Abstract
The puma is an iconic predator that ranges throughout the Americas, occupying diverse habitats. Previous phylogeographic analyses have revealed that it exhibits moderate levels of genetic structure across its range, with few of the classically recognized subspecies being supported as distinct demographic units. Moreover, most of the species’ molecular diversity was found to be in South America. To further investigate the phylogeographic structure and demographic history of pumas we analyzed mtDNA sequences from 186 individuals sampled throughout their range, with emphasis on South America. Our objectives were to refine the phylogeographic assessment within South America and to investigate the demographic history of pumas using a coalescent approach. Our results extend previous phylogeographic findings, reassessing the delimitation of historical population units in South America and demonstrating that this species experienced a considerable demographic expansion in the Holocene, ca. 8,000 years ago. Our analyses indicate that this expansion occurred in South America, prior to the hypothesized re-colonization of North America, which was therefore inferred to be even more recent. The estimated demographic history supports the interpretation that pumas suffered a severe demographic decline in the Late Pleistocene throughout their distribution, followed by population expansion and re-colonization of the range, initiating from South America.
Keywords: conservation genetics, mammals, mitochondrial DNA, molecular time estimate, phylogeography
Introduction
The puma (Puma concolor Linnaeus, 1771) is a large felid that ranges throughout the Americas, exhibiting the broadest latitudinal distribution of any terrestrial mammal (Nowak, 1999). Pumas have remarkable dispersal capabilities (Maehr et al., 2002; Beier et al., 2003) and successfully occupy a diverse array of habitats, illustrating their potential to adapt to the breadth of environmental conditions occurring across the continent, from tropical forests and marshes to dry scrubland and cold Andean or Patagonian biomes (Redford and Eisenberg, 1992; Nowak, 1999). Pumas are solitary and territorial, with large home ranges. Females tend to occupy smaller areas and disperse shorter distances, thus being more philopatric than males (Logan and Sweanor, 2001; Maehr et al., 2002). Overall, the species’ ecological flexibility and dispersal capabilities have the potential to induce broad genetic connectivity across large geographic areas, unless historical barriers have limited gene flow among populations.
Fossil evidence indicates that pumas were already present in North America 0.4 million years ago (MYA) (Kurtén and Anderson, 1980). In parallel, molecular data (Johnson et al., 2006) have led to an estimate of its divergence from the sister-species P. yagouaroundi of 4.17 MYA (C.I.: 3.16–6.01MYA), suggesting a much longer history as a distinct evolutionary lineage. The speciation event that separated these lineages may have occurred in North or South America, with the molecular dating estimate supporting the former, as it tends to predate the Great American Biotic Interchange (GABI) (ca. 2.5–3.5 MYA) and the implied colonization of South America by any felid (Woodburne, 2010; Eizirik, 2012). However, since the credibility interval of this estimate slightly overlaps the timing of the GABI, this issue is still not fully settled. Interestingly, Barnett et al. (2005) provided molecular evidence indicating that the extinct North American felid Miracinonyx trumani is the puma’s closest relative, with a divergence time estimated at 3.19 MYA. This finding would support the hypothesis of a North American origin for the puma, with subsequent colonization of South America by this species, in parallel with that of the jaguarundi.
In a thorough study of puma phylogeography, Culver et al. (2000) assessed the current and historical genetic diversity present in this species, based on a large sampling of individuals from across its range. That study indicated that most of the 32 classical puma subspecies did not correspond to definable genetic units and reduced the number of recognized subspecies to six. Four of these subspecies were distributed in South America, indicating that most of the species’ historical subdivision occurred in that subcontinent. In addition, the genetic diversity within South America was found to be larger than in Central and North America, suggesting that pumas from the latter subcontinent actually derive from a recent re-colonization event, following extinction in North America in the Late Pleistocene (ca. 10,000–12,000 years ago).
