Abstract
The 18S rDNA phylogeny of Class Armophorea, a group of anaerobic ciliates, is proposed based on an analysis of 44 sequences (out of 195) retrieved from the NCBI/GenBank database. Emphasis was placed on the use of two nucleotide alignment criteria that involved variation in the gap-opening and gap-extension parameters and the use of rRNA secondary structure to orientate multiple-alignment. A sensitivity analysis of 76 data sets was run to assess the effect of variations in indel parameters on tree topologies. Bayesian inference, maximum likelihood and maximum parsimony phylogenetic analyses were used to explore how different analytic frameworks influenced the resulting hypotheses. A sensitivity analysis revealed that the relationships among higher taxa of the Intramacronucleata were dependent upon how indels were determined during multiple-alignment of nucleotides. The phylogenetic analyses rejected the monophyly of the Armophorea most of the time and consistently indicated that the Metopidae and Nyctotheridae were related to the Litostomatea. There was no consensus on the placement of the Caenomorphidae, which could be a sister group of the Metopidae + Nyctorheridae, or could have diverged at the base of the Spirotrichea branch or the Intramacronucleata tree.
Keywords: Caenomorpha, Metopus, Nyctotherus, sensitivity analysis
Introduction
The Class Armophorea Lynn, 2004 is represented by ciliates that live in anoxic environments, have mitochondria that transformed into hydrogenosomes during the course of evolution, and harbor symbiotic methanogenic prokaryotes (Jankowski, 1964a,b; Fenchel and Finlay, 1991; Gijzen and Barugahare, 1992; van Hoek et al., 2000a,b; Lynn, 2008). Many armophoreans occur as free-living forms, such as Brachonella Jankowski, 1964, Caenomorpha Perty, 1852, and Metopus Claparède & Lachmann, 1858, while others, such as Nyctotherus Leidy, 1849 and Nyctotheroides Grassé, 1928, inhabit the digestive system of animals, particularly insects and amphibians (Lynn, 2008). In the past, the armophoreans were classified as heterotrichs based on their morphology (Corliss, 1979). However, phylogenetic analyses of the 18S-rDNA (Embley et al., 1995; Hirt et al., 1995; van Hoek et al., 1998; Shin et al., 2000; Affa’a et al., 2004; Gong et al., 2009; Miao et al., 2009a,b; Vd’acny et al., 2010) and of histone H4 and α-tubulin data (Israel et al., 2002; Katz et al., 2004) have all grouped these organisms outside the Heterotrichea Stein, 1859, and within the Intramacro-nucleata Lynn, 1996. Hence, Armophorea is now considered as a molecular class, sometimes referred to as a “riboclass”, for which morphological synapomorphies are unknown (Lynn, 2008).
The phylogenetic affinities of the armorphoreans to other intramacronucleates remain unclear, with recent classifications suggesting an uncertain placement near the Spirotrichea Bütschli, 1889 or the Litostomatea Small & Lynn, 1981 (Riley and Katz, 2001; Cavalier-Smith, 2004; Lynn, 2008; Katz and Kovner, 2010); this uncertainty reflects divergent competing phylogenetic hypotheses (Shin et al., 2000; Gong et al., 2009; Miao et al., 2009a,b; Li et al., 2010; Vd’acny et al., 2010; Zhang et al., 2010; Lynn and Wright, 2013). In addition, the monophyly of armophoreans is sometimes rejected when 18S data of Caenomorpha are considered (Miao et al., 2009a,b; Lynn and Wright, 2013). Statistical support for the branching pattern of armophoreans and the intervening taxa is also variable.
As indicated by various authors (Rannala et al., 1998; Zwickl and Hillis, 2002; Bergsten, 2005; Heath et al., 2008), the quality of taxon and character sampling is an important factor that interferes with phylogenetic hypotheses. This was shown by Vd’acny et al. (2010), who found that the stability of the Armophorea is to some extent dependent on the number of sequences from representatives of other taxa included in the alignment. However, the sequences of armophorean representatives have never been broadly sampled to adequately evaluate their phylogenetic stability. The sensitivity of data to nucleotide alignment parameters and character weighting (Wheeler, 1995; Morrison and Ellis, 1997; Hall, 2005; Kjer et al., 2007; Goloboff et al., 2008; Dessimoz and Gil, 2010) is related to differences in phylogenetic hypotheses. This has already been demonstrated for ciliates by Kivimaki et al. (2009), although their study did not include data on armophoreans. Inconsistencies among phylogenies may reflect the properties of the analytical frameworks used, i.e., different sets of premises, concepts and processes underlying the phylogenetic analyses (Hillis, 1987; Huelsenbeck and Kirkpatrick, 1996; Bruno and Halpern, 1999; Swofford et al., 2001).
In this study, we examined the phylogenetic relationships of armophoreans based on 18S-rDNA sequences available in the NCBI/GenBank database and used a broad sample that included various sequences from unidentified armophoreans. We also explored the usefulness of two nucleotide alignment criteria and three phylogenetic frameworks, in addition to undertaking a sensitivity analysis. The systematics of the Armophorea is discussed based on these results and data from the literature.
Material and Methods
Sequence acquisition
Since the monophyly of the Armophorea is questionable and the phylogenetic affinities are variable (Shin et al., 2000; Miao et al., 2009a,b; Vd’acny et al., 2010), we broadly sampled ciliophoran 18S sequences to include representatives of all recognized ciliate classes (Lynn, 2008). One hundred and ninety-five ciliate 18S rDNA sequences, including 44 armophorean sequences, were downloaded from the NCBI database (Table S1) and assembled for multiple alignment. To avoid confusion when discussing the systematics of armophoreans, only sequences that were identified at least to the level of family or order were sampled. The armophorean sequences used represented the Families Nyctotheridae Affa’a, 1987 (Order Clevelan-dellida Puytorac & Grain, 1976), Caenomorphidae Poche, 1913 and Metopidae Kahl, 1927 (Order Armophorida Jankowski, 1964). Many of the armorphorean 18S sequences available from the NCBI were partial so that the missing homologous regions were treated as absent.
Sequence alignments
There is generally little agreement on how to treat ‘ambiguously alignable’ regions. Some authors recommend elimination of the nucleotide positions of ambiguous alignments as a means of improving phylogenetic hypotheses (Olsen and Woese, 1993; Swofford et al., 1996; Talavera and Castresana, 2007), while others consider that such positions contain information that is potentially useful for phylogenetic reconstructions (Lutzoni et al., 2000; Aagesen, 2004; Redelings and Suchard, 2009). In this study, we opted to preserve this information and to explore different alignments (Wheeler, 1995; Doyle and Davis, 1998). To assess how different alignment criteria might influence phylogenetic hypotheses obtained from the ciliate 18S data, the sequences were multiple-aligned using the ClustalW algorithm and the 18S rRNA secondary structure. The resulting alignment files were inspected in BioEdit v7.0.5 (Hall, 1999) to code leading and trailing gaps as missing data. The overall and mean p-distances displayed in Tables 1 and 2 were calculated with the program MEGA 5 (Tamura et al., 2011), using pairwise deletion as a treatment for gaps and missing data.
