Abstract
MicroRNAs (miRNAs) have been identified in cells as well as in exosomes in biological fluids such as milk. In mammary gland, most of the miRNAs studied have functions related to immunity and show alterations in their pattern of expression during lactation. In mastitis, the inflammatory response caused by Streptococcus uberis alters the expression of miRNAs that may regulate the innate immune system. These small RNAs are stable at room temperature and are resistant to repeated freeze/thaw cycles, acidic conditions and degradation by RNAse, making them resistant to industrial procedures. These properties mean that miRNAs could have multiple applications in veterinary medicine and biotechnology. Indeed, lactoglobulin-free milk has been produced in transgenic cows expressing specific miRNAs. Although plant and animal miRNAs have undergone independent evolutionary adaptation recent studies have demonstrated a cross-kingdom passage in which rice miRNA was isolated from human serum. This finding raises questions about the possible effect that miRNAs present in foods consumed by humans could have on human gene regulation. Further studies are needed before applying miRNA biotechnology to the milk industry. New discoveries and a greater knowledge of gene expression will lead to a better understanding of the role of miRNAs in physiology, nutrition and evolution.
Keywords: lactation, mastitis, milk, miRNA
Introduction
The mammary gland is an interesting model for studying gene expression since this organ experiences different cycles of differentiation and regression during adult life. In eukaryotes, gene expression is a complex process that involves DNA methylation, chromatin modification, imprinting, and interfering RNA (Mattick and Makunin, 2006). Although Fire and Mello received the Nobel Prize for their discovery of double-strand interfering RNA in Caenorhabditis elegans (Fire et al., 1998), the first miRNA (Lin 4) was in fact discovered years before by two groups and published simultaneously in 1993 (Lee et al., 1993; Wightman et al., 1993). In 2000, a second miRNA (Lin 7) was identified in the same nematode (Reinhart et al., 2000) and soon after in many other species (Pasquinelli et al., 2000). By 2001, it was clear that miRNAs represent a class of small conserved RNAs (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). Three main categories of interfering RNAs have been recognized: microRNA (miRNA), short interfering RNA (siRNA) and piwi interactive RNA (piRNA). These three RNAs are of similar size (∼23 nucleotides in length) but have sequence-specific inhibitory functions (Carthew and Sontheimer, 2009; Ambros, 2010). siRNAs are excised from long, fully complementary double-stranded RNAs (Tomari and Zamore, 2005; Ambros, 2010) and were initially considered to protect the host genome from foreign nucleic acids (such as viruses, transposons and transgenes) but that view changed with the discovery of abundantly expressed endogenous siRNAs in animal cells (Golden et al., 2008). piRNAs are small RNAs that bind the piwi subfamily of argonaute proteins and protect the germline (Ambros and Chen, 2007) and embryonic stem cells (Morin et al., 2008) from invasive transposable elements.
miRNAs regulate gene expression at a post-transcriptional level either by causing RNA degradation or by blocking translation through base-pairing with complementary sequences within mRNA. Since partial miRNA complementarities are enough to target an mRNA, each miRNA has the ability to regulate a large number of genes (Jackson et al., 2003). Another feature is that a conserved miRNA can regulate different genetic pathways and developmental processes in various organisms (Ambros and Chen, 2007).
In recent years, many miRNAs have been identified in plants and animals by experimental and computational approaches. Adequate characterization of the actions of miRNAs in the mammary gland in different physiological states could provide new insights into the regulation of gene expression. The goals of this review are to outline and integrate the experimental and computational studies on miRNA in healthy and mastitic mammary gland and to address the stability of miRNA in milk and possible inter-species transmission.
Biogenesis of miRNA
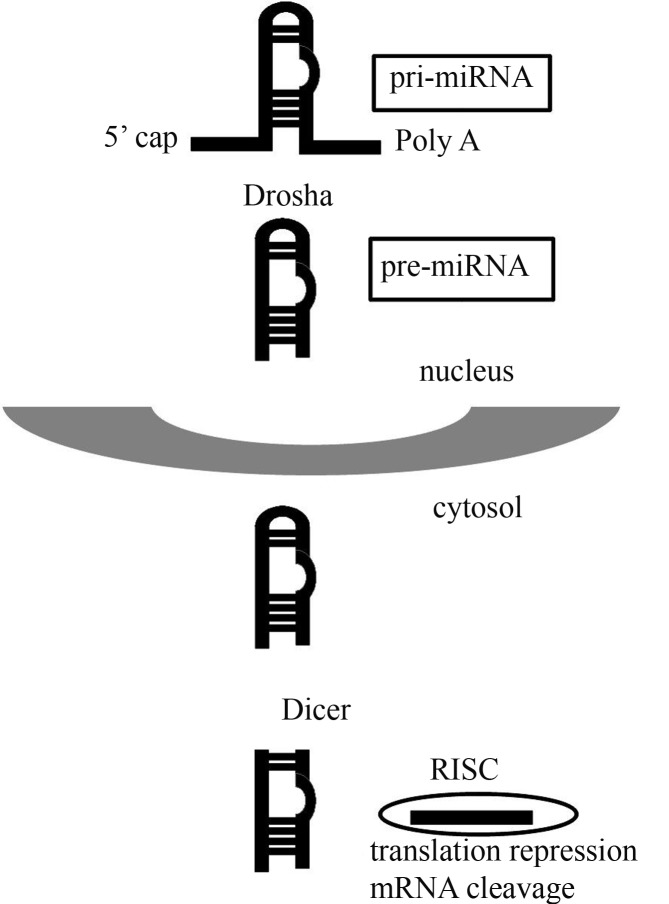
miRNAs are DNA-derived RNA transcripts that are not translated into proteins. miRNA biosynthesis is a complex process that involves several steps. The canonical pathway for miRNA biogenesis requires two RNaseIII enzymes, Drosha and Dicer. In this process, miRNAs are initially transcribed by RNA polymerase II as long transcript hairpin-shaped units. As with mRNA, these molecules are spliced, capped and polyadenylated to produce primary miRNA (pri-miRNA). This precursor is processed by the enzyme Drosha to yield pre-miRNA that is then transported to the cytoplasm where it is cleaved by Dicer to the mature length. The functional strand of mature miRNA is loaded onto an argonaute (Ago) protein, the main constituent of the RNA-induced silencing complex (RISC), while the other strand is degraded (Lee et al., 2003; Du and Zamore, 2005; Shabalina and Koonin, 2008) (Figure 1). Chong et al. (2010) used Dicer- and Drosha-knock-out mice to confirm the requirement of both enzymes for this canonical miRNA biosynthesis. However, not all miRNAs are formed by this pathway. It is now known that there are variations in many of the steps involved and that this can influence the biogenesis results (Okamura et al., 2007; Winter et al., 2009). Deep sequencing technology has revealed marked variability in miRNA biogenesis and has shown that many different sequences can share the same miRNA precursor.
