Abstract
The loss of soybean yield to Brazilian producers because of a water deficit in the 2011–2012 season was 12.9%. To reduce such losses, molecular biology techniques, including plant transformation, can be used to insert genes of interest into conventional soybean cultivars to produce lines that are more tolerant to drought. The abscisic acid (ABA)-independent Dehydration Responsive Element Binding (DREB) gene family has been used to obtain plants with increased tolerance to abiotic stresses. In the present study, the rd29A:AtDREB2A CA gene from Arabidopsis thaliana was inserted into soybean using biolistics. Seventy-eight genetically modified (GM) soybean lines containing 2–17 copies of the AtDREB2A CA gene were produced. Two GM soybean lines (P1397 and P2193) were analyzed to assess the differential expression of the AtDREB2A CA transgene in leaves and roots submitted to various dehydration treatments. Both GM lines exhibited high expression of the transgene, with the roots of P2193 showing the highest expression levels during water deficit. Physiological parameters examined during water deficit confirmed the induction of stress. This analysis of AtDREB2A CA expression in GM soybean indicated that line P2193 had the greatest stability and highest expression in roots during water deficit-induced stress.
Keywords: Arabidopsis, differential expression, genetically modified organism, water deficit
Introduction
Brazil is the world’s second largest soybean producer (Empresa Brasileira de Pesquisa Agropecuária - EMBRA-PA, 2012), and its main consumer market is China, which represents 67% of Brazilian soybean exports (United States Department of Agriculture - USDA, 2011). According to some estimates, exports involving the soybean complex will represent US$30 billion in 2013 (Associação Brasileira das Indústrias de Óleos Vegetais - ABIOVE, 2013).
However, the 2011–2012 harvest yield was reduced by 12.9% compared to the previous year because of severe droughts in southern Brazil, one of the main soybean-producing regions (Companhia Nacional de Abastecimento - CONAB, 2012). The alternatives available to minimize/reduce problems arising from water deficit include the use of biotechnology to develop cultivars more tolerant to drought. To address this problem, a group of researchers from the Japan International Research Center for Agricultural Sciences (JIRCAS) identified, characterized and isolated a family of transcription factors (Dehydration Responsive Element Binding - DREB) from Arabidopsis thaliana that are associated with increased tolerance to abiotic stressors (Shinozaki and Yamaguchi-Shinozaki, 2000).
Transcription factor (TF) AtDREB1A, which is responsive to cold, was inserted into soybean and subjected to molecular, anatomical and physiological analysis by Polizel et al. (2011) using experimental water deficit conditions. Transcription factor DREB2A, which belongs to the same TF family, was also characterized in A. thaliana and responded to drought, high salinity and heat. Overexpression of constitutively active (CA) DREB2A resulted in significant tolerance to drought and heat stress in transgenic Arabidopsis plants (Sakuma et al., 2006a,b). AtDREB2A-homologous genes were studied in maize (Qin et al., 2007), rice (Dubouzet et al., 2003), sunflower (Almogueva et al., 2009), wheat (Terashima and Takumi, 2009) and chrysanthemum (Liu et al., 2008). Recently, Mizoi et al. (2013) identified a soybean DREB2 gene, GmDREB2A;2, and showed that its heterologous expression in Arabidopsis improved the plants tolerance to stress. These findings indicate that plants overexpressing AtDREB2A and DREB2A-like proteins have increased tolerance to abiotic stress, drought and heat, which often occur together under field conditions.
There is a pressing need for soybean cultivars that are more tolerant to drought and heat, particularly since data provided by the IPCC (Intergovernmental Panel on Climate Change, 2007) indicate that the Earth’s average temperature will increase in forthcoming years and lead to a reduction in land area suitable for growing soybean and other crops. The aim of this study was to insert the TF DREB2A isolated from A. thaliana into soybean using biolistics and then examine this genes effect on resistance to drought. The number of transgene copies inserted into the soybean genome was quantified using quantitative polymerase chain reaction (qPCR) and Southern blotting. The transgene expression under drought conditions was analyzed by qPCR in leaves and roots. The results described here may be useful in developing soybean cultivars that are more tolerant to drought and heat.
Materials and Methods
Phylogenetic analysis
The similarity between the sequence of AtDREB2A (the constitutively active form of CA; JIRCAS) and the soybean DREB genes, and the homology of AtDREB2A CA with other genes belonging to the same subfamily in other plant species, were assessed by in silico analyses done using sequences downloaded from the National Center for Biotechnology Information (NCBI) expressed sequence tags (EST) database. The sequences were aligned using Clustal X software and two phylogenetic trees were created using MEGA 4.0 (Molecular Evolutionary Genetics Analysis) software.
Production of genetically modified soybean plants
Embryos from the conventional soybean cultivar BR 16, which is sensitive to drought (Oya et al., 2004), were co-transformed with the genetic constructs rd29A:AtDREB2A CA (Patent no. WO 2006/006236 PCT/JP2004/01003) and ahas:AtAHAS using biolistics, based on the methods of Aragão et al. (2000) and Rech et al. (2008). The genetic construct rd29A:AtDREB2A CA contains the stress-inducible rd29A promoter (Yamaguchi-Shinozaki and Shinozaki, 1994) and the coding region of AtDREB2A is in the CA form, which has a deletion in the negative regulation domain between amino acid residues 136 and 165. The ahas:AtAHAS construct acts as a selection gene that confers tolerance to imazapyr, an imidazolinone herbicide (Aragão et al., 2000).
