Abstract
Gastric cancer is the most common cancer in Asia and most developing countries. Despite the use of multimodality therapeutics, it remains the second leading cause of cancer death in the world. To identify the molecular underpinnings of gastric cancer in the Asian population, we applied an RNA-sequencing approach to gastric tumor and noncancerous specimens, generating 680 million informative short reads to quantitatively characterize the entire transcriptome of gastric cancer (including mRNAs and microRNAs). A multi-layer analysis was then developed to identify multiple types of transcriptional aberrations associated with different stages of gastric cancer, including differentially expressed mRNAs, recurrent somatic mutations and key differentially expressed microRNAs. Through this approach, we identified the central metabolic regulator AMPK-α as a potential functional target in Asian gastric cancer. Further, we experimentally demonstrated the translational relevance of this gene as a potential therapeutic target for early-stage gastric cancer in Asian patients. Together, our findings not only provide a valuable information resource for identifying and elucidating the molecular mechanisms of Asian gastric cancer, but also represent a general integrative framework to develop more effective therapeutic targets.
Keywords: RNA-sequencing, integrative analysis, microRNA, AMPK, gastric cancer
Introduction
Gastric cancer is the fourth most common cancer and the second leading cause of cancer-related deaths in the world (1). The incidence of cancer affecting the distal stomach has significantly declined over the past 40 years, but the incidence of proximal gastric and gastroesophageal junction adenocarcinoma in the United States and Europe has increased at a rate substantially exceeding that of prostate cancer, brain cancer, or melanoma (2). Moreover, the 5-year relative survival rate of patients with gastric cancer has not improved significantly in recent decades, remaining at approximately 20-30%.
The high mortality rate of gastric cancer is due largely to late-stage diagnosis of the cancer and a lack of effective medical treatment options. Treatment often consists of drug combinations that have provided survival advantages for patients with other cancer types (3, 4). Thus, there is clearly a need for new therapies specifically targeting gastric cancer. A comprehensive molecular profile of gastric cancer would provide important information about the disease pathways and targets that could facilitate the development of new therapeutic agents and strategies (5, 6).
So far, most transcriptional profiling studies in gastric cancer have used hybridization microarrays. For example, aberrant microRNA (miRNA) expression signatures in gastric tumor samples from Japanese and Italian patients have been reported using miRNA expression microarrays (7, 8), and mRNA expression signatures from Chinese patients have been recently reported using exon microarrays (9). RNA sequencing (RNA-seq) technology is rapidly supplanting hybridization-based approaches. This approach not only enables investigators to quantify gene expression levels, but to simultaneously assess alternative splicing and gene fusion events and to detect nucleotide variations in transcribed regions (10, 11). Thus, multidimensional datasets from a single platform can generate a rich profile of cancer progression and development. In particular, whole-transcriptome RNA-seq provides a detailed and precise view of the entire spectrum of expressed transcripts for both mRNA and non-coding RNA.
In this study, we generated comprehensive mRNA and miRNA profiles for Asian gastric tumors. First, we performed transcriptome-wide, unbiased analyses of the RNA-seq data to identify different types of transcriptional aberrations (mRNA, miRNA and somatic mutation candidates) to leverage the existing knowledge of the pathogenesis. Second, integrating the results of our multi-layer analyses, we identified a potential role in cancer progression for PRKAA2, which encodes AMPK-α2, a subunit of the AMPK serine/threonine protein kinase complex involved in the regulation of cellular and organismal metabolism. We experimentally validated the expression changes of PRKAA2 between early- and late-stage gastric cancers. Third, through functional studies in gastric cancer cell lines, we demonstrated the translational relevance of PRKAA2 as a potential therapeutic target. Our work provides a valuable information basis for elucidating the molecular mechanisms of gastric cancer progression, and also represents a general framework for the more effective development of disease-focused therapeutic targets.
Methods and Materials
Sample collection and characterization
This is a retrospective study conducted in archival fresh frozen human tissue specimens obtained from the National Research Resource Bank Program of the Korea Science and Engineering Foundation in the Ministry of Science and Technology. Patients consented to the use of the tissue specimens for research purpose, and the Institutional Review Boards of the College of Medicine, Yonsei University (Seoul, Korea), and the University of Texas MD Anderson Cancer Center (Houston, TX, USA) approved the use of the specimens. Histological classification and tumor stage were reviewed by a pathologist at the Gene Bank at Yonsei University Severance Hospital. Among 82 initially enrolled gastric cancer cases, 24 tumors and 6 noncancerous gastric tissues that met the criteria (sufficient amount and quality of RNA) were included in this study. Clinical and histopathological characteristics obtained from the patients are summarized in Supplementary Table 1.
RNA-seq library preparation and SOLiD sequencing
The Miravana Kit (Ambion/Applied Biosystems, Foster City, CA, USA) was used to isolate total RNA according to the vendor’s protocol. The whole-transcriptome-sequencing (WT-seq) and small RNA-seq libraries were prepared using the small RNA expression kit (SREK, PN 4397682) of Applied Biosystems Inc. (ABI), based on SOLiD WT and small RNA standard protocols provided by ABI. The individual prepared “barcode” libraries were quantified and pooled equally together for multiplexing. The sequencing runs were performed on SOLiD v 3.0 for both WT-seq and small RNA-seq. WT-seq samples were sequenced in 1/4 slide per sample using 50-nucleotide (nt) single tags; and small RNA-seq samples were sequenced in 1/10 slide per sample using 35-nt single tags. Detailed information is provided in the Supplementary Materials and Methods.
