Abstract
Although the mechanism of Aβ action in the pathogenesis of Alzheimer's disease (AD) has remained elusive, it is known to increase the expression of the antagonist of canonical wnt signalling, Dickkopf-1 (Dkk1), whereas the silencing of Dkk1 blocks Aβ neurotoxicity. We asked if clusterin, known to be regulated by wnt, is part of an Aβ/Dkk1 neurotoxic pathway. Knockdown of clusterin in primary neurons reduced Aβ toxicity and DKK1 upregulation and, conversely, Aβ increased intracellular clusterin and decreased clusterin protein secretion, resulting in the p53-dependent induction of DKK1. To further elucidate how the clusterin-dependent induction of Dkk1 by Aβ mediates neurotoxicity, we measured the effects of Aβ and Dkk1 protein on whole-genome expression in primary neurons, finding a common pathway suggestive of activation of wnt–planar cell polarity (PCP)–c-Jun N-terminal kinase (JNK) signalling leading to the induction of genes including EGR1 (early growth response-1), NAB2 (Ngfi-A-binding protein-2) and KLF10 (Krüppel-like factor-10) that, when individually silenced, protected against Aβ neurotoxicity and/or tau phosphorylation. Neuronal overexpression of Dkk1 in transgenic mice mimicked this Aβ-induced pathway and resulted in age-dependent increases in tau phosphorylation in hippocampus and cognitive impairment. Furthermore, we show that this Dkk1/wnt–PCP–JNK pathway is active in an Aβ-based mouse model of AD and in AD brain, but not in a tau-based mouse model or in frontotemporal dementia brain. Thus, we have identified a pathway whereby Aβ induces a clusterin/p53/Dkk1/wnt–PCP–JNK pathway, which drives the upregulation of several genes that mediate the development of AD-like neuropathologies, thereby providing new mechanistic insights into the action of Aβ in neurodegenerative diseases.
Keywords: Alzheimer's, amyloid, clusterin, Dickkopf-1, tau, wnt
Introduction
Alzheimer's disease (AD) is characterised by two neuropathological lesions: plaques, composed of aggregated β-amyloid (Aβ) peptides,1 and neurofibrillary tangles, composed of hyperphosphorylated forms of tau.2 It is widely accepted that Aβ is an initiating factor in the pathological process leading to tau pathology, a concept encapsulated in the ‘amyloid cascade hypothesis' formulated over 20 years ago,3 although the details of this cascade remain largely unknown.4, 5
Wnt signalling has been widely implicated in neurodegeneration.6, 7, 8, 9, 10, 11, 12, 13 Canonical wnt/β-catenin signalling promotes cell survival,14 whereas Aβ induces the neuronal expression of the canonical wnt antagonist, Dickkopf-1 (Dkk1).8 Silencing DKK1 protects against Aβ-induced apoptosis and tau phosphorylation,8 and neutralising the Dkk1 protein blocks the deleterious effects of Aβ on synapses.15 Clusterin, the product of CLU, recently identified by large genome-wide association studies to be a susceptibility factor for late-onset AD16, 17 also promotes cell survival,18 whereas both Dkk119 and clusterin20 protein levels are increased in amyloid-based mouse models of AD and clusterin is increased early in disease in blood in humans.20, 21
As both clusterin and Dkk1 are cell survival factors, are implicated in AD pathogenesis and are regulated by wnt signaling,8, 22, 23 we speculated that both lie on a common pathway, most likely to be the amyloid cascade.
Materials and methods
Primary neuronal cultures
Primary neuronal cultures were generated from Sprague Dawley E18 rat and C57BL/6J E16 mouse embryos by papain dissociation according to the manufacturer's instructions (Worthington, Lakewood, NJ, USA) and cultured as described.24
Aβ preparation
Aβ25-35 and Aβ35-25 peptides were solubilised in water at 2 mg ml−1 and incubated at 37 °C for 1 h. Aβ1-42 oligomer preparation has been described.25
siRNA knockdown
Small interfering RNA (siRNA) oligonucleotides were designed as previously described26 with dTdT 3′overhangs and a 5′-thiol modification to the sense strand and coupled to Pen1 as described.27
Western blotting
Western blotting was performed as previously described.28 Antibodies were detected using fluorophor conjugates emitting at 680/700 or 800 nm with an infrared scanner (LI-COR Biosystems, Cambridge, UK). Phosphoepitope immunoreactivity values were divided by non-phospho-dependent immunoreactivity values and are presented as arbitrary units.
Human samples
Human hippocampal brain samples were obtained from the Medical Research Council (MRC) London Neurodegeneration Brain Bank (Denmark Hill, London, UK). Average ages were: Controls, 70.1; AD, 78.4; and frontotemporal dementia (FTD), 72.3 years. Average post-mortem delays were: Control, 33.1 h; AD, 35.3 h; and FTD, 18.2 h. Sexes were similarly distributed among the three groups. The 10 nondemented controls had either no amyloid or tau pathology or a Braak stage score of <2. Of the 10 AD cases, 8 had Braak stage scores of 6 and the remaining 2 cases had Braak scores of 5. Frontotemporal lobar degeneration was the primary diagnosis for all 10 FTD cases. Frozen tissues were thawed and homogenised in Trizol and total RNA extracted according to the manufacturer's instructions (Invitrogen, Paisley, UK).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA (1 μg) was reverse transcribed using random hexamers and a Taqman RT kit (Applied Biosystems, Cheshire, UK) according to the manufacturer's instructions. PCR primers were designed using the Universal Probe Library package (Roche Molecular Biochemicals, Lewes, UK) for rodent genes and Primer Bank (Harvard.edu) for human genes, and used in SYBR Green-based PCR reactions performed on a Bio-Rad Dyad Disciple thermal cycler (Bio-Rad, Bath, UK). Relative quantification of gene expression between samples was determined using the 2-ΔΔCT method as described by Livak and Scmittgen.29 Internal control genes used to normalise for RNA input were HPRT for rodent samples and GAPDH for human samples. All samples were run in triplicate from three independent experiments. The mean crossing threshold (CT) values for both the target and internal control genes in each sample were determined and the 2-ΔΔCT calculations performed. The fold change in the target genes (after normalising to the internal control gene) were calculated for each sample and the mean calculated. Statistical significance was determined by one-way analysis of variance and post hoc t-tests. Data are represented as normalised fold increases over control samples. For human samples, the resultant gene expression data are represented as box-and-whisker plots to show data spread.
Statistical analysis
All routine experiments (for example, cell survival assays and immunblotting) were performed in duplicate or more and repeated a minimum of three times. Unless otherwise stated, the statistical significance of such data was determined by one-way analysis of variance with post hoc t-test or by Student's t-test, using SPSS or Excel, respectively. Values are given as mean±s.e.m., unless otherwise stated. Processing and statistical analyses of microarray and bioinformatics data are described in detail in the appropriate sections of the Supplementary Information.