Table 1.
Informational content of the secondary structure alignment (SSA) and Q-score optimized alignment (QOA).
| Content | SSA | QOA |
|---|---|---|
| Averaged Q-score | 49.7616 | 56.5533 |
| Overall mean p-distance | 0.171 | 0.187 |
| Mean p-distance within Caenomorphidae | 0.045 | 0.109a | 0.081 | 0.135a |
| Mean p-distance within Metopidae | 0.068 | 0.066 a | 0.079 | 0.074 a |
| Mean p-distance within Nyctotheridae | 0.050 | 0.053 |
| Total number of characters | 2,424 | 2,099 |
| Characters informative for parsimony (%) | 58.3 | 62.5 |
| Gaps (%) | 25.3 | 15.6 |
Table 2.
Mean p-distances between armophorean families.
| Families/alignment criteria | 1
|
2
|
|---|---|---|
| SSA | QOA | SSA | QOA | |
| Caenomorphidae | ||
| Metopidae a | 0.194 | 0.211 | |
| Nyctotheridae | 0.198 | 0.224 | 0.099 | 0.106 |
ClustalW alignment
The sequences were aligned with the program ClustalW 1.81 through the CIPRES Science Gateway (Miller et al., 2010). Gap-opening (GOP) and gap-extension (GEP) penalties for multiple-alignment were optimized based on the averaged Q-scores, calculated with the program TuneClustalX (Hall, 2004) and used as a benchmark for alignment accuracy (Hall, 2005). The range explored for the parameters included GOP values of 10, 20, 30 and 40, each combined with integer GEP values that varied evenly from 1 to GOP/2. For comparison, we also included the ClustalW default values, i.e., GOP = 10 and GEP = 0.2, to yield 76 combinations (Figure 1). The alignment of the highest averaged Q-score was inspected and refined by eye in BioEdit as a means of improving the averaged Q-score. This alignment is referred to as the Q-score optimized alignment (QOA).
Figure 1.
Tridimensional bar plot showing the averaged Q-score variation across the explored parameters. The short arrow indicates the global optimum value that was further improved by manually refining the alignment (see text); the long arrow indicates the averaged Q-score obtained for ClustalW default parameters. The scale on the right defines the variation in averaged Q-scores, with darker shading indicating higher values.
Secondary structure alignment
As an alternative approach, the sequences were aligned based on the eukaryotic 18S rRNA secondary structure using the SINA web aligner (Pruesse et al., 2007) with its default settings. The alignment was inspected in BioEdit to remove gap-only columns followed by further refinement by eye that took into account the structural similarity among the sequences. This alignment is referred to as the secondary structure alignment (SSA).
Phylogenetic analyses
Sensitivity analysis
To evaluate whether differences in the GOP-GEP choices in the ClustalW alignment affected the hypotheses for ciliate 18S phylogeny, the BioNJ algorithm (Gascuel, 1997) implemented in the program PAUP* 4b10 (Swofford, 2003) was used to build neighbor-joining (NJ) trees from p-distance matrices of each alignment. For this purpose, the alignments were not refined manually in order to prevent altering the decisions made by ClustalW for each GOP-GEP combination. The resulting trees were gathered with PAUP* and a strict consensus tree was built to show the insensitive branches, with emphasis on ciliate higher taxa (Figure 2).
Figure 2.
Strict consensus of 76 NJ trees, each obtained under a different combination of GOP and GEP.
Analyses of the QOA and SSA data sets
Both data sets were independently analyzed using Bayesian inference (BI), maximum likelihood (ML) and maximum parsimony (MP) frameworks. In all resulting trees, the root was placed a posteriori at the Intramacronucleata-Postciliodesmatophora split (Lynn, 2008). The BI and ML routines employed a GTR + I (= 0.19) + Γ (= 0.6) model, selected based on the Akaike information criterion (AIC) (Akaike, 1974; Bos and Posada, 2005) in MODELTEST 3.7 (Posada and Crandall, 1998). To test whether the optimal trees were significantly different from suboptimal ones, all of the trees were statistically compared based on the data sets and optimality criteria by which they had been generated, with emphasis on the monophyly/non-monophyly of the Armophorea; this comparison was done using the approximately unbiased (AU) test for maximum likelihood and the Templeton test for maximum parsimony (Templeton, 1983; Shimodaira, 2002). The tests were done using the package CONSEL v0.1i (Shimodaira and Hasegawa, 2001; Shimodaira, 2004) and PAUP* 4b10, respectively. The taxonomy of higher taxa displayed in the phylogenetic trees (Figures 2–6) is mostly according to Lynn (2008), although the taxonomy of the Ventrata and Lamellicorticata agrees with Vd’acny et al. (2010).
Figure 6.
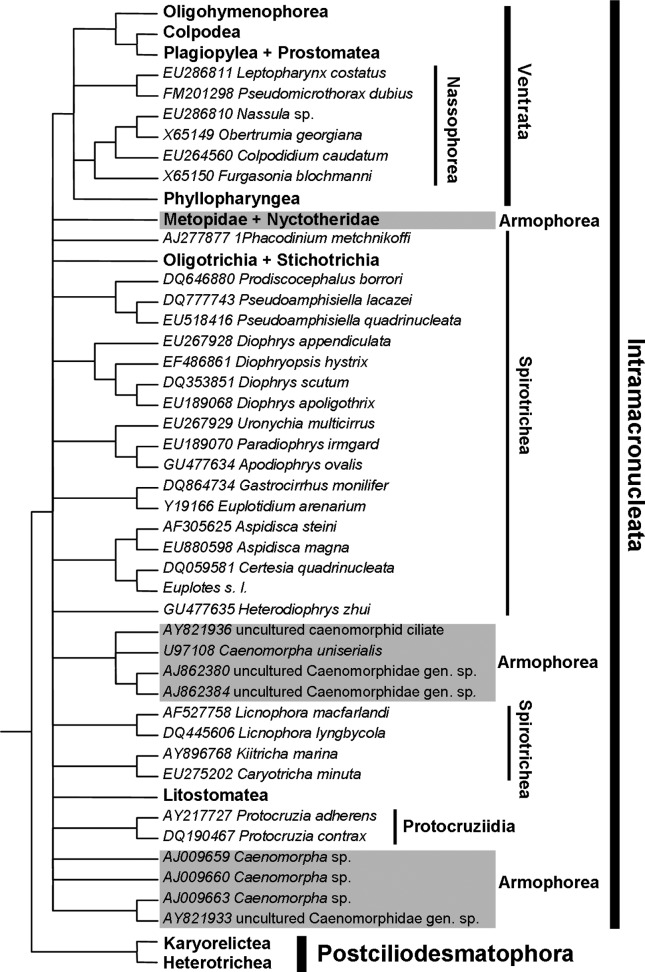
Strict consensus of 10 MP cladograms obtained from the secondary structure alignment (CI = 0.193–0.195; RI = 0.622–0.686). Arrows indicate sequences that supposedly belong to metopids; values associated with nodes are symmetric resampling percentages (values < 50% are omitted); values in balloons represent the number of synapomorphies common to all trees and the total possible synapomorphies of a given node, respectively.