Computational analyses of RNA-seq data
WT-seq short reads were mapped to the human reference genome (hg19) and exon junctions (defined as RefSeq gene annotation) using the ABI Bioscope™ (version 1.21) WT-seq analysis pipeline with default parameters. The reads mapped to the sequences that were not of biological interest, such as rRNAs, tRNAs, and repetitive elements, were first filtered. Then mapped reads with mapping quality ≥10 were defined as uniquely mapped reads and used in the downstream analysis. The SOLiD System Small RNA Analysis Pipeline Tool (corona RNA2MAP version 0.50) was used to analyze small RNA-seq reads: after filtering, the reads were mapped to mature miRNAs in miRBase (verson13.0) (12) and the human reference genome, respectively.
To identify gastric-cancer-related differentially expressed genes, the “reads per kilobase of exon per million mapped sequence reads” (RPKM) (13) values of the human RefSeq genes were calculated using the RNA-seq flow in the Partek® Genomics Suite™ (version 6.5 beta, Partek Inc., St. Louis, MO, USA) and then log transformed. Single-factor analysis of variance (ANOVA) was used to detect differentially expressed genes among 18,890 protein-coding genes: P < 9.5×10−4 (false discovery rate [FDR] < 0.05) was used as the cut-off in the five-group comparison (normal, tumor stage I, II, III or IV); and P < 7×10−4 was used in the four-stage comparison (tumor stage I, II, III or IV). A similar analysis was performed on 2,569 long non-coding RNAs. Cross-platform gene expression comparison was performed with a recent microarray study in gastric cancer (14). A gene ontology analysis was performed using GOminer (15) and a disease association analysis was performed using Ingenuity Pathway Analysis software (version 7.0). Recurrent somatic mutations were identified based on a recent exome-sequencing study on Asian gastric cancer patients (16). MiRNA expression was quantified as reads per million (RPM) of reads mapped to known miRNAs, and key differentially expressed miRNAs were defined based on their differential expression and expression anti-correlation with their potential target genes. Detailed information is provided in the Supplementary Materials and Methods.
Experiments on biological function
Cell culture
Human NCI-N87 and AGS gastric cancer cells were obtained from the American Type Culture Collection (http://www.atcc.org/). The study was conducted within 6 months of resuscitation, and they were cultured in RPMI-1640 (Cellgro) and 10% fetal calf serum (FCS; Hyclone) at 37°C in 5% CO2. ATCC uses short tandem repeat (STR) profiling. For a hypoxia assay, culture flasks were incubated for various times at 37°C in humidified air, 5% (normoxia), or 1% O2, 5% CO2, and 94% N2 (hypoxia) using an in vivo Hypoxia Workstation 500 with Ruskin hypoxic gas mixer (Biotrace International, Bothell, WA, USA). Cells (2.5×105) were seeded and incubated under normoxic conditions to 70% confluence, and then incubated under hypoxic conditions for 18 h in the presence or absence of metformin at 10 mM concentration. The NCI-N87-HRE cells were established according to the manufacturer’s protocol, Cignal HIF Reporter (luc) (SA Biosciences, QIAGEN Co., Frederick, MD, USA). Firefly luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, Sunnyvale, CA, USA) according to the manufacturer’s protocol.
Western blotting
Cells were grown under hypoxic conditions in the presence or absence of 10 mM metformin. The cells were washed twice in a phosphate-buffered saline solution and Western blotting was conducted, as previously described (17).
Real-time reverse transcription-PCR
Total RNA was isolated from cell lysates using the PARIS kit (Ambion/Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. Next, TaqMan quantitative reverse transcription (RT)-PCR was performed on the ABI 7300 system using the TaqMan one-step RT-PCR Master Mix kit and predesigned primer/probe pairs for PRKAA1, PRKAA2, PRKAB1, PRKG1, STK11, HNF4α, and β2-microglobulin (Applied Biosystems, Foster City, CA, USA). Normalization procedures and analyses were carried out with β2-microglobulin using the 2(−ΔΔCt) method as the internal reference (18) and using Applied Biosystems GeneAmp 5700 SDS software. All measurements were performed in triplicate.
Small interfering RNA transfection
Small interfering RNA (siRNA) SMARTpool sequences were obtained from Dharmacon/Thermo Fisher Scientific (Waltham, MA, USA). The cells were transfected with 25 nM siRNA-PRKAA2, siRNA-PRKAB1, siRNA-PRKAG1, siRNA-PRKAG2, or a siRNA non-targeting control using Dharma-FECT 1 lipid transfection reagent. The transfection medium was removed after 24 h and replaced with fresh medium, and the cells were grown in 5% CO2 at 37°C for an additional 48-72 h. RT-PCR and/or Western blot analyses were performed to confirm target knockdown by siRNA. The transfected cells were treated with metformin and cultured under hypoxic conditions for an additional 18 h.
Statistical analysis
For the experiments on individual genes in this section, statistical significance (P < 0.05) was determined using a student’s t test to compare data points with control data.
Results
Overview of the gastric cancer RNA-seq data
Using Life Technologies SOLiD™ sequencing platform, we performed transcriptome-wide profiling of gastric cancer samples from 30 anonymous, unrelated Asians of both sexes. Included were six noncancerous gastric tissue samples and 24 gastric tumor samples that represented stages I through IV of tumor development (Clinical characteristics of patients are shown in Supplementary Table 1). Applying two protocols that complementarily cover RNA fragments of different sizes from each sample resulted in two parallel large-scale datasets that allowed us to simultaneously measure mRNA and miRNA expression.
