Abstract
Tissue-specific gene expression is thought to be one of the major forces shaping mammalian gene order. A recent study that used whole-genome chromosome conformation assays has shown that the mammalian genome is divided into specific topological domains that are shared between different tissues and organisms. Here, we wanted to assess whether gene expression and regulation are involved in shaping these domains and can be used to classify them. We analyzed gene expression and regulation levels in these domains by using RNA-seq and enhancer-associated ChIP-seq datasets for 17 different mouse tissues. We found 162 domains that are active (high gene expression and regulation) in all 17 tissues. These domains are significantly shorter, contain less repeats, and have more housekeeping genes. In contrast, we found 29 domains that are inactive (low gene expression and regulation) in all analyzed tissues and are significantly longer, have more repeats, and gene deserts. Tissue-specific active domains showed some correlation with tissue-type and gene ontology. Domain temporal gene regulation and expression differences also displayed some gene ontology terms fitting their temporal function. Combined, our results provide a catalog of shared and tissue-specific topological domains and suggest that gene expression and regulation could have a role in shaping them.
Keywords: topological domains, RNA-seq, ChIP-seq
Introduction
Gene order was initially assumed to be random. Over time, mounting evidence has shown that gene order is not random, but rather guided by gene expression (Hurst et al. 2004). This has been observed not only in vertebrate genomes but also in plants (Field and Osbourn 2012) and bacteria (Willenbrock and Ussery 2004). Studies show that the genomic location of a gene in linear DNA sequence and its position in the three-dimensional nucleus is important for its regulation (Hurst et al. 2004; Willenbrock and Ussery 2004). Genomic regions that contain the most actively expressed genes may also be those of the highest gene density (Versteeg et al. 2003) and exist in clusters, such as the case of housekeeping genes (Lercher et al. 2002). These clusters could contain genes that are expressed in a tissue-specific (Reymond et al. 2002; Mégy et al. 2003; Hurst et al. 2004) or pathway-specific (Lee and Sonnhammer 2003) manner, or the opposite, with genes being organized in specific domains because of the need to silence them in specific tissues (Lunyak et al. 2002). Genomic location can have important implications on transgene integration (Milot et al. 1996), development (Akashi et al. 2003), or human disease (Ahituv et al. 2005; Kleinjan and van Heyningen 2005; Lettice et al. 2011; Ahituv 2012). For example, human chromosomal aberrations can change the location of functional sequences, either genes and (or) regulatory elements, which can subsequently lead to human disease (Ahituv et al. 2005; Kleinjan and van Heyningen 2005; Lettice et al. 2011; Ahituv 2012).
With advances in sequencing technologies, chromatin conformation capture (3C) assays can now be carried out in a genome-wide manner using technologies such as Hi-C (Lieberman-Aiden et al. 2009) or chromatin interaction analysis followed by paired-end tag sequencing (ChIA-PET) (Fullwood et al. 2009). Using Hi-C assays, universal mammalian topological domains were determined by analyzing mouse embryonic stem (ES) cells, mouse cortex, human ES cells, and human IMR90 fibroblasts (Dixon et al. 2012). This led to the subsequent identification of shared domain boundaries between these different organisms, cell lines, and tissues. The boundaries themselves were found to be enriched for transfer RNAs, short interspersed elements (SINEs), housekeeping genes, and binding of the insulator-associated CCCTC-binding factor (CTCF). These boundaries are thought to separate the genome into megabase-sized regions that have local chromatin interactions and were termed topological domains.
A recent study produced a map of nearly 300 000 murine cis-regulatory sequences for 17 diverse mouse tissues (Shen et al. 2012). These tissues comprise 13 adult tissues (8 weeks old mice) that include bone marrow, cerebellum, cortex, heart, intestine, kidney, liver, lung, olfactory bulb, placenta, spleen, testis, thymus; and 4 embryonic tissues (embryonic day 14.5) that include whole brain, heart, limb, and liver. Using RNA-seq and chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) for RNA polymerase II (polII), the insulator-binding protein CCCTC-binding factor (CTCF), and three chromatin modification marks: histone H3 lysine 4 trimethylation (H3K4me3), histone H3 lysine 4 monomethylation (H3K4me1), and H3 lysine 27 acetylation (H3K27ac) (Shen et al. 2012) the regulatory landscape of these different tissues and time points was determined. These assays provide a unique resource for a deeper examination of gene expression and regulation within these topological domains.
