacology | Neurobiology (45.8) |
| 47 | 1995.42 | 234 | *Cell cycle | *Genes, p53 | Cell Biology (31.9) |
| 48 | 1995.54 | 92 | Photosynthetic Reaction Center, bacterial | *Bacterial proteins | Neurobiology (32.6) |
| 49 | 1995.64 | 150 | *Protein folding | *Protein conformation | Biophysics (69.4) |
| 50 | 1995.65 | 302 | Molecular sequence data | *Genetic vectors | Biochemistry (22.8) |
| 52 | 1996.09 | 156 | Cytotoxicity, immunologic | Killer cells, natural/ *immunology | Immunology (53.4) |
| 53 | 1996.10 | 200 | Lymphocyte transformation | Immunology (33.0) | |
| 57 | 1996.54 | 176 | RNA, messenger/genetics/metabolism | Defensins | Biochemistry (19.3) |
| 59 | 1996.86 | 173 | DNA primers | Tetracycline/*pharmacology | Biochemistry (22.8) |
| 60 | 1997.00 | 82 | cIF-2 kinase | NF-κ B/*antagonists & inhibitors | Immunology (33.3) |
| 61 | 1997.21 | 227 | *DNA repair | Leptin | Medical Sciences (28.8) |
| 62 | 1997.35 | 215 | Protein p53/*metabolism | *Genetics, population | Medical Sciences (24.6) |
| 63 | 1997.45 | 183 | Sirolimus | 1-phosphatidylinositol 3-Kinase/metabolism | Cell Biology (27.3) |
| 64 | 1997.63 | 286 | *Apoptosis | Protooncogene proteins c-bcl-2 | Cell Biology (24.5) |
| 65 | 1997.92 | 139 | Ubiquitins/*metabolism | Multienzyme complexes/*metabolism | Cell Biology (29.6) |
| 66 | 1997.93 | 205 | Models, molecular | Crystallography, x-Ray | Biochemistry (49.0) |
| 67 | 1997.94 | 120 | Neoplasm transplantation | Serine endopeptidases/*metabolism | Medical Sciences (25.0) |
| 68 | 1998.01 | 123 | Adenomatous polyposis coli protein | Genes, APC | Medical Sciences(22.4) |
| 69 | 1998.31 | 222 | Tumor cells, cultured | *Telomere | Biochemistry (31.7) |
| 70 | 1999.55 | 162 | Gene expression profiling | Oligonucleotide array sequence analysis | Genetics (27.1) |
Italics indicate topics with <30% dominance of a cluster.
The high-impact maps of Fig. 5 show two distinct features: a core group of 20 close-knit clusters in the center, and the remaining clusters that are dispersed and focus on individual topics. The central position of the core clusters indicates their centrality to the focus of PNAS over the 20-year period. This core work had much to do with molecular cloning, hybridization, sequencing, and other key techniques during the first 10 years, shifting into more applied work on growth factors, cancers, and gene expression in the middle years (see Fig. 5 and Table 3 to match diagnostic terms to clusters and times). The most recent work in this core area deals with molecular sequencing, RNA, and cell metabolism.
The dispersed clusters do not have a common focus, but most have strong links (through bibliographic coupling) to the core. In general, the shift has been to more applied topics, often using the revolutionary techniques associated with molecular cloning, hybridization, and sequencing, but maintaining a focus on the application. As a result, clusters of activity have focused on such topics as brain-related research, specific gene and protein activity, protein folding, molecular models, and apoptosis, which was identified as a hot topic from the same data by Griffiths and Steyvers (21).
Another interesting shift is shown by the dominant topics in Table 3. One might assume that papers would tend to cluster within PNAS topics, and that authors would cite heavily to papers of the same topic. Over time, this occurrence has proved to be less and less the case. The number of clusters with less than 30% of their papers belonging to a dominant topic has increased over time. This finding indicates either that coupling between PNAS topics is on the increase or that the perceived boundaries between these topics are becoming more fuzzy.
It is also interesting to consider the characteristics of PNAS topics. Topic assignments are made by authors rather than editors, yet both may wish to see characteristics by topic in that it may influence publishing choices. Second-level topics along with their counts and mean percentile rankings are shown in Table 4. The top 14 topics by percentile are all Biological Sciences topics. Medical Sciences and Cell Biology, although being two of the largest categories, rank highest. The largest category, Biochemistry, has a mean percentile of 49. Physical Sciences and Social Sciences categories all have mean percentiles under 50, which is not surprising for a journal centered in biochemistry.