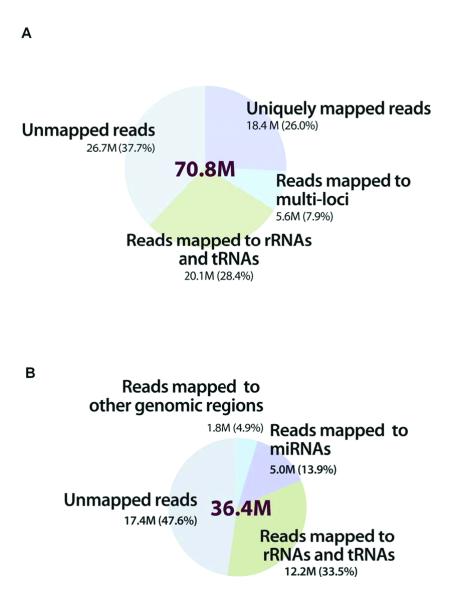
From the WT-seq protocol we generated a WT-seq dataset of 2.1 billion 50-nt short reads from the 30 samples (mean of 70.8 million; Supplementary Table 1). Using the ABI Bioscope™ WT-seq analysis pipeline, 62.3% of the short reads, on average, were mappable: 28.4% were mapped to sequences of no biological interest for this study (e.g., rRNAs and tRNAs), so we removed them from further analysis. On average, 26.0% (18.4 million per sample) of the short reads were uniquely mapped to the human reference genome (hg19) or exon-junction sequences (0.5 million per sample); we used these in the downstream analysis (Fig. 1A and Supplementary Table 1).
Figure 1. Distribution of short reads in the WT-seq and small RNA-seq datasets when mapped to the human reference genome.
(A) WT-seq dataset of 2.1 billion 50-nt short reads from the 30 samples. Using the ABI Bioscope™ (version 1.21) WT-seq analysis pipeline, 62.3% of the short reads were mappable: 28.4% were mapped to the sequences of no biological interest (e.g., rRNAs and tRNAs) and were therefore filtered; and 26.0% (18.4 million per sample) were uniquely mapped to the human reference genome (hg19) or exon junctions. (B) Small RNA-seq dataset of 894 million 35-nt short reads, mean of 36.4 million reads from the 25 samples. Using the SOLiD System Small RNA Analysis Pipeline Tool, 52.4% of the short reads were mappable; 33.6% mapped to sequences of no biological interest were removed from further analysis. On average 13.9% (5.1 million per sample) were mapped to known miRNAs in the miRBase database (version 13.0).
Applying the second small RNA-seq protocol to 19 gastric tumor samples (5 of the original 24 yielded insufficient sample amounts) and 6 noncancerous gastric tissue samples resulted in a small RNA-seq dataset of 894 million 35-nt short reads (mean of 36.4 million; Supplementary Table 1). Using the SOLiD System Small RNA Analysis Pipeline Tool, 52.4% of the short reads, on average, were mappable: 33.6% were mapped to sequences of no biological interest, as in the whole-transcriptome analysis, and we removed them from further analysis. On average, 13.9% (5.1 million per sample) were mapped to known miRNAs in the miRBase database (Fig. 1B and Supplementary Table 1) (version 13.0) (12). To our knowledge, this is one of the largest RNA-seq datasets on human cancer available to date.
Identification of gastric-cancer-related differentially expressed genes
With millions of short reads mapped to the human genome/transcriptome, we quantified the expression levels of known genes in each sample using the conventional parameter, RPKM (13). Among 18,890 annotated RefSeq coding genes, 15,421 genes, on average, per sample had detectable expression (RPKM > 0.05, about one mapped read for a gene with 1kb exons) (Supplementary Table 1). To evaluate the reproducibility of our RNA-seq approach on expression quantification, for a subset of samples with available microarray data from a previous study (14), we compared the gene expression data from the two platforms (microarray and RNA-seq) and found sample-by-sample correlations, RS = 0.73 ± 0.04 (Spearman rank correlation, Supplementary Fig. 1A) that are similar to those reported in the literature (11, 13). The clustering pattern from an unsupervised analysis largely reflected the disease/staging status of the samples under survey (Supplementary Fig. 2). Moreover, a principal component analysis of the global gene expression profiles showed that gastric cancer samples differed from gastric intestinal stromal tumor samples, thereby confirming the pathological classification of our tumor samples (Supplementary Fig. 3).
To identify gastric-cancer-related differentially expressed genes, we performed a single-factor ANOVA on the RPKM (log-scale transformed) data of 18,890 protein-coding genes across five groups (tissue type = normal or stage I, II, III or IV as the independent variable). At FDR < 0.05 (19) (P < 9.5×10−4), we identified 356 differentially expressed genes (Supplementary Table 2). As an independent validation, we performed the same analysis on a published microarray dataset (14) consisting of 83 Asian gastric samples taken from normal tissue and four tumor stages. In that analysis, 78% of the differentially expressed genes identified by our RNA-seq analysis showed significant expression changes in the same direction (tumor vs. normal) based on the microarray data; whereas only 17% would be expected if a same-size gene set were randomly chosen (P < 1×10−4; Supplementary Fig. 1B). In addition, we performed a similar analysis on 2,569 long non-coding RNA genes and identified seven differentially expressed genes (Supplementary Table 2).
