Abstract
From data on allozyme, nuclear DNA and mitochondrial DNA markers, we show that the originally North Pacific/Northwest Atlantic mussel Mytilus trossulus is widespread on North European coasts, earliM. trossuluser thought to be inhabited only by Mytilus edulis. Several local occurrences of , interspersed with a dominant M. edulis, were recorded on the North Sea, Norwegian Sea and Barents Sea coasts of Norway and the Barents and White Sea coasts of Kola Peninsula in Russia. The proportion of M. trossulus genetic background observed at any one site varied from 0 to 95%. These new occurrences are not related to the previously known, introgressed M. trossulus population that occupies the Baltic Sea. The new northern occurrences retain both the F and M M. trossulus mitochondria, which have been lost from the Baltic stock. While hybridization takes place wherever M. trossulus and M. edulis meet, the extent of hybrization varies between the different contact areas. Hybrids are rare, and the hybrid zones are bimodal in the northern areas; more interbreeding has taken place further south in Norway, but even there genotypic disequilibria are higher than those in the steep transition zone between the Baltic mussel and M. edulis: there is no evidence of a collapse toward a hybrid swarm unlike in the Baltic. The Barents and White Sea M. trossulus are genetically slightly closer to the NW Atlantic than NE Pacific populations, while the Baltic mussel has unique features distinguishing it from the others. We postulate that the presence of M. trossulus in Northern Europe is a result of repeated independent inter- or transoceanic cryptic invasions of various ages, up to recent times.
Electronic supplementary material
The online version of this article (doi:10.1007/s00227-010-1609-z) contains supplementary material, which is available to authorized users.
Introduction
Populations of the common mussel Mytilus spp. are key elements of intertidal and subtidal hard-bottom communities throughout the temperate-to-subarctic coasts of the Northern Hemisphere, and also a major object of aquaculture. Over the past decades, molecular studies of these externally relatively uniform bivalves have disclosed a systematic and genetic complexity which is making them model organisms in several branches of evolutionary marine biology, including zoogeography, invasion biology and speciation research. The Mytilus edulis complex consists of three basic taxa, M. edulis, M. galloprovincialis, and M. trossulus, whose distributions evidently reflect a history of vicariance, dispersal, secondary contact and hybridization through the Pleistocene time, which has recently been intensified with (cryptic) trans- and inter-oceanic invasions brought about by human activity.
The current concepts of the diversity and zoogeography of Mytilus arose from studies of allozyme character variation conducted since the 1970s (e.g., Skibinski and Beardmore 1979; Skibinski et al. 1983; Varvio et al. 1988; McDonald and Koehn 1988; McDonald et al. 1991; Väinölä and Hvilsom 1991; Seed 1992), and then continued with DNA markers, both mitochondrial and nuclear (e.g., Inoue et al. 1995; Hilbish et al. 2000). The current distribution of the three taxa is complicated; each of the four northern continental margins is inhabited by more than one of them (e.g., McDonald et al. 1991). There are broad patterns of transition between more southerly and more northerly taxa on each coast, but the ranges are not distinctly parapatric: there is considerable overlap, with regional interdigitation and mosaic hybrid zones (Riginos and Cunningham 2005; Rawson and Harper 2009). In the general framework of the ongoing globalization of marine life, the distributions of the component members of M. edulis complex, and the integrity of the taxa themselves in face of the frequent hybridization, could also be in a state of continuous dynamic change.
Mytilus trossulus is evidently the (only) native taxon of the M. edulis complex in the Northern Pacific, but it is also widespread on the North American coasts of the NW Atlantic, from the Gulf of Maine north (McDonald et al. 1991). In Europe, M. trossulus has been known from the Baltic Sea (Varvio et al. 1988; McDonald and Koehn 1988; Väinölä and Hvilsom 1991; Riginos and Cunningham 2005), whereas M. edulis and M. galloprovincialis are the principal mussel taxa elsewhere in the North-East Atlantic area (Seed 1992; Hummel et al. 2001). A steep intergradation zone in multiple allozyme characters between Baltic M. trossulus and North Sea M. edulis in the Öresund strait was described by Theisen (1978) and Väinölä and Hvilsom (1991). In other genetic characters, the Baltic population, however, appears to be strongly introgressed by parts of the M. edulis genome (e.g., Rawson and Hilbish 1998; Borsa et al. 1999; Riginos and Cunningham 2005; Kijewski et al. 2006; Stuckas et al. 2009). In this, the Baltic mussel makes an evident analogue to another circum-boreal bivalve Macoma balthica, whose Baltic population is also thought to have arisen through amalgamation of distinct Pacific and Atlantic origins, and to represent a rapid genomic reorganization in a new, post-glacially emerged habitat (Väinölä 2003; Nikula et al. 2008). The Pacific Macoma lineage is thought to have reached Europe through the Arctic basin at some point of time and is found in secondary contact with the Atlantic M. b. rubra also elsewhere in Northern Europe, e.g., in the Barents and White seas (Strelkov et al. 2007).
It is notable in this context that while M. trossulus is regarded as a typically northern member of the Mytilus complex, several genetic studies have found M. edulis to be the exclusive Mytilus taxon in the very North of Europe (e.g., Varvio et al. 1988; McDonald et al. 1991; Hummel et al. 1999, 2001; Smietanka et al. 2004; Riginos and Henzler 2008; this includes all localities so far reported from Iceland and the Norwegian, Barents and the White seas). Yet there are previous notions of unexpected systematic heterogeneity of mussels in the Barents Sea sector from DNA-DNA hybridization data (Milyutina and Petrov 1989, 1997) and from shell characters also from the White Sea (Zolotarev and Shurova 1997). More recently a presence of typical molecular characters of M. trossulus have been recorded in some areas of Northwest Europe, i.e., in mussel farms of western Scotland (Beaumont et al. 2008; Zbawicka et al. 2010) and on the Bergen coast of Norway (Ridgway and Nævdal 2004), which contradicts the earlier concept that the Baltic M. trossulus was a unique outpost of this taxon in Europe. The data on systematic heterogeneity from Bergen have been based on two allozyme loci only though, and involved no direct comparison to reference M. trossulus (Ridgway 2001; Ridgway and Nævdal 2004).
The origin (mode and time of immigration) of the European M. trossulus encountered so far remains enigmatic. Both for the Baltic and for the recent Scottish occurrences, a hypothesis of an early post-glacial relict of a wider distribution in Europe has been proposed (Väinölä and Hvilsom 1991; Beaumont et al. 2008). Yet the hypotheses addressed should involve both pre-glacial and Early Holocene immigration to Europe—either from the NW Atlantic or across the Arctic from the Pacific, as well as inadvertent transfer and cryptic invasion with transoceanic navigation in the recent decades or centuries (cf. Väinölä 2003).
Prompted by the emerging indications of M. trossulus genes found outside the Baltic in Europe, by the complex immigration hypotheses arising from inferences from the zoogeographical analogue Macoma balthica (Nikula et al. 2007), by the notion of cryptic invasion biology, and by the indications of the different behavior of different genomic components following secondary contacts in Mytilus and other bivalves (e.g., Riginos and Cunningham 2005; Strelkov et al. 2007), we set out to explore the distribution and genetic background of North European M. trossulus on a broader scale. We employ a wider set of diagnostic characters representing the different genomic components and include direct comparisons with populations from the putatively ancestral distributions. Particularly we have re-assessed the identity of the northern mussels in a survey of samples along the Barents, White and Pechora Sea coasts, including port localities that more likely would be exposed to recent introductions.
In this paper, we shall (1) describe a widespread presence of M. trossulus in samples from North European localities outside the Baltic Sea, based on allozyme and DNA marker data; (2) assess the patterns of interbreeding between the two taxa in their various contact zones, disclosing marked inter-regional variation in levels of hybridization, possibly related to contact age; (3) review the distribution of M. trossulus across the Baltic Sea and its transition zone toward North Sea M. edulis in terms of an allozyme genetic component; and (4) assess the differentiation and relative affinities between the European and American (NW Atlantic, NE Pacific) M. trossulus populations using multilocus allozyme data, and from these data discuss the colonization history of the new occurrences.
Materials and methods
Molecular characters
The diagnosis of M. trossulus by McDonald and Koehn (1988) was initially based on several nearly diagnostic allozyme characters, primarily on the Gpi, Pgm, Mpi and Est loci, and additional partially diagnostic loci including Idh and Odh (octopine dehydrogenase) were described by Väinölä and Hvilsom (1991). Several DNA-based characters have also been established, which diagnostically differentiate the species in putatively pure populations (e.g., Pacific vs. Atlantic). Particularly, these include the two distinct mitochondrial DNAs of Mytilus, independently inherited in the doubly uniparental inheritance system (DUI). The maternally inherited F and paternally inherited M genomes themselves are 20% diverged, and within each the coding genes show strong inter-specific differentiation, which can be addressed by e.g., sequencing or restriction digestion of various gene fragments, following amplification by F or M molecule-specific primers that amplify both species (e.g., Rawson and Hilbish 1995, Rawson et al. 1996). A number of PCR-based nuclear markers are also available (see Bierne et al. 2003; Riginos and Cunningham 2005). While all markers serve concordantly to distinguish pure species populations, their behavior in contact zones may vary, and the information from any single marker type alone should be viewed in that context (e.g., Riginos and Cunningham 2005). This study is primarily based on information from allozyme characters, supplemented for confirmation by a set of commonly used DNA marker loci.
Allozyme electrophoresis
For allozyme assays, mussels had been stored frozen at –80°C. Mixed hepatopancreas and adductor muscle tissue, homogenized in 0.1-% β-mercaptoethanol, were used for the analyses. The general methodology of starch gel electrophoresis was as in Murphy et al. (1996). Assay conditions were basically as described in Väinölä and Hvilsom (1991) for starch gels. In addition to enzymes therein, two further loci, Eno (enolase) and Aap (‘alanine-amino-peptidase’) were assessed in parts of the material. The set of loci used for various subsets of the data varied according to the question addressed.
DNA markers
Markers representing each of the two gender-associated mitochondrial genomes of Mytilus, and two diagnostic nuclear DNA markers ME and ITS were studied in subsets of the material. DNA was isolated from adductor muscle or mantle tissue using a salt-extraction procedure (Aljanabi and Martinez 1997). For an assay of the F mtDNA molecule, a segment of the 16S rRNA gene was specifically amplified using the universal primer 16Sbr combined with the F-specific Pr19, and cut with TruI, which discriminates between the M. trossulus and M. edulis haplotypes (Rawson et al. 1996). The M mtDNA-specific 16S primers Pr17 and Pr18 (Rawson and Hilbish 1995) were used in an attempt to amplify the corresponding gene of the M-genome; no other effort to identify sex was made. The PCR-product from positively scoring individuals was digested with HindIII for species identification. A set of PCR-products of both the F and M mtDNAs were also sequenced to confirm their identities with published M. trossulus/M.edulis mtDNA genomes from other areas, and to corroborate the identifications made by restriction digestion (GenBank accession numbers HQ832566-HQ832573). The length polymorphism of the nuclear ME (or Glu-5) was studied using the primers me15 and me16 of Inoue et al. (1995), and ITS using the PCR-Hin6I-digestion assay of Heath et al. (1995).
