Most snapdragons (Antirrhinum majus) live in scientific obscurity, taking a humble place in springtime gardens or bouquets. However, in the 1980s, molecular geneticist Enrico Coen helped to place these lowly plants in the spotlight. His work at the John Innes Center (JIC) in Norwich, U.K., used snapdragons as a model organism for studying plant development. Coen has identified and cloned several genes that affect flower shape, size, and color (1). Using these snapdragon studies and parallel research on Arabidopsis, he and his colleagues developed a unified model to describe how genes interact to direct flower formation (2–5). These contributions have earned him numerous awards and recognition, such as a 1998 election to the Royal Society in Britain and a 2001 election as a foreign associate of the National Academy of Sciences.
Figure 1.
Enrico Coen (left) and his collaborator, Przemyslaw Prusinkiewicz, seated on a glacier in Calgary, Canada.
In Coen's most recent work, he has strived to understand how patterns of gene activity lead to specific sizes and shapes of plant organs, a feat that combines experimental studies and computer modeling. In his Inaugural Article, published in this issue of PNAS, Coen and his colleagues describe a combination of factors integrated into a recently developed model to explain petal development (6). Similar models could eventually be applied to either plants or animals, aiding the search for key developmental genes.
A Blossoming Career
Coen grew up surrounded by science: his father was a physicist and his mother was a chemist. Drawn to his family's career path, Coen decided to take a somewhat different route by eschewing the physical sciences in favor of the life sciences. He cultivated an interest in biology, stimulated at age 15 by a biochemistry book titled The Chemistry of Life (7). “[Reading the book] was a real eye opener. All of a sudden, I realized you could make sense of the chemical basis of what was going on in living organisms,” he said.
Biochemistry continued to captivate Coen throughout high school and college. However, during his third year at the University of Cambridge, he fretted over which scientific niche to ultimately concentrate on. Attracted to more abstract analysis, he narrowed his choices to either chemistry or genetics, but changed his mind between the two on an almost daily basis. In the end, the tie was broken by surprisingly simple reasons. “I found out that the genetics lectures began at half-past nine in the morning, whereas the chemistry lectures began at nine. Also, the genetics people gave you coffee with your exam, so I thought they were obviously more civilized,” he said.
Thus began his career in genetics. Comfortable at the lab where he had spent much of his undergraduate years, Coen chose to stay at Cambridge after graduation in 1979 to pursue his doctoral degree. Under the mentorship of geneticist Gabriel Dover, he completed his thesis in 1982 on the evolution and function of genes needed to make ribosomal RNA (8). By examining fruit fly lines that had been subject to selection for the number of bristles (sensitive hairs located on the abdomen), Coen found that an exchange of information between ribosomal genes on the X and Y chromosomes had been responsible for a major change in bristle number (9). This was one of the first cases to define the molecular basis of a response to artificial selection.
“We had lovely fields of snapdragons, 99.9% of which were of no interest to us.”
With his Ph.D. completed, Coen chose Cambridge again for his postdoctoral degree. However, disillusioned with some of the competitive politics of mainstream science, Coen decided to pursue a less-developed area of research. He concentrated his efforts on studying the mechanism of “super genes,” putative gene clusters that act together in ways that affect both evolution and development. At the time, one of the best-defined super genes was in primroses, yet few researchers had delved into the molecular aspects of this system. Coen decided to write a proposal for molecular research on primrose super genes, and he was soon accepted as a research fellow in the lab of plant biologist Dick Flavell at the Plant Breeding Institute, Cambridge. Coen fondly remembers this project, which involved hours of sitting in sunny fields collecting primroses with different alleles. However, progress was slow because of the lack of molecular and genetic tools to research this system.
The ABCs of Floral Development
After a year or so, Coen realized that his foray into primrose genetics had perhaps been rather naïve. “Even though I'd now become quite interested in plants, I started to realize that maybe I'd bitten off more than I could chew,” he said. Seeking a different plant system to continue his molecular genetics research, Coen happened upon an open position at JIC, a facility that had studied snapdragon genetics for decades. The head of the program, a plant biologist named Brian Harrison, was about to retire and Coen, together with colleague Cathie Martin, was hired to carry on Harrison's studies.,
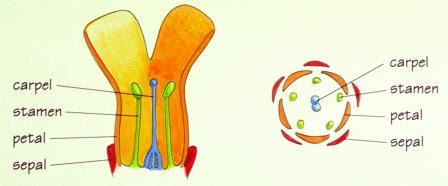
Figure 2.
The four whorls of a wild-type Antirrhinum flower, seen in radial (Left) and longitudinal (Right) cross section. (Courtesy of Enrico Coen; illustration by Nigel Orme.)