Table 4. Summary of properties for PNAS topics, 1997-2001.
| Topic | No. of ALNR | Mean percentile | Times cited | Independence |
|---|---|---|---|---|
| Medical Sciences (BS) | 1,555 | 60.0 | 1,614 | 0.53 |
| Cell Biology (BS) | 1,239 | 57.5 | 1,206 | 0.43 |
| Pharmacology (BS) | 189 | 54.3 | 126 | 0.33 |
| Plant Biology (BS) | 489 | 53.3 | 486 | 0.69 |
| Genetics (BS) | 988 | 51.9 | 986 | 0.47 |
| Microbiology (BS) | 499 | 51.7 | 514 | 0.50 |
| Neurobiology (BS) | 1,358 | 51.5 | 1,098 | 0.72 |
| Physiology (BS) | 341 | 51.2 | 209 | 0.41 |
| Immunology (BS) | 865 | 51.0 | 730 | 0.67 |
| Biochemistry (BS) | 2,586 | 49.0 | 2,521 | 0.64 |
| Developmental Biology (BS) | 372 | 46.6 | 266 | 0.46 |
| Applied Biological Sciences (BS) | 95 | 46.5 | 67 | 0.15 |
| Biophysics (BS) | 640 | 46.3 | 798 | 0.59 |
| Agricultural Sciences (BS) | 44 | 45.3 | 39 | 0.64 |
| Computer Sciences (PS) | 10 | 42.5 | 5 | 0.00 |
| Evolution (BS) | 527 | 42.1 | 470 | 0.61 |
| Chemistry (PS) | 253 | 41.8 | 208 | 0.33 |
| Population Biology (BS) | 43 | 39.4 | 37 | 0.19 |
| Psychology (SS) | 124 | 33.9 | 80 | 0.56 |
| Ecology (BS) | 137 | 33.7 | 49 | 0.80 |
| Applied Physical Sciences (PS) | 42 | 33.3 | 11 | 0.36 |
| Engineering (PS) | 25 | 31.2 | 11 | 0.27 |
| Geophysics (PS) | 26 | 27.5 | 4 | 0.50 |
| Anthropology (SS) | 83 | 25.7 | 74 | 0.57 |
| Social Sciences (SS) | 11 | 25.1 | 4 | 0.75 |
| Geology (PS) | 49 | 24.6 | 9 | 0.44 |
| Statistics (PS) | 20 | 22.5 | 15 | 0.20 |
| Physics (PS) | 46 | 22.3 | 21 | 0.43 |
| Applied Mathematics (PS) | 54 | 16.4 | 22 | 0.50 |
| Astronomy (PS) | 14 | 11.2 | 3 | 1.00 |
| Mathematics (PS) | 42 | 7.0 | 5 | 1.00 |
| Economic Sciences (SS) | 15 | 4.3 | 3 | 0.67 |
BS, Biological Sciences; PS, Physical Sciences; SS, Social Sciences.
Mapping of literatures in the ways shown here: i.e., generation of visual maps, clustering, and analysis of the evolution of topics over time, is amenable to discipline level or structural studies as well as to the single journal study given here.
Import-Export Within PNAS. Diffusion of information between scientific disciplines is a relatively new topic of study. The largest of these studies to date looked at 644,000 articles from the 1999 CDROM version of the SCIE (17). Fifteen broad categories of science were defined (e.g., Basic Life Sciences, Biology, Physics, etc.), and the percentage of references from each category to the others was calculated. Physics was found to be the most independent, whereas Biology was nearly as dependent on Basic Life Sciences as upon itself.
Import and export between fields can also be investigated within a single multidisciplinary journal such as PNAS. Here, we look at diffusion between PNAS topics as defined in Table 4. The normalized (number of citations to topic divided by the number of citations to all topics) diagonal of the citation matrix (data not shown) is defined as an index of independence (17, 51), and is given in Table 4. A higher independence value indicates a larger fraction of references given to papers within topic. Independence is thought to correlate with the basic or applied nature of a field, with high independence indicating a basic science (17). A reordering of Table 4 by independence reveals that, in general, the topics order themselves from basic to applied. Plant Biology, Neurobiology, Biophysics, and Biochemistry are all more basic fields than Genetics, Developmental Biology, Cell Biology, or Physiology. For comparison, Rinia et al. (17) found that for the entire Science Citation Index for 1999, Basic Life Sciences had an independence value of 0.63, whereas the more applied Biology had a value of 0.36. However, they also found that Clinical Life Sciences had an independence of 0.67. The PNAS Medical Sciences topic has a value of 0.53, indicating that PNAS Medical Sciences papers may be more enabling (ability to export) than medical sciences papers overall. The full citation matrix shows that Medical Sciences receives >10% of the citations from 11 of the other PNAS topics, including the nonbiological Computer Sciences and Applied Mathematics. The most enabling topic, receiving large fractions of citations from multiple topics, is Biochemistry, which is consistent with the common perception that it forms the core of PNAS publications. Chemistry is anomalous in that it cites heavily to Biochemistry and Biophysics, with an independence of 0.33. The corresponding value from Rinia et al. (17) is 0.63. Thus, the PNAS Chemistry topic must be an evolved brand of chemistry that has more to do with application of biology than chemistry at large.