Data sets
1. Main data set. To explore the presence of M. trossulus in northernmost Europe, a new set of samples covering much of the White Sea basin and the Russian Barents Sea coast (including the Pechora Sea) was collected mainly in 2001–2004 (Table 1; Fig. 1). This was supplemented by a number of Norwegian samples from the Barents Sea, Norwegian Sea and North Sea coasts. Particularly, we studied material from the Bergen area (Bergen city harbor and Espeland marine biological station with repeated samples from 2001 to 2004, the same sites were studied with two loci by Ridgway 2001; Ridgway and Nævdal 2004). Three samples from within the Baltic Sea and two from the Baltic-North Sea transition zone in the Öresund and Kattegat were included as reference in this data set. Also reference samples from pure North American M. edulis (#4, Searsport, Maine), and North American M. trossulus both from the NW Atlantic (#3, St Andrews, New Brunswick) and the NE Pacific coasts (#1, 2, Puget Sound, Washington). This sample set, of 44 samples and ca 2,500 individuals, was generally genotyped for the set of eight polymorphic allozyme loci Ap, Est, Gpi, Idh-1, Lap, Mpi, Odh and Pgm. The data set was used in the first place for the identification of taxa on European coasts outside the Baltic and for estimating the relative genomic contributions of parental taxa in populations of mixed origin. Analysis of genotypic structure to assess interspecies hybridization in this set was based on four of the most informative loci, Est, Gpi, Odh, Pgm, which feature frequency differences of 0.97, 0.88, 0.77 and 0.77, respectively, between pure reference samples (Electronic supplement 4).
Table 1.
Sample site information for the Main data set analyzed for eight allozyme loci and by PCA
| Sample code | Sampling site | Sea area | Region | Coordinates N/E | Year | |
|---|---|---|---|---|---|---|
| North American M. trossulus reference | ||||||
| 1* | Whidbey Island | NE Pacific | Washington, Puget Sound | 48°13′19″ 122°41′30″W | 1990 | |
| 2* | Driftwood Key | NE Pacific | Washington, Puget Sound | 47°54′20″ 122°35′24″W | 1990 | |
| 3* | St. Andrews | NW Atlantic | New Brunswick, Bay of Fundy | 45°06′07″ 67°03′06″W | 1986 | |
| M. edulis reference | ||||||
| 4 | Searsport | NW Atlantic | Maine, Gulf of Maine | 44°27′20″ 68°55′22″W | 1986 | |
| 6* | Jarfjorden, Storbukta | Barents Sea | Norway, Varangerfjorden | 69°41′ 30°28′ | 1987 | |
| 7 | Hjerting | North Sea | SW Denmark, Wadden Sea | 55°31′20″ 8°21′11″ | 1988 | |
| Baltic transect | ||||||
| 8 | Egense | Kattegat W | Denmark | 56°58′53″ 10°13′33″ | 1988 | |
| 9 | Taarbæk, boat harbor | Öresund | Denmark | 55°47′18″ 12°35′46″ | 1987 | |
| 10 | Sölvesborg | Baltic Sea S | SE Sweden | 56°01′33″ 14°33′45″ | 1987 | |
| 12 | Hanko, Tvärminne | Baltic Sea N | SW Finland | 59°50′50″ 23°16′14″ | 1999 | |
| 13* | Tammisaari, Vitsand | Baltic Sea N | SW Finland | 59°56′14″ 23°22′29″ | 1990 | |
| North Sea/Norwegian Sea | ||||||
| 14 | Bergen, city harbor | North Sea | SW Norway | 60°22′49″ 5°20′08″ | 2001 | |
| 15 | Bergen, city harbor | North Sea | SW Norway | 60°22′49″ 5°20′08″ | 2004 | |
| 16 | Espeland, Marine Biology Station | North Sea | SW Norway | 60°16′10″ 5°13′20″ | 2001 | |
| 17 | Espeland, Marine Biology Station | North Sea | SW Norway | 60°16′10″ 5°13′20″ | 2004 | |
| 18 | Årsetfjorden, N Trondelag | Norwegian Sea | W Norway | 65°02′ 11°51′ | 1999 | |
| 19 | Tromsø, boat harbor | Norwegian Sea | N Norway | 69°39′10″ 18°59′10″ | 2009 | |
| Barents Sea | ||||||
| 20 | Kirkenes harbor | Barents Sea | Norway, Varangerfjorden | 69°43′47″ 30°02′58″ | 2008 | |
| 21 | Pechenga | Barents Sea | Russia, Western Murman | 69°35′07″ 31°15′24″ | 2004 | |
| 22* | Abram Mys | Barents Sea | Murman, Kola Bay | 68°58′55″ 33°01′40″ | 2001 and 2002 | |
| 23* | Mishukovo | Barents Sea | Murman, Kola Bay | 69°02′46″ 33°02′29″ | 2002 | |
| 24 | Tyuva Inlet | Barents Sea | Murman, Kola Bay | 69°10′27″ 33°38′13″ | 2003 | |
| 25 | Tyuva Inlet | Barents Sea | Murman, Kola Bay | 69°10′27″ 33°38′13″ | 2003 | |
| 26 | Kildin, Mogilnaya Inlet (sheltered) | Barents Sea | Murman | 69°19′23″ 34°19′39″ | 2003 | |
| 28 | Kildin Island, Sunduki (exposed) | Barents Sea | Murman | 69°20′11″ 34°24′12″ | 2003 | |
| 29 | Yarnishnaya Inlet, Yarnishni stream | Barents Sea | Eastern Murman | 69°05′14″ 36°03′00″ | 2002 | |
| 30 | Yarnishnaya Inlet, Yarnishni stream | Barents Sea | Eastern Murman | 69°05′14″ 36°03′00″ | 2001 | |
| 31 | Gremikha, Svyatonossky Bay | Barents Sea | Eastern Murman | 68°03′34″ 39°31′13″ | 2002 | |
| 32 | Gremikha, Svyatonossky Bay | Barents Sea | Eastern Murman | 68°03′13″ 39°29′47″ | 2001 | |
| 33 | NW Cape Kanin Nos | Barents/White Sea | Kanin Peninsula | 68°23′31″ 43°58′25″ | 2002 | |
| Pechora Sea | ||||||
| 34 | SE Dolgy Island | Pechora Sea | Dolgy Island | 69°09′10″ 59°13′38″ | 2002 | |
| 35 | Lyamchina Inlet | Pechora Sea | Vaigach Island | 69°53′33″ 59°14′58″ | 2002 | |
| White Sea | ||||||
| 36 | Olenitsa | White Sea | Gulf of Kandalaksha | 66°27′39″ 35°19′35″ | 2002 | |
| 37* | Umba, Malaya Pilskaya Inlet | White Sea | Gulf of Kandalaksha | 66°39′51″ 34°19′34″ | 2002 | |
| 38 | Umba, Malaya Pilskaya Inlet | White Sea | Gulf of Kandalaksha | 66°41′08″ 34°20′09″ | 2004 | |
| 39 | Luvenga | White Sea | Gulf of Kandalaksha | 67°05′56″ 32°42′07″ | 2004 | |
| 40* | Kartesh | White Sea | Gulf of Kandalaksha | 66°20′08″ 33°38′58″ | 1990 | |
| 41 | Matrenin Island | White Sea | Gulf of Kandalaksha | 66°18′34″ 33°37′56″ | 2001 | |
| 42 | Keret, river mouth | White Sea | Gulf of Kandalaksha | 66°16′38″ 33°34′19″ | 1994 | |
| 43 | Rabocheostrovsk | White Sea | Onega Bay | 64°59′30″ 34°47′22″ | 2004 | |
| 44 | Solovetsky Island | White Sea | Onega Bay | 65°02′05″ 35°40′43″ | 2001 | |
| 45 | Tamitsa | White Sea | Onega Bay | 64°08′31″ 37°59′57″ | 2004 | |
| 46 | Una Inlet | White Sea | Dvina Bay | 64°47′52″ 38°23′50″ | 2004 | |
| 47 | Yagri Island | White Sea | Dvina Bay | 64°37′03″ 39°48′44″ | 2002 | |
Samples included also in the Circumboreal multilocus data set are indicated by an asterisk (*)
Fig. 1.
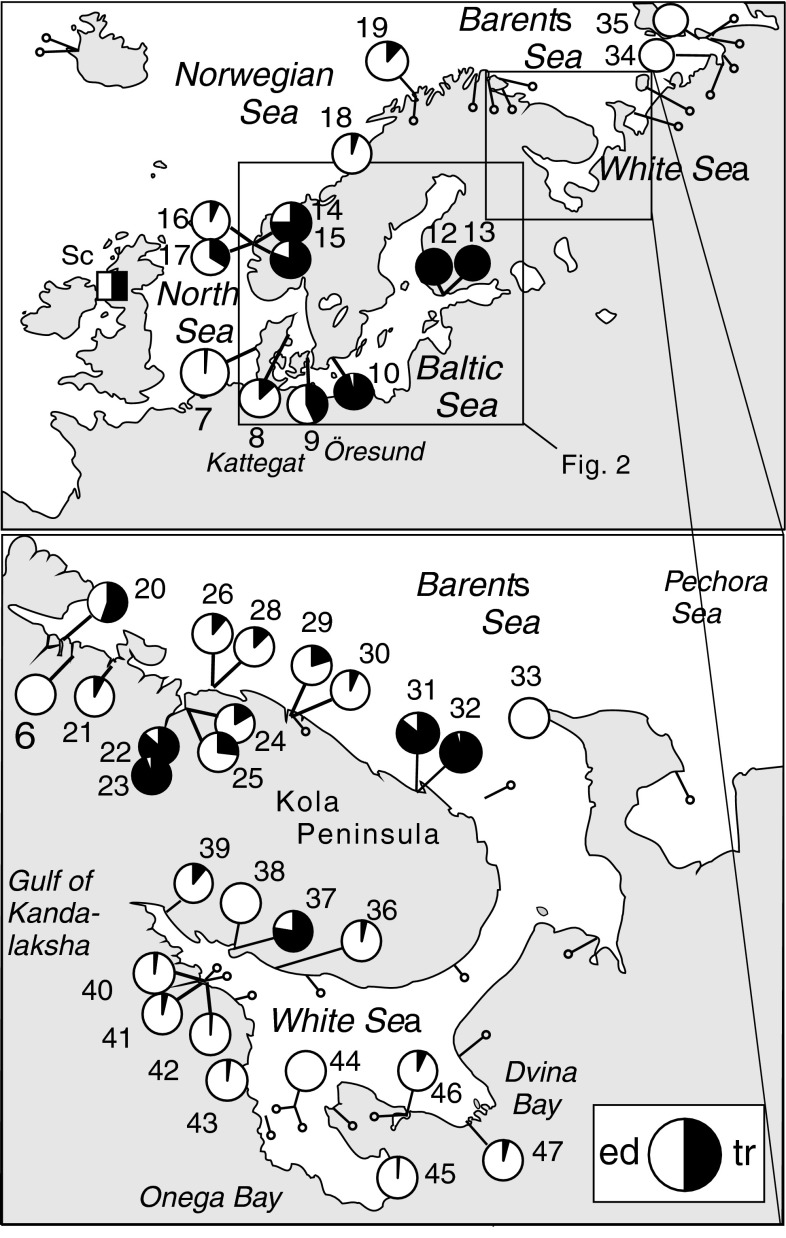
Distribution of the studied samples of the Main data set in Northern Europe. The pie charts indicate the samples studied with the 8-locus allozyme character set, with estimates of M. trossulus (black) versus M. edulis (white) genomic proportions as obtained from the PCA ordination in Fig. 3. The smaller symbols (pins) indicate samples from literature data, from where M. edulis only was previously recorded on genetic grounds (Varvio et al. 1988; McDonald et al. 1991; Hummel et al. 1999, 2001). “Sc” (square symbol) indicates published observations of M. trossulus from a Scottish loch, within the M. edulis/M. galloprovincialis range (Beaumont et al. 2008)
2. DNA character data set. To confirm the congruence of allozyme and of mtDNA and nDNA variation patterns, a subset of the European main data set mussels were analyzed for four diagnostic DNA markers (mtDNA typing and sequencing, nuclear genotyping). The subset was selected mainly to represent populations where M. trossulus and its hybrids were identified by allozymes, and involved 147 specimens (Fig. 6 legend). A number of North Pacific mussels were used as reference.