Results
The induction of Dkk1 by Aβ is neurotoxic and clusterin dependent
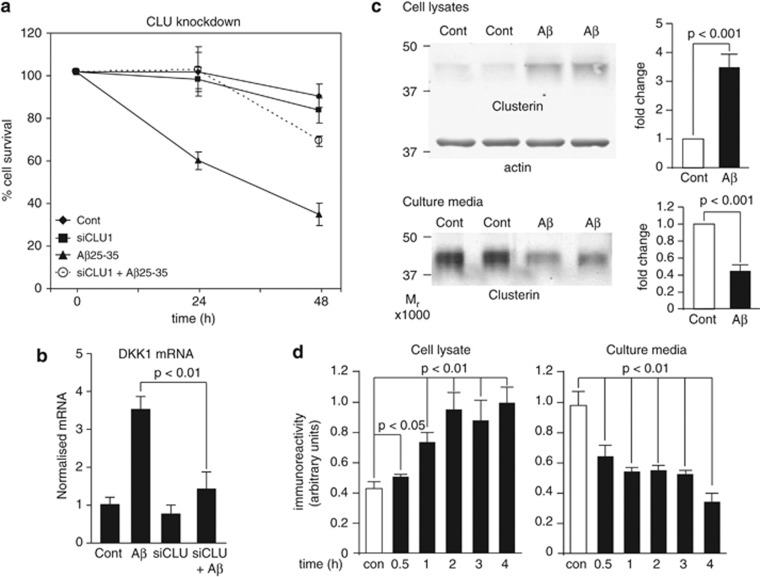
Caricasole et al.8 demonstrated that the Aβ peptide fragment, Aβ25-35, induces neuronal expression of the wnt antagonist Dkk1 and that silencing of DKK1 blocks Aβ neurotoxicity. Using a penetrating peptide (Pen1)-coupled siRNA duplex to the CLU gene, we found that knockdown of clusterin also protects against 20 μM Aβ25-35-induced neurotoxicity (Figure 1a) and furthermore prevents the rapid induction of DKK1 expression by Aβ (Figure 1b). Conversely, a 3 h treatment of neurons with Aβ25-35 resulted in a substantial (∼3.5-fold) increase in intracellular clusterin protein with a concomitant 55% decrease in clusterin in the cell medium (Figure 1c), in line with previous evidence that intracellular clusterin is pro-apoptotic whereas secreted clusterin is cytoprotective.30, 31 These acute changes in clusterin protein (within 30 min; Figure 1d) occur in the absence of changes in clusterin expression and are specific to Aβ as other stressors had little or different effects on clusterin. Thus, both ultraviolet (UV) irradiation (300 Joules m−2) and the proteasome inhibitor MG132 (20 μM) caused significant cell death at 24 h but UV had no detectable effects on clusterin protein, whereas MG132 caused a small decrease in cellular levels, with no effect on extracellular levels (data not shown).
Figure 1.
Aβ induction of Dickkopf-1 (Dkk1) is clusterin dependent. (a) Rat primary cortical neurons were treated with Pen1 small interfering RNA (siRNA) to CLU overnight and subsequently with 20 μM Aβ25-35 for 24 and 48 h and cell survival determined by the nuclear morphology assay. Cont, Control. (b) Neurons were treated as in (a), RNA collected after 3 h of Aβ treatment and qRT-PCR performed (detailed in Supplementary Methods). (c) Neurons were treated for 4 h with 20 μM Aβ25-35 and culture media and total cell lysates were collected and immunoblotted for clusterin. Clusterin in cell lysates was normalised to β-actin levels. (d) Neurons were treated with 20 μM Aβ25-35 for the times indicated and immunoblotted as in (c). Error bars in (c) and (d) show s.d. qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Aβ and Dkk1 induce a common signalling pathway
To explore this neurotoxic signalling pathway further, we performed whole-genome expression analyses of primary neuronal cultures following acute treatments with either Aβ (20 μM Aβ25-35 for 3 h) or recombinant Dkk1 protein (800 ng ml−1 for 2 h). Within the two resultant lists of gene independently responsive to Aβ and Dkk1, 2061 genes were common to both. A test for similarities in ordered gene lists was then performed on the two lists using the OrderedLists software package (http://www.bioconductor.org/packages/release/bioc/html/OrderedList.html) within the Bioconductor R environment. A test of the significance of their ordering by fold change was performed, and in the top 500 genes a P-value of ≤0.000 was obtained (this package returns values to a maximum of three decimal places). When gene expression was ranked in this way, we observed that of the top eight genes, five were common to the two treatments (Table 1), a finding that a hypergeometric distribution calculation indicates is very unlikely to have occurred by chance alone (P<2.2e−16). These observations suggest that Dkk1 mediates the effects of Aβ on gene expression.
Table 1. Aβ and Dkk1 induce a common signalling pathway.
| Aβ treated | Dkk1 treated | |
|---|---|---|
| 1 | EGR1 | EGR1 |
| 2 | IER2 | NAB2 |
| 3 | NAB2 | VCL |
| 4 | CCND1 | KLF10 |
| 5 | FOS | TIMP1 |
| 6 | BAIAP2 | FOS |
| 7 | ATF6B | CCND1 |
| 8 | KLF10 | CCL20 |
| 9 | HNRNPL | TUBB6 |
| 10 | THOC3 | RHOQ |
Microarray data from Aβ25-35-treated (20 μM, 3 h) primary mouse cortical neurons and Dkk1-treated (800 ng ml−1, 2 h) primary rat cortical neurons were processed (Supplementary Information) and ranked by fold change. The top 10 genes from each treatment are shown.
Only one of these common genes, CCND1 (cyclin D1), is a known canonical wnt target,32 with the remaining four encoding transcription factors: EGR1 (early growth response-1), NAB2 (Ngfi-A-binding protein-2), KLF10 (Krüppel-like factor-10) and FOS (FBJ murine osteosarcoma viral oncogene homologue). We confirmed the induction of these genes by both Aβ25-35 and by Dkk1 using qRT-PCR (Supplementary Figure S1a and 1b).
In these experiments we used Aβ25-35 to confirm the observations of Caricasole et al.8 and also noting data suggesting that this peptide contains the ‘active' portion of Aβ and has been detected in human brain.33 However, oligomeric forms of Aβ1-42 are widely accepted to be the physiologically relevant neurotoxic species of Aβ, and we therefore assessed the effects of 3 μM oligomeric Aβ1-42 (Aβ1-42(olig)), finding that these Aβ species also increased the expression of all five Aβ/Dkk1 target genes (Supplementary Figure S1c) and of DKK1 (Supplementary Figure S1d) and had very similar effects on clusterin (data not shown). Thus, in our hands, the effects of Aβ25-35 and Aβ1-42(olig) were largely indistinguishable, in agreement with findings from other laboratories.34 The oligomeric nature of the Aβ1-42(olig) preparation is shown in Supplementary Figure S1e.
Aβ-induced gene expression is dependent on p53 and is necessary and sufficient for neurotoxicity
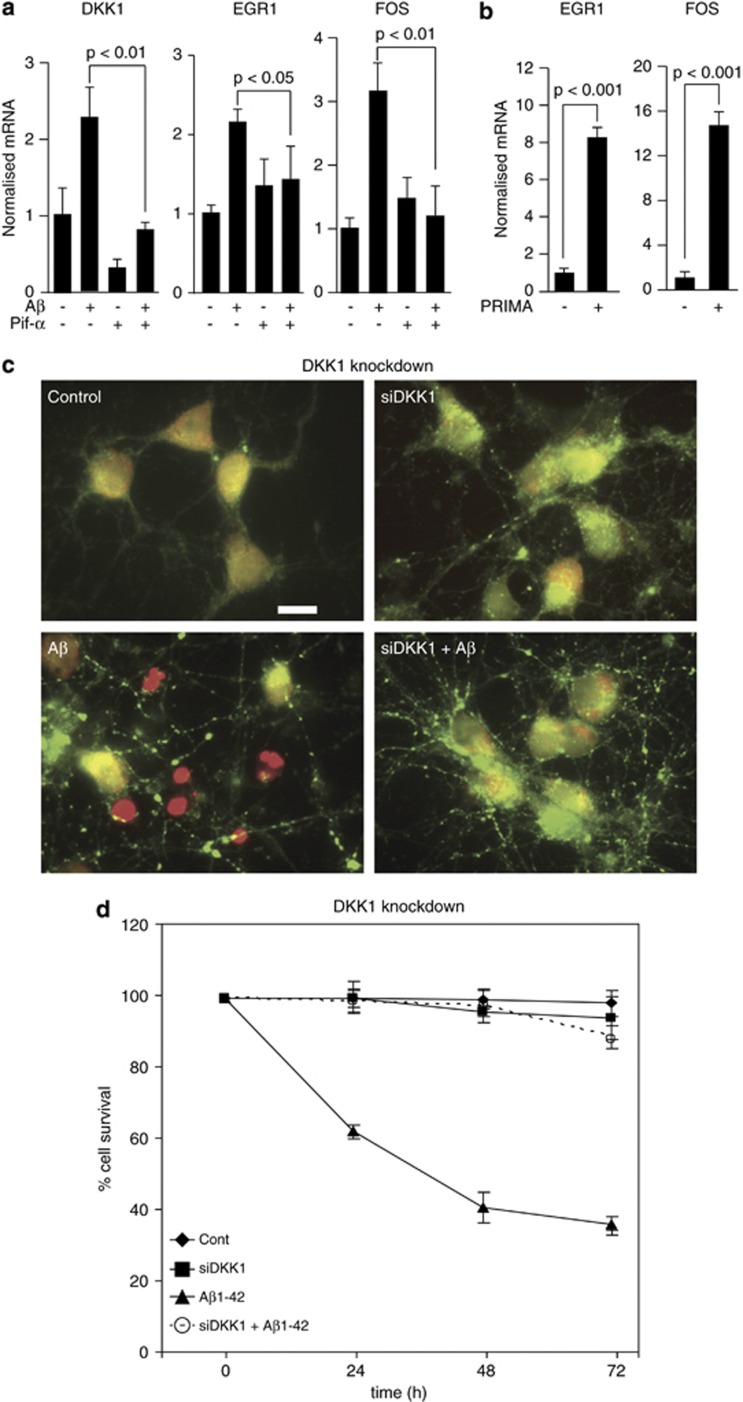
Investigating the molecular mechanism by which Aβ drives gene expression, we first verified the p53 dependency of DKK1 induction using a small-molecule inhibitor of p53 transcriptional activity, pifithrin-α. This blocked the Aβ1-42(olig) induction of DKK1 and also of the Aβ/Dkk1 target genes EGR1 and FOS (Figure 2a). Conversely, activating p53 with PRIMA1 induced DKK1 (data not shown) and target gene expression (Figure 2b). A penetrating siRNA duplex was then generated to target DKK1. Silencing of DKK1 in primary rodent neurons also blocked Aβ1-42(olig) induction of the target genes (data not shown), together confirming that Aβ induction of Dkk1 is p53 dependent,8 and that Aβ induction of the target genes is p53 and Dkk1 dependent.