Figure 1.
A schematic representation of the canonical biogenesis of miRNA. Initially, a long hairpin-shaped (pri-miRNA) is transcribed by RNA polymerase II and then cleaved by Drosha (to yield pre-miRNA) prior to leaving the nucleus; the molecule is subsequently cleaved by a Dicer enzyme to yield double-stranded mature miRNA. Finally, miRNA is incorporated into the RNA-induced silencing complex (RISC), thereby allowing separation of the functional strand that interferes with mRNA by repressing translation or cleaving mRNA.
Identification of miRNA in mammary gland and milk
miRNAs have been identified in cells and in fluids such as saliva, amniotic fluid, blood, urine and milk. In fluids, miRNAs are enclosed in exosomes (Zhou et al., 2012). The number of miRNAs reported for mammary gland varies considerably (Table 1). Chen at al. (2010) used high-throughput sequencing technology to search for expressed small RNA in cow colostrum and milk. After classifying the molecules based on size, they found a similar number of reads in both fluids (1,594,965 in colostrum vs 1,418,136 in milk). A search in the miRBase (a database of published miRNA sequences and annotation) resulted in the identification of 230 and 213 known miRNAs in colostrum and mature milk, respectively. Among the differently expressed miRNAs, 108 were up-regulated and only eight were down-regulated in colostrum compared to mature milk. These results indicate a dynamic gene expression during lactation. Izumi et al. (2012) identified a smaller number of miRNAs than Chen et al. (2010), with 100 being unique for colostrum and 53 for mature milk. It should be noted that whereas Chen et al. (2010) prepared RNA from a larger amount of milk and purified small RNA by PAGE, Izumi et al. (2012) started with a smaller amount of milk and used microarray analysis. Li et al. (2012d) reported 11,964,909 and 15,968,116 clean reads from mammary gland tissues in the lactating and non-lactating periods, respectively. After aligning the reads against the Bos taurus genome and miRBase, 885 pre-miRNAs were identified and encoded for 921 miRNAs; ∼60% of these miRNAs were expressed during the lactation and non-lactation period; of these, 248 miRNAs were known, 57 were conserved and 239 were new identifications. In human milk, Zhou et al. (2012) identified exosomes containing pre-miRNA from four libraries. Four of the top ten miRNAs (30b, 182, 200a and 148a) were related to different aspects of the immune system, with the most abundant of them being miRNA 148a, which is also expressed in bovine milk (Chen et al., 2010).
Table 1.
Total number of miRNAs reported in colostrum, milk and mammary gland in cow, goat and sheep.
| Number of miRNAs
|
Methods | Reference | |||
|---|---|---|---|---|---|
| Species | Colostrum | Milk | Mammary gland | ||
| Cow | 230 | 213 | Solexa deep sequencing analysis | Chen et al. (2010) | |
| Cow | 921 | Solexa deep sequencing analysis | Li et al. (2012d) | ||
| Goat | 100 | 53 | Microarray-real time PCR | Izumi et al. (2012) | |
| Goat | 328 | Solexa deep sequencing analysis | Ji et al. (2012) | ||
| Goat | 180 | 441 | Solexa deep sequencing analysis | Li et al. (2012c) | |
| Sheep | 101 | cDNA sequencing | Galio et al. (2012) | ||
In a sheep mammary gland library constructed from early pregnancy tissue, Galio et al. (2012) identified 54 sequences already described in the miRBase; two of these miRNAs (27e and 36e) were identified for the first time in mammals and only miRNA 379 had previously been shown to occur in sheep. In goats, Ji et al. (2012) characterized miRNAs from a pool of five Laoshan breed animals. A total of 18,031,615 read sequences were obtained after discarding ∼2.6% that did not meet the control criteria (i.e., they were of low quality, had contaminants formed by adapter-adapter ligation, and contained reads without insert tags). Of these reads, 9,093,530 had a perfect match to the Ovis aries genome and 305,711 were new sequences. Furthermore, 290 conserved miRNAs and 38 novel miRNAs were identified, and this total of 338 miRNAs was very similar to that reported by Li et al. (2012c) for dairy goats (441 miRNAs).