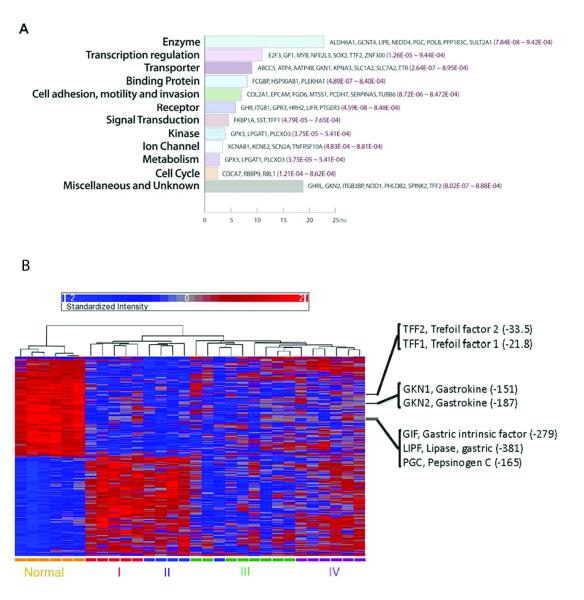
To identify biological characteristics of the differentially expressed genes, we performed a gene ontology analysis using GOminer (15). We found that the 356-gene dataset showed high enrichment of genes involved in the biological processes of digestion and phosphagen metabolism, and the molecular functions of transmembrane transport and ATPase activity (Table 1). Using the Ingenuity Pathway Analysis software, we identified 101 of the 356 genes (28.4%) as associated with cancer (Fisher’s exact test, P = 2.8×10−4), and 46 of them (12.9%) as related to gastrointestinal disease (Fisher’s exact test, P = 1.1×10−4). Fig. 2A categorizes these 356 genes by biological and molecular functions (Supplementary Table 2). Since highly expressed genes tend to be identified in RNA-seq-based differential analysis (20), we used 12,213 genes with the same expression distribution as that of the 356 genes, rather than the whole gene set, as a reference set in the above analyses (see Supplementary Methods and Materials). As shown in Fig. 2B, these differentially expressed genes provided substantial power for classifying normal vs. tumor tissue, as well as for distinguishing different clinical stages of the gastric tumors (although the distinction between stages III and IV became a little fuzzy). In comparison with the expression levels in the normal tissue, genes related to gastrointestinal disease were downregulated in four tumor stages (Fig. 2B). For example, loss of expression of gastrokines GKN1 and GKN2 occurs frequently in gastric adenocarcinoma, which is associated with shorter overall survival in the intestinal subtype of distal gastric cancer (21). Our results showed a dramatic tumor-related loss of expression level in both GKN1/2 and trefoil factor family peptides 1/2 (TFF1/2), supporting their potential use as predictive biomarkers (9).
Table 1.
Gene ontology analysis of gastric-cancer-related differentially expressed genes.
| GO term | Description | Enrichment fold |
LOG10 (P) | False discovery rate |
|---|---|---|---|---|
| Biological process | ||||
| GO:0007586 | Digestion | 6.82 | −6.39 | 0.002 |
| GO:0006599 | phosphagen metabolism |
19.35 | −3.55 | 0.059 |
| GO:0006599 | ion transport | 2.02 | −3.80 | 0.071 |
| Molecular function | ||||
| GO:0022891 | substrate-specific transmembrane transporter |
1.98 | −3.66 | 0.052 |
| GO:0022804 | transmembrane transporter |
2.63 | −3.75 | 0.055 |
| GO:0031420 | metal ion binding | 3.11 | −3.79 | 0.061 |
| GO:0015662 | ATPase activity | 6.45 | −3.57 | 0.059 |
Biological characteristics of the 356 differentially expressed genes identified using GOminer. To adjust the bias towards highly expressed genes in detecting differential expression analysis, we used as the reference set 12,213 genes with the same expression distribution as that of the 356 differentially expressed genes.
Figure 2. Biological and molecular characteristics of 356 differentially expressed genes related to gastric cancer.
These genes were identified with a single-factor analysis of variance (ANOVA) on RPKM (log-scale transformed) across five groups (tissue type = normal or stage I, II, III or IV as the independent variable) (at FDR < 0.05 and P < 9.5×10−4). (A) Biological and molecular functions of representative genes known to be related to gastric cancer. Their raw P-values in the ANOVA are shown in red. (B) The clustering heatmap of 30 samples based on the 356 differentially expressed genes, generated with Partek® Genomics Suite™ v 6.5. Each column is labeled with different colors according to the sample type; several key genes related to gastrointestinal disease are highlighted with their P-values and fold changes (FC) for differential expression.
In addition, we performed a similar analysis on the 24 tumor samples using single-factor ANOVA (tissue type = stage I, II, III or IV). In general, the expression variation among tumor samples was much less than that between normal and tumor samples. At P < 7×10−4, we identified 28 genes with significant stage-specific expression change. Based on these genes, the tumor samples clearly clustered according to their stage, with the largest distinction between stages I, II vs. III, IV (Supplementary Fig. 4).
Identification of recurrent somatic mutation candidates in gastric cancer
To take full use of our RNA-seq data, we also made efforts to identify somatic mutation candidates in gastric cancer. Since we did not sequence the normal DNA from the same patients, we had a very limited power to infer somatic mutations based on our RNA-seq data alone. Instead, we took advantage of a recent exome-sequencing study on Asian gastric cancer patients (16) and obtained a list of 2,651 somatic mutations with a potential functional effect (nonsynonymous/nonsense mutations and those at splicing sites). Among these reported somatic mutation positions, we detected the exact mutant alleles at 14 mutation positions in our WT-seq data, suggesting that they are recurrent somatic mutations (Supplementary Table 3). TP53 is the only gene with multiple recurrent mutation candidates (four mutations), consistent with its known high mutation frequency in gastric cancer (22). In addition, we detected 92 potential recurrent coding somatic mutations based on the COSMIC database (23) (although these recurrent mutations may not be specific to gastric cancer) (Supplementary Table 4).