Fig. 6.
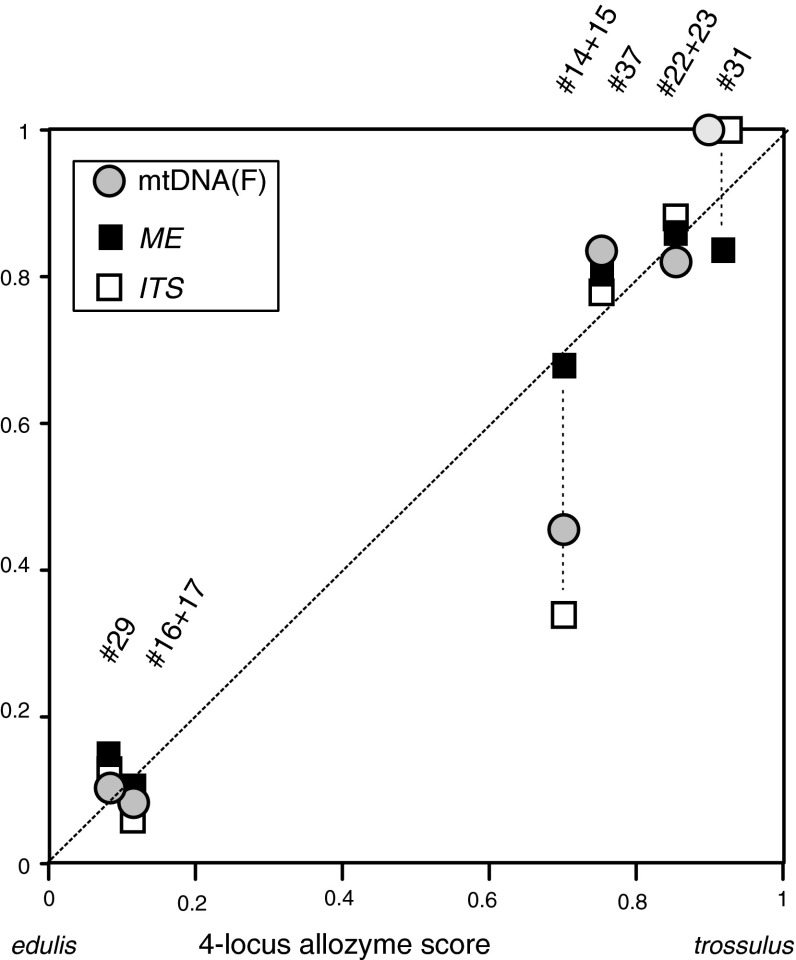
Correspondence of allelic compositions in allozyme versus DNA-based genetic markers in a set of selected regional samples of mixed ancestry. Frequencies of M. trossulus-specific alleles of three different DNA-based markers (Y-axis) are plotted against the scaled M. trossulus score from four nearly diagnostic allozyme loci (calculated from the same individuals, as in Fig. 4). Sample codes refer to Table 1 and Fig. 1, with sample sizes as follows. #14 + 15 Bergen city (N = 37), #16 + 17 Bergen Espeland (N = 22), #22 + 23 inner Kola Bay (N = 35), #29 Yarnishnaya, Murman coast (N = 18), #31 Gremikha, Murman coast (N = 10), #37 Umba, White Sea (N = 25)
3. Circumpolar multilocus data set. To quantify the trans- and inter-oceanic relationships within each of the mussel taxa and to assess the source areas of potentially newly invaded populations, the affinities among Pacific, NW Atlantic, NE Atlantic and Baltic populations were estimated from a broader allozyme character set of 19 loci. Three American and three European M. trossulus populations were included in this analysis, along with one American and two North European populations of pure M. edulis (see Table 3; altogether c. 400 individuals).
Table 3.
Estimates of genetic differentiation among selected circumpolar Mytilus populations, from the 19-locus allozyme data set (Electronic supplement 2)
| Species | M. trossulus | M. edulis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NE Pacific Wa. | Wa. | NW Atlantic N.B. | NE Atlantic Barents Sea | White Sea | N Baltic | NEA White Sea | Barents Sea | NWA Maine | ||
| Sample code | 1 | 2 | 3 | 22+23 | 37 | 13 | 40 | 6 | 4 | |
| M. trossulus | ||||||||||
| 1 | NEP Washington | – | −0.004 | 0.030 | 0.023 | 0.025 | 0.079 | 0.322 | 0.348 | 0.400 |
| 2 | NEP Washington | 0.005 | – | 0.021 | 0.025 | 0.019 | 0.062 | 0.302 | 0.316 | 0.379 |
| 3 | NWA New Brunswick | 0.062 | 0.047 | – | 0.011 | 0.005 | 0.055 | 0.294 | 0.306 | 0.342 |
| 22+23 | NEA Barents Sea | 0.053 | 0.045 | 0.033 | – | −0.009 | 0.055 | 0.297 | 0.316 | 0.360 |
| 37 | NEA White Sea | 0.057 | 0.056 | 0.025 | 0.002 | – | 0.053 | 0.287 | 0.308 | 0.348 |
| 13 | North Baltic | 0.171 | 0.138 | 0.129 | 0.137 | 0.143 | – | 0.259 | 0.283 | 0.347 |
| M. edulis | ||||||||||
| 40 | NEA White Sea | 0.369 | 0.355 | 0.359 | 0.363 | 0.364 | 0.403 | – | 0.003 | 0.027 |
| 6 | NEA Barents Sea | 0.388 | 0.370 | 0.375 | 0.386 | 0.397 | 0.439 | 0.021 | – | 0.024 |
| 4 | NWA Maine | 0.420 | 0.409 | 0.396 | 0.409 | 0.408 | 0.463 | 0.075 | 0.066 | – |
| H T | (gene diversity) | 0.354 | 0.358 | 0.344 | 0.349 | 0.349 | 0.234 | 0.293 | 0.254 | 0.283 |
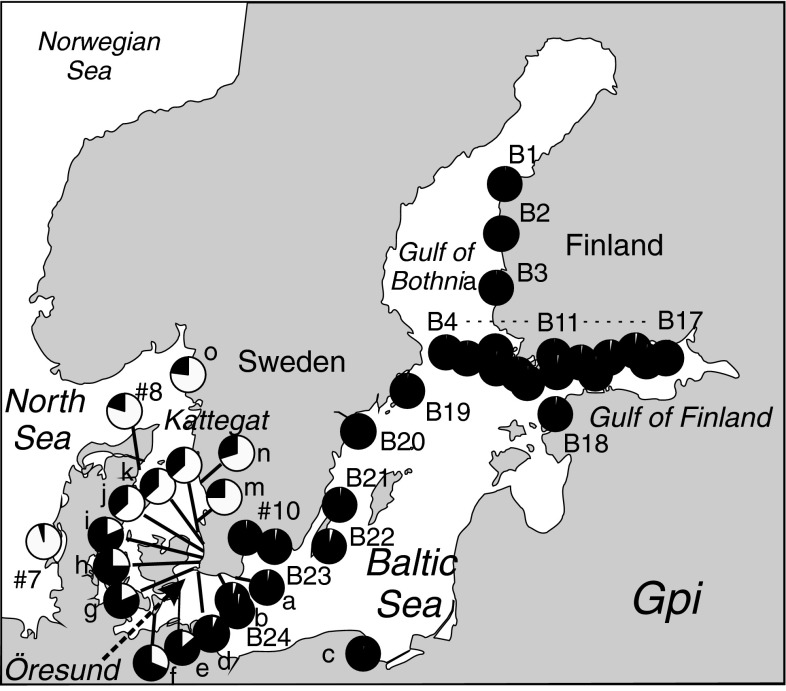
4. Baltic-North Sea data set. The homogeneity of the identity of the Baltic mussel population through the basin, and the genetic transition between the Baltic and the North Sea populations through the Öresund strait, were illustrated on the basis of single-locus data of the diagnostic enzyme GPI. The Gpi locus is a good representative of the general allozyme composition, as demonstrated by the correlation structure in the main data set (1), and data in Väinölä and Hvilsom (1991). The intra-Baltic data comprises 31 geographical samples (total N = 4,881) mainly from the years 1980–1982 (Väinölä 1985), and the Öresund–Kattegat transition zone data 10 samples mainly from the Swedish coast, from a data set of 1986–1987 (N = 1,256) (Fig. 2; Electronic supplement 3).
Fig. 2.
The Baltic-North Sea data set, indicating single-locus allele frequency estimates of the pooled Gpi allele (black), used as a representative of the Baltic allozyme genetic composition (locality and frequency data in Electronic supplement 3). This is a nearly diagnostic M. trossulus marker (sample #7 represents pure North Sea M. edulis)
Data analysis
PCA of population data. Species identification at the individual level is in the first place based on multiple nearly diagnostic characters, each of which similarly discriminates species in pure populations. The presence of mixed systematic or genomic composition at the population level was illustrated by PCA (principal component analysis) ordination of multilocus allele frequency data. This plots populations in the simple Euclidean allele frequency space; the main axis PC1 is expected to reflect the species discrimination, and in the ideal case the relative genomic contributions in mixed populations should be readable directly from their projection on this axis, relative to pure species samples (cf. Strelkov et al. 2007). These scaled PC scores will serve as our operational estimates of the population-level systematic “purity” of the non-reference samples. The PC analysis was performed on a covariance matrix of 8-locus allozyme data using the PAST software (Hammer et al. 2001); one of the most common alleles at each locus was removed to reduce redundancy, and a number of rare alleles were pooled (binned) with adjacent ones to avoid inevitable risks of mis-scoring.
Intra-population structure and identification of hybrids. The extent of hybridization in populations of mixed taxonomic background was assessed by comparing the observed genotypic distributions in the samples to the distributions expected in panmictic populations on the one hand, and in non-hybridizing populations on the other. This was illustrated in terms of the conventional genotypic disequilibrium measures, following the approach in Nikula et al. (2008). Disequilibria (i.e., single-locus heterozygote deficit F IS and the analogous two-locus genotypic correlation measure R’) were calculated as weighted averages over (pairs of) the four loci (Est, Gpi, Odh, Pgm, each coded as two-allelic, T/E) and compared to the maximum values of those measures as expected in hypothetical mixtures of non-interbreeding “pure” taxa, which would have the same average allele frequencies. The observations and expectations were plotted against the population mixing index, for this purpose obtained from a simple sum of the characteristic T-allele frequencies over the four loci, scaled to [0, 1]. In another illustration, the histograms of individual “hybrid indices” of representative intermediate populations were compared with the corresponding predicted distributions in locally panmictic and in non-interbreeding mixture populations having the same average genetic composition (mixture of pure gene pools in proportions of the estimated mixing index for that population). The individual hybrid index used was again the sum of synthetic T alleles of that individual, over the four loci (range 0…8; cf. Riginos and Cunningham 2005).
Large-scale genetic affinities. Genetic distances were assessed from the 19-locus allozyme frequency data of the circumpolar data set (3), in terms of Nei’s “unbiased” standard genetic distance D (e.g., Nei 1987) and the Reynolds et al. (1983) drift-based F ST distance (equivalent to θ of Weir and Cockerham 1984). Affinities were illustrated by (linear) multidimensional scaling and by a neighbor-joining tree from the F ST matrix (NTSYS-software, Rohlf 1990). For the calculation of allele frequencies in samples from areas of potential taxon overlap or hybridization, only individuals judged to be taxonomically pure were included (the operational criterion permitting no more than one potentially introgressed allele at the set of five most diagnostic loci).