The biological material (mammary gland tissue or milk), the amount of sample used, and the species and breed examined can influence the results. Moreover, differences in the quality control criteria used to classify the sequencing products (raw and clean reads) and discrepancies associated with library normalization may also contribute to variations in the number of miRNAs reported. Finally, some degree of variation may be introduced by the mode of sample preparation. Lee et al. (2010) observed no changes in the 3′ end of a 22-nucleotide-long synthetic RNA introduced into RNA samples, but a fraction of the synthetic sequences were truncated at the 5′ end. These authors concluded that the changes in the 5′ end may have been caused by premature termination during the production of synthetic RNA. This finding suggests that at least part of the variation reported for end-region sequences may be linked to sample processing prior to sequence analysis. An additional source of diversity is that many miRNAs may vary from the published reference sequences. To address the latter phenomenon, Morin et al. (2008) proposed the terminology “isomers” to refer to sets of miRNAs that show similarity in their sequences. These authors suggested that isomer variability could be related to variation in the cleavage positions for the enzymes Dicer or Drosha within the pre-miRNA hairpin and showed results in which the variability among isomers influenced the differential expression of miRNA.
Functions of miRNA in mammary gland and milk
An important question in assessing miRNA function is whether the miRNAs present in milk are derived from blood or are specific for mammary gland. To address this issue, Chen et al. (2010) compared the miRNA profile of milk with that of serum from healthy cows and found that the total number of miRNAs in milk was about two-fold higher than in serum; they also identified 47 miRNAs unique to milk. Human breast milk also has a different pattern of miRNA expression compared to blood plasma (Kosaka et al., 2010). These results clearly indicate that mammary alveolar cells express their own miRNAs.
Another question is whether the pattern of miRNA expression in mammary gland is constant throughout the lactation period. Of the 1,692,810 reads described by Li et al. (2012d), 34% were expressed only in the dry period compared to the peak period of milk production. Moreover, analysis of the expression patterns of 173 differentially expressed miRNAs showed that 165 were down-regulated during peak lactation compared to the dry period. Among the sequences reported by Li et al. (2012d), 56 showed significant differences in expression between lactating and non-lactating cows, as assessed using the IDEG6 package (Romualdi et al., 2003); of these, nine were expressed only in lactating animals and six in non-lactating animals. However, 48 of these were confirmed by deep sequencing (Li et al., 2012d), indicating that deep sequencing may be more sensitive and reliable than microarray analysis in identifying differentially expressed miRNAs. Together, these findings indicate that the pattern of miRNA expression varies according to the animals physiological state.
To examine the expression of specific miRNAs associated with cellular proliferation, metabolism and the innate immune response during lactation, Wang et al. (2012) assessed the expression of 13 miRNAs in cows during the dry period (30 d prepartum), fresh period (7 d postpartum) and early lactation (30 d postpartum). Twelve of the miRNAs identified (miRNAs 10a, 15b, 16, 21, 33b, 145, 146b, 155, 181a, 205, 221 and 223) were down-regulated in the dry period compared to during lactation. The exception was miRNA 31, which showed greater expression in early lactation compared to the dry period. Under normal conditions and using bioinformatic assays and biological experiments, Xue et al. (2013) demonstrated that miRNA 31 up-regulated IL-2 (interleukin 2) expression by reducing the levels of the cytokines upstream kinase suppressor, KSR2 (kinase suppressor of ras 2). Interleukins are present in human milk (Bryan et al., 2006) and have an important role in modulating the offsprings immunological system (Aspinall et al., 2011). These findings indicate that miRNA 31 may have an indirect immunological role in the neonate.
Based on microarray analysis, Galio et al. (2012) identified three major patterns of miRNA expression in sheep. In pattern 1, expression was down-regulated during pregnancy, in pattern 2, miRNA expression was induced during pregnancy, and in pattern 3, miRNA expression was induced during lactation. The authors selected one miRNA from each pattern to confirm their expression by RT-qPCR in four animals per pattern. miRNA 21, which is expressed in alveolar epithelial cells, was up-regulated in non-pregnant sheep and at the beginning of pregnancy. This expression profile was attributed to a role for miRNA 21 in adipogenic differentiation. In this regard, Kim et al. (2009) showed that the activity of miRNA 21 in adipogenic tissue was mediated through TGF-β signaling. In contrast, miRNA 205 was expressed mainly in the basement membrane of normal mammary ducts and lobules during the first half of pregnancy and miRNA 200 was expressed in epithelial cells throughout pregnancy but was up-regulated at the end of pregnancy and lactation.
Bioinformatic analysis using the program RNA hybrid identified a miRNA 15a target sequence on the growth hormone receptor (Li et al., 2012a). However, this miRNA was not identified in a sequence search by Chen et al. (2010). To confirm their finding, Li et al. (2012a) transfected this small RNA in mammary epithelial cells and observed a reduction in growth hormone receptor (GHR) transcription and in the expression of β casein. Growth hormone is the most relevant galactopoietic hormone in ruminants (Bauman and Vernon, 1993) and triggers casein expression (Sakamoto et al., 2005). These results therefore indicated that miRNA 15a indirectly decreases milk production by blocking the expression of growth hormone receptor and thus identified a novel regulatory mechanism for GHR.