Identification of key differentially expressed miRNAs related to gastric cancer
In parallel with the analysis of gene expression using the WT-seq dataset, we used the small RNA-seq dataset to quantify the expression levels of known miRNAs in each sample using RPM, a measure analogous to RPKM for coding genes. Among 698 annotated, non-redundant mature miRNAs in the miRBase (version 13.0), on average, ~60% of the miRNAs had mapped reads. The observation that a large proportion of annotated miRNAs (40%) have zero or very low expression levels may be partially due to the inflation of the current miRBase annotation, as suggested by recent studies (24, 25). Therefore, we focused on 402 miRNAs with reliable expression (max RPM > 4 in 25 samples) in the subsequent analysis (see Supplementary Methods and Materials).
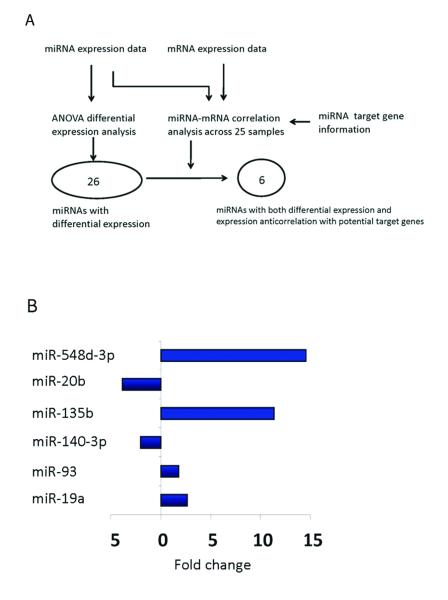
To identify the miRNAs that play a key role in gastric tumor development, we reasoned that (i) the key miRNAs themselves should show significant expression variations across different sample groups; and (ii) they should have detectable repression effects on the expression of their target genes (26). Therefore, we performed a two-step analysis. First, we performed single-factor ANOVA (tissue type as the independent variable) on the RPM data (log-scale transformed) across five groups (normal tissue and four tumor stages). At P < 0.01 (FDR < 0.15), 26 miRNA genes showed significant differential expression: nine upregulated and 17 downregulated (Fig. 3A and Supplementary Table 5). One of the upregulated miRNAs, miR-21 is the most commonly upregulated miRNA in both solid and hematological tumors (8). Seven of the 26 differentially expressed miRNAs (Supplementary Table 5) were also identified by a recent microarray study on the miRNA biomarkers for the progression/prognosis of gastric cancer using samples from Japanese patients (7).
Figure 3. Integrative approach to identify key differentially expressed miRNAs related to gastric cancer.
(A) Two criteria were used to identify key differentially expressed miRNAs: based on the ANOVA on miRNA expression data, at P < 0.01, 26 miRNA genes showed significant differential expression; and integrating miRNA expression, mRNA expression and miRNA target information, at P < 0.01, six out of the 26 miRNAs showed significant anti-correlation with their potential target genes. As a result, the six miRNAs were defined as key differentially expressed miRNAs related to gastric cancer. (B) The expression fold change of the six miRNAs in the tumor samples related to normal samples.
Second, for each of the 26 miRNAs, we used Spearman’s rank correlation (Rs) to quantify its expression correlations with protein-coding genes across 25 samples with both the available coding gene and the miRNA expression data. We then tested whether the Rs values of its potential target genes were significantly lower than those of other genes using Wilcoxon’s rank sum test, resulting in six miRNAs showing significant anticorrelation with their potential targets (Wilcoxon’s rank sum test P < 0.01, see Supplementary Methods and Materials). We defined these six miRNAs as key differentially expressed miRNAs of Asian gastric cancer (Fig. 3B).
Integrative analysis suggests a potential role of PRKAA2, an AMPK activator in early-stage gastric cancer
To identify candidate genes with the highest potential functional impact in gastric tumorigenesis, we surveyed all three types of transcriptional aberrations: (i) differentially expressed genes; (ii) genes related to recurrent somatic mutation candidates; and (iii) potential target genes of key differentially expressed miRNAs. Through a simple scoring analysis, PRKAA2 (AMPK-α2) was the only gene identified by all the three criteria, suggesting that it is a potential key modulator in gastric cancer progression (Supplementary Fig. 5). PRKAA2 is the catalytic subunit of AMP-activated protein kinase, which is a heterotrimer consisting of a catalytic α-subunit and regulatory non-catalytic β- and γ-subunits, each with two or three isoforms (27). AMPK is an energy-sensing enzyme and plays a central role in the regulation of energy homeostasis (28).
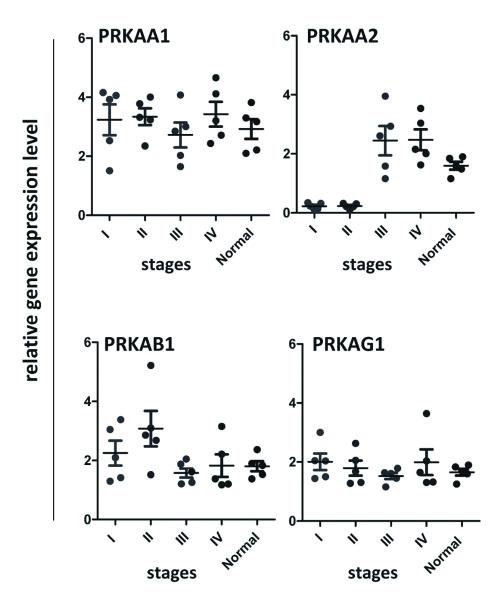
The most compelling evidence is that PRKAA2 is among the most highly differentially expressed genes identified in both 5-group and 4-stage expression analyses (5-group ANOVA, P = 4.65×10−6; 4-stage ANOVA, P = 4.7×10−4; only 22 genes were identified by both analyses). PRKAA2 showed a differential loss of mRNA level in tumor stage I/II relative to normal or tumor stage III/IV (Supplementary Fig. 6). Using qRT-PCR on an independent set of Asian gastric cancer samples (including normal, stage I through IV cases), we validated our findings by comparing the expression patterns of PRKAA2 along with the other three subunits of AMPK, PRKAA1, PRKAB1 and PRKAG1, for which we had not observed changes in the RNA-seq experiment (Fig. 4).