Figure 2.
Aβ-induced gene expression is dependent on p53 and is necessary and sufficient for neurotoxicity. (a) Rat cortical neurons were treated with 10 μM pifithrin-α for 18 h as indicated and subsequently with Aβ1-42(olig) (3 μM, 3 h). The expression levels of DKK1 (Dickkopf-1), EGR1 (early growth response-1) and FOS (FBJ murine osteosarcoma viral oncogene homologue) were determined by qRT-PCR. (b) Neurons were treated as in (b) using 10 μM PRIMA-1 and then with Aβ and qRT-PCR performed. (c) Rat neurons were treated o/n at 7 d.i.c. with control or Pen1 small interfering RNA (siRNA) to DKK1 (160 nM), and then with 3 μM Aβ1-42(olig) for 24 h and cytotoxicity assayed by the live/dead assay. Healthy cells are labelled green and dead cells are red. Scale bar=10 μM. (d) Neurons were treated as in (c) and cell survival determined at 24, 48 and 72 h by the nuclear morphology assay. d.i.c., days in culture; o/n, over night; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
These data therefore demonstrate a pathway whereby Aβ disrupts the balance between intracellular and extracellular clusterin, resulting in a cascade of events including p53-dependent induction of DKK1 expression followed by increased expression of a set of genes including the transcription factors EGR1, NAB2, KLF10 and FOS as well as CCND1. We then investigated the relevance of this pathway to Aβ neurotoxicity, generating Pen1 siRNA duplexes to these genes. First, we pretreated primary neurons with the DKK1 siRNA or the control duplex as above and then subsequently with 3 μM Aβ1-42(olig) for up to 72 h and assessed cell survival. By the live/dead assay, silencing DKK1 afforded almost complete protection from Aβ at 24 h (Figure 2c) and by an assay of nuclear integrity at up to 72 h (Figure 2d). Similar levels of protection were observed by lactate dehydrogenase assay (data not shown), confirming Dkk1 is a necessary mediator of Aβ neurotoxicity.
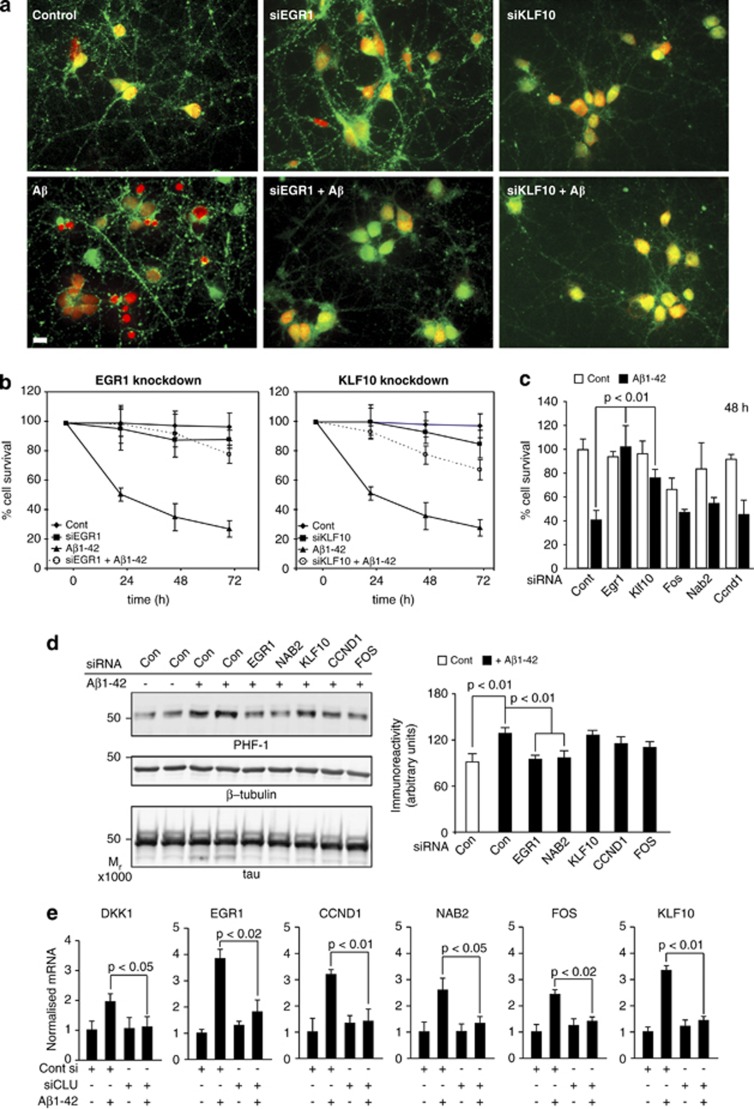
We then individually silenced each of the five genes, treated with Aβ1-42(olig) and measured cell survival as above. Silencing NAB2, FOS and CCND1 afforded no protection against Aβ, but silencing EGR1 or KLF10 each gave substantial protection as measured by the live/dead assay (Figure 3a). Using an assay of nuclear morphology (Figure 3b), EGR1 gave 80% and KLF10 gave 73% protection at 72 h and both were protective as measured by lactate dehydrogenase assay (Figure 3c), demonstrating EGR1 and KLF10 are necessary mediators of Aβ-induced neurotoxicity.
Figure 3.
EGR1 (early growth response-1), KLF10 (Krüppel-like factor-10) and NAB2 (Ngfi-A-binding protein-2) mediate neurotoxicity and tau phosphorylation. (a) Neurons were treated o/n with Pen1 small interfering RNAs (siRNAs) to EGR1, FOS (FBJ murine osteosarcoma viral oncogene homologue), KLF10, NAB2, CCND1 (cyclin D1) and then with 3 μM Aβ1-42(olig) for 24 h and cytotoxicity assayed by the live/dead assay. Protective effects of siRNAs targeting EGR1 and KLF10 are shown. (b) Neurons were treated as in (a) and cell survival determined by the nuclear morphology assay up to 72 h. Significance values (not shown) for the effect of EGR1 and KLF10 siRNA on cell survival at each time point were ≤0.01. (c) Neurons were treated as in (a) and cell survival measured by lactate dehydrogenase (LDH) release. (d) Neurons were treated as in (a) and subsequently with 3 μM Aβ1-42(olig) for 4 h. Total lysates were collected and immunoblotted for phospho-tau using PHF-1. Immunoreactivity values for PHF-1 were normalised to total tau values; densitometric values are shown in the right. (e) Neurons were treated with Pen1-siCLU or control Pen1 siRNA and subsequently with 3 μM Aβ1-42(olig) for 3 h, RNA collected and qRT-PCR performed for DKK1 and the five Aβ/Dkk1 target genes. o/n, over night; qRT-PCR, quantitative reverse transcription polymerase chain reaction.