Bayesian inference was determined with MrBayes 3.1.2 (Ronquist and Huelsenbeck, 2003) and was based on two independent Markov chain Monte Carlo (MCMC) simulations that were run with four chains of 1,000,000 generations. The trees were sampled every 200 generations (temperature of heat chains = 0.2), and the first 100,000 generations were discarded as burn-in. A 50% majority rule consensus of the trees remaining after burn-in was used to calculate the Bayesian posterior probabilities of the recovered kinships; these probabilities were used as node support measures for BI (Schneider, 2007).
For ML, the data sets were analyzed with the program PhyML 3.0 (Guindon and Gascuel, 2003), starting with a BioNJ tree for which the likelihood was improved via SPR branch-swapping to generate the ML tree. Node stability was evaluated via the SH-like aLRT branch support (Anisimova and Gascuel, 2006; Guindon et al., 2010). The MP analyses were done with the program TNT 1.1 (Goloboff et al., 2003, 2008), using a strategy that combined routines of parsimony-ratchet (Nixon, 1999) with tree-drifting, tree-fusing and sectorial searches (Goloboff, 1999) in order to find optimal cladograms. Gaps were coded as a “fifth base” to accommodate their phylogenetic information (Giribet and Wheeler, 1999; Schneider, 2007), and only parsimony-informative characters were analyzed. We agree with Goloboff (1993) and Platnick et al. (1991, 1996) concerning the incompleteness of equal-weighted cladistics and thus applied the implied-weighting approach of Goloboff (1993) to the data. For this, Goloboff’s parameter K was explored at integer intervals varying evenly from 1 to 10. The resulting trees were summarized via strict consensus. The common synapomorphies of the nodes of interest were assessed by optimizing the characters of each data set onto the trees using TNT. Node support was measured via 1,000 symmetric resampling (Goloboff et al., 2003, 2008) replicates, assuming K = 10.
Results and Discussion
Sequences and alignments
The global optimum for the averaged Q-scores distribution was found at GOP = 30, GEP = 14 (Figure 1), and the corresponding alignment was selected as optimal among the GOP-GEP combinations that were explored. However, regions with higher averaged Q-scores may exist for wider GOP-GEP intervals. Manual refinement of the optimal alignment improved the averaged Q-score by ∼ 0.75% (56.5533). The ClustalW default parameters produced a suboptimal result (55.5774) within the GOP-GEP space explored (Figure 1).
The alignment based on the corresponding 18S rRNA secondary structure generated a data set for which the averaged Q-score was 49.7616. Although this value appeared to be considerably suboptimal, it should not be interpreted as strictly in SSA since the Q-score is a measure of alignment quality that reflects the minimization of changes among nucleotides, whereas alignments based on secondary structure tend to minimize the changes among RNA structures (Thompson et al., 1994, Hall, 2005; Kjer et al., 2007).
The G + C content of the ciliate 18S data was 44.5 mol%. QOA yielded 2,099 characters, of which 1,312 (62.5%) were parsimony-informative, 408 were constant and 379 variable, but parsimony-uninformative. On the other hand, SSA yielded 2,424 characters, of which 1,412 (i.e., 58.3%) were parsimony-informative, 455 were constant, and 557 variable, but parsimony-uninformative. Thus, although QOA provided fewer characters it contained slightly more information for parsimony analysis than SSA. The quantitative difference in the number of characters between the two data sets is explained by SSA having more gap entries than QOA (25.6% vs. 15.3%).
The overall mean p-distance of the 18S data was 0.187 for QOA and 0.171 for SSA; the within- and among-group distances also varied according to the alignment criteria. For the armophorean families examined here, the lowest within-group mean distances were found within the Caenomorphidae in SSA whereas this same family had the largest mean distance in QOA (Table 1). Inter-group distance comparisons of the 18S sequences indicated that the Metopidae and Nyctotheridae were genetically closer to each other than to the Caenomorphidae (Table 2).
Sensitivity to GOP-GEP variation
An analysis of sensitivity to GOP-GEP variation (Figure 2) indicated that most higher taxa relationships in the Intramacronucleata (especially those of spirotrich clusters) depended upon how indels were estimated during multiple alignments and the influence of such estimates on the calculation of distance matrices in NJ methods. This situation is aggravated by differences in the placement of ‘difficult’ positions for each combination of parameters. These findings not only corroborate previous observations based on cladistic analyses of implied-aligned matrices of ciliate 18S rRNA (Kivimaki et al., 2009), but also emphasize the need for thorough indel parameter exploration during automated nucleotide alignments (Morrison and Ellis, 1997; Carroll et al., 2006; Smythe et al., 2006; Kjer et al., 2007).
Bayesian inference, Maximum likelihood and Maximum parsimony results
Although a considerable number of sequences from all major ciliate groups was analyzed in this work, the following discussion focuses on the kinships of the Armophorea. BI and ML yielded two topologies (one for each alignment), with the log likelihood of the ML tree obtained with SSA being slightly higher than that for QOA (Figures 3 and 4). In MP analyses, the strict consensus of cladograms resulting from SSA provided more resolution than that from QOA. This finding suggested that the former was slightly more robust to variation in Goloboff’s K parameter than the latter as it dealt with the relationships among higher taxa and affinities within Metopidae and Nyctotheridae (Figures 5 and 6). The matrices resulting from both QOA and SSA showed considerable character incongruence, as indicated by the relatively low ensemble consistency index of their resulting cladograms. On the other hand, the ensemble retention index was relatively high, indicating that most nucleotide primary homologies contributed to synapomorphy. The cladograms from QOA were rather more consistent and showed slightly more homology than those from SSA. Clades representing the main divergences of armophorean lineages hypothesized from the QOA matrix were united by more synapomorphies than those from SSA, except for the Caenomorphidae (Figures 5 and 6).
Figure 3.
BI/ML tree hypothesized from the Q-score optimized alignment (-lnL = 71422.61314). Arrows indicate sequences that supposedly belong to metopids; values associated with nodes are Bayesian posterior probabilities/aLRT support; * = full support; - = support < 50%. Scale bar = 2 substitutions per 10 nucleotide positions.
Figure 4.
BI/ML tree hypothesized from the secondary structure alignment (-lnL = 68901.83179). Arrows indicate sequences that supposedly belong to metopids; values associated with nodes are Bayesian posterior probabilities/aLRT support; * = full support; - = support < 50%. Scale bar = 2 substitutions per 10 nucleotide positions.
Figure 5.
Strict consensus of 10 MP cladograms obtained from the Q-score optimized alignment (CI = 0.194–0.196; RI = 0.728–0.732). Arrows indicate sequences that supposedly belong to metopids; values associated with nodes are symmetric resampling percentages (values < 50% are omitted); values in balloons represent the number of synapomorphies common to all trees and the total possible synapomorphies of a given node, respectively.