Figure 4. qRT-PCR validation of gene expression of AMPK subunits in an independent set of Asian gastric samples.
AMPK is characterized by a heterotrimeric structure, catalytic α1, 2-subunit and regulator β1,2-and γ1,2,3-subunits. Measurement of qRT-PCR expression level of PRKAA1, PRKAA2, PRKAB1 and PRKAG1 between normal tissue and tumors, stages I, II, III and IV, in an independent set of Asian gastric samples (n = 5 per group). Means of the measurements are shown with black lines.
Moreover, through our mRNA:miRNA integrative analysis (Fig. 4), PRKAA2 mRNA expression is inversely correlated with miR-19a expression (Pearson correlation test, P < 0.02), one of the six key differentially expressed miRNAs we identified. PRKAA2 is a strong candidate target gene for miR-19a since its 3′ UTR contains an extremely conserved target site (29).
AMPK activities are closely related to the balances between ATP and AMP, and we found that ATPases were significantly enriched in the differentially expressed genes (P = 4.7×10−4 in Table 1). In addition, PRKAA2 directly regulates TP53 (30, 31), the gene with multiple recurrent somatic mutation candidates detected in our study (although no recurrent mutations were found in PRKAA2). These observations further support the hypothesis that loss of PRKAA2 expression might carry important functional consequences for AMPK activity in the early stages of gastric cancer.
Experimental evaluation of PRKAA2 as a potential functional modulator of energy homeostasis in gastric cancer
The AMPK enzyme occupies a unique position in the signaling pathways that monitor energy consumption and balance between glycolysis and lipid metabolism (32). AMPK directly inhibits hepatocyte nuclear factor 4 alpha (HNF4α), which activates fatty acid and lipid metabolism. Additionally, AMPK inhibits the mTOR pathway, a major cancer growth promoting signaling that regulates hypoxia-inducible factor-1alpha (HIF-1α) (33, 34). Therefore, in order to explore the functional significance of PRKAA2 expression loss, we used both HNF4α and HIF-1α as downstream readouts for AMPK activity.
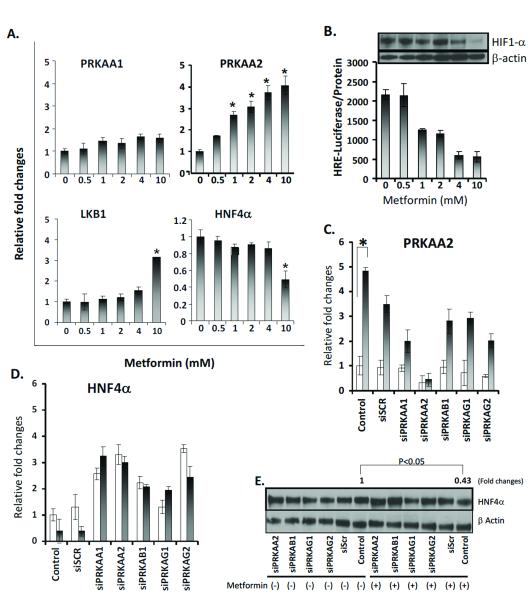
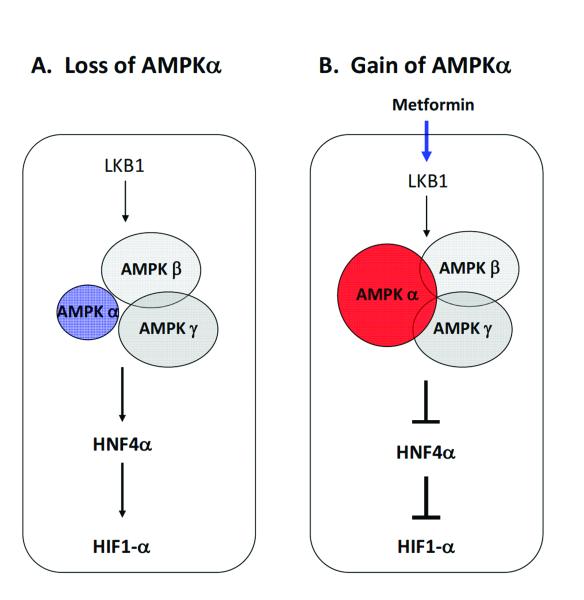
We treated two gastric cancer cell lines (NCI-N87 and AGS) with metformin, an AMPK activator (35). We found increased mRNA expression of LKB1 (liver kinase B1, STK11), which is known to mediate AMPK activity upon metformin treatment (36). As expected, we observed a concentration-dependent increase in PRKAA2 mRNA levels, reflecting the activation of AMPK signaling (Fig. 5A). Importantly, we observed decreased expression of HNF4α (Fig. 5A for NICN87 data; similar results were observed for AGS, data not shown). Meanwhile, we observed clear inhibition of the expression and transactivating activity of HIF-1α by metformin (Fig. 5B, Western blot panel). Furthermore, PRKAA2 knockdown with siRNA inhibited the decrease of HNF4α with metformin treatment at both the mRNA and protein levels (Fig. 5C, D and E). We also observed increases of HNF4α expression for siRNA-PRKAA1, siRNA-PRKAB1, siRNA-PRKAG1, and siRNA-PRKAG2 (the siRNAs targeting the mRNAs of other subunits of AMPK), regardless of metformin treatment (Fig. 5C, D and E). Consistently, in our RNA-seq data, the expression levels of HNF4α and HIF-1α in stage I and II were significantly higher than those in stage III, IV and normal samples (t-test, HNF4α, P < 0.04 and HIF-1α, P < 0.02). Taken together, our results demonstrated the functional relevance of PRKAA2 loss for the AMPK signaling pathway, with downstream consequences that increase both HNF4α and HIF-1α. These data suggest that in early-stage gastric cancer, the loss of PRKAA2 may sustain tumor growth through the activation of HIF-1α (Fig. 6).