Diffusion between PNAS and other journals could also be examined by using a similar analysis on the citations to and from PNAS.
Conclusions
Impact and funding indicators and citation-based maps have been used to provide a characterization of publication in PNAS from 1982 to 2001. The types of maps and analysis shown here can be applied at many levels: single journal, single discipline, groups of disciplines, etc., given appropriate data. Accurate funding data, and especially, accurate records of the relationship between individual grants and papers is needed. Given these data, similar analyses could be performed for large fields of science, or perhaps, even the whole of science. The ultimate goal is to provide an interactive means of exploring and evaluating scientific and technical information (publications, grants, etc.) to help us obtain answers to questions of strategic importance and aid the innovation process.
Acknowledgments
I thank the Laboratory Directed Research and Development Program, Sandia National Laboratories, and Katy Börner, Richard Shiffrin, and several anonymous reviewers for insightful comments and suggestions. This work was supported by the U.S. Department of Energy under Contract DE-AC04-94AL85000.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Mapping Knowledge Domains,” held May 9-11, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Sciences and Engineering in Irvine, CA.
Abbreviations: SCIE, Science Citation Index Expanded; ISI, Institute for Scientific Information; ALNR, articles, letters, notes, and reviews; PI, principal investigator.
Footnotes
These data are extracted from Science Citation Index Expanded [Institute for Scientific Information, Inc. (ISI), Philadelphia, PA; Copyright ISI]. All rights reserved. No portion of these data may be reproduced or transmitted in any form or by any means without the prior written permission of ISI.
National Institute on Aging funding is ≈6% of the National Institutes of Health total annually. The 1,862 National Institute on Aging papers are 7.4% of the total National Institutes of Health papers in the PNAS data set.
References
- 1.Garfield, E. (1955) Science 122, 108-111. [DOI] [PubMed] [Google Scholar]
- 2.Garfield, E. (1970) Nature 227, 669-671. [DOI] [PubMed] [Google Scholar]
- 3.Price, D. J. D. (1963) Little Science, Big Science (Columbia Univ. Press, New York).
- 4.Price, D. J. D. (1965) Science 149, 510-515. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter, M. P. & Narin, F. (1973) J. Am. Soc. Inf. Sci. 24, 425-436. [Google Scholar]
- 6.Börner, K., Chen, C. & Boyack, K. W. (2003) Annu. Rev. Inf. Sci. Technol. 37, 179-255. [Google Scholar]
- 7.Kim, S. K., Lund, J., Kiraly, M., Duke, K., Jiang, M., Stuart, J. M., Eizinger, A., Wylie, B. N. & Davidson, G. S. (2001) Science 293, 2087-2092. [DOI] [PubMed] [Google Scholar]
- 8.Card, S., Mackinlay, J. & Shneiderman, B. (1999) Readings in Information Visualization: Using Vision to Think (Morgan Kaufmann, San Francisco).
- 9.Chen, C. (2003) Mapping Scientific Frontiers: The Quest for Knowledge Visualization (Springer, London).