In all analyses (BI, ML and MP), the Litostomatea was adelphotaxon of some armophoreans, with high support (> 80%) most of the times. However, a completely monophyletic but relatively weakly supported Armophorea was only hypothesized by the BI and ML trees from QOA (Figure 3), which were significantly different (AU test; p < 0.05) from those in which the armophoreans were not monophyletic. For all other trees, the Armophorea were polyphyletic, and Litostomatea was sister to Metopidae + Nyctotheridae. In these trees, the Caenomorphidae branched outside the Lamellicorticata and diverged at the base of Intramacronucleata or near Licnophora spp. and the remaining spirotrich branch (Table 3). These topologies also differed significantly from those in which the armophoreans were monophyletic (AU test, Templeton test; p < 0.05).
Table 3.
Phylogenetic position of the Caenomorphidae branch according to different alignment criteria and analytic frameworks.
| Alignment criterion | Analytic framework | Position of the Caenomorphidae branch | Monophyletic Armophorea? | Number of trees |
|---|---|---|---|---|
| QOA | BI/ML | Adelphotaxon of Metopidae + Nyctotheridae | Yes | 2 |
| SSA | BI/ML | In a trichotomy among Licnophora spp. and the remaining Spirotrichea | No | 2 |
| QOA | MP (K = 1–5) | Diverged at the base of the Intramacronucleata | No | 5 |
| QOA | MP (K = 6–10) | Diverged at the base of a clade formed by Spirotrichea (Ventrata) | No | 5 |
| SSA | MP (K = 1–10) | Diverged at the base of the Intramacronucleata | No | 10 |
BI - Bayesian inference, ML - Maximum likelihood, MP - Maximum parsimony, QOA - Q-score optimized alignment; SSA - secondary structure alignment.
The Caenomorphidae were distributed in a fully supported symmetric group, dichotomized in branches containing four terminals, the position of which varied slightly depending on the alignment criteria and analytic framework. Three Caenomorphid terminals that were classified to family level in NCBI/GenBank, namely AJ009658, AJ009661 and AJ009662, unambiguously branched within the Metopidae (Figures 3–6). Moving these sequences into the main Caenomorphidae branch consistently augmented the mean p-distance of this group but had little effect on that of the Metopidae (Table 1). We therefore suppose that such sequences might belong to actual metopids. They were originally mentioned in a paper by van Hoek et al. (1999) as Caenomorpha-“like” species, so their identity is unknown.
The Metopidae comprised a pectinate line of branches, paraphyletic in relation to the monophyletic Nyctotheridae, and showed the least stable phylogenetic pattern among armophoreans in MP analyses; the latter was seen as polytomies in the consensus trees (Figures 5 and 6), whereas the BI-ML trees of both alignments yielded little inconsistency (Figures 3 and 4). Remarkably, Metopus contortus (Quennerstedt, 1867) Kahl, 1932, always diverged at the base of the Metopidae + Nyctotheridae, and Metopus palaeformis Kahl, 1927, was consistently monophyletic, even though the genus Metopus was not. This situation not only reflects the position of these species in relation to nyctotherids, but also the finding that the branch containing Brachonella arose from within Metopus. The Nyctotheridae were hypothesized to be monophyletic in all analyses, with the affinities of Nyctotherus ovalis Leidy, 1950, sequences varying according to the alignment criteria and phylogenetic framework. The monophyly of Nyctotherus depended on the position of its type species N. velox, which was unstable, although Nyctotheroides was always monophyletic.
Systematics of the Armophorea
Our results rejected the classification of Armophorea within the heterotrichs (Corliss, 1979) and corroborated previous studies based on 18S data (Embley et al., 1995; Hirt et al., 1995; van Hoek et al., 1998; Gong et al., 2009; Miao et al., 2009a,b; Vd’acny et al., 2010) and other molecular markers (Israel et al., 2002; Katz et al., 2004; Lynn, 2008) that indicated their status as a separate class.
Regarding their internal kinships, the monophyly of Armophorea was rejected in all but two analyses (Figures 4–6; Table 3) in which it was weakly supported by the data (Figure 3). Miao et al. (2009b) also found armophoreans to be not monophyletic, but in a different scenario than that hypothesized here. Thus, these authors found Caenomorpha uniserialis to branch off the base of the Litostomatea, while the Metopidae + Nyctotheroidae were a sister group of the protohypotrichs. This contrasts with the study by Shin et al. (2000), in which C. uniserialis branched off the base of a Metopus + Nyctotherus group with moderate to high support (74–98) in distance-based, ML and MP trees.
In his recent classification of the Ciliophora, Lynn (2008) considered the Armophorea to contain two orders (Armophorida and Clevelandellida). The former included the Families Metopidae and Caenomorphidae, while the latter included the Family Nyctotheridae plus five other families that unfortunately were not represented in our study (see Lynn, 2008). The affinity of the Metopidae to the Nyctorheridae contradicts the Order Armophorida proposed by Lynn (2008), who considered metopids to be closely related to caenomorphids. On the other hand, our results seem to fit the system of Jankowski (2007) better, with the caenomorphids, metopids and nyctotherids placed in three separate orders, viz. Armophorida, Metopida Jankowski, 1980, and Clevelandellida, respectively. The sequence of Epalxella antiquorum (Penard, 1922) Corliss, 1960, representative of the Order Odontostomatida Sawaya, 1940, a group traditionally associated with armophoreans (Jankowski, 1964a,b, 2007), clustered consistently with Class Plagiopylea Small & Lynn, 1985 (Ventrata) in all analyses (not shown). These findings corroborate a previous study by Stoeck et al. (2007) who found E. antiquorum to be related to trimyemids and plagiopylids. Lynn (2008) thus tentatively transferred the odontostomatids from the Armophorea to the Plagiopylea, but considered that phylogenetic analyses of further representatives and of other markers were necessary in order to decide on their affinity. The placement of Brachonella within the pectinate assemblage of Metopus terminals casts some doubt on the validity of the former, as it involves their morphological separation (see Esteban et al. (1995) and Foissner and Agatha (1999)).
Although the non-monophyly of armophoreans has been reported in the literature (e.g. Miao et al., 2009a,b), it has never been discussed in detail. The unambiguous proximity of the Metopidae and Nyctotheridae is frequent (e.g., Riley and Katz, 2001; Affa’a et al., 2004; Gong et al., 2009), and their position as an adelphotaxon of Litostomatea agrees with the recent study by Vd’acny et al. (2010), who proposed the name Lamellicorticata for the taxon formed by Armophorea and Litostomatea. Accordingly, one putative morphological synapomorphy of this group is the plate-like organization of the postciliary microtubules that form a layer right and between the ciliary rows (Foissner and Agatha, 1999; Lynn, 2008; Vd’acny et al., 2010).