To classify the aforementioned universal topological domains (Dixon et al. 2012), we analyzed the gene expression (RNA-seq) and gene regulation (ChIP-seq on H3K4me1 and H3K27ac) in each of these domains for these 17 different mouse tissues (Shen et al. 2012). We found different types of domains, such as those that are extremely active with high gene expression and regulation or ones that are inactive with low gene expression and regulation. Several of these domains were shared between tissues, and some were tissue-specific. For the shared active domains, we observed a significant enrichment for housekeeping genes, and for the inactive ones we observed enrichment for DNA repeats and gene deserts. Analysis of similar tissues both in embryonic and developmental time points showed a difference in active domains that somewhat coincided with their temporal function. Our results assign functional annotations for topological domains and suggest that gene expression and regulation might be involved in shaping them.
Materials and methods
Establishment of common tissue domains
We identified common domains by using previously characterized shared Hi-C boundaries (Dixon et al. 2012). From this dataset, we took 1159 shared topological boundaries identified in mESC and mouse cortex Hi-C data. The central point of each boundary was then used to generate the coordinates for these boundaries. It is worth noting, that in these Hi-C domains, telomeric regions are not included and only 97.4% of the genome is covered. In total, 1175 common domains were obtained by the shared boundaries.
RNA-seq analysis
RNA-seq bam files from 17 different mouse tissues was downloaded from http://chromosome.sdsc.edu/mouse/download.html (Shen et al. 2012). We then used Cufflinks (Trapnell et al. 2010), using the mm9 genome assembly and default parameters, to calculate FPKM for each tissue and each replicate. Analyses of the replicates using R for the same tissue showed a good correlation (average correlation coefficient was 0.9706; Pearson test) (supplementary data, Fig. S21). The FPKM for each tissue is calculated as the average of the two replicates.
ChIP-seq analysis
We downloaded H3K27ac and H3K4me1 ChIP-seq data for the 17 mouse tissues from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29184) (Shen et al. 2012). All the reads were aligned to mm9 using Illumina’s ELAND software with a seed length of 25 bases and up to 2 mismatches. Only sequences mapping to exactly one location were used for analysis. If multiple sequences aligned to the same location, all but one of the sequences were discarded (Shen et al. 2012). By counting the number of reads that fell within each 100 base pair bin, the RPKM value was normalized as the tag counts in each bin (Shen et al. 2012). For each tissue we looked for the region that had an RPKM > 0 in both H3K4me1 and H3K27ac and then used the H3K27ac RPKM in those regions for further analysis.
Topological domain scoring
We defined a gene expression or regulation score of each topological domain to be
| (1) |
P(l, λ) is a Poisson distribution parameterized by average coverage in a domain, where
| (2) |
| (3) |
Dw is the width of a current domain, and Gw is the genome effective length for all the domains.
ri are the FPKM or RPKM for the effective region or gene with the width dwi. Given this definition, the scores of a domain represent the negative logarithm of the possibility of enrichment of RPKM and FPKM ChIP-seq and RNA-seq reads to hit a specific location in the genome with equal probability. The higher the score, the less likely the observed profile occurs by chance. We thus obtained two different scores for each tissue: gene expression score and gene regulatory score. We then used a quartile method to categorize each score in every tissue. We defined the lower quartile to be the lower 25% of the data (L) and the top quartile to be the higher 25% (H). Density plots of these scores for all 17 tissues are shown in supplementary Fig. S3.
GO term analysis
All the reference genes in each domain were pooled out and gene ontology (GO) analysis was done using DAVID’s (Huang et al. 2009a, 2009b) default parameters. These parameters generate an EASE score that is based on a Fisher’s exact test, only a bit more stringent. FDR was used to correct for multiple testing. Significant p values for both EASE and FDR were considered as those that were ≤0.05.
Results
Genome-wide analysis of gene expression and regulation in topological domains
We took advantage of topological domain boundaries generated from Hi-C data (Dixon et al. 2012) to partition the genome to universal domains. For this purpose, we used the topological domain boundaries established from Hi-C on mouse embryonic stem cells (mESC) and cortex (Dixon et al. 2012). Using the common boundaries established in this study (see Materials and methods), we defined 1175 common domains with an average length of 2 megabases (Mb) and ranging in size from 40 kilobase to 22 Mb (supplementary Fig. S1).
