Abstract
DNA methylation is mediated by DNA methyltransferases (DNMTs) that add a methyl group to the 5′-carbon of cytosine. The enzyme methylenetetrahydrofolate reductase (MTHFR) catalyzes the reduction of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate in the rate-limiting step of the cycle involving the methyl donor S-adenosyl-L-methionine (SAM). The MTHFR C677T polymorphism results in a thermolabile enzyme with reduced activity that is predicted to influence the DNA methylation status. In this study, we investigated the impact of the MTHFR C677T polymorphism on the global DNA methylation of oral epithelial cells obtained from 54 healthy subjects. There were no significant differences in global DNA methylation among the MTHFR CC, CT and TT genotypes (p = 0.75; Kruskal-Wallis test).
Keywords: DNA methylation, epigenetic, MTHFR C677T, oral epithelial cells, polymorphism
DNA methylation is dependent on a methyl donor and S-adenosyl-L-methionine (SAM) is the primary methyl group donor for most biological methylation reactions (Chiang et al., 1996). SAM is generated by the methionine cycle in which 5-methyltetrahydrofolate transfers single methyl groups to homocysteine in a reaction catalyzed by methionine synthase to produce methionine. After donating the methyl group, 5-methyltetrahydrofolate is converted to tetrahydrofolate and then to 5,10-methylenetetrahydrofolate by serine hydroxymethyltransferase. 5,10-methylenetetrahydrofolate is a key substrate that can be directed towards nucleotide biosynthesis or methionine regeneration. Methylenetetrahydrofolate reductase (MTHFR) catalyzes the irreversible conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate that can be used in the methionine cycle to generate SAM (Schwann and Rozen, 2001). The MTHFR C677T polymorphism leads to the amino acid alanine being replaced by valine (p.Ala222-Val) and the production of a thermolabile variant of MTHFR with 30% less enzyme activity (Sharp and Little, 2004). The MTHFR C677T polymorphism has been suspected to induce hypomethylation and then activate proto-oncogenes, which could explain the association between this polymorphism and some types of cancer (Liu et al., 2012; Saberi et al., 2012; Izmirli, 2013), including oral cancer (Sailasree et al., 2011). This hypomethylation has been suggested to involve the entire genome or specific CpG sites (Matsubayashi et al., 2005; Graziano et al., 2006; Zhu et al., 2011).
Variations in the DNA methylation profile are common among healthy individuals and may be gender-, age-and tissue-specific (El-Maarri et al., 2007; Thompson et al., 2010; Fernandez et al., 2012). Even within the same age group, gender and cell types, inter-individual variations are still observed and are influenced by environmental and genetic factors (Valenza-Schaerly et al., 2001; Weaver et al., 2004; Fraga et al., 2005; Sinclair et al., 2007; Bjornsson et al., 2008).
The aim of this study was to assess the influence of the MTHFR C677T polymorphism on global DNA methylation in oral epithelial cells in young subjects with no systemic disorders or visible alterations in the oral mucosa.
This study was approved by the Institutional Review Board of the Federal University of Paraiba (protocol number 0427/2012). All volunteers were informed about the nature of the proposed research and provided written informed consent. A convenience sample of unrelated male and female subjects > 19 years old was recruited for the study. All subjects were in good general health and had at least 20 healthy teeth. The exclusion criteria included any systemic disorder, pregnancy or lactation, smoking habit and the systemic use of antibiotics or anti-inflammatory medicines within six months prior to baseline. Individuals with a history of oral disease or with clinical signs of damage to the oral mucosa were excluded. The subjects were classified into one of three categories based on their MTHFR genotype, viz. CC, CT and TT.
Oral epithelial cell samples were collected by mouthwash with 3% dextrose and centrifuged at 3000 rpm for 10 min (Trevilatto and Line, 2000). DNA was purified using DNAzol® (Invitrogen) based on the manufacturer’s recommendations. The MTHFR C677T genetic polymorphism was detected by PCR-RFLP, as previously described (Arruda et al., 1997). Global DNA methylation levels were assessed with an ELISA-based commercial kit (MDQ1, Imprint® methylated DNA quantification kit; Sigma Aldrich, St. Louis, MO, USA) according to the manufacturer’s recommendations. The global DNA methylation levels among groups were compared using the Kruskal-Wallis test while the relationship between DNA methylation and age or gender was assessed using analysis of variance (ANOVA) and Students unpaired t-test, respectively. A value of p < 0.05 indicated significance. All data analyses were done using the software package Prism 5.0 (GraphPad Inc., La Jolla, CA, USA).