In a detailed fine structure study of Caenomorpha medusula Perty, 1852 (Figures 7 and 8), Santa-Rosa (1975) found that postciliary microtubules were not developed in the somatic kinetids, thus precluding the organization mentioned above (Figures 7C, D). Consequently, if the Armophorea are to be considered monophyletic, then a loss of the plate-like arrangement of postciliary microtubules is assumed to have occurred after the Caenomorphidae lineage diverged at the base of the armophorean cluster (Figure 3). On the other hand, assuming that caenomorphids are distantly related to armophoreans and that no further traditional armophoreans are found to lack the plate-like arrangement of postciliary microtubules, the presence of such features can be assumed to be a feasibly consistent synapomorphy of the Lamellicorticata ex Caenomorphidae.
Figure 7.
Micrographs of Caenomorpha medusula, from Santa-Rosa (1975). a–b. Protargol impregnated specimens; c–d. Transmission electron microscopy sections. a. Specimen in lateral view showing meridian (bell) kineties (BK); b. Specimen in aboral view showing adoral membranelles (AM) and perizonal stripe kineties (PZ). c. Kinetid organization of a bell kinety. Arrows indicate cathetodesmal fibers departing in opposite directions; d. Kinetid organization in the perizonal stripe. Arrows indicate cathetodesmal fibers. Magnifications: a. 650x; b. 570x; c–d. 30,000x.
Figure 8.
Transmission electron micrographs of Caenomorpha medusula, from Santa-Rosa (1975). a. Tangential section of two adoral membranelles, delimited by brackets at the anterior region and with ciliary rows of one membranelle numbered 1–4. Postciliary microtubules (arrow), transverse microtubules (arrowheads), interkinetosomal connective (double arrowhead; our interpretation), and desmoses (D) are shown; b. Diplostichomonad paroral (P) with peristomial ridge (arrowhead); c. Longitudinal section of the kinetosomes in the perizonal region showing transverse microtubules (arrows; our interpretation), a naked (barren) kinetosome (KSN) and a prokaryote symbiont (B). Magnifications: a. 30,000x; b. 12,000x; c. 36,000x.
The classification of metopids alongside with caenomorphids is generally based in the assumption of homology of the perizonal ciliary stripe by Small and Lynn (1985) and Puytorac (1994), as discussed by Foissner and Agatha (1999). Foissner and Agatha (1999) described and compared the morphogenetic process in Metopus hasei Sondheim, 1929 and M. inversus (Jankowski, 1964) Foissner and Agatha, 1999 to that described for C. medusula by Martin-Gonzalez et al. (1987) and concluded that they have different morphogenetic origin and function (Foissner and Agatha, 1999). Accordingly, the metopid perizonal stripe generates only the paroral for the opisthe, whereas the caenomorphid stripe generates the paroral plus the adoral membranelles and the opisthe’s juvenile perizonal stripe (Martin-Gonzalez et al., 1987; Foissner and Agatha, 1999). Additionally, the kinetome organization of metopids differs from that of caenomorphids (Santa-Rosa, 1975; Sola et al., 1990; Silva-Neto, 1993; Decamp and Warren, 1997; Foissner and Agatha, 1999). In M. hasei and M. inversus the somatic dikinetids have a barren anterior kinetosome, while in perizonal dikinetids both kinetosomes were ciliated (Foissner and Agatha, 1999). On the other hand, in C. medusula all of the kinetosomes in the meridian (bell) kineties are ciliated whereas the posterior kinetosome of perizonal dikinetids is barren (Santa-Rosa, 1975) (Figures 7D and 8C). The three lowermost perizonal dikinetids in M. hasei and M. inversus are not positioned equidistantly, compared to the equidistant placement in C. medusula.
While the foregoing features can be used to support hypotheses that the Caenomorphidae are not closely related to the Metopidae + Nyctotheridae, there is morphologic evidence to support the monophyly of Armophorea, although sometimes ambiguously. The most obvious morphological characteristic is the body torsion present in metopids and caenomorphids (Jankowski, 1964b; Corliss, 1979). Among the metopids, this torsion is conspicuous in the campanulate-shaped representatives of Brachonella Jankowski, 1964. Furthermore, Brachonella darwini (Kahl, 1927) Jankowski, 1964b, exhibits a thorn-like posterior projection resembling those of the Caenomorpha (Jankowski, 1964b). The presence of interkinetosomal connectives jointing adoral membranelles of C. medusula (see Figure 8A) characterizes them as heteromembranelles (Puytorac and Grain, 1976; Lynn, 2008) that also occur in clevelandellids (Lynn, 2008).
Such evidence might be considered support for a close relationship between caenomorphids and clevelan-dellids. However, the presence of heteromembranelles best fits the phylogenetic trees as two independent gains, viz. one in the Caenomorphidae branch and another in the Nyctotheridae (representing the clevelandellids). This also applies even to trees in which Armophorea is monophyletic, given the paraphyly of Metopidae (Figure 3). A diplostichomonad paroral, in which a ridge separates two rows of kinetosomes (Figure 8B) was found in C. medusula by Santa-Rosa (1975), thus matching this structure’s configuration in clevelandellids (Paulin, 1967; Puytorac and Grain, 1976; Takahashi and Imai, 1989; Grim, 1998), but also in the metopid Parametopidium circumlabens (Biggar & Wenrich, 1932) Aescht, 2001 (Silva-Neto, 1993). However, this possibly differs from the seemingly linearly arranged oral dikinetids in Metopus (Esteban et al., 1995; Foissner and Agatha, 1999; Lynn, 2008).
The BI-ML trees inferred from the secondary structure alignment and the MP cladograms (Figures 5 and 6; Table 3) also show the possibility of caenomorphids having either diverged at the base of the Intramacronucleata tree, as suggested by Lynn and Wright (2013) or to be related to Spirotrichea. The phylogeny of ciliates based on other markers exhibit different branching patterns (Lynn, 2008) in which metopids and nyctotherids are closely related to spirotrichs. Based on α-tubulin amino acids, Israel et al. (2002) hypothesized a neighbor-joining cluster formed by M. palaeformis + N. ovalis with the spirotrich Euplotes spp. distantly placed from the litostomatean cluster. Moreover, based on histone H4 data, a neighbor-joining tree was hypothesized by Katz et al. (2004) in which M. palaeformis and N. ovalis branched off a trichotomy formed by Spirotrichea and the remaining ciliate clusters (the rooting method was not specified), except for litostomateans, which were not included (Katz et al., 2004). In any case, α-tubulin and H4 phylogenies must be interpreted cautiously because of paralogy (Israel et al., 2002; Katz et al., 2004). These results corroborate the close affinity of metopids to nyctotherids shown by our analyses. However, further data, especially on caenomorphids, is still required to improve our understanding of armophorean kinships based on α-tubulin and H4 phylogenies.