We next analyzed RNA-seq and ChIP-seq datasets on mouse tissues in these domains. For consistency purposes, we decided to focus on the mouse ENCODE data (Dixon et al. 2012; Shen et al. 2012), owing to it being generated from the same laboratory and having all experiments (RNA-seq and ChIP-seq) preformed on matching tissues (bone marrow, whole brain at embryonic day (E) 14.5, cerebellum, cortex, heart at E14.5, heart, intestine, kidney, limb at E14.5, liver at E14.5, liver, lung, olfactory bulb, placenta, spleen, testis, and thymus). For the RNA-seq datasets, we examined the two available replicates for each tissue (these gave a significant correlation coefficient for all tissues that was on average 0.9706; Pearson test; supplementary Fig. S2). We then calculated expression scores for each topological domain in each tissue, based on its fragments per kilobase of exon per million fragments mapped (FPKM) RNA-seq scores. The total score for each domain was normalized based on domain size and ranked in order; high expressing and low expressing domains in each tissue were defined as those in the top and low quartile, respectively (see Materials and methods; supplementary Fig. S3; supplementary Table S1).
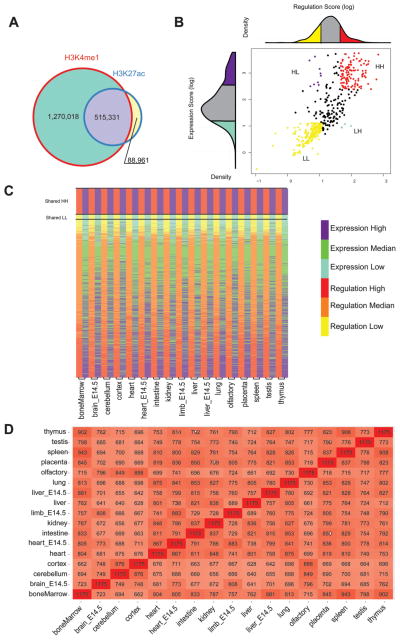
We then calculated the enrichment score of ChIP-seq peaks for the 17 different mouse tissues from two histone modifications that mark active regions, H3K4me1 and H3K27ac. Both H3K4me1 and H3K27ac were shown to mark enhancers (Barski et al. 2007; Heintzman et al. 2009; Ernst et al. 2011). However, owing to previous reports that found H3K27ac to mark active enhancers (Creyghton et al. 2010; Rada-Iglesias et al. 2011), we only considered sequences that had an H3K27ac mark in our analysis. To be even more conservative in our definition of an active ChIP-seq mark, we only analyzed H3K27ac peaks that also overlapped an H3K4me1 peak. Combining all the data from 17 tissues, we observed that 85.23% of the H3K27ac peaks overlap H3K4me1 peaks (Fig. 1A). For each domain in each of the 17 tissues, a regulatory score was calculated based on reads per kilobase per million (RPKM) for H3K27ac overlapping H3K4me1 peaks (see Materials and methods). Similar to gene expression scores, domains were ranked based on their scores; the top and bottom quartiles were considered as having high and low gene regulation, respectively (supplementary Fig. S4; supplementary Table S1).
Fig. 1.
Genome-wide analysis of gene expression and regulation in topological domains. (A) Venn diagram showing the overlap between H3K4me1 and H3K27ac ChIP-seq peaks for all 17 tissues. (B) Graph showing how the olfactory topological domains were separated to four different types based on their gene expression and regulation scores. The regulation score for this tissue is shown above the x axis and the expression score in the y axis. (C) Heat map showing topological domain clusters for each tissue. High regulation scores are shown in red and low in yellow. High expression scores are depicted in purple and low in light blue (D) Overall similarity between domain type and the different tissues. The number of topological domains having a similar domain type between the different tissues is written for each tissue. The darker the color the more similar the tissues are.
Combined, these gene expression and regulation scores provided us with four types of topological domains (Figs. 1B, 1C) that were defined in the following manner: (i) High gene expression and regulation scores were termed HH. (ii) Low gene expression and regulation scores were called LL. (iii) Domains that have high regulatory scores but a low gene expression score were named HL. (iv) Domains that have low regulatory scores but high gene expression scores were termed LH. The quantities and coordinates for these domains can be found in supplementary Table S2. Because the total number of HL and LH domains was extremely low, we focused our subsequent analysis only on HH and LL domains.