Figure 5. Analysis of metformin-activated AMPK function in gastric cancer cells.
NCI-N87 and NCI-N87-HRE cells were cultured for 18 h under hypoxic conditions (1% O2, 5% CO2, 94% N2). (A) NCI-N87 cells were treated in different concentrations of metformin. PRKAA2 expression was measured by RT-PCR and expressed relative to cells without metformin. Values are the means of three measurements. (B) NC-N87 cells transfected with HRE-luciferase treated at different concentrations of metformin. HRE-luciferase was used to measure the HRE response. Values are the means of three measurements. (Panel) Western blot of HIF-1α protein level in NCI-N87, treated at different metformin concentrations, β-actin as loading controls. (C, D) NCI-N87 cells transfected for 72 h with different siRNAs and treated without (open bars) or with 10 mM metformin (filled bars). PRKAA2 and HNF4α expression was measured by RT-PCR and expressed relative to cells treated without metformin (control). Values are the means of three measurements. *P < 0.05 compared to cells without metformin. (E) Western blot of HNF4α protein levels of cells transfected with different siRNAs, treated without or with 10 mM metformin.
Figure 6.
Consequences of activation and signaling by the AMP-activated protein kinase in gastric cancer cells.
Discussion
Compared with previous RNA-seq studies, our whole-transcriptome RNA-seq approach has several merits. First, we employed two protocols that complementarily cover RNA fragments of different sizes in the samples. Thus, we were able to quantify the expression of mRNA, long non-coding RNA and miRNA simultaneously, greatly facilitating the downstream integrative analysis. Second, we sequenced ribosome-depleted RNA samples rather than polyA-enriched RNA samples, generating a less biased view of the population of transcribed molecules (37, 38). As a result of this approach, the percentages of informative reads are relatively low because a considerable proportion of the sequenced short reads came from rRNAs or tRNAs. Nevertheless, due to the large number of total reads per sample, there were still sufficient numbers for our downstream analyses (18.4 million per sample for expression quantification of coding genes; and 5.1 million per sample for expression quantification of miRNAs). Third, our protocols generated strand-specific short reads, which allows for more accurate quantification of gene expression since antisense transcription is widespread in humans (39).
Many challenges exist for the interpretation of transcriptome profiling data, both within and across individual studies. In the present study, we first performed a multidimensional analysis to depict different types of transcriptional aberrations related to gastric cancer. Besides gene expression signatures, we took advantage of recently available somatic mutation data in gastric cancer and used our RNA-seq data to detect potential recurrent somatic mutations in transcribed regions, thereby obtaining additional information from RNA-seq data. Through integrating these analyses, we were able to pinpoint individual key genes for further functional investigation. Our study demonstrates the importance of multi-layer data integration, which may more effectively identify candidate genes than conventional single-dimensional analysis.
While our study provides valuable insights into gastric cancer progression, there are some limitations. First, our RNA-seq data were single-tag reads generated from fragment libraries and the read length is relatively short; therefore, we had limited power to study aberrant splicing and gene fusion events. A key extension to our study will be to perform transcriptome profiling using paired-end and longer reads. This would provide a more comprehensive view of the transcriptional aberrations. Second, a lack of normal tissue samples from the same patients who provided tumor samples limited our ability to detect differentially expressed genes as well as to identify de novo somatic mutations (e.g., distinguishing somatic mutations from polymorphisms and RNA editing changes). Third, our study is based on only Asian patients, so future studies on gastric cancer in other patient populations are needed.
Through a multi-dimensional and integrative analysis of RNA-seq data of Asian patients, we identified a potentially critical role of AMPKα in the early stages of gastric cancer. The reason for different expression levels between stage I/II vs. III/IV is unclear, and we speculate that late-stage tumor development may require higher energy-sensing enzymes. Through our metformin-based functional experiments, we further demonstrated the translational relevance of PRKAA2, which encodes a central component of the energy-sensing AMPK enzyme. Since the expression level of PRKAA2 significantly affects key signaling nodes regulating tumor metabolism and angiogenesis, and shows activation by metformin, a drug widely used to treat type II diabetes, PRKAA2 may represent a promising therapeutic target for early gastric cancer. Our functional evidence supporting an important role of PRKAA2 in gastric cancer is still preliminary, and further functional studies are essential to elucidate how PRKAA2 modulation contributes to gastric cancer progression and to evaluate whether this gene is an effective therapeutic target.
Supplementary Material
Acknowledgements
We thank Keith Baggerly, Ph.D., for statistical suggestions; Julie Izzo, M.D., for reading the manuscript; and LeeAnn Chastain for editorial assistance.