Concluding Remarks
The present study has shown that different nucleotide alignment criteria and the use of different phylogenetic frameworks provided different hypotheses to explain the evolutionary affinities of armophoreans based on the 18S marker. This and the sensitivity of some basal branching patterns of the Intramacronucleata to GOP-GEP variation highlight the importance of explicitness in nucleotide alignment criteria. Whereas the 18S phylogeny of the Armophorea results in an apparently stable placement of metopids and nyctotherids near the litostomateans, the same cannot be said for caenomorphids. Moreover, assumptions regarding the evolution of morphological features based on 18S phylogeny are quite general and ambiguous and must not be over-interpreted since various aspects of the life cycle (which includes morphogenesis) and fine structure of most representatives of Armophorea (Foissner and Agatha, 1999; Lynn, 2008) remain unknown. Improvements in taxon sampling for phylogenetic analyses, the use of additional molecular markers, and advances in our knowledge of the life cycle and fine structure of armophoreans should shed some light on the natural history of these organisms.
Acknowledgments
The authors thank Prof. Dr. Milden Rodrigues de Santa-Rosa for kindly allowing the use of his micrographs of Caenomorpha medusula, and the anonymous reviewers for their comments and suggestions. This study was financed by a post-doctoral fellowship to TSP by CNPq (PDJ) and CAPES (PRODOC) via the project PROTAX (no. 52/2010).
Footnotes
Associate Editor: Guillermo Orti
Supplementary Material
The following supplementary material is available for this article:
Table S1 - The 18S-rDNA sequences used in this study.
This material is available as part of the online version of this article from http://www.scielo.br/gmb.
References
- Aagesen L. The information content of an ambiguously alignable region, a case study of the trnL intron from the Rhamnaceae. Org Divers Evol. 2004;4:35–49. [Google Scholar]
- Affa’a FM, Hickey DA, Struder-Kypke M, Lynn DH. Phylogenetic position of species in the genera Anoplophrya, Plagiotoma, and Nyctotheroides (Phylum Ciliophora), endosymbiotic ciliates of annelids and anurans. J Euk Microbiol. 2004;51:301–306. doi: 10.1111/j.1550-7408.2004.tb00570.x. [DOI] [PubMed] [Google Scholar]
- Akaike HA. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Bos DH, Posada D. Using models of nucleotide evolution to build phylogenetic trees. Dev Comp Immunol. 2005;29:211–227. doi: 10.1016/j.dci.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Bruno WJ, Halpern AL. Topological bias and inconsistency of maximum likelihood using wrong models. Mol Biol Evol. 1999;16:564–566. doi: 10.1093/oxfordjournals.molbev.a026137. [DOI] [PubMed] [Google Scholar]
- Carroll H, Ridge P, Clement M, Snell Q. Effects of gap open and gap extension penalties. In: Clement M, Snell Q, editors. Proceedings of the Third Biotechnology and Bioinformatics Symposium. Brigham Young University; Utah: 2006. pp. 19–23. [Google Scholar]
- Cavalier-Smith T. Chromalveolate diversity and cell megaevolution: Interplay of membranes, genomes and cytoskeleton. In: Hirt RP, Horner DS, editors. Organelles, Genomes and Eukaryote Phylogeny. CRC Press; Boca Raton: 2004. pp. 75–108. [Google Scholar]
- Corliss JO. The Ciliated Protozoa Characterization, Classification and Guide to the Literature. Pergamon Press; Oxford: 1979. p. 445. [Google Scholar]
- Decamp O, Warren A. Observations on the morphology of Caenomorpha uniserialis Levander, 1894 (Ciliophora, Heterotrichida) isolated from a wastewater treatment plant. Acta Protozool. 1997;36:105–110. [Google Scholar]
- Dessimoz C, Gil M. Phylogenetic assessment of alignments reveals neglected tree signal in gaps. Genome Biol. 2010;11:R37. doi: 10.1186/gb-2010-11-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Davis JI. Homology in molecular phylogenetics: A parsimony perspective. In: Solis DE, Soltis PS, Doyle JJ, editors. Molecular Systematics of Plants. II. DNA Sequencing. Kluwer Academic Publishers; Boston: 1998. pp. 101–131. [Google Scholar]
- Embley TM, Finlay BJ, Dyal PL, Hirt RP, Wilkinson M, Williams AG. Multiple origins of anaerobic ciliates with hydrogenosomes within the radiation of aerobic ciliates. Proc R Soc B: Biol Sci. 1995;262:87–93. doi: 10.1098/rspb.1995.0180. [DOI] [PubMed] [Google Scholar]
- Esteban G, Fenchel T, Finlay B. Diversity of free-living morphospecies in the ciliate genus Metopus. Arch Protistenkunde. 1995;146:137–164. [Google Scholar]
- Fenchel T, Finlay BJ. Synchronous division of an endosymbiotic methanogenic bacterium in the anaerobic ciliate Plagiopyla frontata Kahl. J Protozool. 1991;38:22–28. [Google Scholar]
- Foissner W, Agatha S. Morphology and morphogenesis of Metopus hasei Sondheim, 1929 and M. inversus (Jankowski, 1964) nov. comb. (Ciliophora, Metopida) J Euk Microbiol. 1999;46:174–193. [Google Scholar]
- Gascuel O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gijzen HJ, Barugahare M. Contribution of anaerobic protozoa and methanogens to hindgut metabolic activities of the American cockroach, Periplaneta americana. Appl Environ Microbiol. 1992;58:2565–2570. doi: 10.1128/aem.58.8.2565-2570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giribet G, Wheeler WC. On gaps. Mol Phylogenet Evol. 1999;13:132–143. doi: 10.1006/mpev.1999.0643. [DOI] [PubMed] [Google Scholar]
- Goloboff PA. Estimating character weights during tree search. Cladistics. 1993;9:83–91. doi: 10.1111/j.1096-0031.1993.tb00209.x. [DOI] [PubMed] [Google Scholar]
- Goloboff PA. Analyzing large data sets in reasonable times: Solutions for composite optima. Cladistics. 1999;15:415–428. doi: 10.1111/j.1096-0031.1999.tb00278.x. [DOI] [PubMed] [Google Scholar]
- Goloboff PA, Carpenter JM, Arias JS, Esquivel DRM. Weighting against homoplasy improves phylogenetic analysis of morphological data sets. Cladistics. 2008;24:1–6. [Google Scholar]