Characterization of the transgenic plants
Identification of plants containing the AtDREB2A CA gene by conventional PCR
Conventional PCR was used to confirm the positive GM events, chimerization and segregation of the transgene in leaf genomic DNA extracted according to Doyle and Doyle (1987). The primer set used was: forward (F) 5′-GTTTGAACGATCGGGGAAT-3′ and reverse (R) 5′-GGGAAGCTTGCCATAGATGCAATTCAATCAAA CT-3′, which amplified a 1975 bp fragment between the promoter and terminator regions. The reaction mixture consisted of 5 μM of each F and R primer, 2.5 mM dNTPs (Invitrogen, Grand Island, NY, USA), 50 mM magnesium chloride (MgCl2) (Sigma), 2.5 μL of 10X buffer, one unit (U) of Taq DNA polymerase (Invitrogen) and 60 ng of genomic DNA. Amplification was done using an initial denaturation step at 95 °C for 5 min followed by 35 cycles of 95 °C for 1 min, 55 °C for 1 min and 72 °C for 1 min. Once identified, the transgenic plants were transferred to pots in a greenhouse where they were grown and used as described below.
Quantification of copy number by qPCR
Quantitative PCR (qPCR) was used to determine the number of transgenic cassette copies of rd29A:AtDREB2A CA inserted into the soybean genome. Sixteen PCR-positive plants from the T0 generation (P1397, P7186, P7413, P2193, P7195, P7417, P7212, P7418, P7231, P7430, P7256, P7374, P7431, P7174, P7393 and P7531) were analyzed. The endogenous lectin gene GmLec (accession no. K00821) was used as the reference gene for normalization since it is a species-specific gene and there is only a single copy in the soybean haploid genome (Meyer et al., 1994). The primer set used for the target gene was: F5′-CCGAGAGTCAACAAAGTGGTTTT-3′ and R 5′-CGAGCTGAACGGAGGTATTCC-3′, and the probe sequence was 5′-FAMCCGCTACAAAGC-3′. The primer set used for the reference gene was F 5′-TCCCGAGTG GGTGAGGATAG-3′ and R 5′-CATGCGATTCCCCAG GTATG-3′, and the probe sequence was 5′-TETTCTC TGCTGCCACGGGACTCG-3′. The cycling conditions consisted of 50 °C for 2 min, denaturation at 95 °C for 10 min, and 40 cycles of 95 °C for 15 s and 60 °C for 1 min.
To calculate the number of copies inserted into T0 generation plants, the formula of Livak and Schmittgen (2001) was modified for 2−ΔCt since the plants were hemizygous in this generation and the resulting value was multiplied by two to correct for the 2:1 proportion of the endogenous gene corresponding to the transgene. One T0 generation plant containing a large copy number of AtDREB2A CA (P2193) and one containing a low copy number (P1397) were selected for gene expression analysis.
Southern blotting
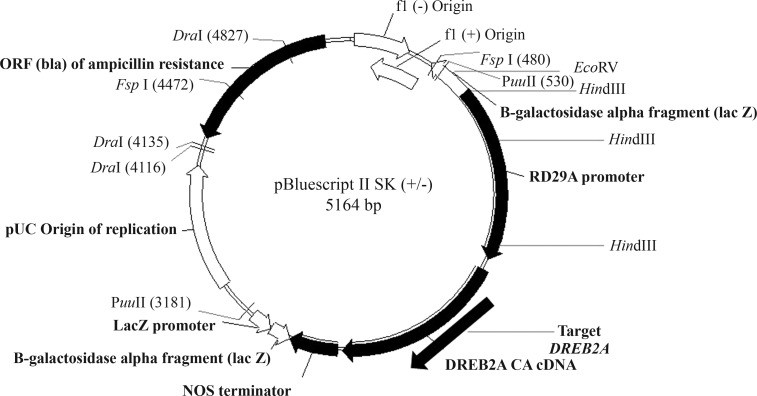
The number of transgenic cassette insertions was assessed by Southern blotting using soybean genomic DNA obtained from T0, T1 and T2 plants of P2193, which had high transgene expression levels. After extraction, the DNA was digested using the restriction enzymes HindIII and EcoRV and then hybridized using a 32P-labeled radioactive probe that targeted AtDREB2A CA (Figure 1). Hybridized bands were detected by autoradiography (Southern, 1974).
Figure 1.
Plasmid containing the transgene rd29A:AtDREB2A CA and the map of restriction enzymes used in this work. The enzymes used in Southern blotting were HindIII and EcoRV.
Leaf dehydration test using a biological oxygen demand (BOD) incubator
GM plants P2193 and P1397 were grown in Typic Haplorthox (Embrapa, 2006) for 60 days in a greenhouse with drip irrigation (twice per day for 5 min each) at 28 ± 3 °C. The experiment was based on the protocol by Kariola et al. (2006), with the following modifications: (a) one leaflet was collected from each plant to be analyzed and divided into three parts, (b) each leaflet part was considered to be one biological replicate, and (c) the initial and final dehydration time points were sampled based on the water loss of the detached leaves. Thirty and 90 min cell dehydration treatments were done by subjecting the detached leaves to a temperature of 30 °C and 60% humidity in a BOD incubator. Leaflets of the non-transformed, conventional soybean cultivar BR 16 were used as negative controls.
Leaf and root dehydration under hydroponic conditions
To simulate a water deficit in the roots of P2193 plants, a hydroponic experiment was done according to the protocol of Martins et al. (2008). At the V3 vegetative growth stage, dehydration treatments lasting zero (control - without stress), 30, 60 and 90 min under stress conditions were applied in triplicate, with each plant being considered as a biological replicate. Triplicate measurements of the photosynthetic rate, stomatal conductance, transpiration and leaf-air temperature difference were obtained during the treatments using an LI-6400 portable photosynthesis system. The data were analyzed statistically using Duncan’s test at 5% significance (p < 0.05) (SAS software).
Extraction of total RNA and cDNA synthesis
GM plants P1397 and P2193 and the conventional cultivar BR 16 (non-transgenic control plant) were subjected to dehydration experiments after which total RNA was extracted using TRIzol® reagent (Invitrogen, Grand Island, NY, USA) according to the manufacturer’s instructions. For cDNA synthesis, 5 μg of total RNA treated with DNase was transcribed using M-MLV reverse transcriptase according to the manufacturer’s recommendations. The cDNA was quantified spectrophotometrically (NanoDrop 1000, Thermo Scientific, Wilmington, USA) and its intactness was analyzed electrophoretically in a 1% agarose gel with sodium borate (SB) buffer.