Mitochondrial DNA (mtDNA) markers have been extensively used in evolutionary studies of felids (e.g. Eizirik et al., 1998, 2001; Trigo et al., 2008). Although limited because they reveal only matrilineal history and contain a single realization of the stochastic processes of coalescence and lineage sorting (Avise, 1998), they are still powerful molecular tools to investigate recent phylogeographic patterns and demographic history. Given that felids tend to have male-biased gene flow (Pusey and Packer, 1987; Smith, 1993; Logan and Sweanor, 2001), mtDNA assessments of phylogeography are expected to provide more refined patterns of geographic structuring due to historical processes than nuclear markers. They may therefore be expected to be relevant benchmarks for maximum historical structuring, serving as baselines against which nuclear data sets may be compared. In addition, due to their four-fold lower effective population sizes (Ne) relative to autosomal markers, mtDNA segments should be more sensitive to the effect of historical genetic drift. In combination with their relatively high substitution rates, these features render these markers potentially more sensitive to historical processes of population subdivision than equivalent segments of the nuclear genome.
Within the mtDNA, the NADH dehydrogenase subunit 5 (ND5) gene has been successfully used in phylogenetic and phylogeographic studies of felids and other carnivores (e.g. Culver et al., 2000; Trinca et al., 2012). In a previous study focusing on pumas (Culver et al., 2000), three mtDNA segments were employed (ND5, 16S and ATP8). Of these, ND5 showed the highest polymorphic content in this species, based on a segment spanning 318 bp. A new primer set for this gene was designed specifically for carnivores (Trigo et al., 2008), amplifying a longer fragment (ca. 750 bp) and exhibiting successful amplification across several families (e.g. Felidae, Mustelidae, Mephitidae, Procyonidae [unpublished data]).
In the present study we employ this longer ND5 segment to investigate the evolutionary history of P. concolor, with emphasis on South American populations, which were previously found to harbor high levels of diversity and inferred to have played a key role in the historical demography of this species (Culver et al., 2000). Given that the geographic sampling of South American pumas was limited in that first study, we aimed here to expand the representation of the various regions of this sub-continent, so as to allow refined inferences of population structure, maternal gene flow and demographic history. In addition to expanding the geographic coverage of South American regions to refine inferences on patterns of matrilineal subdivision, we have performed novel analyses on puma demographic history, which revealed consistent evidence of a recent population expansion in South America, prior to re-colonization of North America.
Materials and Methods
Sample collection and laboratory procedures
We obtained blood and tissue samples from 77 pumas including wild individuals captured during field-ecology projects, caught by farmers or road-killed, as well as captive animals with known geographic origin (Table S1). In addition, we also collected data from 109 additional individuals whose DNA was already available in the participant laboratories, some of which had been used in earlier genetic studies employing different markers (Culver et al., 2000; Castilho et al., 2011; Miotto et al., 2011, 2012). Therefore, as a whole we collected novel data from a total of 186 individuals. These samples were originated from a diverse array of geographic regions throughout most of the puma range, with greater emphasis on South America (see Table S1). Two samples of Puma yagouaroundi were also included to be used as outgroups in some analyses.
To select a marker that would provide suitable information levels, we initially examined the mtDNA fragments used in previous studies, especially those involving Neotropical felids (e.g. Eizirik et al., 1998, 2001; Johnson et al., 1998, 1999; Culver et al., 2000). We selected the ca. 750 bp-long fragment of the ND5 gene reported by Trigo et al. (2008), thus considerably increasing the information content derived from this marker relative to the previous phylogeographic study of the puma (Culver et al., 2000). Finally, the availability of ND5 sequences for the extinct felid Miracinonyx trumani (Barnett et al., 2005) was an additional asset of this segment, allowing the inclusion of this fossil taxon in some analyses.
In the case of new biological samples, DNA was extracted using either a standard phenol-chloroform approach (Sambrook et al., 1989) or a CTAB-based protocol (Doyle and Doyle, 1987). For all samples, a fragment of the ND5 gene was amplified by the Polymerase Chain Reaction (PCR) (Saiki et al., 1985) using the primers ND5-DF1 and ND5-DR1 (Trigo et al., 2008). PCR reactions were performed in a total volume of 20 μL containing 2 μL of empirically diluted DNA, 1X buffer, 1.5 mM MgCl2, 0.2 μM of each primer, 0.2 mM dNTPs and 0.5 U Platinum Taq DNA polymerase (Invitrogen), with a thermocycling profile as described by Tchaicka et al. (2007). PCR products were visualized on a 1% agarose gel stained with GelRed (Biotium), purified with the enzymes Exonuclease I and Shrimp Alkaline Phosphatase (Exo-SAP, GE Healthcare), sequenced in both directions using the DYEnamic ET Dye Terminator cycle sequencing kit (GE Healthcare) and read in a MegaBacE 1000 automated DNA sequencer.