- Gong J, Stoeck T, Yi Z, Miao M, Zhang Q, Roberts DM, Warren A, Song W. Small subunit rRNA phylogenies show that the Class Nassophorea is not monophyletic (Phylum Ciliophora) J Euk Microbiol. 2009;56:339–347. doi: 10.1111/j.1550-7408.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- Grim JN. A comparison of three populations of the ciliate genus, Paracichlidotherus Grim, 1992. New fish hosts, and biogeography; Revised genus description. J Euk Microbiol. 1998;45:40–44. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hall BG. Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol Biol Evol. 2005;22:792–802. doi: 10.1093/molbev/msi066. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Series. 1999;41:95–98. [Google Scholar]
- Heath TA, Hedtke SM, Hillis DM. Taxon sampling and the accuracy of phylogenetic analyses. J Syst Evol. 2008;46:239–257. [Google Scholar]
- Hillis DM. Molecular vs. morphological approaches to systematics. Annu Rev Ecol Syst. 1987;18:23–42. [Google Scholar]
- Hirt RP, Dyal PL, Wilkinson M, Finlay BJ, Roberts DM, Embley TM. Phylogenetic relationships among karyorelictids and heterotrichs inferred from small subunit rRNA sequences: Resolution at the base of the ciliate tree. Mol Phylogenet Evol. 1995;4:77–87. doi: 10.1006/mpev.1995.1008. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Kirkpatrick M. Do phylogenetic methods produce trees with biased shapes? Evolution. 1996;50:1418–1424. doi: 10.1111/j.1558-5646.1996.tb03915.x. [DOI] [PubMed] [Google Scholar]
- Israel RL, Pond SLK, Muse SV, Katz LA. Evolution of duplicated alpha-tubulin genes in ciliates. Evolution. 2002;56:1110–1122. doi: 10.1111/j.0014-3820.2002.tb01425.x. [DOI] [PubMed] [Google Scholar]
- Jankowski AW. Morphology and evolution of Ciliophora. I. The new system of sapropelebiotic Heterotrichida. Zoologichesky Zhurnal. 1964a;43:503–517. [Google Scholar]
- Jankowski AW. Morphology and evolution of Ciliophora. III. Diagnoses and phylogenesis of 53 sapropelebionts, mainly of the Order Heterotrichida. Arch Protistenkunde. 1964b;107:185–294. [Google Scholar]
- Jankowski AW. Phylum Ciliophora Doflein, 1901. In: Krylow MV, Frolov AO, editors. Protista: Handbook on Zoology, Part 2. Nauka; St. Petersburg: 2007. pp. 415–976. [Google Scholar]
- Katz LA, Kovner AM. Alternative processing of scrambled genes generates protein diversity in the ciliate Chilodonella uncinata. J Exp Zool, Part B, Mol Develop Evol. 2010;314:480–488. doi: 10.1002/jez.b.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, Bornstein JG, Lasek-Nesselquist E, Muse SV. Dramatic diversity of ciliate histone H4 genes revealed by comparisons of patterns of substitutions and paralog divergences among eukaryotes. Mol Biol Evol. 2004;21:555–562. doi: 10.1093/molbev/msh048. [DOI] [PubMed] [Google Scholar]
- Kivimaki KL, Bowditch BM, Riordan GP, Lipscomb DL. Phylogeny and systematic position of Zosterodasys (Ciliophora, Synhymeniida): A combined analysis of ciliate relationships using morphological and molecular data. J Euk Microbiol. 2009;56:323–338. doi: 10.1111/j.1550-7408.2009.00403.x. [DOI] [PubMed] [Google Scholar]
- Kjer KM, Gillespie JJ, Ober KA. Opinions on multiple sequence alignment, and an empirical comparison of repeatability and accuracy between POY and structural alignment. Syst Biol. 2007;56:133–146. doi: 10.1080/10635150601156305. [DOI] [PubMed] [Google Scholar]
- Li LF, Stoeck T, Shin MK, Al-Rasheid KAS, Al-Khedhairy BA, Song W. Protocruzia, a highly ambiguous ciliate (Protozoa; Ciliophora): Very likely an ancestral form for Heterotrichea, Colpodea or Spirotrichea? With reevaluation of its evolutionary position based on multigene analyses. Science China (Life Sci) 2010;53:131–138. doi: 10.1007/s11427-010-0012-9. [DOI] [PubMed] [Google Scholar]
- Lutzoni F, Wagner P, Reeb V, Zoller S. Integrating ambiguously aligned regions of DNA sequences in phylogenetic analyses without violation positional homology. Syst Biol. 2000;49:628–651. doi: 10.1080/106351500750049743. [DOI] [PubMed] [Google Scholar]
- Lynn DH. The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature. 3rd edition. Springer; Dordrecht: 2008. p. 606. [Google Scholar]
- Lynn DH, Wright AD. Biodiversity and molecular phylogeny of Australian Clevelandella species (Class Armophorea, Order Clevelandellida, Family Clevelandellidae), intestinal endosymbiotic ciliates in the wood-feeding roach Panesthia cribrata Saussure, 1864. J Euk Microbiol. 2013;60:335–341. doi: 10.1111/jeu.12037. [DOI] [PubMed] [Google Scholar]
- Martin-Gonzalez A, Serrano S, Fernandez-Galiano D. Cortical morphogenesis and conjugation process in Caenomorpha medusula (Ciliophora, Heterotrichida) Eur J Protistol. 1987;23:111–121. doi: 10.1016/S0932-4739(88)80054-3. [DOI] [PubMed] [Google Scholar]
- Miao M, Shao C, Jiang J, Li L, Stoeck T, Song W. Caryotricha minuta (Xu et al., 2008) nov. comb., a unique marine ciliate (Protista, Ciliophora, Spirotrichea), with phylogenetic analysis of the ambiguous genus Caryotricha inferred from the small-subunit rRNA gene sequence. Int J Syst Evol Microbiol. 2009a;59:430–438. doi: 10.1099/ijs.0.65855-0. [DOI] [PubMed] [Google Scholar]
- Miao M, Song W, Clamp JC, Al-Rasheid KAS, Al-Khedhairy AA, Al-Arifi S. Further consideration of the phylogeny of some “traditional” heterotrichs (Protista, Ciliophora) of uncertain affinities, based on new sequences of the small subunit rRNA gene. J Euk Microbiol. 2009b;56:244–250. doi: 10.1111/j.1550-7408.2009.00391.x. [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop; New Orleans: IEEE; 2010. pp. 1–8. [Google Scholar]
- Morrison DA, Ellis JT. Effects of nucleotide sequence alignment on phylogeny estimation: A case study of 18S rDNAs of Apicomplexa. Mol Biol Evol. 1997;14:428–441. doi: 10.1093/oxfordjournals.molbev.a025779. [DOI] [PubMed] [Google Scholar]
- Nixon KC. The Parsimony Ratchet, a new method for rapid Parsimony analysis. Cladistics. 1999;15:407–414. doi: 10.1111/j.1096-0031.1999.tb00277.x. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR. Ribosomal RNA: A key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- Paulin JJ. The fine structure of Nyctotherus cordiformis (Ehrenberg) J Protozool. 1967;14:183–196. doi: 10.1111/j.1550-7408.1967.tb01981.x. [DOI] [PubMed] [Google Scholar]
- Platnick NI, Coddington JA, Forster RR, Griswold CE. Spinneret morphology and the phylogeny of haplogyne spiders (Araneae, Araneomorphae) Am Mus Novitat. 1991;3016:1–73. [Google Scholar]