Quantification of the relative gene expression by qPCR
The gene expression levels in transgenic plants P1397 and P2193 and the negative control BR 16 were assessed by quantitative PCR (qPCR). GmrRNA18S (accession no. X02623.1) and Gmβ_actin (Stolf-Moreira et al., 2011) were used as reference genes for normalization. The primers and probes used were: (a) F 5-GGTTGACAGACTG AGAGCTCTTTC-3 and R 5-CAAATCGCTCCAACTAAGAAC-3 and probe 5-VICCACCACCCATAGAAT CAA-3 for GmrRNA18S, (b) F 5-TTGCCTGATGGACA GGTCATT-3 and R 5-CAATCAGAGAAGGCTGGA AC AG-3, and probe 5-AGGTTCTGAGAGATTCCGTTGC CCAGA-3 for Gmβ_actin, (c) F 5-CCGAGAGTCAACA AAGTGGTTTT-3 and R 5-GGGAAGCTTGCCATAGA TGCAATTCAATCAAACT-3, and probe 5-FAMCCGCT ACAAAGCCTCAACTACGGAATACCT-3 for Primer AtDREB2A, and (d) F 5-CAGGCCCGAGAGTCAACA AA-3 and R 5-GAGGTATTCCGTAGTTGAGGCTTT-3, and probe 5-FAMTAGCGGATCAAAACCAC-3 for Assay AtDREB2A. Gmβ_actin was not used in the cell dehydration experiment with the T0 generation.
The reactions were done in biological and technical triplicates using a TaqMan® PreAmp Master mix kit (Applied Biosystems) and a 7300 Real Time System thermocycler (Applied Biosystems) under the following cycling conditions: 50º C for 2 min, denaturation at 95º C for 10 min, followed by 40–45 cycles of 95 °C for 15 s and 60 °C for 1 min. The Relative Expression Software tool (RESTmsc software; Pfaffl et al., 2002) was used to analyze the gene expression data.
Results
Sequence alignment
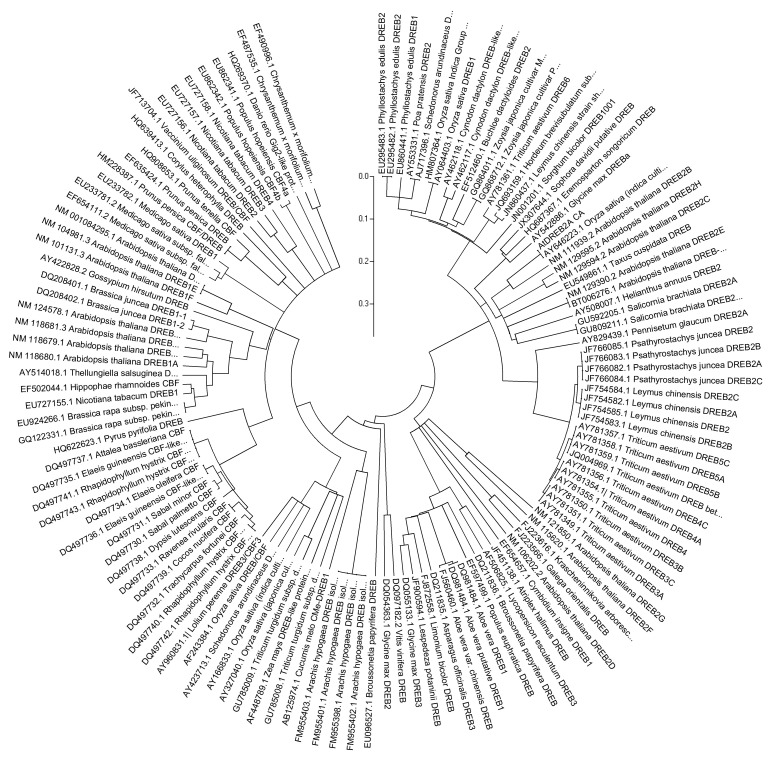
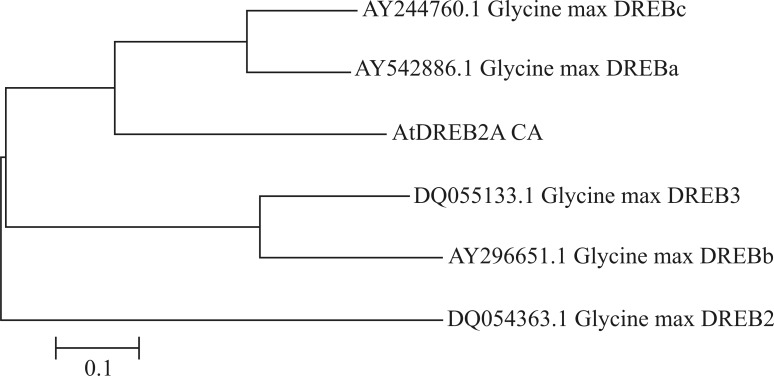
The analysis of various sequences of DREB-homologous genes yielded a single phylogenetic tree (Figure 2). Sequence alignment indicated conservation of the DREB genes in the species investigated, which suggested that the A. thaliana DREB2A CA gene inserted into soybean could be expressed to activate the endogenous genes responsive to water deficit stress. This conclusion was supported using the phylogenetic tree constructed with soybean DREB genes (Figure 3).
Figure 2.
Phylogenetic tree generated from the DREB-coding DNA sequence (CDS) from various plant species. The sequences were obtained from the NCBI website and were aligned and clustered using Clustal X and MEGA 4.0, respectively.
Figure 3.
Phylogenetic tree generated from CDS sequences of the soybean DREB gene. The sequences were obtained from the NCBI website and were aligned and clustered using Clustal X and MEGA 4.0, respectively.