We next analyzed the various domain types for their overall similarity between the different tissues (Fig. 1D). We found several tissues to have corresponding domain types. For example, the bone marrow, spleen, and thymus had the largest number of domains that were identical in their domain type (HH, LL, HL, or LH), probably because of their immunological role. The tissues that were the least similar were the embryonic brain (E14.5 brain) and the adult liver, having only 641 similar domain types. The brain tissues, E14.5 brain, cerebellum, and cortex, were the tissues that were the most different from all other tissues, having an average of 709 domains with a similar type to other tissues (supplementary Fig. S5). The placenta was the tissue that was most similar to all other tissues with an average of 790 domains matching another tissue (supplementary Fig. S5).
Domains with high gene expression and regulation (HH)
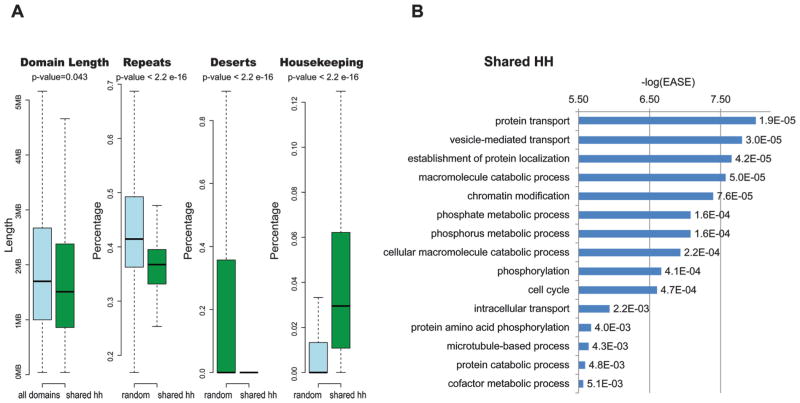
We next analyzed domains that had high gene expression and regulation scores, which we termed HH. We observed that these domains could be divided into two different classes: (i) domains that are shared across all tissues (shared HH) and (ii) domains that are tissue-specific. For the first class of shared HH domains, we found 162 domains that were shared across the different tissues (supplementary Table S2). These domains were significantly shorter than the other domains (p value = 0.043; Wilcoxon test) (Fig. 2A). In addition, they had less repeats (p value < 2.2 × 10−16; Kolmogorov–Smirnov test) and less gene deserts (p value < 2.2 × 10−16; Kolmogorov–Smirnov test), as defined by Ovhcarenko et al. (Ovcharenko et al. 2005) (Fig. 2A), when compared with 162 randomly picked domains of similar length tested over 1000 times.
Fig. 2.
Tissue-shared topological domains with high gene expression and regulation (HH). (A) Boxplots comparing HH tissue shared domains with all other domains for length, and prevalence of repeats, gene deserts, and housekeeping genes compared 1000 times with 162 random domains with a similar length. p values above the boxplots are based on a Wilcoxon test for the domain length and a Kolmogorov–Smirnov test for the other comparisons. (B) Enriched GO terms in shared HH domains. −log(EASE) p values generated by DAVID (Huang et al. 2009a, 2009b) are shown in the x axis and to the right of each bar are its FDR p values.
We then analyzed the shared HH domains for enrichment of housekeeping genes. This was done using a dataset of previously defined ubiquitously expressed human genes (Tu et al. 2006). This allowed us also to check the validity of our domain types, expecting domains with high gene expression scores to be enriched for these ubiquitously expressed housekeeping genes. We found that the shared HH domains showed a significant enrichment for housekeeping genes compared 1000 times with 162 randomly picked domains of similar length (p value < 2.2 × 10−16; Kolmogorov–Smirnov test) (Fig. 2A). Analysis of the gene ontology (GO) terms using DAVID (Huang et al. 2009a, 2009b), after correcting for multiple testing using a false discovery rate (FDR), found that these domains are enriched for GO terms associated with protein transport function, metabolic and catabolic processes, chromatin modification, phosphorylation, and others (Fig. 2B; supplementary Table S3). Combined, these results suggest that shared HH domains have high gene expression and regulation scores owing to them encompassing housekeeping genes, which falls in line with previous reports showing that housekeeping genes are located in clusters within the genome (Lercher et al. 2002).