Results
Mytilus trossulus in the North Sea, Barents Sea and White Sea: allozyme data
Allozyme assessment of the multilocus genetic composition of individual mussels in the first place disclosed the presence of both Mytilus trossulus, M. edulis and their hybrids at several North European localities outside the Baltic Sea. In direct multilocus comparisons with reference populations, we found pure M. trossulus (i.e., specimens with no diagnostic/characteristic M. edulis alleles), or traces of its genome (multiple characteristic M. trossulus alleles) to be present in the White Sea and on the Barents Sea coast of the Kola Peninsula and NE Norway, as well as the eastern Norwegian Sea (Tromsø). Likewise, the data comprising multiple diagnostic characters confirmed the genetically peculiar North Sea occurrences in and near Bergen as representing true M. trossulus. The relative representation of M. trossulus (or its genome) in these samples varied from ca 80 to 95% at sites of the inner Kola Bay near Murmansk (#22, 23), Gremikha in the NE Kola Peninsula (31, 32) and at Umba (#37) on the south coast of the Kola Peninsula in the White Sea, to 10–25% at most other Murman coast samples (#24–30), and to just a few hybrid or backcross individuals (<5% trossulus contribution) in most White Sea samples. Several samples still contained only M. edulis (e.g., #6 Jarfjord of the Barents Sea, #44 the Solovetsky Islands and other sites in the west and south of the White Sea, and the easternmost samples from the Pechora Sea, #33, 34). Below, we shall refer to the newly identified European M. trossulus as the Northeast Atlantic or NEA populations, to a distinction from the Baltic M. trossulus (“Baltic mussel”) and the reference NWA (Northwest Atlantic) and NEP (Northeast Pacific) M. trossulus.
The estimated genomic contributions are illustrated in the PCA ordination based on the 8-locus data, where both the European and the North American reference material are included. The first axis PC1, accounting for 89% of variance, shows the conventional pure M. edulis and M. trossulus at the opposite ends, independent of their geographical origins (Fig. 3). The new occurrences with M. trossulus influence in the NEA fall in between the pure populations, and so do the Öresund and Kattegat populations (#8, 9) from the Baltic Sea-North Sea hybrid zone. This axis is strongly correlated with the frequencies of the major alleles of most loci included, often with an r > 0.95, and with r > 0.85 for all loci but Ap (Table 2). The estimates of X (=M. trossulus genomic proportion) from the 8-locus data are defined as the relative position of the sample on PC1 between reference pure M. edulis (from average of samples #4, 6, 7) and the NW Atlantic M. trossulus (#3) and are also summarized on the maps as pie-diagrams (Fig. 1a, b).
Fig. 3.
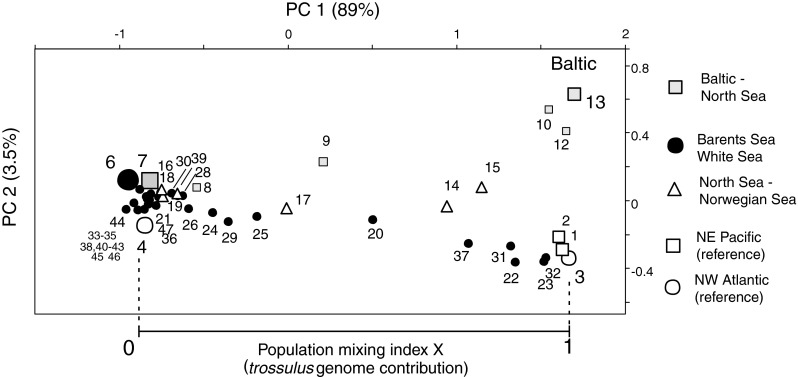
PCA ordination of the samples based on multilocus allele frequency data from 8 allozyme loci in the Main data set (Table 1, Electronic supplement 1). The plot shows the distribution of the new North European material between reference samples of pure European M. edulis (represented by samples #6 and #7) and North American M. trossulus (#3; St Andrews, New Brunswick). The relative placement of each sample on the PC1 axis between these points [0, 1] is taken as an estimate of the M. trossulus genomic contribution at the population level (population mixing index X; cf. Strelkov et al. 2007). Different symbols indicate different geographical regions, as indicated on the right. The axes are drawn to be proportional to the square root of the explained allele frequency variance in the analysis
Table 2.
Component correlations (correlation between allele frequencies and PC1 and PC2 scores) at the 8 loci included in the PCA of Fig. 3 (Main data set)
| Locus | Allele | Component correlation | |
|---|---|---|---|
| PC1 | PC2 | ||
| Est | 90* | 0.99 | – |
| 95* | 0.56 | 0.63 | |
| 100 | −0.99 | – | |
| 110 | −0.39 | – | |
| Odh | 90 | −0.37 | – |
| 95 | 0.28 | – | |
| 100 | −0.97 | – | |
| 105* | 0.98 | – | |
| 108 | – | – | |
| 110 | −0.30 | – | |
| Pgm | 90 | −0.58 | – |
| 95 | −0.41 | – | |
| 100 | −0.97 | – | |
| 110* | 0.36 | −0.58 | |
| 120* | 0.97 | – | |
| 125* | 0.82 | – | |
| Gpi | 86* | 0.64 | −0.52 |
| 93* | 0.67 | −0.40 | |
| 96* | 0.61 | −0.53 | |
| 98* | 0.95 | – | |
| 100 | – | −0.52 | |
| 102 | −0.89 | – | |
| 105 | −0.62 | – | |
| 107 | −0.96 | – | |
| 110 | −0.59 | – | |
| Idh | 93 | −0.90 | – |
| 100 | 0.89 | – | |
| 102 | −0.50 | – | |
| 107 | – | −0.42 | |
| 114 | – | – | |
| Ap | 90 | – | – |
| 95 | 0.39 | −0.57 | |
| 100 | – | 0.59 | |
| 105 | – | −0.41 | |
| 110 | – | −0.34 | |
| Lap | 92 | 0.62 | −0.55 |
| 94 | 0.77 | −0.43 | |
| 96 | −0.62 | – | |
| 98 | −0.87 | – | |
| 100 | 0.90 | 0.84 | |
| Mpi | 87 | 0.47 | 0.51 |
| 90 | – | – | |
| 94 | 0.99 | – | |
| 100 | −0.99 | – | |
The second axis PC2 only accounts for 3.5% of the allele frequency variance and serves to separate the North American M. trossulus (of both coasts) from the Baltic mussel. It also makes some amphi-Atlantic difference within M. edulis, i.e., between the Maine (#4) and the European references (#6, 7). This “trans-Atlantic axis” mainly reflects the variation of Lap and Ap loci, but also of minor alleles at Est and Gpi (Table 2). Neither of the first two components (in all 93%) does make a difference between the Pacific and Atlantic (NWA) M. trossulus populations.
On the PC1–PC2 plane, the Russian populations with a M. trossulus influence (black dots) fall clearly on the line between the American M. trossulus and European M. edulis, whereas the samples from the North Sea–Baltic transition (squares) fall rather on a line between the North Sea–inner Baltic populations. The positions of the mixed North Sea–Norwegian Sea samples (triangles) appear somewhat intermediate even on PC2.
Homogeneity of the Baltic mussel (M. trossulus)
For the Baltic Sea–North Sea area, the location of the M. trossulus–M. edulis transition and the stability of the intra-Baltic structure are presented in terms of the Gpi allozyme frequencies in Fig. 2 (a nearly diagnostic M. trossulus marker). In terms of this character, populations representing pure Baltic M. trossulus occupy the Baltic coasts from Southern Sweden up to the Bothnian Sea and the Gulf of Finland. There are little data from the eastern and southern Baltic coasts, but a sample from the Gulf of Gdansk (c) is classified as pure M. trossulus in these data. The transition across the Öresund along the Swedish coast is relatively smooth. Kattegat-Skagerrak populations still bear a considerable introgressive M. trossulus contribution (as also seen in sample #8 in the main multilocus data set). It should be noted that in this transect other, DNA-based marker characters show different patterns of introgression, and a different picture would be obtained from them (e.g., Kijewski et al. 2006; Stuckas et al. 2009).
Intra-population structure and extent of hybridization
From direct examination of the genotypes in mixed populations, some hybridization between the taxa is generally inferred wherever they co-occur. Considerable variation in the extent of this hybridization in the different regions is however evident from the quantitative assessment of genotypic disequilibria (Figs. 4, 5). The pattern appears to vary between the various contact areas and possibly with regard to the direction of introgression.
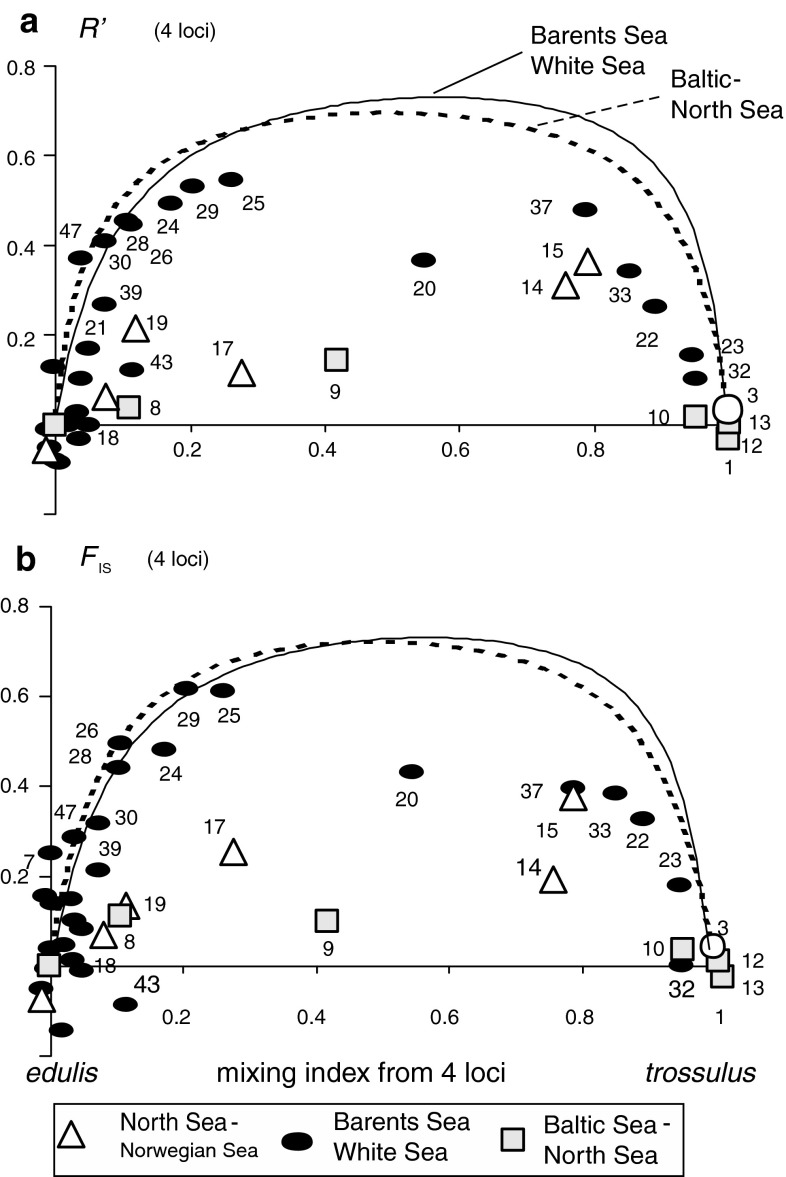
Fig. 4.