Sequence electropherograms were visually inspected and edited using Chromas Lite 2.01 or FinchTV 1.4.0. Sequences were aligned with the CLUSTALW algorithm (Higgings et al., 1996) implemented in MEGA 4 (Tamura et al., 2007), followed by manual verification and editing. The resulting novel puma sequences were deposited in GenBank (accession numbers KF460496-KF460523).
Data analysis
Nucleotide and haplotype (gene) diversity indices were estimated with DnaSP 5.10 (Librado and Rozas, 2009) for the total sample, as well as for a sample set restricted to South America. In addition, we estimated diversity indices for geographic subsets identified in this study as distinct historical units (see below).
To investigate the evolutionary relationships among puma mtDNA lineages, haplotype networks were built using the median-joining approach (Bandelt et al., 1999), as implemented in the software Network 4.5.1.6. We explored two outgroup options for rooting the network, one with the P. yagouaroundi sequences generated here, and the other employing M. trumani sequences (Barnett et al., 2005).
We also performed an Analysis of Molecular Variance (AMOVA) (Excoffier et al., 1992) with Arlequin 3.5.1.2 (Excoffier and Lischer, 2010) to assess the differentiation among geographic populations, using samples from 174 individuals. Twelve samples were excluded (see Table S1) due to insufficient information on local provenance, i.e. cases where the original geographic information was absent (the case of some zoo animals) or too broad, such as a large country. This subset was also used in all other analyses that required precise geographic provenance, such as all AMOVAs, Mantel tests, spatial autocorrelation, regional neutrality tests, mismatch distribution and Bayesian skyline plots (see below for additional details).
AMOVAs were performed using Φst computed from a pairwise matrix based on p-distances. Statistical significance of Φst values was tested using 10,000 permutations. To minimize the effect of prior assumptions on puma geographic subdivision we tested a variety of different scenarios attempting to identify the best possible way to represent historical population structure in this species (i.e. the one that maximizes the estimated Φst). These AMOVAs were based on two sample sets: (1) South America (SA) only: and (2) the full sample of SA, Central America (CA) and North America (NA).
Within sample set 1, we assessed the following scenarios: 1a) the four SA subspecies suggested by Culver et al. (2000); 1b) grouping samples according to their biomes of origin (adapted from IBGE (2010) and Olson et al. (2001)) while sequentially joining adjacent populations that exhibited non-significant Φst values; 1c) a sequential approach in which samples were initially grouped by immediate geographic proximity, then merged with other groups when they exhibited non-significant p-values for their pairwise Φst, and subsequently joined to additional contiguous populations with which the Φst estimate was low. For strategies 1b and 1c, the overall Φst value was monitored at every sequential step and the successive grouping strategy was maintained until a peak was observed. In this process, when we observed non-significant p-values between population groups, we still tested the effect of changing individuals from one adjacent unit to the other, so as to fine-tune the estimate that maximized the overall Φst value for this sample set.
In the second round of AMOVAs, based on sample set 2 (full sample), we employed the following strategies: 2a) the six subspecies suggested by Culver et al. (2000); 2b) incorporating the best grouping for SA identified in strategy 1c and adding the CA and NA samples as additional populations; 2c) testing the overall division into three subcontinents, including an additional adjustment separating Central America into two subgroups (northern and southern); 2d) a two-level hierarchical analysis considering subcontinents (best scenario found in 2c) as broader groups, within which we placed the regional populations identified with strategy 1c.
A third round of AMOVAs was performed to test the hypotheses raised by Culver et al. (2000), who postulated that major South American rivers could act as historical barriers to gene flow in this species. This was performed separately for the Amazon and Paraná rivers, by considering populations on either side of each river as distinct geographic units. Only populations located near the respective river were considered here. Given the available sampling on both sides of each river, we established an arbitrary cutoff of ≤ 610 km on each side of the Paraná, and ≤ 1340 km for the Amazon, so as to include a reasonable number of individuals and localities in both assessments.