Some miRNAs have been suggested to have immunosuppressive roles. In silico analyses of two members of the miRNA 30 family (miRNAs 30a-5p and 30d-5p) predicted binding sites in several suppressors of cytokine signaling inhibitors of the JAK/STAT pathway that regulate IL-10 transcription (Gaziel-Sovran et al., 2011). This miRNA has also been implicated in the formation of the adipose pad in mammary gland. Le Guillou et al. (2012) constructed transgenic mice over-expressing miRNA 30b and observed (based on histological analysis) that these animals had acinar structures with abnormally small lumens. Even when there were no differences in the concentration of major milk proteins, the number of lipid droplets was smaller and did not show the spherical shape seen in the wild type. Microarray analyses of animals that did not express miRNA 30b showed that 164 genes were up-regulated and 56 genes were down-regulated. All of the up-regulated genes were associated with tissue development, except for seven that were involved in the inflammatory response. It is remarkable that the blockade of just a single miRNA altered the expression levels of 222 genes.
Further studies are needed to understand the biological roles of most of the reported miRNAs. After identifying miRNAs in mammary gland by sequencing procedures or computational searches, it is important to validate the results in functional experiments and to study their expression pattern in physiological and pathological conditions (Huang et al., 2010). The purpose of expression studies is to compare patterns between groups, e.g., disease vs. healthy and lactation vs. dry. The subsequent variation observed between experimental groups should reflect differences in expression between the groups and not be attributable to other sources of variation such as sampling methods, stabilization procedures and extraction methods. A normalizer or internal control should be used. This control is generally an RNA that exhibits invariant expression across all samples, is expressed along with the target miRNA in the cells of interest, and demonstrates equivalent storage stability and efficiency of extraction and quantification as the target miRNA of interest (Vandesompele et al., 2002; Peltier and Latham, 2008).
Selecting an optimal normalizer (an aspect that is frequently undervalued) may help to avoid inconsistent results. Gu et al. (2012) proposed a set of miRNAs for porcine milk studies. These authors compared the expression of six porcine milk miRNAs from different lactation periods and proposed three of them (miRNAs 17, 107 and 103) as internal controls because they were stabled throughout the periods studied (1 h and 3, 7, 14, 21 and 28 days postpartum). In cow, Izumi et al. (2012) normalized the samples using a synthetic cel-miRNA 39. Naeem et al. (2012) normalized the data using miRNA 320 and miRNA U6. The usefulness of the latter miRNA has been questioned because of its stability in serum (Chen et al., 2008). For the next generation of sequencing technology, normalization will be challenging because different sequencing experiments may generate quite different total numbers of reads (Li et al., 2012b). The total count normalization is inadequate for data generated by new generation sequencing technologies (Garmire and Subramaniam, 2012). To overcome this limitation various statistical models have been proposed for the normalization of data. Although there is still no general agreement about the most adequate internal controls and the best methods for normalizing data, it is nevertheless essential to establish criteria for selecting which controls should be used for each species and tissue and which method should be used to normalize the data in order to decrease the false discovery rate.
Functions of miRNA in mastitis
Mastitis, or inflammation of the mammary gland, is one of the most prevalent and costly diseases in dairy animals (for specific reviews see Halasa et al., 2007; Akers and Nickerson, 2011; De Vliegher et al., 2012). Intra-mammary infection occurs when bacteria cross the teat sphincter and reach the alveolar lumen after passing through the teat and gland cisterns. The first response of the immune system is a neutrophil influx via chemotaxis to establish the inflammatory process (Akers and Thompson, 1987). Understanding the molecular mechanisms involved in mastitis would be helpful in developing new strategies to prevent and treat this condition. Streptococcus uberis is one of the major etiological agents of mastitis. This Gram-positive bacterium can cause contagious or environmental mastitis (Rato et al., 2008). Two recent studies examined the miRNA expression pattern in S. uberis-induced mastitis: one focused on in vivo infection and miRNA expression at 12 h post-infection while the other examined the expression pattern at different times in cultured mammary epithelial cells. Both studies provided interesting insights into the rapid and diverse response triggered after infection.
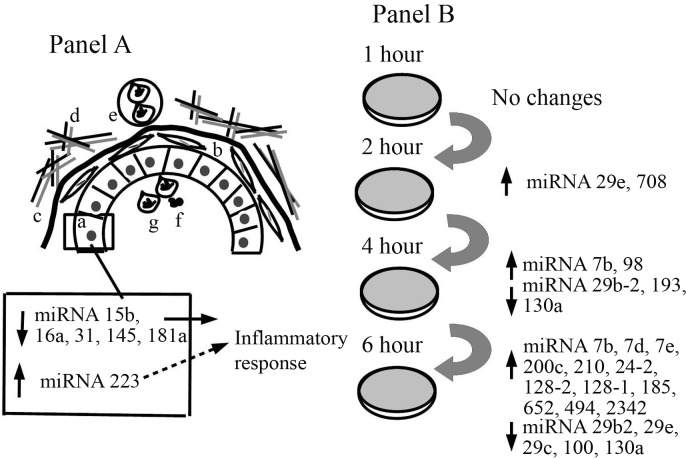
In the first of these studies, Naeem et al. (2012) examined the expression pattern of 14 miRNAs in mammary gland 12 h after a challenge with S. uberis. The resulting data plus the microarray gene expression patterns of 2,102 genes were used in bioinformatic analyses to identify miRNA targets and the biological pathways involved. The results showed down-regulation of miRNAs 15b, 16a, 31, 145 and 181a, and up-regulation of miRNA 223 in mastitis compared with healthy control animals (Figure 2, panel A). The target genes identified were mostly associated with immunological regulation, metabolic processes and cellular proliferation/differentiation. The change in miRNA 16a expression was associated with the up-regulation of some interleukins (IL-6, IL-8 and IL-10). The authors suggested that miRNA 16a might control the level of key inflammatory components in bovine mammary gland and could play a role in regulating the response to mastitis. Mastitis caused the down-regulation of miRNA 181a which has a role in the immune system (such as an increase in toll-like receptor and B cell receptor signaling). However, the decrease in miRNA 181a was the opposite of that observed in mice with an acute inflammatory response (Xie et al., 2013). In the latter case, the levels of miRNA 181a increased within 2 h after the induction of inflammation and remained high for up to 6 h post-treatment; however, 12 h later the expression was lower than in control animals. The discrepancy between these two studies may be related to the sampling intervals since Naeem et al. (2012) obtained the mammary biopsies 20 h after the bacterial challenge. However, differences among species should also be considered. The only miRNA that was up-regulated in udder mastitis was miRNA 223, which can inhibit several cellular signaling pathways mediated via the down-regulation of IGF1R (insulin growth factor 1 receptor). Negative modulation of the immune response may be required to avoid damage to the host tissue.