We then examined the role of the five genes in another aspect of neurotoxicity, Aβ-induced tau phosphorylation. Neurons were pretreated with the five siRNA duplexes as above and protein lysates collected after 4 h Aβ1-42(olig) treatments. Penetrating siRNAs against KLF10, FOS and CCND1 had little effect on Aβ-induced tau phosphorylation, whereas those against EGR1 or NAB2 significantly reduced Aβ-induced increases in PHF-1 immunoreactivity (Figure 3d), demonstrating that Egr1 and Nab2 are mediators of Aβ-induced tau phosphorylation at this phosphoepitope. Of note, Egr1 has been shown to drive tau phosphorylation at the PHF-1 epitope and other sites in rat brain via the activation of p35/cdk5,35 whilst, Nab2 is a regulator of Egr1 activity.36, 37
Given that silencing CLU protected against Aβ neurotoxicity in a cell survival assay and also blocked the induction of DKK1 (Figure 1 above), we asked if silencing CLU would also block the downstream target genes including the identified mediators of toxicity and increase tau phosphorylation. Rat primaries were pretreated with the si-control or siCLU and treated next day with 3 μM Aβ1-42(olig) for 3 h, RNA collected and qRT-PCR performed. Aβ induction of DKK1 and all five of the common genes, including EGR1, NAB2 and KLF10, was significantly blocked by the silencing of CLU (Figure 3e), substantiating further that CLU lies on the Aβ neurotoxic pathway and is a necessary component of the signalling cascade.
Aβ induced gene expression in amyloidopathy, but not tauopathy, brain
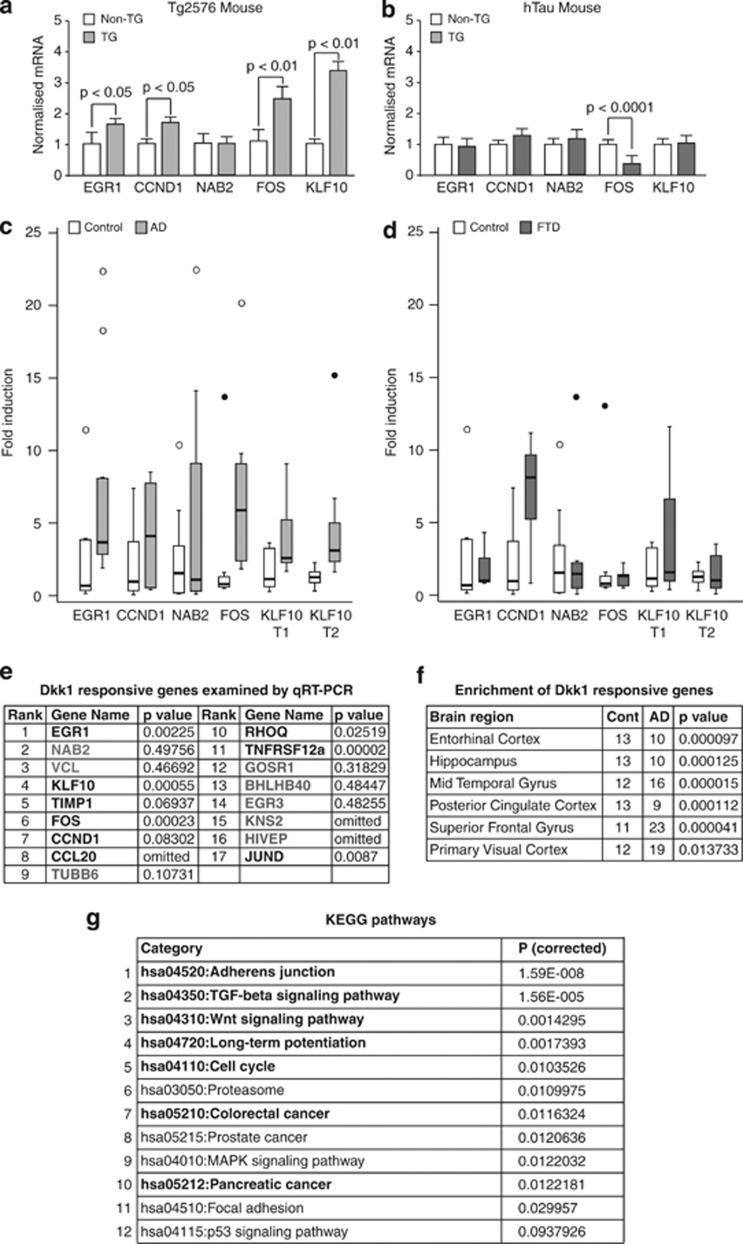
Moving from rodent primary neurons to in vivo mouse models, we measured the expression levels of the five common genes in cortex from amyloid-based (Tg257638) and tau-based (hTau39) lines and their respective nontransgenic littermates. In Tg2576 mice, four of the five genes were significantly upregulated (Figure 4a). None were upregulated in hTau mice but FOS was significantly downregulated (Figure 4b). Moving to humans, we assessed gene expression in hippocampal tissues from AD and FTD cases as both feature tau pathology but FTD lacks amyloid pathology. In AD cases, EGR1, KLF10 and FOS were significantly (P<0.01) upregulated and CCND1 showed an increased trend (P=0.083; Figure 4c). None of the genes were altered in FTD (Figure 4d). Examining a further nine of the neuronal Dkk1-responsive genes, we found JUND, RHOQ and TNFRSF12A to be significantly (P<0.05) upregulated and TIMP1 to have an increased trend (P=0.069) in AD hippocampus (see table in Figure 4e). We then looked for enrichment of the top 50 most significantly responsive Dkk1 genes in transcriptomic data sets from control and AD brain. In a large data set in which data from several cortical regions had been pooled,42 we found significant (P<0.001) gene enrichment in AD cases. In data sets from separate brain regions,40 we found enrichment in AD in all regions but with greater significance in those regions most affected by the disease (for example, mid-temporal gyrus; P=0.000015) and least in a relatively spared region (primary visual cortex P=0.014; Figure 4f), indicating that Dkk1 contributes to the expression pattern of genes in the AD brain.
Figure 4.
Aβ induced gene expression in amyloidopathy, but not tauopathy, brain. (a–d) Expression levels of the five common Aβ/Dickkopf-1 (Dkk1)-responsive genes were measured by qRT-PCR in total RNA from: cortex of (a) 12-month-old Tg2576 mice (n=9) and their non-TG littermate controls (n=7) and (b) 12-month-old hTau mice (n=5) and their non-TG littermate controls (n=5); and from hippocampi of (c) Alzheimer's disease (AD; n=10) and age-matched controls (n=9) and (d) frontotemporal dementia (FTD; n=9) and age-matched controls (n=9). TG, transgenic. Mouse data were normalised to HPRT and human data to GAPDH by the 2-ΔΔCT method. Significance was determined by one-way analysis of variance (ANOVA) and post hoc t-tests. Data are represented as normalised fold increases over control. Significance values of gene changes in human samples are given in table (e). Outlier values (between 1.5 and 3 times the interquartile range) and extreme values (>3 times the range) are shown as circles and filled circles, respectively. (e) Dkk1-responsive genes examined in AD hippocampus. Nonsignificant and omitted genes are shown in grey. (f) Human brain transcriptome data sets were mined with the top 50 most significant Dkk1-responsive genes. Significance of Dkk1 gene enrichment in AD in the six brain regions examined by Liang et al.40 are shown. Significance was determined by asymptotic globaltest (see Supplementary Information for full description). (g) KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis of human homologues of rat Dkk1-responsive genes. The pathway identifier, name and P-values after correcting for multiple testing by the method of Benjamini are shown. Pathways in bold have been associated with disease in AD brain expression data by Huang et al.41 qRT-PCR, quantitative reverse transcription polymerase chain reaction.