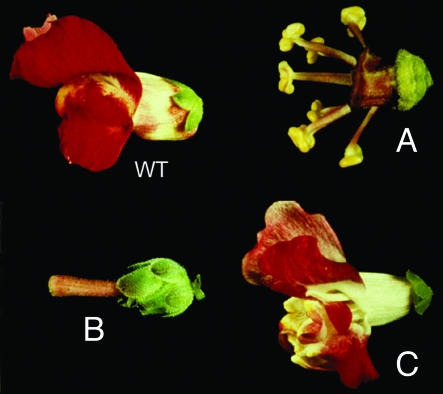
Figure 3.
Antirrhinum flowers of wild-type and organ identity mutants. (Courtesy of Enrico Coen.)
“I knew zero about snapdragons,” said Coen. However, he was aware that snapdragons have transposons, rare genetic elements that give rise to several types of mutations. Researchers have long known that transposons, also known as “jumping genes,” move to different positions in the genome of a single cell. Geneticist Barbara McClintock, who won the 1983 Nobel Prize in physiology or medicine, discovered transposons in corn in the 1940s. At the time Coen began working at JIC, most transposon research elsewhere continued McClintock's work on corn. However, Harrison saw definite advantages in using snapdragons for genetics. “Snapdragons are smaller plants, and they are nice and easy to grow. You can grow large numbers in a smaller space than corn, which is a bit of a colossal beast,” Coen said. Using some of Harrison's material, collaborators Heinz Saedler and Hans Sommer at the Max Planck Institute in Cologne had already cloned several active snapdragon transposons in the early 1980s, ushering in the molecular era of snapdragon research.
Upon Coen's first visit to JIC, Harrison and his research technician, Rosemary Carpenter, showed him their extensive collection of mutant snapdragons. Although Harrison and Carpenter's work concentrated mostly on color mutations, such as variegated flowers, the pair had several snapdragon specimens with mutant bloom morphologies. Coen immediately surmised that because transposons may sometimes land within genes important for floral development, they could give researchers a novel way to isolate and study developmental genes in flowering plants. Together with Carpenter, he began screening snapdragons for developmental mutants caused by transposon insertions. To locate and classify these naturally occurring mutants, Carpenter and Coen had to carefully comb through extensive snapdragon plantings, searching for anything out of the ordinary. “We had lovely fields full of flowering snapdragons, 99.9% of which were of no interest to us whatsoever. We were looking for the rare exceptions,” said Coen.
Along the radial axis, normal snapdragon flowers have four types of organs arranged in concentric whorls: sepals on the outside, then petals, stamens, and finally carpels inside. Within his first few years at JIC, Coen saw flowers with one of three morphological mutations, later known as types A, B, and C. The B type, which Harrison and Carpenter showed Coen on his original tour of the lab, had sepals instead of petals and carpels instead of stamens. The A type, which he and Carpenter identified later, had carpel-like organs instead of sepals in the flower's outermost whorl and stamen-like organs instead of petals. The C type mutant had a repetition of sepal- or petal-like organs, rather than stamens and carpels, in its two innermost whorls. Coen was puzzled about why these three mutations took place, but sensed that they were related.
After a long day looking at snapdragon flowers, Coen came to a sudden conclusion. “The idea involved a combination of gene activities: that by having certain combinations, you could account for the particular organ identities observed in the mutants,” he said.
Coen eventually discovered that three classes of genes (A, B, and C), later christened “organ identity genes,” worked in various combinations to control development of each whorl in wild-type snapdragons: class A controlled sepal identity, A and B worked together to define petal identity, B and C established stamen identity, and C alone specified carpel identity. Coen's simple model, which he presented at a conference in 1989 and subsequently published in 1990, explained how the functional loss of any one of these gene classes was responsible for the mutations he had observed earlier (2). A few months after presenting his model, Coen heard a talk by a postdoc in the lab of plant geneticist Elliot Meyerowitz of the California Institute of Technology and realized that the model Meyerowitz's group had developed for the flowering plant Arabidopsis was strikingly similar (10). Coen and Meyerowitz later published a joint paper that reviewed these two models and discussed the idea that a similar mechanism underlies flower development in Antirrhinum and Arabidopsis, which likely diverged from a common plant ancestor about 100 million years ago (3). The discovery suggested that “floral development has some unity and logic to it as opposed to each species having its own collection of mutant forms,” said Coen. Isolation of the genes involved, by the labs of Coen, Meyerowitz, and of Zsuzsanna Schwarz-Sommer and Hans Sommer in Cologne, showed that these similarities also extended to the molecular level (4, 11).
The Genetics of Geometry
In the 1990s, Coen and his colleagues at JIC made great strides in identifying genes controlling several other aspects of snapdragon development, such as flower asymmetry and inflorescence architecture (12–15). However, he gradually became puzzled by another mystery: how interaction between genes and cells affects the development of organ shapes. For example, how do the five petals of a snapdragon grow to form the flower's distinctively shaped “mouth”?