Figure 2.
Diagram of the mammary epithelial cell response to infection by Streptococcus uberis in vivo and in vitro. (A) Mammary alveolus showing (a) epithelial cells, (b) myoepithelial cells, (c) basement membrane, (d) extracellular matrix and (e) capillary. In the alveolar lumen: (f) bacterial infiltration and (g) neutrophil infiltration. The accompanying box shows that 12 h after inoculation with S. uberis there was a decrease in the expression of miRNAs 15b, 16a, 31, 145 and 181a and an increase in miRNA 223 to modulate the inflammatory response (Naeem et al., 2012). (B) Mammary epithelial cells inoculated with S. uberis. No changes were observed at 1 h but there was an increase in miRNAs 29e and 708 at 2 h, an increase in miRNAs 7b and 98 and a decrease in miRNAs 29b-2, 193 and 130a at 4 h and, finally, an increase in 12 miRNAs (7b, 7d, 7e, 200c, 210, 24-2, 128-2, 128-1, 185, 652, 494 and 2342) concomitantly with a decrease in miRNA 29b2, 29e, 29c, 100 and 130a at 6 h (Lawless et al., 2013).
In the second study, Lawless et al. (2013) identified 15 miRNAs that showed altered expression in cultured bovine mammary epithelial cells challenged with S. uberis. No changes were observed in the first hour after inoculation but at 2 h, miRNA 29e and miRNA 708 were up-regulated. This was followed by the up-regulation of miRNA 7b and miRNA 98 at 4 h post-challenge. At 6 h, 12 miRNAs were up-regulated (miRNAs 7b, 7d, 7e, 24-2, 128-1, 128-2, 185, 200c, 210, 494, 652 and 2342). Down-regulation was first observed only at 4 h and involved miRNAs 29b-2, 193a and 130a. Two miRNAs (29b-2 and 130a) were down-regulated at 4 h and 6 h (Figure 2, Panel B). Although a large number of 5′ isomers were identified, they were expressed at a low rate. The prediction of target genes showed that only the miRNAs down-regulated at 4 h and 6 h post-inoculation were significantly enriched in genes with a role in innate immunity. Table 2 summarizes the different miRNAs reported in mammary gland and milk and their level of expression in cow and sheep.
Table 2.
Summary of miRNA expression in mammary gland, mammary epithelial cell lines and milk in cow and sheep.
| miRNA | Tissue or biological fluid | Species | Expression level | Reference |
|---|---|---|---|---|
| miRNA 7b | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 7d | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 7e | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 10a | MGT | Cow | Elevated in lactation | Wang et al. (2012) |
| miRNA 15a | MECL | Cow | Elevated in transfected cells | Li et al. (2012a) |
| miRNA 15b | MGT | Cow | Elevated in lactation and reduced in mastitis | Wang et al. (2012); Naem et al. (2012) |
| miRNA 16 | MGT | Cow | Elevated in lactation | Wang et al. (2012) |
| miRNA 16a | MGT | Cow | Reduced in mastitis | Naem et al. (2012) |
| miRNA 21 | MGT | Cow/sheep | Elevated in cow during lactation and in sheep during early pregnancy | Galio et al. (2012); Wang et al. (2012) |
| miRNA 24-2 | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 29-b2 | Milk | Cow | Reduced in mastitis | Lawless et al. (2013) |
| miRNA 29c | Milk | Cow | Reduced in mastitis | Lawless et al. (2013) |
| miRNA 29e | Milk | Cow | Elevated and then reduced in mastitis | Lawless et al. (2013) |
| miRNA 31 | MGT/milk | Cow | Reduced in lactation and mastitis | Wang et al. (2012); Naem et al. (2012); Lawless et al. (2013) |
| miRNA 33b | MGT | Cow | Elevated in lactation | Wang et al. (2012) |
| miRNA 98 | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 100 | Milk | Cow | Reduced in lactation | Lawless et al. (2013) |
| miRNA 128-1 | Milk | Cow | Elevated in lactation | Lawless et al. (2013) |
| miRNA 128-2 | Milk | Cow | Elevated in lactation | Lawless et al. (2013) |
| miRNA 130a | MGT | Cow | Reduced in mastitis | Lawless et al. (2013) |
| miRNA145 | MGT | Cow | Elevated in lactation and reduced in mastitis | Wang et al. (2012); Naem et al. (2012) |
| miRNA 146b | MGT | Cow | Elevated in lactation | Wang et al. (2012) |
| miRNA 148a | Milk | Cow | Elevated in lactation | Chen et al. (2010) |
| miRNA 155 | MGT | Cow | Elevated in lactation | Wang et al. (2012) |
| miRNA 181a | MGT | Cow | Elevated in lactation and reduced in mastitis | Wang et al. (2012); Naem et al. (2012) |
| miRNA 185 | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 193a | MGT | Cow | Reduced in mastitis | Lawless et al. (2013) |
| miRNA 200 | MGT | Sheep | Elevated in lactation | Galio et al. (2012) |
| miRNA 200c | MGT/milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 205 | MGT | Cow/sheep | Elevated in cow lactation and mastitis and second half of sheep pregnancy | Galio et al. (2012); Wang et al. (2012) |
| miRNA 210 | MGT/milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 221 | MGT | Cow | Elevated in lactation | Wang et al. (2012) |
| miRNA 223 | MGT | Cow | Elevated in lactation and mastitis | Wang et al. (2012); Naem et al. (2012) |
| miRNA 494 | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 652 | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 708 | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
| miRNA 2342 | Milk | Cow | Elevated in mastitis | Lawless et al. (2013) |
ECL: mammary epithelial cell line, MGT: mammary gland tissue.