People with Trisomy 21 (Down's syndrome (DS)) have a high incidence of dementia almost certainly because of the presence of an additional copy of the APP gene, resulting in the excess generation of Aβ. This is reflected in the almost invariable presence of AD pathology in post-mortem brain from people with DS surviving to mid-life. Interrogating a publically available whole-genome expression data set from DS brain,43 we found highly significant enrichment of Dkk1-responsive genes (P<0.0027), adding further to the evidence that this gene signature is driven by Aβ.
Next, we performed further bioinformatics analyses on the neuronal Dkk1-responsive genes. Interrogating the Ingenuity database with the 100 most significant Dkk1-responsive genes, the top pathway identified contained EGR1, NAB2 and KLF10, the second CCND1 and FOS, from which EGR1, NAB2 and KLF10 were absent, indicating the two gene sets may serve separate functions. We then mapped the genes to a human protein–protein interaction network and used jActiveModules (Ryan Kelley, UCSD, USA) to identify sub-networks of interacting proteins that collectively show significant changes in expression.44 One key differentially expressed functional module was identified within which a number of KEGG pathways were significantly over-represented (Figure 4g). Interestingly, 7 of these top 10 Dkk1-driven pathways also appear in the top 10 disease-associated pathways (shown in bold in Figure 4g) identified by Huang et al.41 in human AD brain expression data, indicating Dkk1 affects cell signalling in the AD brain.
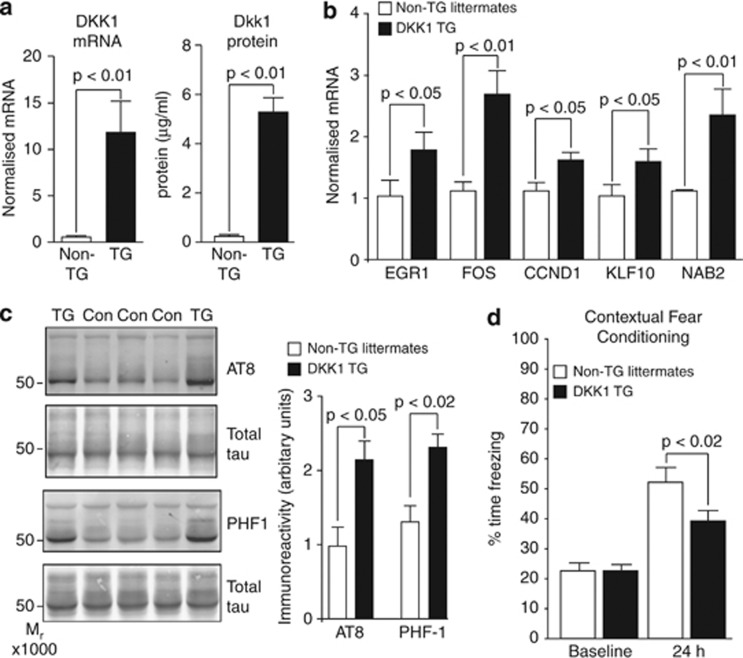
Transgenic overexpression of DKK1 induces ‘Aβ neurotoxicity pathway genes', tau phosphorylation and cognitive deficits
To model pathway activation downstream of Aβ we generated transgenic mice overexpressing murine Dkk1 in neurons. DKK1 transgenics had increased expression of Dkk1 (Figure 5a) and of the five common Aβ/Dkk1 genes in neonatal temporal cortex (Figure 5b). Examining tau phosphorylation in mice aged 20 to 25 months, we found a significant increase in hippocampus with AT8 and PHF-1 (Figure 5c), antibodies recognising phospho-tau epitopes increased in AD.45 Minor and nonsignificant increase in tau phosphorylation was observed at these epitopes from 6 to 9 months of age. To determine the effects on cognition, contextual fear conditioning was used as a measure of amygdala- and hippocampal-dependent memory recall. In young adults, no significant differences were observed. However, in older animals (14–16 months), significant (P<0.02) impairment in memory recall was observed in DKK1 transgenics compared with their littermate controls (Figure 5d). Thus, it appears that chronic overexpression of Dkk1 in brain leads to an age-dependent increase in tau phosphorylation and the appearance of cognitive deficits.
Figure 5.
Transgenic overexpression of Dickkopf-1 (DKK1) induces ‘Aβ neurotoxicity pathway genes', tau phosphorylation and cognitive deficits. (a) DKK1 expression was determined by qRT-PCR (left bar graph) and enzyme-linked immunosorbent assay (ELISA; (right bar graph) in temporal cortex of neonatal DKK1 transgenic (TG) mice (n=6) and their non-transgenic (non-TG) littermates (n=5). (b) Neonatal expression of the five Aβ/Dkk1 genes determined by qRT-PCR. (c) Immunoblots of hippocampal lysates from 18- to 24-month-old DKK1 TG (n=7) and non-TG (n=7) mice using antibodies AT8 and PHF-1. Phosphoimmunoreactivity values were normalised to total tau values, within blot (bar charts, right). (d) The 14–16-month-old Dkk1 TG (n=17) and non-TG littermates (n=16) were subjected to contextual fear conditioning. Time spent freezing upon placement in the conditioning apparatus at baseline and at 24 h after training are shown. qRT-PCR, quantitative reverse transcription polymerase chain reaction.
The Aβ neurotoxicity pathway is the wnt–planar cell polarity pathway
Having determined that Aβ induction of DKK1 is dependent upon a rapid effect on clusterin protein and that Dkk1 then drives the expression of genes mediating key AD-like pathologies in primary neurons and mouse models and would also appear to affect the transcriptome and signalling pathways active in the AD brain, we began to determine in more detail the mechanism by which Dkk1 drives the transcription of these genes.
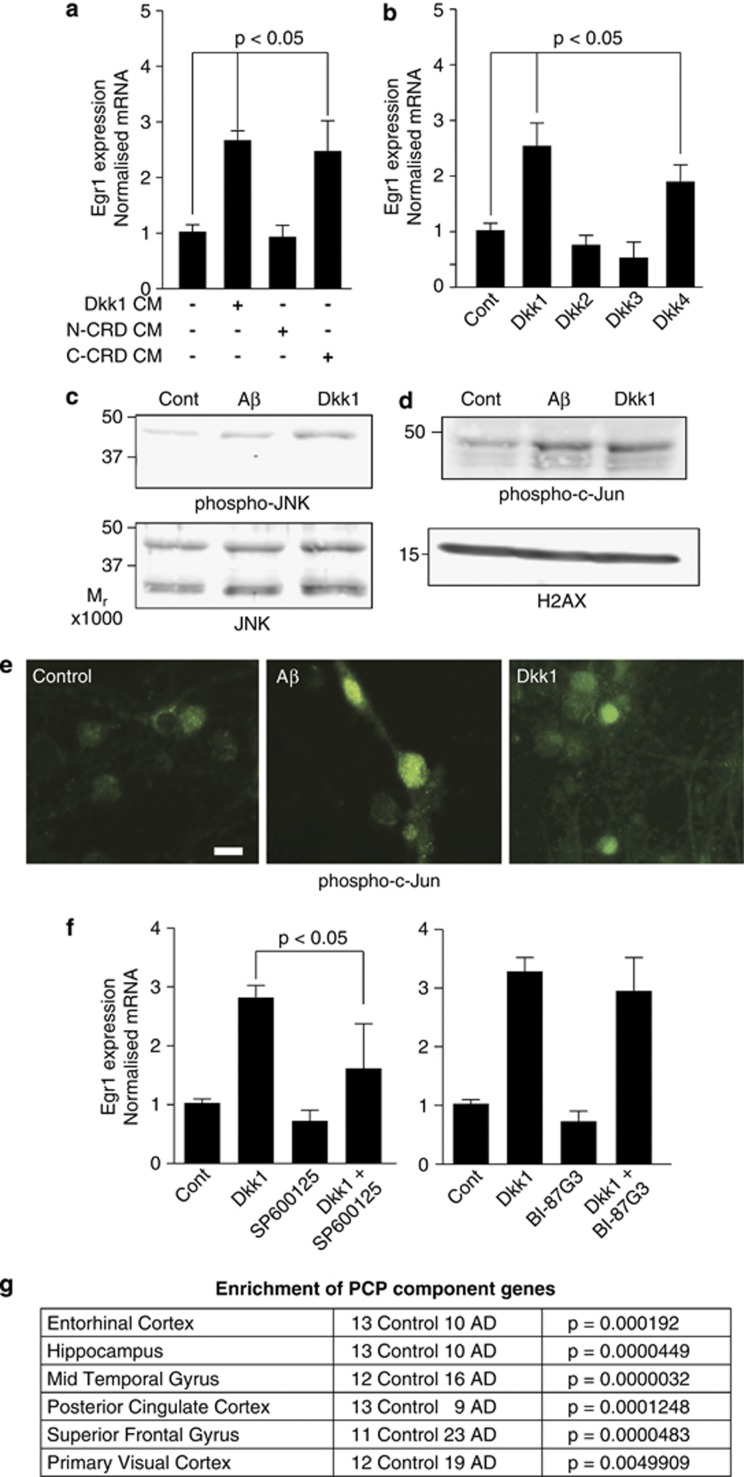
Given that the Aβ/Dkk1 genes are not recognised canonical wnt targets and as the Dkk1 protein contains N- and C-terminal cysteine-rich domains, with the C-terminal one being required for the antagonism of canonical wnt and the N-terminal one possessing wnt-independent activities,46 we generated conditioned media containing full-length or just the C-terminal or N-terminal portions of Dkk1. Treating neurons with full-length and C-terminal conditioned media activated EGR1 (Figure 6a) and FOS expression (data not shown), whereas N-terminal conditioned media did not. Primary neurons were then treated with recombinant Dkk1, Dkk2, Dkk3 and Dkk4 proteins. Dkk4 and Dkk1 induced EGR1 (Figure 6b) and FOS (data not shown) whereas Dkk2 and Dkk3 did not, mirroring the abilities of the Dkk1 family to antagonise canonical wnt.47 This indicates that the Dkk1-dependent transcriptional effects of Aβ are not via a wnt-independent activity of Dkk1, but that they do involve wnt signalling, possibly via effects on one of the noncanonical wnt pathways.