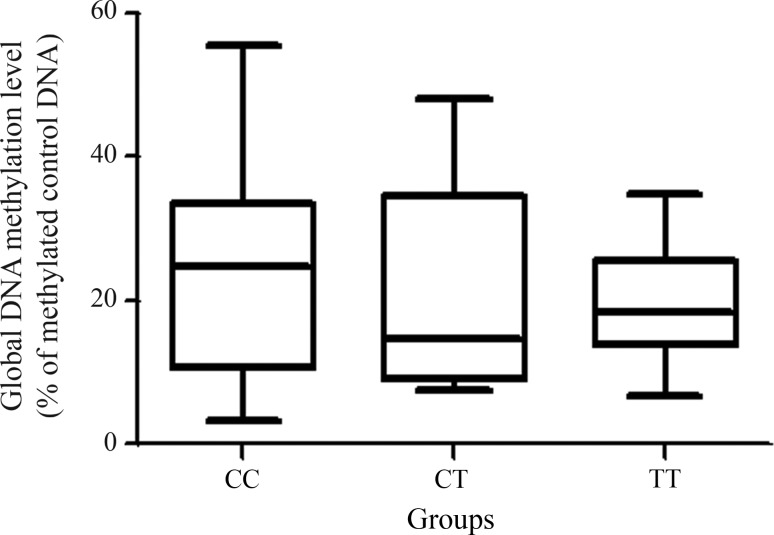
Fifty-four subjects 19–25 years old were enrolled in the study. Of these, 17 were CC (four males and 13 females; mean age: 21.4 ± 2.9 years; mean ± SD), 19 were CT (five males and 14 females; mean age: 21.8 ± 2.1 years) and 18 were TT (11 males and seven females; mean age; 22.7 ± 2.8 years) based on their MTHFR genotype. There were no significant differences in the global DNA methylation levels (expressed as a percentage) among the CC, CT and TT genotypes (p = 0.75; Kruskal-Wallis test) (Figure 1). In addition, the percentage of global DNA methylation was unrelated to age (p = 0.07; ANOVA) or gender (p = 0.30; Students unpaired t-test).
Figure 1.
Global DNA methylation of oral epithelial cells from healthy subjects based on the MTHFR genotypes. The data are shown as the median, minimum and maximum values for 17 (CC), 19 (CT) and 18 (TT) individuals (p = 0.75; Kruskal-Wallis test).
Studies of the effect of the MTHFR C677T polymorphism on DNA methylation have focused primarily on the pathological implications of this phenomenon. Few studies have examined the influence of this polymorphism on DNA methylation in healthy subjects. This is the first study to evaluate this association in oral epithelial cells and no significant differences were detected among the genotypes. A previous study of normal colonic mucosa found no influence of the MTHFR C677T polymorphism on global DNA methylation in this tissue, although a tendency towards a decrease in global methylation was observed in smokers with low folate levels who carriedtheC677T substitution (Pufulete et al., 2005). In addition, studies using lymphocytes found no correlation between the MTHFR C677T polymorphism and decreased DNA methylation in the genome overall (Narayanan et al., 2004; Ono et al., 2012) or at specific CpG sites (Hirsch et al., 2008; Wernimont et al., 2011), regardless of the folate concentration.
However, the existence of this association remains controversial because some studies detected a positive correlation when folate levels were low (Stern et al., 2000; Friso and Choi, 2002; Friso et al., 2002). Folate consumption stimulates the metabolic pathway leading to the biosynthesis of homocysteine that is subsequently converted to its active form, tetrahydrofolate, which is a substrate for the enzyme MTHFR. MTHFR plays a central role in folate metabolism by irreversibly converting 5,10-methylenetetrahydrofolate to 5-methylenetetrahydrofolate, the primary circulating form of folate. This product provides methyl groups for methionine synthesis, which in turn is required for the synthesis of SAM, the primary methyl group donor (Fox and Stover, 2008).
In addition to aging (Bjornsson et al., 2008; Grönniger et al., 2010), the influence of environmental effects on DNA methylation is a confounding factor in establishing a correlation between polymorphisms and modifications of the DNA methylation profile. Indeed, environmental factors such as diet (Heijmans et al., 2008; McKay et al., 2012), alcohol consumption (Philibert et al., 2012; Zhang et al., 2013), tobacco use (Flom et al., 2011; Wangsri et al., 2012), radiation exposure (Grönniger et al., 2010; Chaudhry and Omaruddin, 2012) and air pollution (Tarantini et al., 2009; Salam et al., 2012) have been shown to modulate DNA methylation.