Additionally, to investigate spatial patterns of genetic structure in pumas we assessed the correlation between genetic and geographic distances for all pairs of sampled individuals using a Mantel test (Mantel, 1967). This was performed with the program Alleles in Space (AIS) (Miller, 2005), employing the proportion of differences (i.e. a p-distance equivalent) to generate the genetic distance matrix and 10,000 permutations to assess statistical significance. We also employed AIS to perform a spatial autocorrelation analysis of the data set, using 11 distance classes, equalized sample sizes across classes, and 10,000 permutations to assess significance.
Finally, we conducted a set of analyses to investigate the demographic history of pumas. We performed neutrality tests (Tajima’s D, Fu & Li’ D* and F*, Fu’s Fs) with DnaSP, as well as a mismatch distribution analysis (Rogers and Harpending, 1992; Schneider and Excoffier, 1999) with Arlequin. In addition, we used the program Beast 1.6.1 (Drummond and Rambaut, 2007) to perform estimates of coalescence times and past demography. We defined the best model of nucleotide substitution for our data set, which was the HKY (Hasegawa et al., 1985) + G model, using the Akaike Information Criterion (AIC, Akaike, 1974) implemented in jModelTest 0.1 (Posada, 2008). Our first set of Beast runs was aimed at estimating the molecular clock rate for our segment in puma lineages. We assumed a Yule Process for the tree prior and incorporated an uncorrelated lognormal relaxed molecular clock. In order to better fit the expectations of the Yule process we only included five divergent puma haplotypes, along with the two jaguarundi sequences generated here. To calibrate the molecular clock we assumed that pumas and jaguarundis diverged between 3.16 and 6.01 MYA (95% HPD of the estimate reported by Johnson et al. (2006) using a nuclear supermatrix). We applied a uniform prior using these ages as conservative minimum and maximum boundaries, respectively. The final run was based on 108 MCMC generations, with samples taken every 104 steps, and the first 104 steps removed as burn-in.
The second set of Beast analyses aimed to infer the time of the most recent common ancestor (tMRCA) of different groups of samples, as well as to reconstruct the demographic history of pumas by estimating past fluctuations in population size via the Bayesian Skyline Plot (BSP) approach (Drummond et al., 2005). We assumed a Bayesian skyline tree prior and a strict molecular clock, whose evolutionary rate was based on the estimate derived from the first round of analyses. We assessed the tMRCA for four different sample sets: (i) complete sample; (ii) South America only; (iii) North and Central Americas (NA+NCA); and (iv) a subset of 24 NA+NCA samples that formed a regionally endemic subclade in the haplotype network, suggestive of autochthonous diversification (see Results). The BSP analyses were performed only for sets (i) and (ii), as their larger sample size allowed for more robust demographic inferences. The MCMC parameters were the same as in the first set of Beast analyses.
Results
After removal of incorporated primer sequences and low-quality fragment edges, double-stranded sequences for a 669-bp segment of the mtDNA ND5 gene were obtained for 156 puma individuals sampled in South America, 17 from Central America, and 13 from North America (Table S1, Figure 1). Additional sequences of the same fragment were obtained for two P. yagouaroundi individuals.