The mammary alveolus cell is a three-dimensional structure in which the extracellular matrix plays an active role in epithelial function such as casein expression (Katz and Streuli, 2007). Therefore, the time intervals studied and the biological model used (in vivo vs. cell culture) could account for the divergent miRNA expression reported in these studies. Nevertheless, the results clearly suggest that S. uberis can coordinate different processes by regulating target miRNAs. Another aspect to emphasize is the variability in miRNA expression and their rapid and dynamic temporal expression patterns that can modulate the ability of mammary epithelial cells to mobilize the innate immune system.
miRNA and biotechnology
Lactoglobulin (LGB) is the major whey protein in ruminants whereas human milk contains no LGB (Anderson et al., 1982). This difference in milk composition accounts for some of the milk allergy problems in infants. LGB intolerance in infants has been known since 1965 (Davidson et al., 1965). One strategy to reduce the allergenic potential of milk is to produce LGB-free milk. In pigs, Merkl et al. (2011) reported the effectiveness of short hairpin RNAs and artificial miRNAs in blocking LGB. Jabed et al. (2012) designed a successful strategy to block LGB synthesis by constructing a transgenic cow that expressed a tandem miRNAs construct (miRNA 6 and miRNA 4) against bovine LGB (see Figure 3). By controlling the artificial miRNAs through a lactation-specific-promoter it is possible to express miRNAs only during the lactation period when LGB is being produced. The analysis of milk samples by SDS-PAGE and HPLC showed that milk from miRNA 4 and 6 calves contained no LGB, but there was a strong, compensatory effect on the levels of other milk proteins. Consequently, artificial miRNAs could be an alternative for abolishing the production of specific proteins (Jabed et al., 2012).
Figure 3.
Bos taurus lactoglobulin B (LGB) mRNA sequence (GenBank accession number BC108213.1) showing the positions of miRNA6 and miRNA4 that targeted LGB, as designed by Jabed et al. (2012). The numbers refer to the nucleotide positions.
Stability of miRNA in milk
One of the interesting features of miRNAs is their stability. miRNAs are resistant to acidic conditions, digestion by RNAse, incubation at room temperature and various freeze/thaw cycles (Hata et al., 2010; Kosaka et al., 2010; Gu et al., 2012; Izumi et al.; 2012; Zhou et al., 2012). In milk, this resistance to degradation is explained by the fact that miRNAs are contained in exosomes or microvesicles. Treatment with Triton-X, a detergent that disrupts lipid membranes, results in the degradation of miRNAs by RNAse (Zhang et al., 2010). The resistance to acidic conditions ensures passage through the stomach and absorption into the bloodstream, and this in turn allows the exchange of genetic information between mother and offspring.
An important recent finding is the isolation of miRNAs from industrially processed foods for infants (Izumi et al., 2012). This raises new questions about the possible role that cow miRNAs could have in the end-consumer, especially children. Could cross-species miRNAs have an epigenetic role? In this regard, the study by Zhang et al. (2012) have provided interesting results regarding cross-kingdom transmission and regulation since these authors isolated a plant miRNA (miRNA168a) from the blood of mice fed with rice. They also demonstrated that this miRNA could bind to low density lipoprotein receptor adapter 1 mRNA in liver and that this binding led to a decrease in low density proteins in mouse plasma. This plant-derived miRNA was also identified in people consuming a rice-based diet. Even though plant and animal miRNAs have undergone independent evolutionary adaptations and are unrelated (Ambros, 2011), these findings show that plant miRNA can still exert a cross-kingdom effect.
The identification of cross-kingdom miRNA transmission raises significant questions. To what extent can miRNAs present in the diet regulate mammalian genes? How many miRNAs are incorporated through a normal diet? What are the effects of these miRNAs in health and disease? Milk provides a variety of bioactive components such as lactoferrin, defensin and immunoglobulin. In this regard, miRNAs represent yet another group of molecules transported by milk that could influence the immunological system in neonates.