- Platnick NI, Humphries CJ, Nelson GJ, Williams DM. Is Farris optimization perfect? Three-taxon statements and multiple branching. Cladistics. 1996;12:243–252. doi: 10.1111/j.1096-0031.1996.tb00011.x. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: Testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puytorac P de. Phylum Ciliophora Doflein, 1901. Traité Zoologie. 1994;2:1–15. [Google Scholar]
- Puytorac P de, Grain J. Ultrastructure du córtex buccal et evolution chez les cilies. Protistologica. 1976;12:49–67. [Google Scholar]
- Rannala B, Huelsenbeck JP, Yang Z, Nielsen R. Taxon sampling and the accuracy of large phylogenies. Syst Biol. 1998;47:702–710. doi: 10.1080/106351598260680. [DOI] [PubMed] [Google Scholar]
- Redelings BD, Suchard MA. Robust inferences from ambiguous alignments. In: Rosenberg M, editor. Sequence Alignment: Methods, Concepts, and Strategies. University of California Press; Berkeley: 2009. pp. 209–270. [Google Scholar]
- Riley JL, Katz LA. Widespread distribution of extensive genome fragmentation in ciliates. Mol Biol Evol. 2001;18:1372–1377. doi: 10.1093/oxfordjournals.molbev.a003921. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Santa-Rosa MR de. Université de Clermont-Ferrand; 1975. Contribution a l’Ultrastructure Comparee de Quelques Especes de Cilies Appartenant a Divers Orders. PhD Thesis. [Google Scholar]
- Schneider H. Métodos de Análise Filogenética - Um Guia Prático. 3rd edition. SBG and Holos; Ribeirão Preto: 2007. p. 200. [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. Approximately unbiased tests of regions using multistep-multiscale bootstrap resampling. Ann Stat. 2004;32:2616–2641. [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shin MK, Hwang UW, Kim W, Wright ADG, Krawczyk C, Lynn DH. Phylogenetic position of the ciliates Phacodinium (Order Phacodiniida) and Protocruzia (Subclass Protocruziidia) and systematic of the spirotrich ciliates examined by small subunit ribosomal RNA gene sequences. Eur J Protistol. 2000;36:293–302. [Google Scholar]
- Silva-Neto ID da. Universidade de São Paulo; 1993. Estrutura e ultraestrutura de cinco espé-cies de ciliados heterotríqueos e um estudo comparativo das estruturas infraciliares corticais e bucais da classe Heterotrichea Stein, 1859. PhD thesis. [Google Scholar]
- Small EB, Lynn DH. Phylum Ciliophora Doflein, 1901. In: Lee J, Hutner SH, Bovee EC, editors. An Illustrated Guide to the Protozoa. Allen Press; Kansas: 1985. pp. 393–575. [Google Scholar]
- Smythe AB, Sanderson MJ, Nadler SA. Nematode small subunit phylogeny correlates with alignment parameters. Syst Biol. 2006;55:972–992. doi: 10.1080/10635150601089001. [DOI] [PubMed] [Google Scholar]
- Sola A, Guinea A, Longás JE, Fernandez-Galiano D. Nouvelles données sur l’infraciliature somatique et buccale de Caenornorpha uniserialis Levander, 1894 (Ciliophora, Heterotrichida) Arch Protistenkunde. 1990;138:233–238. [Google Scholar]
- Stoeck T, Bruemmer F, Foissner W. Evidence for local ciliate endemism in an alpine anoxic lake. Microb Ecol. 2007;54:478–486. doi: 10.1007/s00248-007-9213-6. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (* and Other Methods) Version 4. Illinois Natural History Survey; Champaign: 2003. p. 179. [Google Scholar]
- Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Sinauer Associates; Sunderland: 1996. pp. 407–514. [Google Scholar]
- Swofford DL, Waddell PJ, Huelsenbeck JP, Foster PG, Lewis PO, Rogers JS. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst Biol. 2001;50:525–539. [PubMed] [Google Scholar]
- Takahashi EI, Imai S. Light and scanning electron microscopy of Nyctotherus kyphodes (Ciliophora, Plagiotomidae) from the Galapagos giant tortoise, Testudo sp. Bull Nippon Vet Zootechnical College. 1989;38:9–15. [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the humans and apes. Evolution. 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoek AHAM, van Alen TA, Sprakel VSI, Hackstein JHP, Vogels GD. Evolution of anaerobic ciliates from the gastrointestinal tract: Phylogenetic analysis of the ribosomal repeat from Nyctotherus ovalis and its relatives. Mol Biol Evol. 1998;15:1195–1206. doi: 10.1093/oxfordjournals.molbev.a026027. [DOI] [PubMed] [Google Scholar]
- van Hoek AHAM, Sprakel VSI, van Alen TA, Theuvenet APR, Vogels GD, Hackstein JHP. Voltage-dependent reversal of anodic galvanotaxis in Nyctotherus ovalis. J Euk Microbiol. 1999;46:427–433. doi: 10.1111/j.1550-7408.1999.tb04623.x. [DOI] [PubMed] [Google Scholar]
- van Hoek AHAM, Akhmanova AS, Huynen MA, Hackstein JHP. A mitochondrial ancestry of the hydrogenosomes of Nyctotherus ovalis. Mol Biol Evol. 2000a;17:202–206. doi: 10.1093/oxfordjournals.molbev.a026234. [DOI] [PubMed] [Google Scholar]
- van Hoek AHAM, van Alen TA, Sprakel VSI, Leunissen JAM, Brigge T, Vogels GD, Hackstein JHP. Multiple acquisition of methanogenic archaeal symbionts by anaerobic ciliates. Mol Biol Evol. 2000b;17:251–258. doi: 10.1093/oxfordjournals.molbev.a026304. [DOI] [PubMed] [Google Scholar]
- Vd’acny P, Orsi W, Foissner W. Molecular and morphological evidence for a sister group relationship of the Classes Armophorea and Litostomatea (Ciliophora, Intra-macronucleata, Lamellicorticata infraphyl. nov.), with an account on basal haptorid litostomateans. Eur J Protistol. 2010;46:298–309. doi: 10.1016/j.ejop.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Wheeler WC. Sequence alignment, parameter sensitivity, and the phylogenetic analysis of molecular data. Syst Biol. 1995;44:321–331. [Google Scholar]
- Zhang Q, Yi Z, Song W, Al-Rasheid KAS, Warren A. The systematic position of Paraspathidium Noland, 1937 (Ciliophora, Litostomatea?) inferred from primary SSU rRNA gene sequences and predicted secondary rRNA structure. Eur J Protistol. 2010;46:280–288. doi: 10.1016/j.ejop.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ, Hillis DM. Increased taxon sampling greatly reduces phylogenetic error. Syst Biol. 2002;51:588–598. doi: 10.1080/10635150290102339. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Goloboff P, Farris J, Nixon K. T.N.T.: Tree Analysis Using New Technology. 2003 2012 Nov 13; http://www.zmuc.dk/public/phylogeny/TNT.
- Hall BG. Tune ClustalX. Computer software and manual. 2004 2012 Nov 15; http://homepage.mac.com/barryghall/TuneClustalX.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 - The 18S-rDNA sequences used in this study.