Production of GM soybean plants
A total of 6,537 embryos from cultivar BR 16 were bombarded with the genetic construct of interest. Nine hundred and twenty seedlings from 4,185 multiple shoots were screened for the presence of AtDREB2A CA by conventional PCR using a specific primer set, and 78 of these were PCR-positive and produced seeds.
Number of transgene copies in GM soybean plants
The number of copies of the AtDREB2A CA transgene was assessed in the T0 generation of GM soybean plants and ranged from 2–17 (Table 1). No chimeras of P2193 and P1397 were detected, and the number of copies in T1 generation plants indicated segregation in the transition from T0 to T1: plants 1 and 2 of P1397 lost all copies and eight copies, respectively, and plants 1 and 2 of P2193 lost all copies and 48 transgene copies, respectively (Table 1).
Table 1.
Estimation of AtDREB2A copy number in generations T0 and T1 using qPCR and the 2−ΔCt/2 method. The first column displays the selected events, the second and third columns display the results from the independent experiments, the fourth column lists the average of the two experiments, and the fifth, sixth and ninth columns show the standard deviation (SD), coefficient of variation (CV%) and estimated copy number, respectively.
| Sample | First experiment | Second experiment | Average 2−ΔCt | SD 2−ΔCt | CV% 2−ΔCt | SD+ | SD- | Nº of copies |
|---|---|---|---|---|---|---|---|---|
| P 1397 | 15.95 | 10.35 | 13.15 | 3.96 | 30.15 | 17.11 | 9.18 | 9 to 17 |
| P 7186 | 8.39 | 5.73 | 7.06 | 1.88 | 26.60 | 8.94 | 5.18 | 5 to 9 |
| P 7413 | 4.95 | 3.98 | 4.47 | 0.69 | 15.45 | 5.16 | 3.78 | 4 to 5 |
| P 2193 | 966.38 | 225.89 | 596.14 | 523.61 | 87.83 | 1119.74 | 72.53 | High** |
| P 7195 | 9.36 | 6.74 | 8.05 | 1.85 | 23.00 | 9.91 | 6.20 | 6 to 10 |
| P 7417 | 4.05 | 2.92 | 3.49 | 0.80 | 22.95 | 4.29 | 2.69 | 3 to 4 |
| P 7212 | 6.84 | 5.09 | 5.97 | 1.24 | 20.75 | 7.21 | 4.73 | 5 to 7 |
| P 7418 | 7.9 | 5.66 | 6.78 | 1.58 | 23.44 | 8.37 | 5.19 | 5 to 8 |
| P 7231 | 6.23 | 4.25 | 5.24 | 1.40 | 26.72 | 6.64 | 3.84 | 4 to 7 |
| P 7430 | 4.33 | 3.2 | 3.77 | 0.80 | 21.13 | 4.56 | 2.97 | 3 to 5 |
| P 7256 | 9.44 | 4.88 | 7.16 | 3.22 | 45.12 | 10.39 | 3.93 | 4 to 10 |
| P 7374 | 3 12 | 2.32 | 2.72 | 0.57 | 20.86 | 3.29 | 2.15 | 2 to 4 |
| P 7431 | 14 | 8.56 | 11.28 | 3.84 | 34.07 | 15.12 | 7.44 | 7 to 15 |
| P 7174 | 6.17 | 3.88 | 5.03 | 1.62 | 32.26 | 6.65 | 3.41 | 3 to 7 |
| P 7393 | 4.61 | 3.15 | 3.88 | 1.03 | 26.62 | 4.91 | 2.85 | 3 to 5 |
| P 7531 | 9.23 | 5.91 | 7.57 | 2.34 | 30.94 | 9.91 | 5.23 | 5 to 10 |
| P 1397.1 (T1)* | 0.07 | 0.05 | 0.06 | 0.01 | 20.41 | 0.07 | 0.05 | Negative*** |
| P 1397.2 (T1) | 2.94 | 1.7 | 2.32 | 0.88 | 37.85 | 3.20 | 1.44 | 1 to 3 |
| P 2193.1 (T1) | 0.76 | 0.53 | 0.65 | 0.16 | 25.29 | 0.81 | 0.48 | Negative |
| P 2193.2 (T1) | 4.14 | 2.12 | 3.13 | 1.43 | 45.68 | 4.56 | 1.70 | 2 to 5 |
Samples identified with T1 descended from the T0 generation (i.e., generation T1). The copy number in generation T1 was divided by two because these plants were hemizygous. The number followed by a dot in T1 plants indicates the two descendants analyzed of each planta (P 1397 and P 2193), here denominated dot 1 and dot 2.
High fluorescence values caused signal saturation that led to imprecise results; in this case, the plants were considered to have > 50 copies.
Samples with values < 1 were considered to be negative.
Analysis in T0, T1, and T2 generations of P1397 and P2193
Leaf dehydration experiment
Gene expression in GM plants was compared to that in the conventional cultivar BR 16. P2193 showed high transcript levels of AtDREB2A CA in the T0 generation, with 725-, 535- and 146-fold greater expression after zero (control), 30 and 90 min of leaf dehydration in a BOD incubator, although only the 30 and 90 min treatments were significantly different from the levels in BR 16 (p < 0.004 and p < 0.003, respectively). In P1397, the highest levels of AtDREB2A CA transcripts were observed after 30 min of exposure to stress (89-fold, p < 0.11), but none of the values was statistically significant at any dehydration time point. When T1 generation GM plants were assessed, high levels of AtDREB2A CA transcripts were detected at time zero, which was 11-fold higher in P1397 (p < 0.072) and 111-fold greater in P2193 (p < 0.081). In P1397, the transgene expression levels were reduced, which suggests possible repression of gene expression at the 90 min treatment interval (0.001-fold, p < 0.022).
When the results for the T0 and T1 generations were compared, P2193 plants showed the greatest gene expression in both cases (Table 2). This finding most likely indicates greater stability of the transgene rd29A:AtDREB2A CA expression in P2193 compared to P1397, which only exhibited significant expression in control plants from T1. Transgene instability in P1397 was further demonstrated by the possible repression of gene expression after a 90 min exposure to stress (Table 2).