This problem required an understanding not only of genetics, but of geometry as well. Together with computer scientists knowledgeable in biological development, such as Przemyslaw Prusinkiewicz (pictured above) of the University of Calgary (Canada) and Andrew Bangham of the University of East Anglia (Norwich, U.K.), Coen's lab has spent the last decade developing computer modeling techniques that relate gene activity to patterns of cell division and growth. Last year, Coen, Bangham, and graduate student Anne-Gaelle Rolland-Lagan published a paper in Nature on one such model that analyzed snapdragon petal growth (16). After genetically marking cells in a young white flower bud so that the cells and their progeny appeared red, Coen's group watched the flowers blossom into a spotted or stripy appearance in adulthood. Although they could not observe individual marked cells or control the exact starting location with this technique, the researchers were able to ascertain how the petals grew by analyzing and comparing thousands of spots on mature flowers by using a computer. The resulting model suggested that two genetic forces controlled the resulting shape: a signal that promotes growth running from the base to the tip of the flower, and signals causing different regions of petal tube to grow more than others.
The model described in the Nature paper (16) combined several conceptual and experimental approaches that Coen's group had developed to predict biological shape change over time. In his Inaugural Article, published on page 4728 of this issue of PNAS, Coen and his colleagues describe factors taken into account to create such models, using their snapdragon petal model as a case study. The article discusses how certain parameters of cell growth, such as rate, preferred direction of growth, and anisotropy, are affected by various genes and morphogens. By accurately calculating the complex interplay between these factors, the authors suggest that many biological shape changes over time could be successfully modeled.
Although most of his experience thus far has focused on analyzing plant morphological development, Coen stresses that the basic principles he has learned from developing plant models could eventually be applied to animals. Thus, modeling could help speed the search for key developmental genes in any organism, including humans. This broad application keeps him deeply interested and invested in the outcome of his work. “An apple is very different from a planet in lots of ways, but at the end of the day, the great thing about Newton was he realized that apples and planets follow the same laws of gravity. There will certainly be differences when you're dealing with plant and animal development, but there will be common principles as well. I'm interested in exploring both of those things,” he said.
This is a Biography of a recently elected member of the National Academy of Sciences to accompany the member's Inaugural Article on page 4728.
References
- 1.Coen, E. S. (1996) EMBO J. 15, 6777–6788. [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, R. & Coen, E. S. (1990) Genes Dev. 4, 1483–1493. [DOI] [PubMed] [Google Scholar]
- 3.Coen, E. S. & Meyerowitz, E. M. (1991) Nature 353, 31–35. [DOI] [PubMed] [Google Scholar]
- 4.Weigel, D. & Meyerowitz, E. (1994) Cell 78, 203–209. [DOI] [PubMed] [Google Scholar]
- 5.Coen, E. (2000) The Art of Genes (Oxford Univ. Press, Oxford).
- 6.Coen, E., Rolland-Lagan, A.-G., Matthews, M., Bangham, A. & Prusinkiewicz, P. (2004) Proc. Natl. Acad. Sci. USA 101, 4728–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose, S. (1966) The Chemistry of Life (Penguin, Toronto).
- 8.Coen, E. S., Thoday, J. M. & Dover, G. (1982) Nature 295, 564–568. [DOI] [PubMed] [Google Scholar]
- 9.Coen, E. S. & Dover, G. A. (1983) Cell 33, 849–855. [DOI] [PubMed] [Google Scholar]
- 10.Bowman, J. L., Smyth, D. R. & Meyerowitz, E. M. (1991) Development (Cambridge, U.K.) 112, 1–20. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz-Sommer, Z., Davies, B. & Hudson, A. (2003) Nat. Rev. Genet. 4, 655–664. [DOI] [PubMed] [Google Scholar]
- 12.Luo, D., Carpenter, R., Vincent, C., Copsey, L. & Coen, E. (1996) Nature 383, 794–799. [DOI] [PubMed] [Google Scholar]
- 13.Luo, D., Carpenter, R., Copsey, L., Vincent, C., Clark, J. & Coen, E. (1999) Cell 99, 367–376. [DOI] [PubMed] [Google Scholar]
- 14.Bradley, D., Carpenter, R., Copsey, L., Vincent, C., Rothstein, S. & Coen, E. (1996) Nature 379, 791–797. [DOI] [PubMed] [Google Scholar]
- 15.Coen, E. S., Romero, J. M., Doyle, S., Elliott, R., Murphy, G. & Carpenter, R. (1990) Cell 63, 1311–1322. [DOI] [PubMed] [Google Scholar]
- 16.Rolland-Lagan, A.-G., Bangham, J. A & Coen, E. (2003) Nature 422, 161–163. [DOI] [PubMed] [Google Scholar]