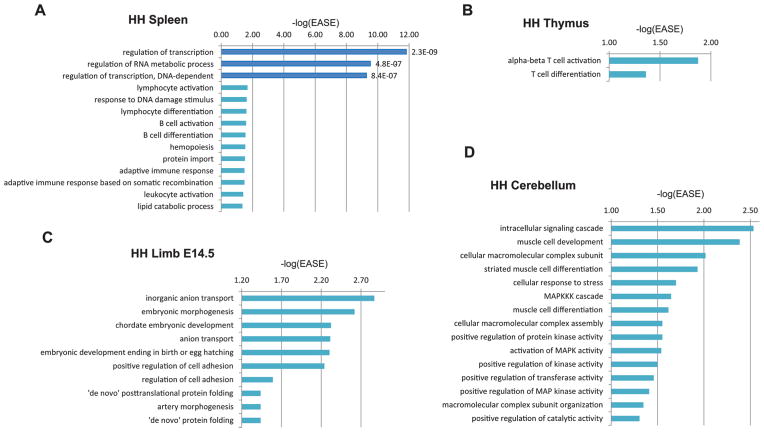
We next analyzed the tissue-specific HH domains. On average, each tissue had 4.8 HH domains that were specific to that tissue (supplementary Table S2). Examination of the GO terms for the genes in these tissue-specific domains, after correcting for multiple testing using FDR, only found the spleen to have significant GO terms. These were regulation of transcription, regulation of RNA metabolic process, and regulation of transcription, DNA-dependent, which are general terms that could fit any tissue (Fig. 3A). However, it is worth noting that if we look at GO terms that have a significant DAVID EASE score (Huang et al. 2009a, 2009b), which is based on a Fisher’s exact test, but without correcting for multiple testing, we found an overall correlation with the tissue and gene function for most of the tissues (supplementary Table S3). For example, for the spleen we observed, in addition to the previous terms, lymphocyte activation, lymphocyte differentiation, B cell activation, B cell differentiation, hemopoiesis, and others (Fig. 3A). For the thymus, we observed terms such as alpha-beta T cell activation and T cell differentiation (Fig. 3B). For the E14.5 limb we observed enrichment for GO terms such as embryonic morphogenesis or chordate embryonic development (Fig. 3C). However, tissue function was not correlated in all tissues for GO terms fitting that specific tissue using these EASE scores. The most distant, based on our data, was the cerebellum, where for example muscle associated terms such as muscle cell development or muscle cell differentiation were observed (Fig. 3D). Combined, our results hint to the potential existence of tissue-specific HH domains that have correlated tissue functions, but do not pass significance upon multiple testing.
Fig. 3.
Tissue-specific topological domains with high gene expression and regulation (HH). (A) Enriched GO terms in spleen tissue-specific HH domains. (B) Enriched GO terms in thymus tissue-specific HH domains. (C) Enriched GO terms in E14.5 limb tissue-specific HH domains. (D) Enriched GO terms in cerebellum tissue-specific HH domains. Dark blue bars represent GO terms that have an FDR ≤ 0.05. −log(EASE) p values generated by DAVID (Huang et al. 2009a, 2009b) are shown in the x axis and to the right of each dark blue colored bar are its FDR p values.
Domains with low gene expression and regulation (LL)
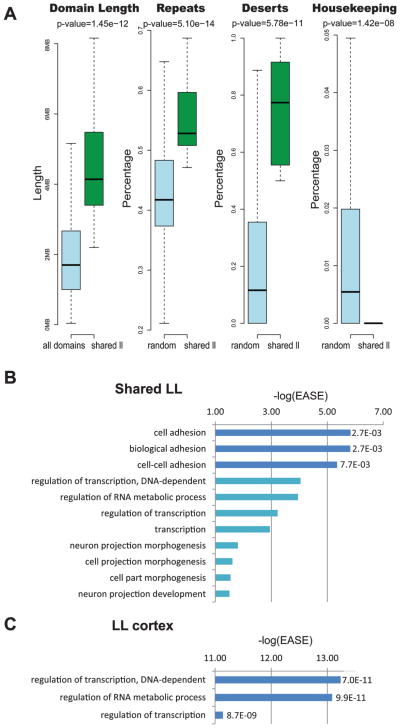
We found 29 domains that were shared across the different tissues that had low gene expression and regulation scores, termed LL (supplementary Table S2). Examination of these 29 shared LL domains found them to be significantly longer than all other domains (p value = 1.45 × 10−12; Wilcoxon test) (Fig. 4A). In addition, these domains were found to have a significantly higher occupancy of DNA repeats (p value = 5.10 × 10−14; Kolmogorov–Smirnov test) and gene deserts (p value = 5.78 × 10−11; Kolmogorov–Smirnov test) and less housekeeping genes (p value = 1.42 × 10−8; Kolmogorov–Smirnov test) when compared with 29 randomly picked domains of similar length over 1000 times (Fig. 4A). We next carried out a GO term analysis of these shared LL domains and found that they are enriched, after correcting for multiple testing using FDR, for various terms that are associated with cell adhesion (Fig. 4B, supplementary Table S3). Analysis of tissue-specific LL domains found only an average of 0.6 per tissue (supplementary Table S2). Because the low number of domains per tissue, only the cortex had GO terms that passed significance (supplementary Table S3), and these were associated with transcriptional regulation or RNA metabolic processes (Fig. 4C).