Table 2.
Relative expression of the AtDREB2A transgene in GM plants P1397 and P2193 in response to dehydration.
| Dehydration experiment | GM plant | Treatment (min) | Expression | Expression profile* | SE | p |
|---|---|---|---|---|---|---|
| T0 Generation | P1397 | 0 | 1.99 | - | ±2.3 | 0.559 |
| 30 | 89.2 | - | ±138.1 | 0.110 | ||
| 90 | 5.1 | - | ±9.8 | 0.241 | ||
| P 2193 | 0 | 725 | - | ±1744.8 | 0.144 | |
| 30 | 535.2 | Up | ±1308.7 | 0.004 | ||
| 90 | 146.1 | Up | ±203.0 | 0.003 | ||
|
| ||||||
| T1 Generation | P 1397 | 0 | 11.3 | Up | ±12.9 | 0.072 |
| 30 | 0.9 | - | ±1.1 | 0.965 | ||
| 90 | 0.001 | Down | ±0.002 | 0.022 | ||
| P 2193 | 0 | 111.1 | Up | ±247.1 | 0.081 | |
| 30 | 0.28 | - | ±0.31 | 0.545 | ||
| 90 | 8.3 | - | ±10.5 | 0.141 | ||
The standard error (SE) was calculated for median values, with p < 0.1.
The values for up-regulated (UP) profiles were significantly different from the control group when the expression profile during water stress diverged from the control at 10% probability (p < 0.1) upon randomization testing (n = 2,000 iterations). All statistical analyses were done using RESTmsc software.
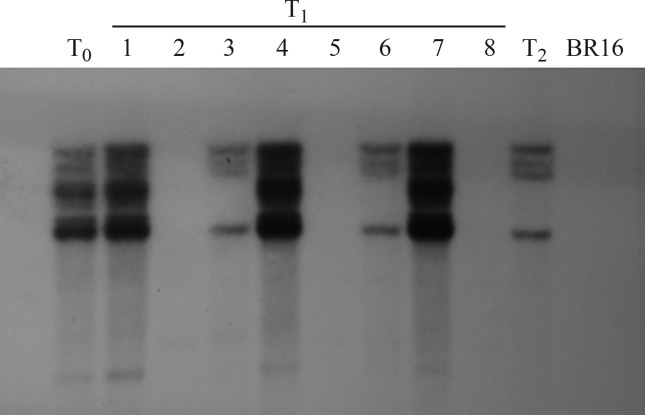
Segregation test and copy number analysis by Southern blotting in P2193
The results of the transgene segregation test in P2193 complied with the proportion expected for the T1 generation according to Mendel’s first law; however, the transgene segregation did not comply with Mendel’s first law in plants P2193.2, P2193.6 and P2193.7 from the T2 generation. Since most P2193 descendants segregated independently, Southern blot analysis was used to determine the number of inserts. In T0, the number of inserts detected for the AtDREB2A CA gene was lower than the copy number detected by qPCR. In T1, however, the results of both methods were similar and indicated three homozygous, two heterozygous and three negative plants. T2 generation plants had the same insert number as T2 plants of P2193.3 and P2193.6, indicating that the transgene behaves in a stable manner (Figure 4).
Figure 4.
Southern blot for generations T0, T1 and T2 of genetically modified P2193 containing the transcription factor AtDREB2A. Lanes 1–8 represent T1 generation plants. BR16 - control (normal) soybean cultivar.
Hydroponic experiment with the T2 generation of P2193
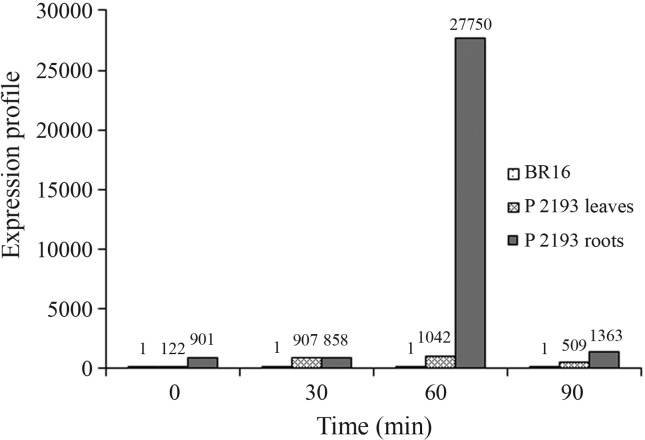
Compared to gene expression in the non-transformed, conventional cultivar BR 16, the results of the hydroponic experiment with the T2 generation of P2193 showed that the AtDREB2A CA transgene was overexpressed in leaf tissue by 122-, 907-, 1,042- and 509-fold (p < 0.001) at zero (control), 30, 60 and 90 min after treatment, respectively (Table 3). Higher levels of gene expression were detected in roots, with the transgene showing 901-, 858- and 1,363-fold higher expression at zero, 30 and 90 min after treatment (p < 0.001), respectively, and a peak increase of 27,750-fold after 60 min (Table 3, Figure 5). Thus, in leaves and roots, the highest levels of transgene expression were detected after 60 min, as indicated above.
Table 3.
Relative gene expression of the AtDREB2A transgene in the leaves and roots of T2 generation, GM plants P2193, grown under hydroponic conditions.
| Organ | Treatment (min) | Expression | Expression profile* | SE | p |
|---|---|---|---|---|---|
| Leaves | 0 | 121.7 | Up | ± 114.5 | 0.001 |
| 30 | 907.5 | Up | ± 146.8 | 0.001 | |
| 60 | 1042.0 | Up | ± 145.2 | 0.001 | |
| 90 | 508.8 | Up | ± 497.9 | 0.001 | |
|
| |||||
| Roots | 0 | 901.3 | Up | ± 111.4 | 0.001 |
| 30 | 857.8 | Up | ± 120.0 | 0.001 | |
| 60 | 27749.7 | Up | ± 137.8 | 0.001 | |
| 90 | 1363.2 | Up | ± 182.8 | 0.001 | |
The data were analyzed using RESTmsc software, which performs random comparisons of each treatment with its corresponding control. The standard error (SE) was calculated for median values, with p < 0.1.