Estimates of average genotypic disequilibria in North European samples studied by multilocus enzyme electrophoresis. On the x-axis, the samples are plotted according to the estimated M. trossulus genomic contribution, obtained as a (scaled) average of the single-locus estimates for each of the four loci Est, Gpi, Odh and Pgm, included in analysis; each locus is treated as bi-allelic (pooled T and E alleles, Table 2, Electronic supplement 4). Disequilibria are expressed in terms of the genotypic correlation measures R′ (two-locus disequilibrium, upper panel) and F IS (single-locus heterozygote deficit, lower). The values are averages over the four strongly differentiated loci, or over all of their six pair-wise comparisons. The curves show the expectations in hypothetical non-interbreeding mixtures of the putative parental populations (“maximum disequilibria”; the expectations are calculated separately for the NEA samples (i.e., North, Norwegian, Barents and White seas; parental frequencies from samples #3 and #6 as given in Electronic supplement 4; continuous line) and for the North Sea–Baltic transect (parental frequencies from #7 and #13, interrupted line). Details of calculation and averaging were as in Nikula et al. (2008)
Fig. 5.
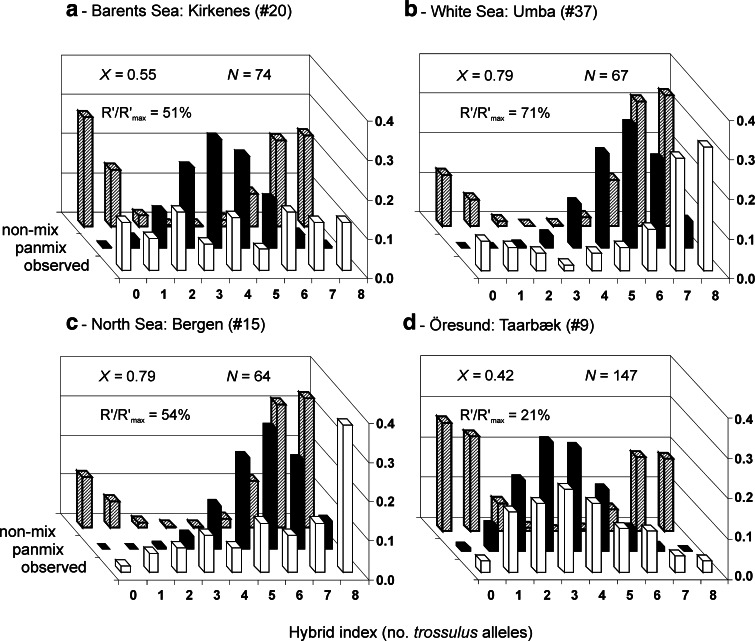
Observed multilocus genotypic distributions in some populations of mixed composition (white, front row), compared with expectations in local panmictic equilibrium populations (black, middle row) and in non-interbreeding mixtures of two taxa (shaded, back row). The picture shows the distribution of the observed hybrid index defined as the total count of M. trossulus-specific alleles (T, see Table 2) at the four strongly differentiated allozyme loci Est, Gpi, Odh and Pgm. The expected distributions are calculated in a hypothetical mixture of the parental taxa, defined by the 4-locus allele frequencies (bi-allelic pooling) and the estimated mixing index X as in Fig. 4, and represent the extreme unimodal and bimodal contact zone structures, respectively. Frequencies of each distribution (Y-axis) sum to 1. X, population-level mixing index. R′/R′max, average two-locus genotypic disequilibrium as a proportion of that in a non-interbreeding mixture (from Fig. 4, cf. Nikula et al. 2008), N, sample size
Comparing the observed inter- and intra-locus correlations R’ and F IS to the expectations under a non-interbreeding situation (curves in Fig. 4), the local disequilibria are generally clearly positive and mostly higher than 50–80% of the maximum level expected in a non-interbreeding situation, particularly in the northern contacts along the Barents and White Sea coasts. A large proportion of individuals appear relatively pure M. trossulus or M. edulis; multilocus hybrid genotypes are rare compared to that expected in a panmictic population. This is illustrated also in the relatively bimodal distribution of the individual hybrid indices for the White Sea situation (4-locus trossulus-allele counts Fig. 5b). The disequilibria however are considerably weaker in the Norwegian samples from the Bergen area (#15–17) and from Tromsø and Kirkenes (#19, 20), where genotypes are distributed more evenly among heterozyogsity classes (Fig. 5a, c), and from which genotypic deviations around 50% of the maximum were estimated. Further, the sample from the Öresund strait (#9), from the middle of the cline connecting the Baltic mussel and North Sea M. edulis, showed still lower disequilibria, only 10–20% of the maxima, and a markedly unimodal genotypic distribution (Figs. 4, 5d). Even this sample was far from a panmictic population structure however.
Moreover, the disequilibrium measures suggest a difference in the level of interbreeding depending on the dominant species in the mixture. For the White Sea–Barents Sea populations where M. edulis remains dominant, the disequilibria are close to the maximum (left end of Fig. 4, black dots), whereas when M. trossulus dominates (right end), the disequilibria are closer to 50%, proving considerable introgression.
Evidence from mitochondrial and nuclear DNA markers
Diagnostic mtDNA and nDNA characters were typed in altogether 147 individuals from nine samples representing six populations, where allozyme evidence of M. trossulus was observed. These included subsamples from Bergen (#14 + 15) and the adjacent Espeland (#16 + 17), pooled Kola Bay populations (#22 + 23), Yarnishnaya (#29) and Gremikha (#31) on the Barents Sea coast of Kola peninsula, and further Umba (#37) from within the White Sea.
In this material, diagnostic M. trossulus alleles (T) of the ME and ITS nuclear markers and of the F mtDNA were found concordantly with the allozyme data. The concordance holds for the relative frequencies of alleles among populations (Fig. 6) and for the distribution of characters in individuals within populations—i.e., in scoring mussels similarly as pure M. trossulus, M. edulis or hybrids, whether using either marker class. The correlation is particularly clear at the Russian sites. Locus-specific deviations from the 1:1 relationship were however also seen, at the Bergen site (#14 + 15) where mtDNA and ITS showed relative under-representation of M. trossulus alleles.
The identity of the F mitochondrial lineage of M. trossulus, as detected by the 16S PCR–RFLP assay, was verified by sequencing two PCR products and comparing with North American GenBank data. The absence of M. trossulus mtDNA in the Baltic mussel has been reported earlier (e.g., Rawson and Hilbish 1998; Kijewski et al. 2006) and was not further explored here.
As to the paternally inherited M mtDNA, the PCR assay by the M-specific 16S primers gave positive results generally in less than 50% of individuals of any sample, and this proportion varied widely between samples. In the HindIII restriction assay, both trossulus and edulis M genomes were recovered in the northern populations of mixed composition. Altogether 20 M. trossulus and 40 M. edulis M mtDNAs were thus identified, and the identities were again confirmed by sequencing several individuals. Yet in individual samples, the E:T M mtDNA frequencies sometimes deviated from those of other markers; thus, no T type M was scored from the Bergen area (#14–17), whereas it was common in the Russian samples. As these results could have been affected by differential amplification success of E vs. T M mtDNAs depending on e.g., life cycle stage or tissue quality (muscle/mantle), or by unequal sex ratios, they are not treated further here.
Broad-scale affinities among circumpolar M. trossulus
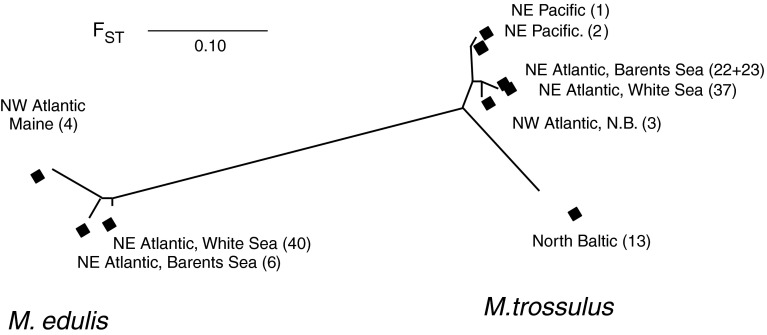
The assessment of the relationships among the M. trossulus populations from the various circumpolar sea regions, using a wider set of 19 allozyme loci (Electronic Supplement 2), involved putatively pure species samples from the NE Pacific, NWAtlantic, Baltic, and NE Atlantic (Kola Bay of the Barents Sea and Umba of the White Sea; not enough pure specimens were identified from the Bergen area). Genetic distances of the new M. trossulus occurrences of the NEA White and Barents seas were slightly closer to the NW Alantic than to the NE Pacific reference material (Nei’s D = 0.008 vs. 0.023, F ST = 0.029 vs. 0.053; Table 3, Fig. 7). The difference in the affinities is primarily based on frequencies at the Ark, Odh, Lap, and Gpi loci. The trans-Atlantic difference in M. edulis was similar to the inter-oceanic NEP-NWA difference in M. trossulus (D = 0.025, F ST = 0.070).
Fig. 7.
Affinities among pure M. edulis, M. trossulus and Baltic mussel samples, based on 19-locus allozyme data. The figure displays a 2-dimensional ordination of the samples by linear multidimensional scaling based on F ST distance matrix in Table 3, and a superimposed neighbor-joining tree from the same distances
The Baltic mussel in turn was more distant from the cluster of the NEA, NWA and NEP M. trossulus populations and barely closer to NEA and NWA than to NEP mussels (average D = 0.06, F ST = 0.14). This distinction is based on several loci and generally reflects a reduced level of allelic variation in the Baltic. The estimate of gene diversity for the Baltic mussel (H E = 0.24) was 30% lower than for the other M. trossulus samples (NEA 0.35, NWA 0.34, NEP 0.36). Diversity of M. edulis from these loci was estimated at H E = 0.25–0.29 (Table 3).
The average interspecies Nei’s distance (M. trossulus vs. M. edulis) from these data was D = 0.32.
Discussion
Earlier data on mussel distribution in Europe have showed a relatively clear partitioning of the Mytilus range between a southern and western M. galloprovincialis and a northwestern and northern M. edulis, apart from an outpost occurrence of M. trossulus in the Baltic Sea (Seed 1992; Hummel et al. 2001). Figure 1 shows, in addition to the new data, also the previously existing gene-marker-based information on mussel identity in Barents-White Sea sector, from which only M. edulis has so far been reported on these grounds (e.g., Varvio et al. 1988; Hummel et al. 1999, 2001). In stark contrast to that concept, the current data, along with those of Dias et al. (2009), show that populations of M. trossulus, or with a strong M. trossulus genomic influence, occur widely throughout the northern parts of M. edulis range, from the North Sea (Bergen) and Western Scotland to the Barents Sea coast of Norway and Russia. The species is also present locally within the White Sea. Still M. edulis appears to be the majority taxon on the northern coasts, often also found alone, whereas where M. trossulus is present, it is found together with M. edulis.
The genetic constitution at the new M. trossulus sites varies with respect to the relative representation of the two mussel taxa (or their genomes) and to the degree of interbreeding (Figs. 1, 3, 4). At any rate, the new findings indicate the presence of a relatively undisturbed M. trossulus genome in Northern Europe, similar to the North American reference samples, seeming to be at the initial stages of mixing with M. edulis. This is in contrast to the earlier established Baltic mussel occurrence, where extensive mixing of the M. trossulus and M. edulis genomes is inferred to have taken place, involving complete replacement of the mtDNA genomes and an established hybrid swarm equilibrium structure through most of the basin.