Conclusion
The identification of small non-coding RNAs has provided new insights into cell regulation and intercellular communication. Studies of the mammary gland in lactation and mastitis show that the temporal expression of miRNAs regulates the innate immune system. The discovery of cross-species and cross-kingdom miRNA regulation provides a basis for new research into the regulatory mechanisms involved and the impact of epigenetic regulation. As stated by Zhang et al. (2012): “like vitamins, minerals and other essential nutrients derived from food sources, plant (or milk) miRNAs may serve as a novel functional component of food and make a critical contribution to maintaining and shaping animal body structure and function”. We are in an exciting time of new discoveries and a new understanding of gene regulation and epigenetic effects. Future research will improve our understanding of the role of miRNAs in health and disease and their importance as food resources.
Footnotes
Associate Editor: Carlos F.M. Menck
References
- Akers RM, Thompson W. Effect of induced leucocyte migration on mammary cell morphology and milk component biosynthesis. J Dairy Sci. 1987;70:1685–1695. doi: 10.3168/jds.S0022-0302(87)80197-2. [DOI] [PubMed] [Google Scholar]
- Akers RM, Nickerson SC. Mastitis and its impact on structure and function in the ruminant mammary gland. J Mammary Gland Biol Neoplasia. 2011;16:275–289. doi: 10.1007/s10911-011-9231-3. [DOI] [PubMed] [Google Scholar]
- Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- Ambros V. MicroRNAs: G enetically sensitized worms reveal new secrets. Curr Biol. 2010;20:R598–R600. doi: 10.1016/j.cub.2010.05.054. [DOI] [PubMed] [Google Scholar]
- Ambros V. microRNAs and developmental timing. Curr Opin Genet Dev. 2011;21:511–517. doi: 10.1016/j.gde.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NG, Powers MT, Tollaksen SL. Proteins of human milk. I. Identification of major components. Clin Chem. 1982;28:1045–1055. [PubMed] [Google Scholar]
- Aspinall R, Prentice AM, Ngom PT. Interleukin 7 from maternal milk crosses the intestinal barrier and modulates T-cell development in offspring. PLoS One. 2011;6:e20812. doi: 10.1371/journal.pone.0020812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman DE, Vernon RG. Effects of exogenous bovine somatotropin on lactation. Annu Rev Nutr. 1993;13:437–461. doi: 10.1146/annurev.nu.13.070193.002253. [DOI] [PubMed] [Google Scholar]
- Bryan DL, Forsyth KD, Gibson RA, Hawkes JS. Inter-leukin-2 in human milk: A potential modulator of lymphocyte development in the breastfed infant. Cytokine. 2006;33:289–2893. doi: 10.1016/j.cyto.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:1–10. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chen X, Gao C, Li H, Huang L, Sun Q, Dong Y, Tian C, Gao S, Dong H, Guan D, et al. Identification and characterization of microRNAs in raw milk during different periods of lactation, commercial fluid, and powdered milk products. Cell Res. 2010;20:1128–1137. doi: 10.1038/cr.2010.80. [DOI] [PubMed] [Google Scholar]
- Chong MM, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson M, Burnstine RC, Kugler MM, Bauer CH. Malabsorption defect induced by ingestion of beta lactoglobulin. J Pediatr. 1965;66:545–554. doi: 10.1016/s0022-3476(65)80119-6. [DOI] [PubMed] [Google Scholar]
- De Vliegher S, Fox LK, Piepers S, McDougall S, Barkema HW. Mastitis in dairy heifers: Nature of the disease, potential impact, prevention, and control. J Dairy Sci. 2012;95:1025–1040. doi: 10.3168/jds.2010-4074. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nat Genet. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Galio L, Droineau S, Yeboah P, Boudiaf H, Bouet S, Truchet S, Devinoy E. microRNA in the ovine mammary gland during early pregnancy: spatial and temporal expression of miR-21, miR-205 and miR-200. Physiol Genomics. 2012;45:151–161. doi: 10.1152/physiolgenomics.00091.2012. [DOI] [PubMed] [Google Scholar]
- Garmire LX, Subramaniam S. Evaluation of normalization methods in mammalian microRNA-Seq data. RNA. 2012;18:1279–1288. doi: 10.1261/rna.030916.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immuno-suppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Mol Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Li M, Wang T, Liang Y, Zhong Z, Wang X, Zhou Q, Chen L, Lang Q, He Z, et al. Lactation-related microRNA expression profiles of porcine breast milk exosomes. PloS One. 2012;7:e43691. doi: 10.1371/journal.pone.0043691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halasa T, Huijps K, Østerås O, Hogeveen H. Economic effects of bovine mastitis and mastitis management: A review. Vet Q. 2007;1:18–31. doi: 10.1080/01652176.2007.9695224. [DOI] [PubMed] [Google Scholar]
- Hata T, Murakami K, Nakatani H, Yamamoto Y, Matsuda T, Aoki N. Isolation of bovine milk-derived micro-vesicles carrying mRNAs and microRNAs. Biochem Biophys Res Commun. 2010;396:528–533. doi: 10.1016/j.bbrc.2010.04.135. [DOI] [PubMed] [Google Scholar]
- Huang Y, Zou Q, Song H, Song F, Wang L, Zhang G, Shen X. A study of miRNAs targets prediction and experimental validation. Protein Cell. 2010;1:979–986. doi: 10.1007/s13238-010-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci. 2012;95:4831–4841. doi: 10.3168/jds.2012-5489. [DOI] [PubMed] [Google Scholar]