The values for up-regulated (Up) profiles were significantly different from the control group when the expression profile during water stress diverged from the control at 10% probability (p < 0.1) upon randomization testing (n = 2,000 iterations).
Figure 5.
Relative gene expression of the AtDREB2A CA transgene in leaves and roots of the T2 generation of P2193 during stress induced by a water deficit. Transgene expression was normalized to the reference genes GmrRNA18S and Gmβ_actin and then calibrated with the corresponding time points and tissue from control plants (cultivar BR 16) (p < 0.001). Statistical analyses were done using RESTmsc software and all treatments of leaves and roots were significantly higher than the control.
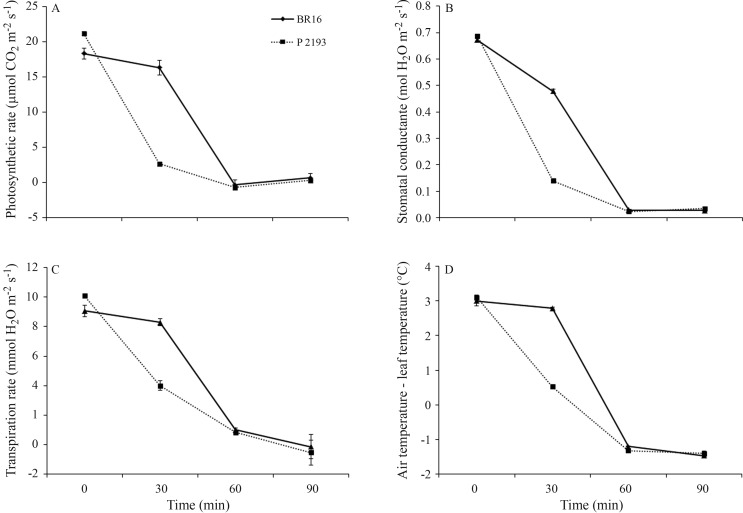
During water stress in the hydroponic system the photosynthetic rate (CER), stomatal conductance, transpiration, and the leaf-air temperature difference decreased in BR 16 and P2193 plants during all treatments. GM plants showed a decline in these parameters after 30 min of dehydration, whereas the greatest decrease in the non-transformed control BR 16 occurred after 60 min of dehydration (Figure 6). During dehydration, the CER and stomatal conductance differed significantly between cultivar BR 16 and GM P2193 at time zero and 30 min, with the value at zero min being higher for P2193 compared to BR 16. Intragroup analysis of BR 16 and P2193 showed that the values for both groups of plants differed from each other in the first 60 min, but were similar after 60–90 min of treatment (Duncan’s test, p ≤ 0.05) (Figure 6).
Figure 6.
Physiological responses in control plants (cultivar BR 16) and transgenic P2193 plants during stress for 30, 60 and 90 min. (A) Photosynthetic rate, (B) Stomatal conductance, (C) Transpiration rate and (D) Air-leaf temperature. The points are the mean ± SE of three determinations. The non-overlapping points at 30 min were significantly different from each other (p ≤ 0.05, Duncan’s test).
Discussion
The phylogenetic tree generated by aligning homologous DREB2A sequences from various plant species indicated minor differences in non-conserved regions (Sakuma et al., 2002) of DREB family genes and grouped species that were phylogenetically close to each other. DREB2A has highly conserved regions, including a DNA-binding domain known as AP2, which classifies A. thaliana DREB2A in the DREB subfamily that contains AP2 and a subgroup referred to as A-2 (Sakuma et al., 2002). As also observed by Li et al. (2005), AtDREB2A CA clustered with the GmDREBa and GmDREBc DREB genes in soybean, thereby corroborating the similarity among DREB genes (Figure 3).
As with TF AtDREB1A (Polizel at al., 2011), TF AtDREB2A CA from A. thaliana was successfully inserted into soybean by biolistics. Chimeric plants are a common occurrence in biolistic-based transformations but were not observed in GM plants P1397 or P2193. According to Rech et al. (2008), use of the herbicide imazapyr to select transformed plants reduces chimerization because of translocation events and decreases the concentration of herbicide in the apical meristematic region of the embryonic axes. Thus, imazapyr hindered the development of cells that did not receive the insert.
The number of transgene copies (2–17) of AtDREB2A CA inserted into the T0 generation of GM plants is characteristic of genetic transformations by biolistics and corroborated the independence of the events and the chance insertions into the receptor genome (Kohli et al., 2010). For P2193, the AtDREB2A CA transgene insertions occurred at a single locus since the data from generations T1 and T2 showed a pattern of segregation that followed Mendel’s first law. This conclusion agrees with the findings of Rech et al. (2008), who showed that transgenic plants transformed using biolistics follow the Mendelian model.
Southern blot data for P2193 indicated a large number of inserts in the T0 generation, a loss of transgene inserts in the following generation and subsequent stability of the transgene, as demonstrated by the finding that generations T1 and T2 had the same number of inserts. P2193 had a high number of copies in the T0 generation, whereas 0–5 copies were detected in samples from T1 (segregation of non-linked inserts). For P1397, 9–17 transgene copies were detected in T0 plants and 0–3 copies were observed in T1 plants.
Loss of the transgene of interest is a common phenomenon during gamete segregation and may become more evident when a construct that includes modifications/deletions is inserted (Kohli et al., 2010). Possible explanations of these losses include mechanisms such as intrachromosomal recombination, gene instability arising from manipulation during tissue culture, patterns of DNA methylation and co-elimination of transgenes activated by a plant genome defensive process (Romano et al., 2005). Nevertheless, as Kohli et al. (2006) observed, multiple transgene copies may result in greater transcript expression compared to events with only a single copy. This situation occurred in P2193, which had a high copy number and high levels of gene expression in T0, T1 and especially T2. This finding suggests that some transgene copies may have been inserted in a completely stable manner.