Financial Support This work was supported by the National Institutes of Health [grant numbers CA 95060, CA129616, CA 17094 and CA 98920, and a grant from the Perot Foundation (GP); CA143883 and CA016672 (JNW and HL)]; UTMDACC – G.S. Hogan Gastrointestinal Research Fund (HL); the University of Texas MD Anderson Research Trust, and by the Center for Targeted Therapy at MD Anderson Cancer Center (XL, CL).
Footnotes
Conflict of Interest The authors declare no conflict of interest related to this work.
References
- 1.Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–50. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- 2.Brown LM, Devesa SS, Chow WH. Incidence of adenocarcinoma of the esophagus among white Americans by sex, stage, and age. J Natl Cancer Inst. 2008;100:1184–7. doi: 10.1093/jnci/djn211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto C, Di Fabio F, Siena S, Cascinu S, Llimpe FLR, Ceccarelli C, et al. Phase II study of cetuximab in combination with FOLFIRI in patients with untreated advanced gastric or gastroesophageal junction adenocarcinoma (FOLCETUX study) Annals of Oncology. 2007;18:510–7. doi: 10.1093/annonc/mdl459. [DOI] [PubMed] [Google Scholar]
- 4.Shah MA, Ramanathan RK, Ilson DH, Levnor A, D’Adamo D, O’Reilly E, et al. Multicenter phase II study of irinotecan, cisplatin, and bevacizumab in patients with metastatic gastric or gastroesophageal junction adenocarcinoma. J Clin Oncol. 2006;24:5201–6. doi: 10.1200/JCO.2006.08.0887. [DOI] [PubMed] [Google Scholar]
- 5.Sotiriou C, Piccart MJ. Opinion - Taking gene-expression profiling to the clinic: when will molecular signatures become relevant to patient care? Nature Reviews Cancer. 2007;7:545–53. doi: 10.1038/nrc2173. [DOI] [PubMed] [Google Scholar]
- 6.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16:559–66. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- 7.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–46. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui J, Chen Y, Chou WC, Sun L, Chen L, Suo J, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger MF, Levin JZ, Vijayendran K, Sivachenko A, Adiconis X, Maguire J, et al. Integrative analysis of the melanoma transcriptome. Genome Res. 2010;20:413–27. doi: 10.1101/gr.103697.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 14.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–7. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, et al. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome Biol. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011 doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 17.Kim YH, Coon A, Baker AF, Powis G. Antitumor agent PX-12 inhibits HIF-1alpha protein levels through an Nrf2/PMF-1-mediated increase in spermidine/spermine acetyl transferase. Cancer Chemother Pharmacol. 2010;68:405–13. doi: 10.1007/s00280-010-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 19.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 20.Oshlack A, Robinson MD, Young MD. From RNA-seq reads to differential expression results. Genome Biol. 2010;11:220. doi: 10.1186/gb-2010-11-12-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss SF, Lee JW, Sabo E, Rubin AK, Rommel J, Westley BR, et al. Decreased expression of gastrokine 1 and the trefoil factor interacting protein TFIZ1/GKN2 in gastric cancer: influence of tumor histology and relationship to prognosis. Clin Cancer Res. 2008;14:4161–7. doi: 10.1158/1078-0432.CCR-07-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uchino S, Noguchi M, Ochiai A, Saito T, Kobayashi M, Hirohashi S. p53 mutation in gastric cancer: a genetic model for carcinogenesis is common to gastric and colorectal cancer. Int J Cancer. 1993;54:759–64. doi: 10.1002/ijc.2910540509. [DOI] [PubMed] [Google Scholar]
- 23.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, et al. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr Protoc Hum Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10: Unit 10 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, Baek D, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang H, Li WH. Lowly expressed human microRNA genes evolve rapidly. Mol Biol Evol. 2009;26:1195–8. doi: 10.1093/molbev/msp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisslthaler B, Fleming I. Activation and signaling by the AMP-activated protein kinase in endothelial cells. Circ Res. 2009;105:114–27. doi: 10.1161/CIRCRESAHA.109.201590. [DOI] [PubMed] [Google Scholar]
- 28.Nader N, Ng SS, Lambrou GI, Pervanidou P, Wang Y, Chrousos GP, et al. AMPK regulates metabolic actions of glucocorticoids by phosphorylating the glucocorticoid receptor through p38 MAPK. Mol Endocrinol. 2010;24:1748–64. doi: 10.1210/me.2010-0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 30.Okoshi R, Ozaki T, Yamamoto H, Ando K, Koida N, Ono S, et al. Activation of AMP-activated protein kinase induces p53-dependent apoptotic cell death in response to energetic stress. J Biol Chem. 2008;283:3979–87. doi: 10.1074/jbc.M705232200. [DOI] [PubMed] [Google Scholar]
- 31.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–93. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 32.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20:427–34. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Jalving M, Gietema JA, Lefrandt JD, de Jong S, Reyners AK, Gans RO, et al. Metformin: taking away the candy for cancer? Eur J Cancer. 2010;46:2369–80. doi: 10.1016/j.ejca.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Boyle JG, Salt IP, McKay GA. Metformin action on AMP-activated protein kinase: a translational research approach to understanding a potential new therapeutic target. Diabet Med. 2010;27:1097–106. doi: 10.1111/j.1464-5491.2010.03098.x. [DOI] [PubMed] [Google Scholar]
- 36.Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–75. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, et al. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods. 2009;6:647–9. doi: 10.1038/nmeth.1360. [DOI] [PubMed] [Google Scholar]
- 38.Wu Q, Kim YC, Lu J, Xuan Z, Chen J, Zheng Y, et al. Poly A- transcripts expressed in HeLa cells. PLoS One. 2008;3:e2803. doi: 10.1371/journal.pone.0002803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–7. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.