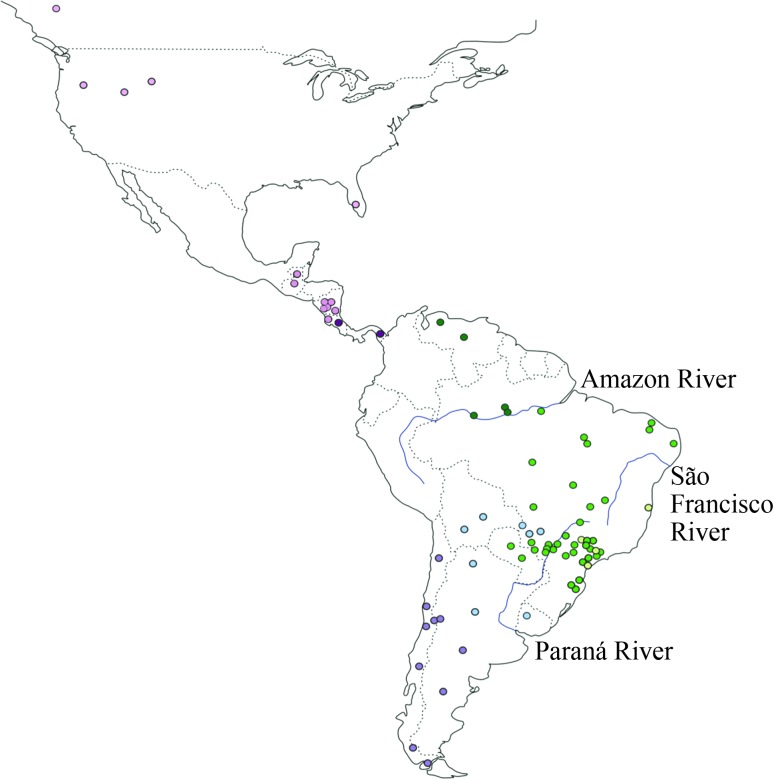
Figure 1.
Geographic distribution of the Puma concolor samples analyzed here. Blue lines indicate three major rivers. The different colors indicate the seven groups identified with the AMOVA (see text and Table 3 for details): Northern Central America + North America (NCA+NA, in light pink), Southern Central America (SCA, in dark pink), Northern South America (NSA, in dark green), Central-North-Eastern South America (CNESA, in medium green), Eastern South America (ESA, in light green), Central-Southern South America (CSSA, in light blue) and Southwestern South America (SWSA, in purple).
The puma data set contained 33 variable sites (32 transitions and one transversion), 25 of which were phylogenetically informative (Table 1). Overall, there were 28 haplotypes, 22 of which occurred in South America (H01–H22), one of them (H09) being shared with Central America (Figure 2A). Moderate to high levels of haplotype diversity were observed, while nucleotide diversity was rather low (Table 2).
Table 1.
Variable sites (33) observed among the 28 haplotypes detected in a 669-bp fragment of the mtDNA ND5 gene in Puma concolor samples (n = 174), along with their respective geographic groups of occurrence as defined by the AMOVA.
| Haplotype | Variable sitesa
|
N | Geographic groupsb | ||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 087* | 115* | 131* | 173* | 183* | 198 | 204* | 273* | 318* | 322 | 327* | 335* | 336* | 339* | 378* | 396 | 399 | 411* | 429* | 442* | 454* | 459* | 465* | 498* | 513 | 531* | 534* | 540* | 552* | 574 | 606 | 637 | 669* | |||
| H01 | T | A | C | A | C | C | G | A | T | C | A | G | C | T | C | C | T | T | G | A | T | C | G | T | C | G | C | C | C | C | C | C | C | 15 | CNESA (15) |
| H02 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 34 | CNESA (34) |
| H03 | . | . | . | . | . | . | A | G | . | . | . | A | . | . | . | . | . | C | . | . | . | . | . | C | . | . | . | . | T | . | . | . | T | 7 | CNESA (1), ESA (6) |
| H04 | . | . | . | . | . | . | . | . | . | T | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 1 | CNESA (1) |
| H05 | . | . | T | . | . | . | . | . | . | . | . | . | . | C | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | T | 2 | CSSA (2) |
| H06 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 4 | CNESA (3), CSSA (1) |
| H07 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | T | 9 | CNESA (4), CSSA (3), SWSA (2) |
| H08 | . | . | . | . | . | T | . | . | . | . | . | . | . | C | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | T | 1 | CSSA (1) |
| H09 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | T | 17 | SCA (3), CNESA (8), CSSA (6) |
| H10 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | T | . | . | . | . | . | . | . | . | . | . | T | 12 | CNESA (12) |
| H11 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | A | . | . | T | 1 | CNESA (1) |
| H12 | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | C | . | G | . | T | . | . | . | . | . | . | . | . | . | . | T | 2 | CSSA (2) |
| H13 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 2 | CNESA (2) |
| H14 | . | . | . | T | . | . | . | . | . | . | . | T | . | T | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 2 | CNESA (2) | |
| H15 | . | . | . | . | T | . | . | . | . | . | . | . | . | . | T | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 8 | CNESA (8) |
| H16 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | G | . | T | . | . | . | . | . | . | . | . | . | . | T | 6 | NSA (5), CNESA (1) |