Figure 6.
The Aβ neurotoxicity pathway is the wnt–planar cell polarity (PCP) pathway. (a) Neurons were treated for 2 h with full-length, N-Terminal CRD or C-terminal CRD containing conditioned media (CM) and qRT-PCR performed. (b) Neurons were treated as in (c) with recombinant Dickkopf-1 (Dkk1), Dkk2, Dkk3 and Dkk4 proteins at 800 ng ml−1 and expression measured. (c) Neuronal lysates were immunoblotted for phospho-Thr183/Tyr185-SAPK/JNK and total SAPK/JNK following 3 h of treatments with 3 μM Aβ1-42(olig) or 800 ng ml−1 Dkk1. (d) Nuclear fractions were prepared from neurons treated as in (e) and immunoblotted for phospho-Serine63-c-Jun. Equal loading was determined using anti-H2A.X. (e) Neurons were treated as in (c), fixed and stained for phospho Ser63-c-Jun. Scale bar=10 μm. (f) Neurons were pre-treated with the c-Jun N-terminal kinase (JNK) inhibitors SP600125 and BI-87G3, each at 10 μM for 3 h, and then with 800 ng ml−1 recombinant Dkk1 for 2 h and qRT-PCR was performed. (g) Human brain transcriptomic data were mined with the wnt–PCP pathway component genes. Significance of differences in Alzheimer's disease (AD) in the six brain regions examined by Liang et al.40 are shown. CRD, cysteine rich domain; qRT-PCR, quantitative reverse transcription polymerase chain reaction; SAPK, stress activated protein kinase.
To signal via the canonical pathway, the Frizzled (Fzd) family receptors must interact with their coreceptor proteins, LRP5 or LRP6.48, 49, 50 Dkk1 antagonises canonical wnt by blocking the Fzd–LRP5/6 interaction and, in doing so, allows Fzd to drive the wnt–Ca2+ and wnt–planar cell polarity (PCP) pathways.46, 51 As PCP signalling induces gene expression via c-Jun N-terminal kinase (JNK) and its target, the transcription factor c-Jun,52, 53, 54 we looked for evidence of their activation in primary neurons following Aβ1-42(olig) and Dkk1 treatments. By western blot analysis, an increase in activated JNK1 was observed (Figure 6c). Subcellular fractionation (Figure 6d) and immunofluorescence microscopy (Figure 6e) demonstrated a concomitant increase in c-Jun activity. Examining target gene expression by qRT-PCR, the use of SP600125, an inhibitor with >20-fold selectivity for all three JNK isoforms over a range of other kinases, blocked induction by recombinant Dkk1 protein, whereas BI-87G3, an inhibitor more selective towards JNK2 and JNK3 than JNK1 at the dose used, did not (Figure 6f). Thus, the transcriptional effects of Aβ appear to be JNK1 dependent and likely because of Dkk1 activation of wnt–PCP signalling.
Finally, to investigate the relevance of PCP in AD in man, a list of wnt–PCP pathway component genes was compiled (Supplementary Figure S1f) and again used to mine the AD expression data sets.40, 42 PCP component genes were significantly (P<0.0005) altered in AD cases in the combined brain region data set,42 and in data sets from separate brain regions,40 again most significantly in AD in mid-temporal gyrus (P<3.2 × 10−6) and least in primary visual cortex (P<0.005; Figure 6g), demonstrating that wnt–PCP signalling is altered in AD brain.
Discussion
We report two novel and important findings regarding a signalling pathway through which Aβ appears to exert its neurotoxic effects. First, we have uncovered a previously unrecognised effect of Aβ on clusterin protein. Although clusterin has long been implicated in AD, and has more recently been identified as a risk gene for the sporadic form of the disease,17 its contribution to disease pathology has remained enigmatic. We demonstrate that Aβ rapidly targets the clusterin protein, causing its intracellular accumulation possibly by blocking its exit from neurons. This effect appears to be specific to Aβ, not being observed with other cytotoxic agents, and is a necessary step in the mechanism by which Aβ exerts its neurotoxic properties. Given that clusterin is known to bind Aβ, an alternative plausible explanation is that Aβ–clusterin complexes form and are internalised and this is responsible for neurotoxicity.
Second, we demonstrate that Aβ-induced changes in clusterin distribution has acute Dkk1-dependent effects on neuronal gene transcription and have identified three of its target genes, EGR1, NAB2 and KLF10, as necessary mediators of Aβ-induced neurotoxicity and its ability to drive tau phosphorylation. These transcriptional effects are mediated not by antagonism of the canonical wnt pathway, as previously thought, but by activation of noncanonical wnt signalling, specifically the wnt–PCP pathway, leading to the activation of JNK/c-Jun.
Data derived from multiple animal models and from post-mortem tissue in humans confirm this pathway is Aβ driven and is not a nonspecific result of cell stress or toxicity. Remarkably, the pathway appears to be active in DS brain too, again indicating that it is Aβ driven and lending further support to the amyloid cascade hypothesis. The mediator of this Aβ/clusterin/Dkk1-induced pathway is p53, reinforcing its emerging role in AD pathogenesis.55 EGR1, the gene most responsive to both Aβ and to Dkk1, is a necessary downstream component of Aβ/clusterin/Dkk1-induced neurotoxicity and tau phosphorylation. In a study of inducible transcription factors, only Egr1 and c-Jun, which we show likely drives EGR1 expression, were found elevated in AD brain.56 Hippocampal expression levels of Egr1 have been shown to positively correlate with disease progression in AD,57 whereas the overexpression of EGR1 in rat brain induces tau phosphorylation via its target, and regulator of cdk5, p35.35 Other Egr1 targets include the major sporadic AD risk genes APOE17 and CLU.58 Although we found no effect on CLU mRNA with acute Aβ treatments, it remains plausible that chronic Aβ exposure could increase CLU expression, contributing to a pathogenic cycle.