The levels of transgene expression in P2193 and P1397 during cell dehydration in a BOD incubator indicated that A. thaliana TF DREB2A was activated in soybean. In addition, the high transgene levels detected in T0 plants, particularly in P2193 after 30 and 90 min of treatment, may be indicative of a defensive response by soybean to the imposed environmental stress, as previously observed by Yamaguchi-Shinozaki and Shinozaki (2006).
Gene activation and expression triggered by the stress-inducible rd29 promoter was observed in A. thaliana (Quist et al., 2009), Brassica napus (Dalal et al., 2009), Arachis hypogaea L. (Bhatnagar-Mathur et al., 2007) and Glycine max L. Merril (Polizel et al., 2011). In the latter study, the investigators inserted and characterized the rd29A:AtDREB1A cassette in soybean and described the activation of the rd29 promoter during leaf water stress. The AtDREB1A transgene levels were approximately 2-, 7- and 3.4-fold after 20, 34 and 41 days of water deficit, respectively. Additionally, genes in the DREB1A-activated cascade, such as GmPI-PLC, GmSTP, GmGR-RBP and GmLEA14, exhibited high expression levels during severe stress (Polizel et al., 2011).
High transgene expression was also detected during dehydration in the hydroponic system. Transgene expression levels in leaves and roots peaked after 60 min of water stress, with increases of 1,042- and 27,750-fold, respectively, compared to the controls. This finding suggested differential AtDREB2A CA expression in organs of soybean plants. Similar findings have been reported in studies of roots containing DREB2A homologous genes and the constitutive 35S promoter in maize (ZmDREB2A) (Qin et al., 2007), A. thaliana (Liu et al., 1998) and Glycine max (GmDREBc) (Li et al., 2005). In soybean, transcription factors may have an indirect action in maintaining leaf and root cells during water stress by triggering the expression of genes associated with the protection of cell structures against dehydration. Zhong et al. (2008) demonstrated that transcription factors may be induced by specific conditions, including the duration of experimental stress.
The rate of photosynthesis, stomatal conductance, transpiration and leaf-air temperature difference in AtDREB2A CA transgenic plants differed from those of the control cultivar BR 16 for each treatment. These findings confirmed the occurrence of stress in the hydroponic system, with a tendency to reduce the rates of stomatal conductance, photosynthesis and transpiration (Jaleel et al., 2009).
The levels of gene expression and copy numbers of AtDREB2A CA in GM plants indicated that this transgene was recognized by the cell enzymatic machinery and resulted in its insertion and expression, most likely in euchromatic regions where transgene methylation and/or transgene silencing did not occur (Kohli et al., 2010). For P2193, the transgene was differentially expressed in three generations (T0, T1 and T2) and in leaves and roots during a water deficit. Future investigations should address the mechanisms involved in this response in P2193 more thoroughly, particularly with regard to the genes associated with the cascade regulated by AtDREB2A CA. In addition, the introduction of AtDREB2A CA into breeding programs aimed at developing soybean cultivars that are more tolerant to drought and heat should also be considered.
Acknowledgments
This work was supported by the Science and Technology Research Partnership for Sustainable Development (SATREPS) of the Japan Science and Technology Agency/Japan International Cooperation Agency. We thank the Plant Biotechnology and Physiology laboratories at Embrapa Soybean (Londrina, PR, Brazil) help in molecular and gas exchange analysis, respectively.
Footnotes
Associate Editor: Dario Grattapaglia
References
- Almogueva C, Prieto-Dapena P, Díaz-Martín J, Espinosa JM, Carranco R, Jordano R. The HaDREB2 transcription factor enhances basal thermotolerance and longevity of seeds through functional interaction with HaHSFA9. BMC Plant Biol. 2009;9:1–12. doi: 10.1186/1471-2229-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragão FJL, Sarokin L, Vianna GR, Rech EL. Selection of transgenic meristematic cells utilizing a herbicidal molecule results in the recovery of fertile transgenic soybean [Glycine max (L.) Merril] plants at a high frequency. Theor Appl Genet. 2000;101:1–6. [Google Scholar]
- Bhatnagar-Mathur P, Devi MJ, Reddy DS, Lavanya M, Vadez V, Serraj R, Yamaguchi-Shinozaki K, Sharma KK. Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep. 2007;26:2071–2082. doi: 10.1007/s00299-007-0406-8. [DOI] [PubMed] [Google Scholar]
- Dalal M, Tayal D, Chinnusamy V, Bansal KC. Abiotic stress and ABA-inducible Group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotechnol. 2009;17:137–145. doi: 10.1016/j.jbiotec.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encoded transcription activators that function in drought, high-salt and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Embrapa . Sistema Brasileiro de Classificação de Solos. 2nd edition. Embrapa; Rio de Janeiro: 2006. p. 306. [Google Scholar]
- Intergovernmental Panel on Climate Change - IPCC . Contribution of working Group I to the fourth assessment report of the intergovernmental panel on climate change. In: Solomon S, Qin D, Manning M, Marquis M, Averyt K, Tignor MMB, Miller HL Jr, Chen Z, editors. Climate Change 2007: The Physical Science Basis. Cambridge University Press; Cambridge: 2007. p. 996. [Google Scholar]
- Jaleel CA, Manivannan P, Wahid A, Farooq M, Al-Juburi HJ, Somasundaram R, Panneerselvam R. Drought stress in plants: A review on morphological characteristics and pigments composition. Int J Agric Biol. 2009;11:100–105. [Google Scholar]