Acknowledgments
We thank the volunteers who donated oral mucosa samples. ITSA was supported by a fellowship from Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Footnotes
Associate Editor: Angela M. Vianna-Morgante
References
- Arruda VR, Von Zube PM, Chiparini LC, Annichino-Bizzacchi JM, Costa FF. The mutation ala677val in the MTHFR gene: A risk factor for arterial disease and venous thrombosis. J Thomb Haemost. 1997;77:818–821. [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, et al. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2888. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry MA, Omaruddin RA. Differential DNA methylation alterations in radiation-sensitive and -resistant cells. DNA Cell Biol. 2012;31:908–916. doi: 10.1089/dna.2011.1509. [DOI] [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-adenosylmethionine and methylation. FASEB J. 1996;10:471–480. [PubMed] [Google Scholar]
- El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: A tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC, Galm O, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22:407–419. doi: 10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JT, Stover PJ. Folate-mediated one-carbon metabolism. Vitam Horm. 2008;79:1–44. doi: 10.1016/S0083-6729(08)00401-9. [DOI] [PubMed] [Google Scholar]
- Flom JD, Ferris JS, Liao Y, Tehranifar P, Richards CB, Cho YH, Gonzalez K, Santella RM, Terry MB. Prenatal smoke exposure and genomic DNA methylation in a multi-ethnic birth cohort. Cancer Epidemiol Biomarkers Prev. 2011;20:2518–2523. doi: 10.1158/1055-9965.EPI-11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso S, Choi SW. Gene-nutrient interactions and DNA methylation. J Nutr. 2002;132:2382S–2387S. doi: 10.1093/jn/132.8.2382S. [DOI] [PubMed] [Google Scholar]
- Friso S, Choi SW, Girelli D, Mason JB, Dolnikowski GG, Bagley PJ, Olivieri O, Jacques PF, Rosenberg IH, Corrocher R, et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci USA. 2002;99:5606–5611. doi: 10.1073/pnas.062066299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano F, Kawakami K, Ruzzo A, Watanabe G, Santini D, Pizzagalli F, Bisonni R, Mari D, Floriani I, Catalano V, et al. Methylenetetrahydrofolate reductase 677C/T gene polymorphism, gastric cancer susceptibility and genomic DNA hypomethylation in an at-risk Italian population. Int J Cancer. 2006;118:628–632. doi: 10.1002/ijc.21397. [DOI] [PubMed] [Google Scholar]
- Grönniger E, Weber B, Heil O, Peters N, Stäb F, Wenck H, Korn B, Winnefeld M, Lyko F. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genetics. 2010;6:e1000971. doi: 10.1371/journal.pgen.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch S, Ronco AM, Guerrero-Bosagna C, de la Maza MP, Leiva L, Barrera G, Llanos M, Alliende MA, Silva F, Bunout D. Methylation status in healthy subjects with normal and high serum folate concentration. Nutrition. 2008;24:1103–1109. doi: 10.1016/j.nut.2008.05.018. [DOI] [PubMed] [Google Scholar]
- Izmirli M. A literature review of MTHFR (C677T and A1298C polymorphisms) and cancer risk. Mol Biol Rep. 2013;40:625–637. doi: 10.1007/s11033-012-2101-2. [DOI] [PubMed] [Google Scholar]
- Liu XM, Liu FH, Tang Y, Li Q. MTHFR C677T polymorphism and pancreatic cancer risk: A meta-analysis. Asian Pac J Cancer Prev. 2012;13:3763–3766. doi: 10.7314/apjcp.2012.13.8.3763. [DOI] [PubMed] [Google Scholar]
- McKay JA, Groom A, Potter C, Coneyworth LJ, Ford D, Mathers JC, Relton CL. Genetic and non-genetic influences during pregnancy on infant global and site-specific DNA methylation: Role for folate gene variants and vitamin B12. PLoS One. 2012;7:e33290. doi: 10.1371/journal.pone.0033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi H, Skinner HG, Iacobuzio-Donahue C, Abe T, Sato N, Riall TS, Yeo CJ, Kern SE, Goggins M. Pancreaticobiliary cancers with deficient methylenetetrahydrofolate reductase genotypes. Clin Gastroenterol Hepatol. 2005;3:752–760. doi: 10.1016/s1542-3565(05)00359-9. [DOI] [PubMed] [Google Scholar]
- Narayanan S, McConnell J, Little J, Sharp L, Piyathilake CJ, Powers H, Basten G, Duthie SJ. Associations between two common variants C677T and A1298C in the methylenetetrahydrofolate reductase gene and measures of folate metabolism and DNA stability (strand breaks, misincorporated uracil and DNA methylation status) in human lymphocytes in vivo. Cancer Epidemiol Biomarkers Prev. 2004;13:1436–1443. [PubMed] [Google Scholar]