Fig. 4.
Topological domains with low gene expression and regulation (LL). (A) Boxplots comparing LL tissue-shared domains with all other domains for length, and prevalence of repeats, gene deserts, and housekeeping genes compared 1000 times with 29 random domains with a similar length. p values above the boxplots are based on a Wilcoxon test for the domain length and a Kolmogorov–Smirnov test for the other comparisons. (B) Enriched GO terms in shared LL domains. (C) Enriched GO terms in cortex tissue-specific LL domains. Dark blue bars represent GO terms that have an FDR ≤ 0.05. −log(EASE) p values generated by DAVID (Huang et al. 2009a, 2009b) are shown in the x axis and to the right of each dark blue colored bar are its FDR p values.
Active and inactive domain temporal changes in the same tissue type
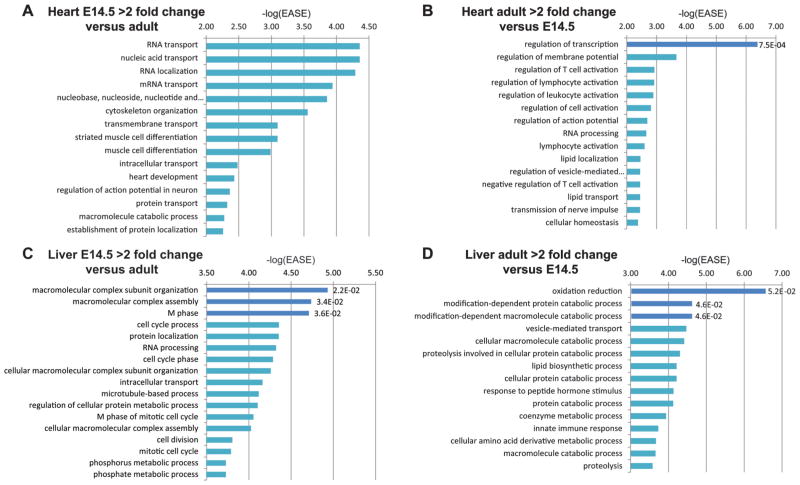
We next wanted to assess whether there are temporal differences in the activation or inactivation of domains in the same tissue. To do this, we took advantage of the liver and heart datasets, as both had embryonic (E14.5) and adult time points. We compared each domain in these tissues for their domain type in both time points and identified domains that have both a gene expression and regulation score that is greater than two-fold in the embryonic liver and heart, respectively, and vice versa (supplementary Table S2). We then carried out a GO term analysis of these domains. For the 77 domains that had two-fold higher gene expression and regulation scores in the embryonic heart versus the adult heart, we did not observe significant GO terms after correcting for multiple testing (supplementary Table S4). For the 59 domains that were two-fold higher in the adult heart versus E14.5 heart, we only found regulation of transcription to be significant (supplementary Table S4). However, if we just look at significant DAVID EASE score (Huang et al. 2009a, 2009b), without correction for multiple testing, we did observe terms that correlated with temporal function. For example, in domains that showed two-fold higher expression and regulation scores in E14.5 heart versus adult, we observed striated muscle cell differentiation, muscle cell differentiation, or heart development (Fig. 5A; supplementary Table S4). For domains that were two-fold higher in the adult heart compared with the embryonic one, we had regulation of membrane potential as the second highest term (Fig. 5B; supplementary Table S4).
Fig. 5.