- Jabed A, Wagner S, McCracken J, Wells DN, Laible G. Targeted microRNA expression in dairy cattle directs production of β-lactoglobulin-free, high-casein milk. Proc Natl Acad Sci USA. 2012;109:16811–16816. doi: 10.1073/pnas.1210057109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Ji Z, Wang G, Xie Z, Zhang C, Wang J. Identification and characterization of microRNA in the dairy goat (Capra hircus) mammary gland by Solexa deep-sequencing technology. Mol Biol Rep. 2012;39:9361–9371. doi: 10.1007/s11033-012-1779-5. [DOI] [PubMed] [Google Scholar]
- Katz E, Streuli CH. The extracellular matrix as an adhesion checkpoint for mammary epithelial function. Int J Biochem Cell Biol. 2007;39:715–726. doi: 10.1016/j.biocel.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Hwang SJ, Bae YC, Jung JS. miR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27:3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Izumi H, Sekine K, Ochiya T. microRNA as a new immune-regulatory agent in breast milk. Silence. 2010;1:1–7. doi: 10.1186/1758-907X-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lawless N, Foroushani ABK, McCabe MS, O’Farrelly C, Lynn DJ. Next generation sequencing reveals the expression of a unique miRNA profile in response to a Gram-positive bacterial infection. PLoS One. 2013;8:e57543. doi: 10.1371/journal.pone.0057543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guillou S, Sdassi N, Laubier J, Passet B, Vilotte M, Castille J, Laloe D, Polyte J, Bouet S, Jaffrezic F, et al. Over-expression of miR-30b in the developing mouse mammary gland causes a lactation defect and delays involution. PLoS One. 2012;7:e45727. doi: 10.1371/journal.pone.0045727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LW, Zhang S, Etheridge A, Ma L, Martin D, Galas D, Wang K. Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA. 2010;16:2170–2180. doi: 10.1261/rna.2225110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with anti-sense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Li H-M, Wang C-M, Li Q-Z, Gao XJ. MiR-15a decreases bovine mammary epithelial cell viability and lactation and regulates growth hormone receptor expression. Molecules. 2012a;17:12037–12048. doi: 10.3390/molecules171012037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Witten DM, Johnstone IM, Tibshirani R. Normalization, testing, and false discovery rate estimation for RNA-sequencing data. Biostatistics. 2012b;13:523–538. doi: 10.1093/biostatistics/kxr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Lan X, Guo W, Sun J, Huang Y, Wang J, Huang T, Lei C, Fang X, Chen H. Comparative transcriptome profiling of dairy goat microRNAs from dry period and peak lactation mammary gland tissues. PLoS One. 2012c;7:e52388. doi: 10.1371/journal.pone.0052388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liu H, Jin X, Lo L, Liu J. Expression profiles of microRNAs from lactating and non-lactating bovine mammary glands and identification of miRNA related to lactation. BMC Genomics. 2012d;13:731. doi: 10.1186/1471-2164-13-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- Merkl C, Leuchs S, Saalfrank A, Kind A, Schnieke A. RNA interference in pigs: comparison of RNAi test systems and expression vectors. Mol Biotechnol. 2011;48:38–48. doi: 10.1007/s12033-010-9346-6. [DOI] [PubMed] [Google Scholar]
- Morin RD, O’Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, Zhao Y, McDonald H, Zeng T, Hirst M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naeem A, Zhong K, Moisá SJ, Drackley JK, Moyes KM, Loor JJ. Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with Streptococcus uberis 1. J Dairy Sci. 2012;95:6397–6408. doi: 10.3168/jds.2011-5173. [DOI] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rato MG, Bexiga R, Nunes SF, Cavaco LM, Vilela CL, Santos-Sanches I. Molecular epidemiology and population structure of bovine Streptococcus uberis. J Dairy Sci. 2008;91:4542–4551. doi: 10.3168/jds.2007-0907. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Romualdi C, Bortoluzzi S, D’Alessi F, Danieli GA. IDEG6: A web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 2003;12:159–162. doi: 10.1152/physiolgenomics.00096.2002. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Komatsu T, Kobayashi T, Rose MT, Aso H, Hagino A, Obara Y. Growth hormone acts on the synthesis and secretion of alpha-casein in bovine mammary epithelial cells. J Dairy Res. 2005;72:264–270. doi: 10.1017/S0022029905000889. [DOI] [PubMed] [Google Scholar]
- Shabalina AS, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008;23:578–587. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Moisá S, Khan MJ, Wang J, Bu D, Loor JJ. MicroRNA expression patterns in the bovine mammary gland are affected by stage of lactation. J Dairy Sci. 2012;9:6529–6535. doi: 10.3168/jds.2012-5748. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- Xie W, Li Z, Li M, Xu N, Zhang Y. miR-181a and inflammation: miRNA homeostasis response to inflammatory stimuli in vivo. Biochem Biophys Res Commun. 2013;430:647–652. doi: 10.1016/j.bbrc.2012.11.097. [DOI] [PubMed] [Google Scholar]
- Xue F, Li H, Zhang J, Lu J, Xia Y, Xia Q. miR-31 regulates interleukin 2 and kinase suppressor of ras 2 during T cell activation. Genes Immun. 2013;14:127–131. doi: 10.1038/gene.2012.58. [DOI] [PubMed] [Google Scholar]
- Zhang L, Hou D, Chen X, Li D, Zhu L, Zhang Y, Li J, Bian Z, Liang X, Cai X, et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell Endocrinol. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, Zhou X, Wang X, Gao X, Li X. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]