Hypotheses of invasion history
From the current understanding of zoogeography, M. trossulus represents a basically North Pacific lineage within the M. edulis complex, evolved in vicarious isolation from the Atlantic M. edulis–M. galloprovincialis through most of the Pleistocene (e.g., Väinölä 2003; Riginos and Cunningham 2005; Rawson and Harper 2009). The discovery of several North Atlantic occurrences of this taxon brings up the questions of the uniqueness, geographical origins, routing and timing of the inter- and transoceanic invasion and colonization events. The time scales of invasions to be considered include pre-glacial (e.g., Eemian, 120 kyr ago), early post-glacial or mid-Holocene (11–5 kyr), early human-influenced (1,000–100 year), or recent human-influenced (<100 year) periods (cf. Väinölä 2003). The origins of the European population(s) could have been directly either from the Pacific or from the NW Atlantic (or vice versa, origin of NWA populations from Europe instead directly from the Pacific; cf. Vermeij 2005); the NEA populations could be post-glacially derived from the Baltic, possibly with human activity, or the Baltic populations serially from the NEA; or the various European occurrences might have independent and plausibly asynchronous trans- or inter-oceanic origins. All these hypotheses cannot be resolved from the present data but some of them can be reasonably rejected.
In the multilocus allozyme data, the NEA, NWA and NEP M. trossulus populations all are closely related, but among these the NEA (Barents and White Sea) and NWA are the closest (Table 3; Fig. 7). Such a sister relationship of the amphi-Atlantic populations suggests that their common ancestor first invaded form the Pacific, and the populations then split by a trans-Atlantic dispersal event. The direction of such dispersal is not resolvable from these data alone (e.g., there is no indication of loss of variation on one or the other coast). Regarding the age of events, there are two informative points of comparison. From mtDNA data, Rawson and Harper (2009) judged that even the NEP-NWA divergence (Pacific-to-Atlantic invasion) most likely post-dates the previous interglacial and might rather be post-glacial (c. 10,000 years). From this, also the closer trans-Atlantic relationship is most likely post-glacial and probably relatively recent. The trans-Atlantic allozyme divergence in M. trossulus is also clearly smaller than the trans-Atlantic divergence within the sister taxon M. edulis, which on the other hand is more similar to the inter-oceanic NEP-NWA M. trossulus divergence, and again suggests a very recent NEA-NWA relationship in M. trossulus. From mtDNA data in turn, the age of amphi-Atlantic distribution of M. edulis is thought to be of the order 105 year at least (Wares and Cunningham 2001; Riginos and Henzler 2008), and the closer allozyme relationship in M. trossulus then again points to a post-glacial invasion. Considering the concentration of new finds in harbor areas, a hypothesis of recent human-mediated trans-Atlantic invasion of M. trossulus from the NWA to the NEA then seems most reasonable, at least for the northerly Barents Sea–White Sea populations.
The close trans-Atlantic affinity of the new North European M. trossulus occurrences stands in contrast to the distinction of the Baltic mussel, which appears to have been modified by a long-lasting introgression process. Particularly, the earlier hypotheses of a recent common ancestry of the Baltic and the North Sea M. trossulus occurrence in Bergen (Ridgway and Nævdal 2004; Riginos and Cunningham 2005) are disputed by the evidence of the retention of the truly “native” M. trossulus (F)-mtDNA in the NEA populations, which is lost in the Baltic. Likewise, a practically fixed trossulus composition at nuclear DNA loci in some populations (Fig. 6) shows they are not recently derived from the Baltic, where the composition is strongly diluted by introgression of edulis alleles (e.g., Kijewski et al. 2006). A similar hypothesis for the Scottish loch occurrence (cf. Beaumont et al. 2008) was similarly refuted by mtDNA evidence of Zbawicka et al. (2010).
Nevertheless, even the relatively greater differentiation of the Baltic mussel in allozyme characters cannot be interpreted in the same terms as that among the other M. trossulus stocks, of time-dependent divergence only. The Baltic population shows a reduced level of variation suggestive of a bottleneck history (30% lower allozyme gene diversity than in other M. trossulus; Table 3), but may also have been influenced by introgression from the neighboring M. edulis (as amply documented in DNA markers, e.g., Stuckas et al. 2009). Moreover, it shows unique differences such as the high frequency of the Lap 100 allele, potentially resulting from selection in the unique environment (Väinölä and Hvilsom 1991). The generation of such a unique structure in itself suggests a considerable independent age. In view of the basin history, a post-glacial process since the origin and colonization of the brackish-water basin still seems the most likely scenario.
So far, there is plenty of evidence on human-mediated range expansions from other mussel taxa. Particularly, the European M. galloprovincialis has efficiently colonized both coasts of the Northern Pacific and partly replaced the resident M. trossulus there (see Brannock et al. 2009 for references). Transfer of mussels has been hypothesized in ballast water and intentionally for aquaculture, but can occur on ship hulls, which mussels may occupy en masse as fouling organisms (e.g., Apte et al. 2000; Lee and Chown 2007).
Remarkably, the new North European finds of M. trossulus are mostly from harbors or harbor-related areas, whereas the sites of pure M. edulis from these and earlier data are largely from less urban environments. On the Norwegian Barents Sea coast, we found M. trossulus in the major Kirkenes harbor (#19) but not in the adjacent, relatively pristine Jarfjorden (#20) or at several sites (including local fishing harbors) of the Varanger peninsula studied by Varvio et al. (1988). Likewise, the three Russian areas with M. trossulus prevalence (Kola Bay #22–25, Gremikha #31, 32, Umba #37) are related to active navigation and international traffic. Among these, Murmansk in the Kola Bay has been a major port of the Russian North since the early the twentieth century; particularly, it was a trans-Atlantic supply port during WWII and continues to involve important bases for the northern naval fleet. M. trossulus influence is currently seen even at sites outside the entrance to the Bay. Important ports have also functioned in Gremikha (military) and Umba (timber export), although that of Umba was abandoned by the end of the century. The latter harbors could also plausibly have been serially colonized from Murmansk, to which they were connected by regular coastal ship traffic. On the other hand, M. trossulus was not found in samples from the vicinity of some other White Sea ports (e.g., #43, 45, 47). Nevertheless, direct White Sea ocean traffic has been mainly to and from the Arkhangelsk harbor (near #47), which was a major commercial and military port even before Murmansk. Notably however, the Arkhangelsk harbor at the mouth of Northern Dvina, as well as that of Onega at the mouth of the Onega river (near #45), are located within the freshwater downstream portions of the rivers themselves, which would make them fatal for incoming mussels.
Given that M. edulis has been a dominant benthic organism on the Northeast Atlantic coasts through millennia, the inferred recent and independent colonization of several bays and harbors by M. trossulus suggests a competitive advantage in the respective habitats. This could be related to a more efficient reproductive output or settlement of the incoming M. trossulus. In North American mixed M. trossulus/M. edulis localities, M. trossulus generally seems to be dominant in spat and younger age classes, whereas edulis will take over with age (Toro et al. 2004), in line with such a hypothesis.
The ability of M. trossulus to invade habitats of the Russian Barents and White Sea coasts could also relate to the peculiar dynamics of the natural mussel populations in those areas. Particularly on the Murman coast and in the Kola Bay, a dramatic collapse of mussel populations took place in the 1960s to early 1970s, plausibly connected with a long climatic deterioration that affected native reproduction (Antipova et al. 1984). This left mussel habitats vacant for colonization, which could have provided an opportunity for transoceanic invaders to take over large areas, such as those in the inner and middle Kola Bay (Golikov and Averintsev 1973). In the White Sea in turn, a general multidecadal cyclicity of local mussel beds has been recorded, involving local extinctions; this has no evident relationship to the dynamics of neighboring populations or to the environment however (Lukanin et al. 1986). Such metapopulation dynamics could likewise have periodically facilitated access to invader spat.
Contrasting patterns of hybridization in different contact zones
Earlier studies of M. trossulus × M. edulis contact zones have drawn attention to a difference between the NW Atlantic, where hybridization is limited and the zone structures thus bimodal (i.e., parental forms remain predominant), versus the Baltic Sea contact, where extensive introgression at a number of loci has resulted in amalgamation of the genetic material into a unimodal hybrid swarm structure, in which the ancestries of individual mussels effectively do not differ in regard to average parental contributions (review in Riginos and Cunningham 2005). Similarly, context-specific differences in hybridization occur in M. trossulus × M. galloprovincialis zones (Brannock et al. 2009). Among the new Northeast Atlantic M. trossulus × M. edulis contact sites, there seems to be a range of genotypic structures ranging from bimodality—parallel to the NW Atlantic zones—toward more advanced mixing, although nowhere approaching a hybrid swarm situation. At least four successive patterns can be seen.
First, while evidence of hybridization was always seen in direct examination of individual genotypes, in several instances the estimates of genotypic disequilibria were very close to the pure mixing situation, indicating an extreme of bimodal zone structure; this was seen particularly at the northern sites with low M. trossulus contribution (e.g., # 24–30, 39, 47). Plausibly such situations represent mixing from a broader mosaic-type of population structure, where M. trossulus breed in relatively pure but so far undetected patches, perhaps in an ecologically peculiar environment, and settle among a dominating M. edulis. The possible production of interspecies hybrids makes an insignificant contribution to the settling larval pool.
Second, in those samples where M. trossulus genome was dominant, the disequilibria appeared weaker, about 50% of the maxima (Fig. 4, #22, 23, 33, 37), a level comparable to what would occur e.g., if an initially non-interbred mix would have undergone one generation of random mating. Speculatively, while M. trossulus in these situations has gained a local dominance, larval flow from a regionally prevalent M. edulis simultaneously keeps the population mixed so that production of a pure M. trossulus cohort offspring is not feasible. At any rate, the parental forms still make a major part of these populations and directly contribute to the process of hybridization; backcrossing remains relatively rare. Overall, the contrast in the observed initial level of hybridization/introgression at M. trossulus versus M. edulis dominated sites makes an interesting parallel to the similar asymmetry of introgression in the NWA hybrid zones of the Canadian maritimes (see Riginos and Cunningham 2005).
Third, in the Norwegian contact populations both from the Bergen and Kirkenes regions, interbreeding appears to be more extensive than in the Russian sector. Measures of disequilibria are <50% maximum even in strongly mixed populations (whether with M. trossulus or M. edulis dominance; Fig. 4), and the mixing is illustrated by the hybrid-index histograms which display relatively even representation of various hybrid classes, and little indication of bimodality (Fig. 5).
Fourth, a further level of mixing is seen in the contact zone between the North Sea and Baltic mussels, for which a large sample from the middle of the relatively steep Öresund cline was examined (#9). The genotypic structure here is definitely unimodal (Fig. 5d), indicating advanced interbreeding between the parental forms. Two notions should however be made when placing this in the framework of the unimodality vs bimodality scale of mussel hybrid zones discussed by Riginos and Cunningham (2005):
Thus, when describing the Baltic M. edulis–M. trossulus mix as an archetype of a unimodal zone, Riginos and Cunningham (2005) indeed did not refer to the current Baltic-North Sea contact zone, but to what we consider as the pure Baltic mussel, from within the Baltic. Within that “zone” our allozyme markers are nearly fixed for T alleles (and thus not informative for its disequilibrium structure), whereas the introgressed, polymorphic DNA markers exhibit a panmictic equilibrium distribution of the unimodal type (Riginos and Cunningham 2005; Kijewski et al. 2006). In effect, the inner Baltic populations represent an independent hybrid swarm that has lost its contact nature. But for a comparison of genotypic structures in different dynamic inter-taxon contact zones, the NWA and NEA situations should thus rather be contrasted with the contact between the Baltic swarm and the North Sea M. edulis, not with the intra-Baltic mixture itself. In this context, the unimodality in the swarm-M. edulis contact in Fig. 5c is even the more remarkable.