Analysis of temporal differences in heart and liver topological domains. (A) Enriched GO terms in domains that have two-fold higher gene expression and regulation scores in the embryonic heart versus the adult heart. (B) Enriched GO terms in domains that have two-fold higher gene expression and regulation scores in the adult heart versus the embryonic heart. (C) Enriched GO terms in domains that have two-fold higher gene expression and regulation scores in the embryonic liver versus the adult liver. (D) Enriched GO terms in domains that have two-fold higher gene expression and regulation scores in the adult liver versus the embryonic liver. Dark blue bars represent GO terms that have an FDR ≤ 0.05. −log(EASE) p values generated by DAVID (Huang et al. 2009a, 2009b) are shown in the x axis and to the right of each dark blue colored bar are its FDR p values.
We carried out a similar analysis on the liver. We found only significant terms that fit with general functions (supplementary Table S4). For example, for the 179 domains that were two-fold higher in the embryonic liver versus adult, we observed the following significant GO terms: macromolecular complex subunit organization, macromolecular complex assembly, and M phase (Fig. 5C). For the 106 domains that were two-fold higher in the adult liver, we found significant association with terms such as oxidation reduction, modification-dependent protein catabolic process, and modification-dependent macromolecule catabolic process (Fig. 5D). If we just look at significant DAVID EASE score, as we did for the heart above, we do observe GO terms that fit their temporal function in domains that are two-fold higher in the adult liver. For example, lipid biosynthetic process, innate immune response, and various metabolic and catabolic processes (Fig. 5D; supplementary Table S4).
Discussion
By analyzing gene expression and regulation within topological domains, we were able to catalog them into different domain types. We found 162 domains that had high gene expression and regulation (HH) in all 17 tissues and are enriched for housekeeping genes (Fig. 2A), fitting with an earlier report that showed that these genes tend to reside in clusters (Lercher et al. 2002). Further analysis of these domains found them to be significantly shorter, have less repeats, and gene deserts (Fig. 2A), fitting with their potential housekeeping functions. In contrast, we identified 29 domains that were shared across tissues and had low gene expression and regulation scores (LL) with significantly less housekeeping genes (Fig. 4A). Also contrary to the HH shared domains, these shared LL domains were significantly longer and had more repeats and gene deserts (Fig. 4A). We did not find any shared domains with high gene regulation and low gene expression scores (HL) and vice versa (LH), but did find a few tissue-specific ones. This observation could likely be due to our selection process, choosing only domains from the top and low quartile of expression and regulation scores.
GO term analysis of tissue-specific HH domains only had significant results for spleen, whose function was not specific for that tissue. However, if we do not correct for multiple testing and just look at significant DAVID EASE scores, we observed some agreement with gene enrichment and tissue-specific function (supplementary Table S3), suggesting that domains with tissue-specific function could exist in the genome. However, it is worth noting that we were only able to interrogate 17 tissues and within those observed an average of 4.8 HH tissue-specific domains from the 1175 total domains. Taking these numbers into account, we assume that when similar data becomes available for additional tissues, the number of HH tissue-specific domains will reduce even further. In addition, as more tissue-specific Hi-C datasets become available, topological domains may be further refined. It is also worth noting that the tissues themselves are, on the majority, a mixture of cell types and not pure cell populations, which could also skew our results.
In our GO term comparison between domains that had a two-fold difference in gene expression and regulation scores between embryonic and adult heart and liver, we observed partial enrichment for terms that had biological function matching their respective time point only when using DAVID EASE scores without correcting for multiple testing. We saw several developmental-associated GO terms in domains that had higher scores in E14.5 heart versus the adult heart (supplementary Table S4) and terms that fit an adult heart in domains that were higher in that tissue compared with the embryonic time point. Our temporal analysis of the liver, even when using just DAVID EASE scores, was less successful. Although we found GO terms that fit with the role of adult liver in domains that had higher scores in adult compared with E14.5 liver, the opposite did not show a good temporal match.
Combined, our results suggest that gene expression and regulation might be one of the forces shaping these topological domains. We observed that housekeeping genes tend to reside in clusters within domains that are shared between all our analyzed tissues that have high gene expression and regulation scores. Tissues with low gene expression and regulation scores tend to be longer and have more repeats and gene deserts. Our tissue-specific domain analysis was limited because of the low number of these domains. As more tissue-specific expression and regulation datasets become available along with Hi-C data for those tissues, the classification of these tissue-specific domains could be expanded. These domains could provide us with a better understanding of how genomic location can influence chromatin organization, transgene or viral integration, development, and human disease.