The deleterious synaptic effects of Aβ have recently shown to be Dkk1 dependent15 and attributed to its antagonism of canonical wnt signalling. Although inhibition of canonical wnt no doubt contributes to the synaptic effects of Aβ, and likely to its neurotoxicity too, it is worth noting that aberrant Rho signalling also causes synaptic defects and cognitive impairment59 and that the wnt–PCP pathway not only drives gene transcription via JNK but also regulates structural changes via Rho and ROCK.60 Thus, Aβ activation of the wnt–PCP pathway may also contribute to the synaptic effects of Aβ, which now warrants further investigation.
To summarise, we show that Aβ-induced neurotoxicity, including tau phosphorylation at specific epitopes, is via the CLU-dependent induction of Dkk1, with Dkk1 then driving wnt–PCP signalling to increase expression of genes that we have identified and shown to be necessary mediators of these pathological processes. In elucidating this, we have positioned a number of components in a previously unrecognised molecular pathway that mediates Aβ toxicity in rodent primary neurons. These data are confirmed in vivo in both animal models of amyloidopathy and in AD and DS brain in humans, adding considerably to the hypothesis that this pathway contributes to disease pathogenesis. Together, this further reinforces the idea that it is Aβ that drives AD pathology, a long-standing hypothesis that is now coming under scrutiny given the failure of drugs aimed at lowering brain amyloid burden to ameliorate the disease. That this pathway is dependent on the sporadic AD risk gene, CLU, and that modelling it in vivo by chronically overexpressing Dkk1 in the mouse brain gives rise to age-dependent increases in tau phosphorylation, and cognitive impairment further indicates that the cascade may be part of the pathway driving AD neuropathology in humans. It also suggests that blocking the effect of Aβ on clusterin or the ability of Dkk1 to drive wnt–PCP signalling are likely to be fruitful areas in which novel targets could be identified against which therapeutic strategies aiming to ameliorate or even halt AD could be developed.
Acknowledgments
This study was funded by the Wellcome Trust, the NIHR BRC for Mental Health at the South London and Maudsley NHS Foundation Trust, the Alzheimer's Society/BUPA Foundation, Alzheimer's Research UK, the Fundacion Alfonso Martin Escudero and the Royal Free Hampstead NHS Trust. We thank the MRC London Neurodegeneration Brain Bank and Brains for Dementia Research and Dr Michael Minchin (of Astellas Pharma Europe (UK), formerly Yamanouchi UK) and Professor Daniel Geshwind for their advice on the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Supplementary Material
References
- Glenner GG, Wong CW. Alzheimer's disease and Down's syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun. 1984;122:1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- Brion JP, Flament-Durand J, Dustin P. Alzheimer's disease and tau proteins. Lancet. 1986;2:1098. doi: 10.1016/s0140-6736(86)90495-2. [DOI] [PubMed] [Google Scholar]
- Hardy J, Allsop D. Amyloid deposition as the central event in the aetiology of Alzheimer's disease. Trends Pharmacol Sci. 1991;12:383–388. doi: 10.1016/0165-6147(91)90609-v. [DOI] [PubMed] [Google Scholar]
- Hardy J. The amyloid hypothesis for Alzheimer's disease: a critical reappraisal. J Neurochem. 2009;110:1129–1134. doi: 10.1111/j.1471-4159.2009.06181.x. [DOI] [PubMed] [Google Scholar]
- Kremer A, Louis JV, Jaworski T, F VanLeuven. GSK3 and Alzheimer's disease: facts and fiction. Front Mol Neurosci. 2011;4:17. doi: 10.3389/fnmol.2011.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DE, Soriano S, Xia X, Eberhart CG, De Strooper B, Zheng H, et al. Presenilin couples the paired phosphorylation of beta-catenin independent of axin: implications for beta-catenin activation in tumorigenesis. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- Alvarez AR, Godoy JA, Mullendorff K, Olivares GH, Bronfman M, Inestrosa NC. Wnt-3a overcomes beta-amyloid toxicity in rat hippocampal neurons. Exp Cell Res. 2004;297:186–196. doi: 10.1016/j.yexcr.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Caricasole A, Copani A, Caraci F, Aronica E, Rozemuller AJ, Caruso A, et al. Induction of Dickkopf-1, a negative modulator of the Wnt pathway, is associated with neuronal degeneration in Alzheimer's brain. J Neurosci. 2004;24:6021–6027. doi: 10.1523/JNEUROSCI.1381-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo EM, Inestrosa NC.Activation of Wnt signaling by lithium and rosiglitazone reduced spatial memory impairment and neurodegeneration in brains of an APPswe/PSEN1DeltaE9 mouse model of Alzheimer's disease Mol Psychiatry 201015272–285.,228. [DOI] [PubMed] [Google Scholar]
- De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, Major MB, et al. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer's disease. Proc Natl Acad Sci USA. 2007;104:9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudher A, Chapman S, Richardson J, Asuni A, Gibb G, Pollard C, et al. Dishevelled regulates the metabolism of amyloid precursor protein via protein kinase C/mitogen-activated protein kinase and c-Jun terminal kinase. J Neurosci. 2001;21:4987–4995. doi: 10.1523/JNEUROSCI.21-14-04987.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick R, Pollard CC, Asuni AA, Mudher AK, Richardson JC, Rupniak HT, et al. Presenilin 1 independently regulates beta-catenin stability and transcriptional activity. J Biol Chem. 2001;276:48554–48561. doi: 10.1074/jbc.M108332200. [DOI] [PubMed] [Google Scholar]
- Rosen EY, Wexler EM, Versano R, Coppola G, Gao F, Winden KD, et al. Functional genomic analyses identify pathways dysregulated by progranulin deficiency, implicating Wnt signaling. Neuron. 2011;71:1030–1042. doi: 10.1016/j.neuron.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Guttridge DC, You Z, Zhang Z, Fribley A, Mayo MW, et al. Wnt-1 signaling inhibits apoptosis by activating beta-catenin/T cell factor-mediated transcription. J Cell Biol. 2001;152:87–96. doi: 10.1083/jcb.152.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purro SA, Dickins EM, Salinas PC. The Secreted Wnt antagonist Dickkopf-1 is required for amyloid beta-mediated synaptic loss. J Neurosci. 2012;32:3492–3498. doi: 10.1523/JNEUROSCI.4562-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch-Brandt C, Morgans C. Clusterin: a role in cell survival in the face of apoptosis. Prog Mol Subcell Biol. 1996;16:130–149. doi: 10.1007/978-3-642-79850-4_8. [DOI] [PubMed] [Google Scholar]
- Rosi MC, Luccarini I, Grossi C, Fiorentini A, Spillantini MG, Prisco A, et al. Increased Dickkopf-1 expression in transgenic mouse models of neurodegenerative disease. J Neurochem. 2010;112:1539–1551. doi: 10.1111/j.1471-4159.2009.06566.x. [DOI] [PubMed] [Google Scholar]
- Thambisetty M, Simmons A, Velayudhan L, Hye A, Campbell J, Zhang Y, et al. Association of plasma clusterin concentration with severity, pathology, and progression in Alzheimer disease. Arch Gen Psychiatry. 2010;67:739–748. doi: 10.1001/archgenpsychiatry.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrijvers EM, Koudstaal PJ, Hofman A, Breteler MM. Plasma clusterin and the risk of Alzheimer disease. JAMA. 2011;305:1322–1326. doi: 10.1001/jama.2011.381. [DOI] [PubMed] [Google Scholar]
- Schepeler T, Mansilla F, Christensen LL, Orntoft TF, Andersen CL. Clusterin expression can be modulated by changes in TCF1-mediated Wnt signaling. J Mol Signal. 2007;2:6. doi: 10.1186/1750-2187-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Sancho JM, Aguilera O, Garcia JM, Pendas-Franco N, Pena C, Cal S, et al. The Wnt antagonist DICKKOPF-1 gene is a downstream target of beta-catenin/TCF and is downregulated in human colon cancer. Oncogene. 2005;24:1098–1103. doi: 10.1038/sj.onc.1208303. [DOI] [PubMed] [Google Scholar]