- 10.Newman, M. E. J. (2001) Proc. Natl. Acad. Sci. USA 98, 404-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glanzel, W. (2001) Scientometrics 51, 69-115. [Google Scholar]
- 12.White, H. D. & McCain, K. W. (1998) J. Am. Soc. Inf. Sci. 49, 327-356. [Google Scholar]
- 13.Salton, G., Yang, C. & Wong, A. (1975) Comm. ACM 18, 613-620. [Google Scholar]
- 14.Deerwester, S., Dumais, S. T., Landauer, T. K., Furnas, G. W. & Harshman, R. A. (1990) J. Am. Soc. Inf. Sci. 41, 391-407. [Google Scholar]
- 15.Landauer, T. K., Laham, D. & Derr, M. (2004) Proc. Natl. Acad. Sci. USA 101, 5214-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bassecoulard, E. & Zitt, M. (1999) Scientometrics 44, 323-345. [Google Scholar]
- 17.Rinia, E. J., van Leeuwen, T. N., Bruins, E. E. W., van Vuren, H. G. & van Raan, A. F. J. (2002) Scientometrics 54, 347-362. [Google Scholar]
- 18.Callon, M. & Law, J. (1983) Social Science Information 22, 191-235. [Google Scholar]
- 19.Leydesdorff, L. (1997) J. Am. Soc. Inf. Sci. 48, 418-427. [Google Scholar]
- 20.Noyons, E. C. M., Moed, H. F. & Luwel, M. (1999) J. Am. Soc. Inf. Sci. 50, 115-131. [Google Scholar]
- 21.Griffiths, T. L. & Steyvers, M. (2004) Proc. Natl. Acad. Sci. USA 101, 5228-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erosheva, E., Fienberg, S. & Lafferty, J. (2004) Proc. Natl. Acad. Sci. USA 101, 5220-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Batagelj, V. & Mrvar, A. (1998) Connections 21, 47-57. [Google Scholar]
- 24.Lin, X. (1997) J. Am. Soc. Inf. Sci. 48, 40-54. [Google Scholar]
- 25.Boyack, K. W., Wylie, B. N. & Davidson, G. S. (2002) J. Am. Soc. Inf. Sci. Technol. 53, 764-774. [Google Scholar]
- 26.Wise, J. A. (1999) J. Am. Soc. Inf. Sci. 50, 1224-1233. [Google Scholar]
- 27.Morris, S. A., Yen, G., Wu, Z. & Asnake, B. (2003) J. Am. Soc. Inf. Sci. Technol. 54, 413-422. [Google Scholar]
- 28.Chen, C. & Kuljis, J. (2003) J. Am. Soc. Inf. Sci. Technol. 54, 453-446. [Google Scholar]
- 29.Godin, B. (2003) Res. Policy 32, 679-691. [Google Scholar]
- 30.National Science Board. (2002) Science and Engineering Indicators 2002 (National Science Foundation, Arlington, VA).
- 31.King, J. (1987) J. Inf. Sci. 13, 261-276. [Google Scholar]
- 32.Martin, B. R. & Irvine, J. (1983) Res. Policy 12, 61-90. [Google Scholar]
- 33.Irvine, J. & Martin, B. R. (1984) Foresight in Science: Picking the Winners (Frances Pinter Publications, London).
- 34.Frame, J. D. & Narin, F. (1976) Fed. Proc. 35, 2529-2532. [PubMed] [Google Scholar]
- 35.McAllister, P. R. & Narin, F. (1983) J. Am. Soc. Inf. Sci. 34, 123-131. [DOI] [PubMed] [Google Scholar]
- 36.Lewison, G. & Dawson, G. (1998) Scientometrics 41, 17-27. [Google Scholar]
- 37.Lewison, G. (1998) Gut 43, 288-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler, L. (2001) Res. Eval. 10, 59-65. [Google Scholar]
- 39.Boyack, K. W. & Börner, K. (2003) J. Am. Soc. Inf. Sci. Technol. 54, 447-461. [Google Scholar]
- 40.Ingwersen, P. & Christensen, F. H. (1997) J. Am. Soc. Inf. Sci. 48, 205-217. [Google Scholar]
- 41.Hood, W. W. & Wilson, C. S. (2001) J. Am. Soc. Inf. Sci. Technol. 52, 1242-1254. [Google Scholar]
- 42.Seglen, P. (1997) Allergy (Copenhagen) 52, 1050-1056. [DOI] [PubMed] [Google Scholar]
- 43.Seglen, P. (1997) Br. Med. J. 314, 498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narin, F. & Hamilton, K. S. (1996) Scientometrics 36, 293-310. [Google Scholar]
- 45.Lewison, G., Dawson, G. & Anderson, J. (1995) in 5th International Conference of the International Society for Scientometrics and Informetrics, eds. Koenig, M. E. D. & Bookstein, A. (Learned Information, Medford, NJ), pp. 255-263.
- 46.Scheffé, H. (1953) Biometrika 40, 87-104. [Google Scholar]
- 47.Kessler, M. M. (1963) Am. Doc. 14, 10-25. [Google Scholar]
- 48.Small, H. (1997) Scientometrics 38, 275-293. [Google Scholar]
- 49.Davidson, G. S., Wylie, B. N. & Boyack, K. W. (2001) in 7th IEEE Symposium Inform Visualization (InfoVis 2001), eds. Andrews, K., Roth, S. & Wong, P. C., (IEEE Computer Society, Los Alamitos, CA), pp. 23-30.
- 50.Davidson, G. S., Hendrickson, B., Johnson, D. K., Meyers, C. E. & Wylie, B. N. (1998) J. Intell. Inform. Syst. 11, 259-285. [Google Scholar]
- 51.Urata, H. (1990) Scientometrics 18, 309-319. [Google Scholar]