- Kariola T, Brader G, Helenius E, Li J, Heino P, Palva ET. Early responsive to dehydration 15, a negative regulator of abscisic acid responses in Arabidopsis. Plant Physiol. 2006;142:1559–1573. doi: 10.1104/pp.106.086223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A, Melendi PG, Abranches R, Capell T, Stoger E, Christou Paul. The quest to understand the basis and mechanisms that control expression of introduced transgenes in crop plants. Plant Signal Behav. 2006;1:185–195. doi: 10.4161/psb.1.4.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli A, Miro B, Twyman RM. Transgenic integration, expression and stability in plants: Strategies for improvements. In: Kole C, Michler CH, Abbott AG, Hall TC, editors. Transgenic Crop Plants: Principles and Development. Springer-Verlag; Berlin: 2010. pp. 201–237. [Google Scholar]
- Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, Chen SY. Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet. 2005;110:1355–1362. doi: 10.1007/s00122-004-1867-6. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought and low-temperature responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhu K, Yang Y, Wu J, Chen F, Yu D. Molecular cloning, expression profiling and trans-activation property studies of a DREB2-like gene from chrysanthemum (Dendranthema vestitum) J Plant Res. 2008;121:215–226. doi: 10.1007/s10265-007-0140-x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Martins PK, Jordão BQ, Yamanaka N, Farias JRB, Beneventi MA, Binneck E, Fuganti R, Stolf R, Nepomuceno AL. Differential gene expression and mitotic cell analysis of the drought tolerant soybean (Glycine max L. Merrill Fabales, Fabaceae) cultivar MG/BR46 (Conquista) under two water deficit induction systems. Genet Mol Biol. 2008;31:512–521. [Google Scholar]
- Meyer R, Candrian U, Lüthy J. Detection of pork in heated meat products by polymerase chain reaction. J AOAC Int. 1994;77:617–622. [PubMed] [Google Scholar]
- Mizoi J, Ohori T, Moriwaki T, Kidokoro S, Todaka D, Maruyama K, Kusakabe K, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. GmDREB2A;2, a canonical dehydration-responsive element binding protein2-type transcription factor in soybean, is posttranslationally regulated and mediates dehydration-responsive element-dependent gene expression. Plant Physiol. 2013;161:346–361. doi: 10.1104/pp.112.204875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oya T, Nepomuceno AL, Neumaier N, Farias JRB, Tobita S, Ito O. Drought tolerance characteristics of Brazilian soybean cultivars - Evaluation and characterization of drought tolerance of various of Brazilian soybean cultivars in the field. Plant Prod Sci. 2004;7:129–137. [Google Scholar]
- Pfaffl MM, Horgan GW, Dempfle L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:1–10. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizel AM, Medri ME, Nakashima K, Yamanaka N, Farias JRB, Oliveira MCN, Marin SRR, Abdelnoor RV, Marcelino-Guimarães FC, Fuganti R, et al. Molecular, anatomical and physiological properties of a genetically modified soybean line transformed with rd29A:AtDREB1A for the improvement of drought tolerance. Genet Mol Res. 2011;10:3641–3656. doi: 10.4238/2011.October.21.4. [DOI] [PubMed] [Google Scholar]
- Qin F, Kakimoto M, Sakuma Y, Maruyama K, Osakabe Y, Tran LSP, Shinozaki K, Yamaguchi-Shinozaki Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 2007;50:54–69. doi: 10.1111/j.1365-313X.2007.03034.x. [DOI] [PubMed] [Google Scholar]
- Quist TM, Sokolchik I, Shi H, Joly RJ, Bressan RA, Maggio A, Narsimhan M, Li X. HOS3, an ELO-Like gene, inhibits effects of ABA and implicates an S-1-P/ceramide control system for abiotic stress responses in Arabidopsis thaliana. Mol Plant. 2009;2:138–151. doi: 10.1093/mp/ssn085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech EL, Vianna GR, Aragão FJL. High-efficiency transformation by biolistics of soybean, common bean and cotton transgenic plants. Nat Protoc. 2008;3:410–418. doi: 10.1038/nprot.2008.9. [DOI] [PubMed] [Google Scholar]
- Romano A, Van Der Plas LHW, Witholt B, Eggink G, Mooibroek H. Expression of poly-3-(R)-hydroxyalkanoate (PHA) polymerase and acyl-CoA-transacylase in plastids of transgenic potato leads to the synthesis of a hydrophobic polymer, presumably medium-chain-length PHAs. Planta. 2005;220:455–464. doi: 10.1007/s00425-004-1349-8. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M, Shinozaki K, Yamaguchi Shinozaki K. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006a;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA. 2006b;103:18822–18827. doi: 10.1073/pnas.0605639103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Southern EM. An improved method for transferring nucleotides from electrophoresis strips to thin layers of ion-exchange cellulose. Anal Biochem. 1974;34:317–318. doi: 10.1016/0003-2697(74)90395-9. [DOI] [PubMed] [Google Scholar]
- Stolf-Moreira R, Lemos EGM, Abdelnoor RV, Beneventi MA, Rolla AAP, Pereira SS, Oliveira MCN, Nepomuceno AL, Marcelino-Guimarães FC. Identification of reference genes for expression analysis by real-time quantitative PCR in drought-stressed soybean. Pesqui Agropecu Bras. 2011;46:58–65. [Google Scholar]
- Terashima A, Takumi S. Allopolyploidization reduces alternative splicing efficiency for transcripts of the wheat DREB2 homolog, WDREB2. Genome. 2009;52:100–105. doi: 10.1139/G08-101. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell. 1994;6:251–264. doi: 10.1105/tpc.6.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Zhou J, McCarthy RL, Ye ZH. A battery of transcription factors involved in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell. 2008;20:2763–2782. doi: 10.1105/tpc.108.061325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- Associação Brasileira das Indústrias de Óleos Vegetais (ABIO-VE) 2013 May 20; http://www.abiove.com.br.
- Companhia Nacional de Abastecimento (CONAB) 2012 May 8; http://www.conab.gov.br.
- Empresa Brasileira de Pesquisa Agropecuária (EMBRAPA) 2012 May 10; http://www.cnpso.embrapa.br.
- United States Department of Agriculture (USDA) 2011 Jul 6; http://www.usda.gov/wps/portal/usda/usdahome.