- Ono H, Iwasaki M, Kuchiba A, Kasuga Y, Yokoyama S, Onuma H, Nishimura H, Kusama R, Ohnami S, Sakamoto H, et al. Association of dietary and genetic factors related to one-carbon metabolism with global methylation level of leukocyte DNA. Cancer Sci. 2012;103:2159–2164. doi: 10.1111/cas.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philibert RA, Plume JM, Gibbons FX, Brody GH, Beach SR. The impact of recent alcohol use on genome wide DNA methylation signatures. Front Genet. 2012;3:54. doi: 10.3389/fgene.2012.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufulete M, Al-Ghnaniem R, Rennie JA, Appleby P, Harris N, Gout S, Emery PW, Sanders TA. Influence of folate status on genomic DNA methylation in colonic mucosa of subjects without colorectal adenoma or cancer. Br J Cancer. 2005;92:838–842. doi: 10.1038/sj.bjc.6602439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saberi S, Zendehdel K, Jahangiri S, Talebkhan Y, Abdirad A, Mohajerani N, Bababeik M, Karami N, Esmaili M, Oghalaie A, et al. Impact of methylenetetrahydrofolate reductase C677T polymorphism on the risk of gastric cancer and its interaction with Helicobacter pylori infection. Iran Biomed J. 2012;16:179–184. doi: 10.6091/ibj.1102.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailasree R, Nalinakumari KR, Sebastian P, Kannan S. Influence of methylenetetrahydrofolate reductase polymorphisms in oral cancer patients. J Oral Pathol Med. 2011;40:61–66. doi: 10.1111/j.1600-0714.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- Salam MT, Byun HM, Lurmann F, Breton CV, Wang X, Eckel SP, Gilliland FD. Genetic and epigenetic variations in inducible nitric oxide synthase promoter, particulate pollution, and exhaled nitric oxide levels in children. J Allergy Clin Immunol. 2012;129:e7. doi: 10.1016/j.jaci.2011.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwann B, Rozen R. Polymorphisms in the methylenetetrahydrofolate reductase gene: Clinical consequences. Am J Pharmacogenomics. 2001;1:189–201. doi: 10.2165/00129785-200101030-00004. [DOI] [PubMed] [Google Scholar]
- Sharp L, Little J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: A huge review. Am J Epidemiol. 2004;159:423–443. doi: 10.1093/aje/kwh066. [DOI] [PubMed] [Google Scholar]
- Sinclair KD, Allegrucci C, Singh R, Gardner DS, Sebastian S, Bispham J, Thurston A, Huntley JF, Rees WD, Maloney CA, et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal peri-conceptional B vitamin and methionine status. Proc Natl Acad Sci USA. 2007;104:19351–19356. doi: 10.1073/pnas.0707258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern LL, Mason JB, Selhub J, Choi SW. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol Biomarkers Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- Tarantini L, Bonzini M, Apostoli P, Pegoraro V, Bollati V, Marinelli B, Cantone L, Rizzo G, Hou L, Schwartz J, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–222. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RF, Atzmon G, Gheorghe C, Liang HQ, Lowes C, Greally JM, Barzilai N. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell. 2010;9:506–518. doi: 10.1111/j.1474-9726.2010.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevilatto PC, Line SR. Use of buccal epithelial cells for PCR amplification of large DNA fragments. J Forensic Odontostomatol. 2000;18:6–9. [PubMed] [Google Scholar]
- Valenza-Schaerly P, Pickard B, Walter J, Jung M, Pourcel L, Reik W, Gauguier D, Vergnaud G, Pourcel C. A dominant modifier of transgene methylation is mapped by QTL analysis to mouse chromosome 13. Genome Res. 2001;11:382–388. doi: 10.1101/gr.163801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangsri S, Subbalekha K, Kitkumthorn N, Mutirangura A. Patterns and possible roles of LINE-1 methylation changes in smoke-exposed epithelia. PLoS One. 2012;7:e45292. doi: 10.1371/journal.pone.0045292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Wernimont SM, Clark AG, Stover PJ, Wells MT, Litonjua AA, Weiss ST, Gaziano JM, Tucker KL, et al. Folate network genetic variation, plasma homocysteine, and global genomic methylation content: A genetic association study. BMC Med Genet. 2011;12:150. doi: 10.1186/1471-2350-12-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Herman AI, Kranzler HR, Anton RF, Zhao H, Zheng W, Gelernter J. Array-based profiling of DNA methylation changes associated with alcohol dependence. Alcohol Clin Exp Res. 2013;37:E108–E115. doi: 10.1111/j.1530-0277.2012.01928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Jin Z, Yuan Y, Lu Q, Ge D, Zong M. Impact of MTHFR gene C677T polymorphism on Bcl-2 gene methylation and protein expression in colorectal cancer. Scand J Gastroenterol. 2011;46:436–445. doi: 10.3109/00365521.2010.537682. [DOI] [PubMed] [Google Scholar]