For the other notion, despite the unimodality in the Öresund contact (#9), in the general context of hybrid zones where taxa interact (e.g., Barton and Hewitt 1985), the disequilibria estimated in the middle of the Baltic-North Sea zone here (R′/R′max = 21%) and similarly in Väinölä and Hvilsom (1991) (R′/R′max = 36%) are still remarkably high. They imply strong selection and limited dispersal, if these are the forces that retain a stability of the zone (Väinölä and Hvilsom 1991). The apparent stability of the zone is seen from the successive studies in Theisen (1978), Väinölä and Hvilsom (1991), and Fig. 2 here.
Factors that could contribute to the differences in levels and patterns of hybridization in the various contacts could involve (1) the different ages of the contacts, particularly a recentness of the new NEA contacts and insufficient time for amalgamation there, (2) ecological differences between the taxa that contribute to the retention of pure stocks in alternative habitats, and (3) differences in the selection against hybrids which prevents the propagation of further generation hybrid classes. The simple time hypothesis (1) arises from the presumed age difference between the Baltic-North Sea and the other European zones (NEA). However, the genotypic structures of the NEA zones rather resemble that of the NWA zones, which presumably are relatively old, i.e., have existed through the post-glacial without leading to fusion (Rawson and Harper 2009); therefore, the alternative reasons should also be considered.
On the other hand, the Bergen samples (#14, 15), which appear intermediate in terms of the genotypic disequilibria (Fig. 4), also show other features that would place them between the North Russian and the Baltic contact zones and could indicate their intermediate age. The Bergen material shows clear differences in the estimates of genetic mixing based on different markers (Fig. 6). This situation may be compared to the more extreme one in the presumably older Baltic swarm, where different genes have been either fixed to alternative species backgrounds, or retained a polymorphic state (e.g., Riginos and Cunningham 2005). Specifically, the Bergen populations of mixed composition show a relatively higher frequency of the edulis F mitochondrion (which in the Baltic has eventually been fixed on a predominant M. trossulus background). Further, no trossulus M mitochondrion was so far seen (verification needed). Even the estimate of the nuclear ITS mixing proportion markedly deviated from the allozyme estimate. Such a deviation is however in part (only) expected for ITS, which strictly does not represent a single locus: it is part of a multicopy operon, and in backcrossing situations would show an excess of heterozygote phenotypes (cf. Kijewski et al. 2006).
Characteristics and identity of the Baltic mussel
The new Northeast Atlantic M. trossulus occurrences described here, and those from Western Scotland in Beaumont et al. (2008) and Zbawicka et al. (2010), all seem to retain a relatively pure component of the North American M. trossulus, characterized by the original parental gene combinations, in at least a part of each population. As noted, that is in contrast to the mussels of the inner Baltic waters, where different loci (or genomes) exhibit different affinities to the parental taxa, but on average the relationships of different individuals to the parental taxa do not essentially differ: populations are in local genotypic equilibrium. While not primarily based on current data, this conclusion deserves clarification, since some confusion seems to remain about the number and identity of the mussel taxa within the Baltic Sea.
The initial characterization of the Baltic mussel as a close relative of the Pacific M. trossulus was on the basis of the very allozyme characters that were used in the original M. trossulus diagnosis (Varvio et al. 1988; McDonald and Koehn 1988; Väinölä and Hvilsom 1991). The current data corroborate a basin-wide genetic purity and uniformity of the Baltic populations from southern Sweden to the northern distribution limits, in terms of a representative allozyme character Gpi. Meanwhile, data on a number of PCR-based nuclear DNA markers show that populations from various parts of the Baltic do indeed have a mixed genetic background of M. trossulus and M. edulis alleles, but in a genetic equilibrium situation that can be characterized as a hybrid swarm (Bierne et al. 2003; Riginos and Cunningham 2005; Kijewski et al. 2006). Further, other characters, particularly the F mtDNA, are completely derived from M. edulis. Overall, the genomes of the Baltic mussel populations thus represent a mixture of genes of the two systematic ancestries, with different loci either predominantly derived from one or the other species (allozymes vs. mtDNA), or almost equally represented (nDNA markers). This can be characterized as a mosaic genomic composition (cf. Nikula et al. 2008). The peculiar mosaic nature is further emphasized by the composition at another set of characters which seem to be unique to the Baltic mussel, and thus most likely represent post-colonization evolution. These involve the Lap allozyme locus, which has a unique dominant allele in the Baltic (Lap 100, Väinölä and Hvilsom 1991), and the M mitochondrial genome, which seems to be a novel recombinant product of the edulis M and F mtDNAs, not found anywhere else (Zbawicka et al. 2007).
Regarding the systematic treatment of the Baltic mussel, there are three points to be stressed: First, while the presence of both M. trossulus and M. edulis genomic influence is evident in mussels throughout the basin, there is no evidence of two taxa of Mytilus being present in the inner parts of the basin, beyond the immediate neighborhood of the main transition zone in the Öresund. The Baltic mussel represents a single, locally uniform and geographically relatively homogeneous gene pool.
Second, in the North European context, the outstanding property of the Baltic Mytilus is its distinction from the neighboring “pure” M. edulis of the North Sea and the NE Atlantic, at a level comparable to the distinction between the well-recognized taxa M. edulis and M. galloprovincialis. A naming convention is needed to reflect this feature even in routine ecological studies, which often imply a comparability of their results to mussel data from other regions. If a simple binomial (either M. edulis or M. trossulus) must be used, the choice should be M. trossulus since (1) this involves the essential indication of distinction from the North Sea mussels and allows no confusion with it, (2) on average from the so far studied characters the genetic affinity to M. trossulus is closer than that to M. edulis (3) the initial taxonomic diagnosis of M. trossulus by McDonald and Koehn (1988) was exclusively based on allozyme characters, and by that criterion the Baltic mussels would score as unequivocal M. trossulus. Nevertheless, more correctly, a label that also highlights the uniqueness of the Baltic stock with respect to the (putatively) ancestral North American M. trossulus should be employed: the informal “Baltic mussel”, “Baltic M. trossulus”, or “[Baltic] M. trossulus × M. edulis” may be recommended at this stage.
Third, in evolutionary terms, the Baltic mussel most likely represents an occasion of rapid evolution of a new local taxon, by means of combining genetic elements from different ancestral sources and of rapid spread of unique characters (such as Lap 100), plausibly as an adaptation to the peculiar Baltic environment or through associated genetic hitch-hiking. Given the information on the origin and colonization history of this peculiar environment, much of this evolution probably took place within the past 8,000 years. In all, the structure and status of the Baltic mussel make a parallel to those of the other Baltic dominant bivalve Macoma balthica, which similarly seems to represent a post-glacial mixture of Atlantic and Pacific genomic elements (Väinölä 2003; Nikula et al. 2008). The Baltic mussel also shows interesting parallels to a number of other recently evolved animal taxa which feature mosaic hybrid genomes, turning up even among vertebrates including fish and mammals (Nolte et al. 2005; Larsen et al. 2010). Ultimately, the Baltic mussel may deserve its own formal taxonomic designation.
Conclusion
We describe the widespread presence of Mytilus trossulus on North European coasts of Norway and Russia, locally along with the dominant Mytilus edulis. The new occurrences are genetically distinct from the Baltic mussel which also has been classified as M. trossulus, but which rather represents a plausibly stable M. trossulus × M. edulis hybrid swarm of mosaic genomic composition, and exhibits an equilibrium genetic structure throughout the basin. Unlike the Baltic mussel, the new northern occurrences retain the original M. trossulus mitochondrial genomes and full M. trossulus nuclear composition at both allozyme and nuclear DNA loci. Also in the new contacts, the two types of mussel hybridize, but this hybridization is less extensive than in the contact zone between the Baltic mussel and North Sea M. edulis in the Danish straits. Judged from the relative genotypic disequilibria, even the various new contact areas show different patterns of genomic mixing. By far at the Russian sites, a bimodal genotypic structure with dominance of parental forms was seen, whereas in Norway, the hybridization has been more extensive.
From allozyme data, the new European finds of M. trossulus are genetically very close to those in the Northwest Atlantic, which in turn have been argued to be derived from NE Pacific ancestors early after the Last Glacial Maximum (ca 10 kyr ago). We hypothesize that the new finds represent a wave of relatively recent trans-Atlantic cryptic invasions of the North American M. trossulus to North European harbors with ship traffic, plausibly during the twentieth century. This adds a new component to the evidence of the geographical and genetic mixing processes of the Northern Hemisphere littoral mollusks, whose systematic complexity has been shaped by repeated inter- and transoceanic dispersal events and vicariance periods through the Plio-Pleistocene, and is now undergoing a new round of reshuffling.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank all who participated in mussel collection, including M.V. Katolikova, G. Ridgway, L.A. Basova, D.V. Belyaeva, V.J. Berger, E.N. Bufalova, K.V. Galaktionov, A.V. Grishankov, K. Keynäs, V.V. Klimenko, H. Laakkonen, D.L. Lajus, H. Lehväslaiho, R. Nikula, G.G. Paskerova, A.V. Poloskin, D.V. Redkin, H. Rockas, N.N. Shunatova, S.N. Shtinnikov, A.A. Sukhotin and S. Varvio. We are also grateful to M.V. Katolikova, H. Mäkinen, H. Nousianen, K. Nylund, P. Seppä and S. Varvio for their contributions to the laboratory work. Over the years, parts of the study have been supported by grants from the Academy of Finland (1011698, 47618, 127471, and BIREME collaborative grant 207862), CIMO, the Emil Aaltonen Foundation, the Nessling Foundation, and the Russian Fund for Basic Research (04-04-57808-AF2004a and 08-04-01315-a).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aljanabi SM, Martinez I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acid Res. 1997;25:4692–4693. doi: 10.1093/nar/25.22.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipova TA, Guerasimova OV, Panasenko LD, Sennikov AM (1984) Quantitative distribution of commercially important invertebrates along Murman coast. In: Benthos of the Barents Sea: distribution, ecology and population structure. USSR Acad Sci, Kola Scientific Centre, MMBI, Apatity, pp 113–123 (in Russian)