Supplementary Material
Acknowledgments
We want to thank H. Di and I. Ovcharenko (NCBI) for providing us with the mouse gene desert dataset and the Ahituv laboratory for helpful comments on the manuscript. This work was supported by the National Institute of Child & Human Development grant number R01HD059862, by the National Institute of Neurological Disorders & Stroke grant number 1R01NS079231, and by the National Human Genome Research Institute (NHGRI) grant numbers 1R01HG006768 and R01HG005058. N.A. is also supported in part by the National Institute of General Medical Sciences (GM61390), National Institute of Diabetes & Digestive & Kidney Diseases (1R01DK090382), and the Simons Foundation (SFARI #256769).
Footnotes
Supplementary data are available with the article throught the journal Web site at http://nrcresearchpress.com/doi/suppl/10.1139/gen-2013-0111.
Contributor Information
Jingjing Zhao, Department of Bioengineering and Therapeutic Sciences, University of California San Francisco, San Francisco, CA 94158, USA; Institute for Human Genetics, University of California San Francisco, San Francisco, CA 94158, USA; Key Laboratory of Advanced Control and Optimization for Chemical Processes of the Ministry of Education, East China University of Science and Technology, 200237, Shanghai, China.
Hongbo Shi, Key Laboratory of Advanced Control and Optimization for Chemical Processes of the Ministry of Education, East China University of Science and Technology, 200237, Shanghai, China.
Nadav Ahituv, Department of Bioengineering and Therapeutic Sciences, University of California San Francisco, San Francisco, CA 94158, USA; Institute for Human Genetics, University of California San Francisco, San Francisco, CA 94158, USA.
References
- Ahituv N. Gene regulatory sequences and human disease. Springer; New York: 2012. [Google Scholar]
- Ahituv N, Prabhakar S, Poulin F, Rubin EM, Couronne O. Mapping cis-regulatory domains in the human genome using multi-species conservation of synteny. Hum Mol Genet. 2005;14(20):3057–3063. doi: 10.1093/hmg/ddi338. [DOI] [PubMed] [Google Scholar]
- Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, et al. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101(2):383–389. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn A. Order in the playground: Formation of plant gene clusters in dynamic chromosomal regions. Mobile Genetic Elements. 2012;2(1):46–50. doi: 10.4161/mge.19348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459(7243):108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pál C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nat Rev Genet. 2004;5(4):299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76(1):8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Sonnhammer EL. Genomic gene clustering analysis of pathways in eukaryotes. Genome Res. 2003;13(5):875–882. doi: 10.1101/gr.737703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 2002;31(2):180–183. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- Lettice LA, Daniels S, Sweeney E, Venkataraman S, Devenney PS, Gautier P, et al. Enhancer-adoption as a mechanism of human developmental disease. Hum Mutat. 2011;32(12):1492–1499. doi: 10.1002/humu.21615. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298(5599):1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Mégy K, Audic S, Claverie JM. Positional clustering of differentially expressed genes on human chromosomes 20, 21 and 22. Genome Biol. 2003;4(2):1. doi: 10.1186/gb-2003-4-2-p1. [DOI] [PubMed] [Google Scholar]
- Milot E, Strouboulis J, Trimborn T, Wijgerde M, de Boer E, Langeveld A, et al. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87(1):105–114. doi: 10.1016/s0092-8674(00)81327-6. 10.1016/S0092-8674(00) 81327-6. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15(1):137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Marigo V, Yaylaoglu MB, Leoni A, Ucla C, Scamuffa N, et al. Human chromosome 21 gene expression atlas in the mouse. Nature. 2002;420(6915):582–586. doi: 10.1038/nature01178. [DOI] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Z, Wang L, Xu M, Zhou X, Chen T, Sun F. Further understanding human disease genes by comparing with housekeeping genes and other genes. BMC Genomics. 2006;7:31. doi: 10.1186/1471-2164-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg R, van Schaik BD, van Batenburg MF, Roos M, Monajemi R, Caron H, et al. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 2003;13(9):1998–2004. doi: 10.1101/gr.1649303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willenbrock H, Ussery DW. Chromatin architecture and gene expression in Escherichia coli. Genome Biol. 2004;5(12):252. doi: 10.1186/gb-2004-5-12-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.