- Theuns J, Remacle J, Killick R, Corsmit E, Vennekens K, Huylebroeck D, et al. Alzheimer-associated C allele of the promoter polymorphism -22C>T causes a critical neuron-specific decrease of presenilin 1 expression. Hum Mol Genet. 2003;12:869–877. doi: 10.1093/hmg/ddg098. [DOI] [PubMed] [Google Scholar]
- Tizon B, Ribe EM, Mi W, Troy CM, Levy E. Cystatin C protects neuronal cells from amyloid-beta-induced toxicity. J Alzheimers Dis. 2010;19:885–894. doi: 10.3233/JAD-2010-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Davidson TJ, Harel S, Arboleda VA, Prunell GF, Shelanski ML, Greene LA, et al. Highly efficient small interfering RNA delivery to primary mammalian neurons induces MicroRNA-like effects before mRNA degradation. J Neurosci. 2004;24:10040–10046. doi: 10.1523/JNEUROSCI.3643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killick R, Scales G, Leroy K, Causevic M, Hooper C, Irvine EE, et al. Deletion of Irs2 reduces amyloid deposition and rescues behavioural deficits in APP transgenic mice. Biochem Biophys Res Commun. 2009;386:257–262. doi: 10.1016/j.bbrc.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Trougakos IP, So A, Jansen B, Gleave ME, Gonos ES. Silencing expression of the clusterin/apolipoprotein j gene in human cancer cells using small interfering RNA induces spontaneous apoptosis, reduced growth ability, and cell sensitization to genotoxic and oxidative stress. Cancer Res. 2004;64:1834–1842. doi: 10.1158/0008-5472.can-03-2664. [DOI] [PubMed] [Google Scholar]
- Bertram L, Tanzi RE. Alzheimer disease: new light on an old CLU. Nat Rev Neurol. 2010;6:11–13. doi: 10.1038/nrneurol.2009.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper C, Killick R, Fernandes C, Sugden D, Lovestone S. Transcriptomic profiles of Wnt3a and insulin in primary cultured rat cortical neurones. J Neurochem. 2011;118:512–520. doi: 10.1111/j.1471-4159.2011.07349.x. [DOI] [PubMed] [Google Scholar]
- Millucci L, Ghezzi L, Bernardini G, Santucci A. Conformations and biological activities of amyloid beta peptide 25-35. Curr Protein Pept Sci. 2009;11:54–67. doi: 10.2174/138920310790274626. [DOI] [PubMed] [Google Scholar]
- Abeti R, Abramov AY, Duchen MR. Beta-amyloid activates PARP causing astrocytic metabolic failure and neuronal death. Brain. 2011;134 (Pt 6:1658–1672. doi: 10.1093/brain/awr104. [DOI] [PubMed] [Google Scholar]
- Lu Y, Li T, Qureshi HY, Han D, Paudel HK. Early growth response 1 (Egr-1) regulates phosphorylation of microtubule-associated protein tau in mammalian brain. J Biol Chem. 2011;286:20569–20581. doi: 10.1074/jbc.M111.220962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumbrink J, Kirsch KH, JP Johnson. EGR1, EGR2, and EGR3 activate the expression of their coregulator NAB2 establishing a negative feedback loop in cells of neuroectodermal and epithelial origin. J Cell Biochem. 2010;111:207–217. doi: 10.1002/jcb.22690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Sevetson BR, Golda T, Stanton JJ, Swirnoff AH, Milbrandt J. Novel mutants of NAB corepressors enhance activation by Egr transactivators. EMBO J. 1998;17:6010–6019. doi: 10.1093/emboj/17.20.6010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin S, et al. Correlative memory deficits, Abeta elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- Andorfer C, Kress Y, Espinoza M, de Silva R, Tucker KL, Barde YA, et al. Hyperphosphorylation and aggregation of tau in mice expressing normal human tau isoforms. J Neurochem. 2003;86:582–590. doi: 10.1046/j.1471-4159.2003.01879.x. [DOI] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Ramsey K, et al. Altered neuronal gene expression in brain regions differentially affected by Alzheimer's disease: a reference data set. Physiol Genomics. 2008;33:240–256. doi: 10.1152/physiolgenomics.00242.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Sun X, Hu G. An integrated genetics approach for identifying protein signal pathways of Alzheimer's disease. Comput Methods Biomech Biomed Engin. 2011;14:371–378. doi: 10.1080/10255842.2010.482525. [DOI] [PubMed] [Google Scholar]
- Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, et al. Genetic control of human brain transcript expression in Alzheimer disease. Am J Hum Genet. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockstone HE, Harris LW, Swatton JE, Wayland MT, Holland AJ, Bahn S. Gene expression profiling in the adult Down syndrome brain. Genomics. 2007;90:647–660. doi: 10.1016/j.ygeno.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18 (Suppl 1:S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- Bertrand J, Plouffe V, Senechal P, Leclerc N. The pattern of human tau phosphorylation is the result of priming and feedback events in primary hippocampal neurons. Neuroscience. 2010;168:323–334. doi: 10.1016/j.neuroscience.2010.04.009. [DOI] [PubMed] [Google Scholar]
- Korol O, Gupta RW, Mercola M. A novel activity of the Dickkopf-1 amino terminal domain promotes axial and heart development independently of canonical Wnt inhibition. Dev Biol. 2008;324:131–138. doi: 10.1016/j.ydbio.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupnik VE, Sharp JD, Jiang C, Robison K, Chickering TW, Amaravadi L, et al. Functional and structural diversity of the human Dickkopf gene family. Gene. 1999;238:301–313. doi: 10.1016/s0378-1119(99)00365-0. [DOI] [PubMed] [Google Scholar]
- Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, et al. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, He X. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- Caneparo L, Huang YL, Staudt N, Tada M, Ahrendt R, Kazanskaya O, et al. Dickkopf-1 regulates gastrulation movements by coordinated modulation of Wnt/beta catenin and Wnt/PCP activities, through interaction with the Dally-like homolog Knypek. Genes Dev. 2007;21:465–480. doi: 10.1101/gad.406007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros M, Paricio N, Strutt DI, Mlodzik M. Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell. 1998;94:109–118. doi: 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- Yamanaka H, Moriguchi T, Masuyama N, Kusakabe M, Hanafusa H, Takada R, et al. JNK functions in the non-canonical Wnt pathway to regulate convergent extension movements in vertebrates. EMBO Rep. 2002;3:69–75. doi: 10.1093/embo-reports/kvf008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur P, Lasche M, Eisenberg LM, Kuhl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- Checler F, Dunys J, Pardossi-Piquard R, Alves da Costa C. p53 is regulated by and regulates members of the gamma-secretase complex. Neurodegener Dis. 2010;7:50–55. doi: 10.1159/000283483. [DOI] [PubMed] [Google Scholar]
- MacGibbon GA, Lawlor PA, Walton M, Sirimanne E, Faull RL, Synek B, et al. Expression of Fos, Jun, and Krox family proteins in Alzheimer's disease. Exp Neurol. 1997;147:316–332. doi: 10.1006/exnr.1997.6600. [DOI] [PubMed] [Google Scholar]
- Gomez Ravetti M, Rosso OA, Berretta R, Moscato P. Uncovering molecular biomarkers that correlate cognitive decline with the changes of hippocampus' gene expression profiles in Alzheimer's disease. PLoS One. 2010;5:e10153. doi: 10.1371/journal.pone.0010153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criswell T, Beman M, Araki S, Leskov K, Cataldo E, Mayo LD, et al. Delayed activation of insulin-like growth factor-1 receptor/Src/MAPK/Egr-1 signaling regulates clusterin expression, a pro-survival factor. J Biol Chem. 2005;280:14212–14221. doi: 10.1074/jbc.M412569200. [DOI] [PubMed] [Google Scholar]
- Tolias KF, Duman JG, Um K. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog Neurobiol. 2011;94:133–148. doi: 10.1016/j.pneurobio.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Crenshaw EB, Kelley MW. Noncanonical Wnt signaling and neural polarity. Annu Rev Neurosci. 2006;29:363–386. doi: 10.1146/annurev.neuro.29.051605.112933. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.