- Apte S, Holland BS, Godwin LS, Gardner JPA. Jumping ship: a stepping stone event mediating transfer of a non-indigenous species via a potentially unsuitable environment. Biol Invasions. 2000;2:75–79. doi: 10.1023/A:1010024818644. [DOI] [Google Scholar]
- Barton NH, Hewitt GM (1985) Analysis of hybrid zones. Ann Rev Ecol Syst 16:113–148
- Beaumont AR, Hawkins MP, Doig FL, Davies IM, Snow M. Three species of Mytilus and their hybrids identified in a Scottish Loch: relicts, endemics and invaders? J Exp Mar Biol Ecol. 2008;367:100–110. doi: 10.1016/j.jembe.2008.08.021. [DOI] [Google Scholar]
- Bierne N, Daguin C, Bonhomme F, David P, Borsa P. Direct selection on allozymes is not required to explain heterogeneity among marker loci across a Mytilus hybrid zone. Mol Ecol. 2003;12:2505–2510. doi: 10.1046/j.1365-294X.2003.01936.x. [DOI] [PubMed] [Google Scholar]
- Borsa P, Daguin C, Caetano SR, Bonhomme F. Nuclear-DNA evidence that northeastern Atlantic Mytilus trossulus mussels carry M. edulis genes. J Mollusc Stud. 1999;65:504–507. doi: 10.1093/mollus/65.4.504. [DOI] [Google Scholar]
- Brannock PM, Wethey DS, Hilbish TJ. Extensive hybridization with minimal introgression in Mytilus galloprovincialis and M. trossulus in Hokkaido, Japan. Mar Ecol Prog Ser. 2009;383:161–171. doi: 10.3354/meps07995. [DOI] [Google Scholar]
- Dias PS, Dordor S, Tulett D, Piertney S, Davies IM, Snow M. Survey of mussel (Mytilus) species at Scottish shellfish farms. Aquac Res. 2009;40:1715–1722. doi: 10.1111/j.1365-2109.2009.02274.x. [DOI] [Google Scholar]
- Golikov AN, Averintsev VG (1973) Ecological description of some bottom communities of the top of Kola Bay and shallows near Cape Zhelaniya (Novaya Zemlya). In: Composition distribution and ecology of the bottom fauna of the Barents Sea. PINRO, Murmansk, pp 16–19 (in Russian)
- Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4(1):9. [Google Scholar]
- Heath DD, Rawson PD, Hilbish TJ. PCR-based nuclear markers identify alien blue mussel (Mytilus spp.) genotypes on the west coast of Canada. Can J Fish Aquat Sci. 1995;52:2621–2627. doi: 10.1139/f95-851. [DOI] [Google Scholar]
- Hilbish TJ, Mullinax A, Dolven SI, Meyer A, Koehn RK, Rawson PD (2000) Origin of the antitropical distribution pattern in marine mussels (Mytilus spp): routes and timing of transequatorial migration. Mar Biol 136:69–77
- Hummel H, Strelkov PP, Sukhotin AA, Naumov AD, Berger VJ, Koukina SE, Bek TA, Novikov GG, Lubin PA, Denisenko SG, Larroux C, Amiard-Triquet C, Calafat-Frau AM, Regoli F, Orlando E, Palerud R (1999). Biodiversity and adaptation strategies of Arctic coastal marine benthos. NIOO-report 1999-2, Yerseke, the Netherlands, ISSN 1381-6519, 98 pp
- Hummel H, Colucci F, Bogaards RH, Strelkov P. Genetic traits in the bivalve Mytilus from Europe, with an emphasis on Arctic populations. Polar Biol. 2001;24:44–52. doi: 10.1007/s003000000174. [DOI] [Google Scholar]
- Inoue K, Waite JH, Matsuoka M, Odo S, Harayama S. Interspecific variations in adhesive protein sequences of Mytilus edulis, M. galloprovincialis, and M. trossulus. Biol Bull. 1995;189:370–375. doi: 10.2307/1542155. [DOI] [PubMed] [Google Scholar]
- Kijewski TK, Zbawicka M, Väinölä R, Wenne R. Introgression and mitochondrial DNA heteroplasmy in the Baltic populations of mussels Mytilus trossulus and M. edulis. Mar Biol. 2006;149:1371–1385. doi: 10.1007/s00227-006-0316-2. [DOI] [Google Scholar]
- Larsen PA, Marchán-Rivadeneira MR, Baker RJ. Natural hybridization generates mammalian lineage with species characteristics. Proc Natl Acad Sci USA. 2010;107:11447–11452. doi: 10.1073/pnas.1000133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Chown SL. Mytilus on the move: transport of an invasive bivalve to the Antarctic. Mar Ecol Prog Ser. 2007;339:307–310. doi: 10.3354/meps339307. [DOI] [Google Scholar]
- Lukanin VV, Naumov AD, Fedyakov VV (1986) Dynamics of size-structure in a population of the White Sea mussels (Mytilus edulis L.). In: Fedyakov VV, Lukanin VV (eds) Ecological investigations of the benthic organisms of the White Sea. Zoological Institute of the USSR Academy of Sciences, Leningrad, pp 50–63 (in Russian)
- McDonald JH, Koehn RK. The mussels Mytilus galloprovincialis and M. trossulus on the Pacific coast of North America. Mar Biol. 1988;99:111–118. doi: 10.1007/BF00644984. [DOI] [Google Scholar]
- McDonald JH, Seed R, Koehn RK. Allozyme and morphometric characters of three species of Mytilus in the northern and southern hemispheres. Mar Biol. 1991;111:323–335. doi: 10.1007/BF01319403. [DOI] [Google Scholar]
- Milyutina IA, Petrov NB. Divergence of unique DNA sequences in bivalve mollusks of subfamily Mytilinae (Bivalvia Mytilidae) Mol Biol (Moscow) 1989;23:1373–1381. [PubMed] [Google Scholar]
- Milyutina IA, Petrov NB. Interpopulation divergence of unique DNA sequences of the mussel Mytilius edulis. Biol Morya. 1997;23:319–324. [Google Scholar]
- Murphy RW, Sites JW, Buth DG Jr, Haufler CH (1996) Proteins: isozyme electrophoresis. In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics, 2nd edn. Sinauer Associates, Sunderland, Massachusetts, pp 51–120
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Nikula R, Strelkov P, Väinölä R. Diversity and trans-Arctic invasion history of mitochondrial lineages in the North Atlantic Macoma balthica complex (Bivalvia: Tellinidae) Evolution. 2007;61:928–941. doi: 10.1111/j.1558-5646.2007.00066.x. [DOI] [PubMed] [Google Scholar]
- Nikula R, Strelkov P, Väinölä R. A broad transition zone between an inner Baltic hybrid swarm and a pure North Sea subspecies of Macoma balthica (Mollusca, Bivalvia) Mol Ecol. 2008;17:1505–1522. doi: 10.1111/j.1365-294X.2007.03688.x. [DOI] [PubMed] [Google Scholar]
- Nolte AW, Freyhof J, Stemshorn KC, Tautz D. An invasive lineage of sculpins, Cottus sp. (Pisces, Teleostei) in the Rhine with new habitat adaptations has originated from hybridization between old phylogeographic groups. Proc R Soc B Biol Sci. 2005;272:2379–2387. doi: 10.1098/rspb.2005.3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawson PD, Harper FM. Colonization of the northwest Atlantic by the blue mussel, Mytilus trossulus postdates the last glacial maximum. Mar Biol. 2009;156:1857–1868. doi: 10.1007/s00227-009-1218-x. [DOI] [Google Scholar]
- Rawson PD, Hilbish TJ. Distribution of male and female transmitted mitochondrial lineages in the mussels Mytilus trossulus and M. galloprovincialis on the west coast of North America. Mar Biol. 1995;124:245–250. doi: 10.1007/BF00347128. [DOI] [Google Scholar]
- Rawson PD, Hilbish TJ. Asymmetric introgression of mitochondrial DNA among European populations of blue mussels (Mytilus spp.) Evolution. 1998;52:100–108. doi: 10.2307/2410924. [DOI] [PubMed] [Google Scholar]
- Rawson PD, Secor CL, Hilbish TJ. The effects of natural hybridization on the regulation of doubly uniparental mtDNA inheritance in blue mussels (Mytilus spp.) Genetics. 1996;144:241–248. doi: 10.1093/genetics/144.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics. 1983;105:767–779. doi: 10.1093/genetics/105.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway G. Interpopulation variation in blue mussels Mytilus edulis L., over short distances. Sarsia. 2001;86:157–161. [Google Scholar]
- Ridgway G, Nævdal G. Genotypes of Mytilus from waters of different salinity around Bergen, Norway. Helgoland Mar Res. 2004;58:104–109. doi: 10.1007/s10152-004-0175-5. [DOI] [Google Scholar]
- Riginos C, Cunningham CW. Local adaptation and species segregation in two mussel (Mytilus edulis × Mytilus trossulus) hybrid zones. Mol Ecol. 2005;14:381–400. doi: 10.1111/j.1365-294X.2004.02379.x. [DOI] [PubMed] [Google Scholar]
- Riginos C, Henzler CM. Patterns of mtDNA diversity in North Atlantic populations of the mussel Mytilus edulis. Mar Biol. 2008;155:399–412. doi: 10.1007/s00227-008-1038-4. [DOI] [Google Scholar]
- Rohlf FJ. NTSYS-pc. Numerical taxonomy and multivariate analysis system. V 1.60. Exeter Software. New York: Setauket; 1990. [Google Scholar]
- Seed RS. Systematics, evolution and distribution of mussels belonging to the genus Mytilus: an overview. Am Malacol Bull. 1992;9:123–137. [Google Scholar]
- Skibinski DOF, Beardmore JA. A genetic study of intergradation between Mytilusedulis and Mytilusgalloprovincialis. Experientia. 1979;35:1442–1444. doi: 10.1007/BF01962773. [DOI] [PubMed] [Google Scholar]
- Skibinski DOF, Beardmore JA, Cross TF. Aspects of the population genetics of Mytilus (Mytilidae; Mollusca) in the British Isles. Biol J Linn Soc. 1983;19:137–183. doi: 10.1111/j.1095-8312.1983.tb00782.x. [DOI] [Google Scholar]
- Smietanka B, Zbawicka M, Wołowicz M, Wenne R. Mitochondrial DNA lineages in the European populations of mussels (Mytilus spp.) Mar Biol. 2004;146:79–92. doi: 10.1007/s00227-004-1418-3. [DOI] [Google Scholar]
- Strelkov P, Nikula R, Väinölä R. Macoma balthica in the White and Barents Seas: properties of a widespread marine hybrid swarm. Mol Ecol. 2007;16:4110–4127. doi: 10.1111/j.1365-294X.2007.03463.x. [DOI] [PubMed] [Google Scholar]
- Stuckas H, Stoof K, Quesada H, Tiedemann R. Evolutionary implications of discordant clines across the Baltic Mytilus hybrid zone (Mytilus edulis and Mytilus trossulus) Heredity. 2009;103:146–156. doi: 10.1038/hdy.2009.37. [DOI] [PubMed] [Google Scholar]
- Theisen BF. Allozyme clines and evidence of strong selection in three loci in Mytilus edulis L. (Bivalvia) from Danish waters. Ophelia. 1978;17:135–142. [Google Scholar]
- Toro J, Innes DJ, Thompson RJ. Genetic variation among life-history stages of mussels in a Mytilus edulis–M. trossulus hybrid zone. Mar Biol. 2004;145:713–725. [Google Scholar]
- Väinölä R (1985) Sinisimpukan geenifrekvenssivaihtelu Itämeressä. M.Sc. thesis, University of Helsinki
- Väinölä R. Repeated trans-Arctic invasions in littoral bivalves: molecular zoogeography of the Macoma balthica complex. Mar Biol. 2003;143:935–946. doi: 10.1007/s00227-003-1137-1. [DOI] [Google Scholar]
- Väinölä R, Hvilsom MM. Genetic divergence and a hybrid zone between Baltic and North Sea Mytilus populations (Mytilidae; Mollusca) Biol J Linn Soc. 1991;43:127–148. doi: 10.1111/j.1095-8312.1991.tb00589.x. [DOI] [Google Scholar]
- Varvio SL, Koehn RK, Väinölä R. Evolutionary genetics of the Mytilus edulis complex in the North Atlantic region. Mar Biol. 1988;98:51–60. doi: 10.1007/BF00392658. [DOI] [Google Scholar]
- Vermeij GJ. From Europe to America: Pliocene to Recent trans-Atlantic expansion of cold-water North Atlantic mollusks. Proc R Soc B Biol Sci. 2005;272:2545–2550. doi: 10.1098/rspb.2005.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wares JP, Cunningham CW. Phylogeography and historical ecology of the North Atlantic intertidal. Evolution. 2001;55:2455–2469. doi: 10.1111/j.0014-3820.2001.tb00760.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.2307/2408641. [DOI] [PubMed] [Google Scholar]
- Zbawicka M, Burzyński A, Wenne R. Complete sequences of mitochondrial genomes from the Baltic mussel Mytilus trossulus. Gene. 2007;406:191–198. doi: 10.1016/j.gene.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Zbawicka M, Burzyński A, Skibinski D, Wenne R. ScottishMytilus trossulusmussels retain ancestral mitochondrial DNA: complete sequences of male and female mtDNA genomes. Gene. 2010;456:45–53. doi: 10.1016/j.gene.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Zolotarev VN, Shurova VN. Relations of prismatic and nacreous layers in the shells of the mussel Mytilus trossulus. Biol Morya. 1997;23:26–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.