| H17 | . | . | . | . | T | . | . | . | . | . | . | . | . | . | T | . | C | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 1 | CNESA (1) |
| H18 | C | . | . | . | . | . | . | . | . | . | G | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | . | . | . | . | T | 11 | SWSA (11) |
| H19 | . | . | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | T | 7 | SWSA (7) |
| H20 | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | T | 1 | NSA (1) |
| H21 | C | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | . | T | 3 | CNESA (3) |
| H22 | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | C | . | G | . | . | . | . | . | . | . | . | . | . | . | T | T | 1 | CNESA (1) |
| H23 | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | A | . | . | . | . | . | . | . | T | T | . | . | . | . | T | 18 | NCA+NA (17), SCA (1) |
| H24 | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | . | C | . | . | . | . | A | . | . | A | . | . | T | . | . | . | T | 3 | NCA+NA (1), SCA (2) |
| H25 | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | A | . | . | . | . | . | T | . | T | T | . | . | . | . | T | 1 | NCA+NA (1) |
| H26 | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | A | . | C | . | . | . | . | . | T | T | . | . | . | . | T | 3 | NCA+NA (3) |
| H27 | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | T | . | C | A | . | . | . | . | . | . | . | T | T | . | . | . | . | T | 1 | NCA+NA (1) |
| H28 | . | G | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | C | A | . | . | . | . | . | . | . | T | T | . | . | T | . | T | 1 | NCA+NA (1) |
Asterisks indicate phylogenetically informative sites.
Geographic group codes are as follows: NCA+NA: Northern Central America + North America; SCA: Southern Central America; NSA: Northern South America; CNESA: Central-North-Eastern South America; ESA: Eastern South America; CSSA: Central-Southern South America; SWSA: Southwestern South America (see Figure 1 and Supplementary Table S2 for details on the distribution of each group).
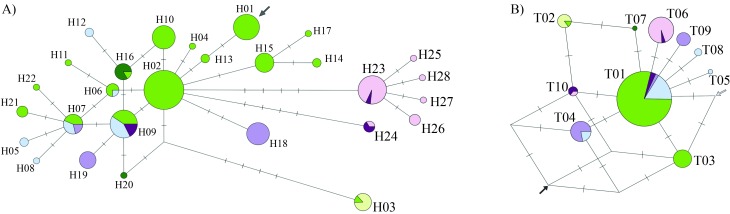
Figure 2.
Median-joining networks based on puma ND5 haplotypes. The area of each circle is proportional to the haplotype frequency. The relative frequency of populations in each haplotype is indicated by the proportion of different colors (coded as in Figure 1). A) Analysis of the full data set (669 bp). The arrow indicates the point where the outgroup P. yagouaroundi is connected. B) Analysis of a subset of the nucleotide sites that overlap with the Miracinonyx trumani sequence (286 bp). Arrows indicate the points where the outgroup taxa join the network: P. yagouaroundi (in gray; 32 mutational steps from the connection point), and M. trumani (in black; 19 steps from the connection point).
Table 2.
Nucleotide and gene diversity levels observed in Puma concolor ND5 sequences. Geographic groups were defined on the basis of the AMOVA results (see text and Tables 1 and 3).
| Sub-Continent | Nb | Nucleotide Diversity (±SE) per site | Gene diversity (±SE) | Haplotypes |
|---|---|---|---|---|
| North America | 13 | 0.00217 (0.00158) | 0.295 (0.156) | H23-H25 |
| Central America | 17 | 0.00523 (0.00114) | 0.794 (0.075) | H09, H23, H24, H26–H28 |
| South America | 156 | 0.00428 (0.00027) | 0.904 (0 011) | H01–H22 |
| Total | 186 | 0.00547 (0.00029) | 0.920 (0.008) | |
|
| ||||
| Geographic regiona | ||||
|
| ||||
| NA+NCA | 24 | 0.00179 (0.00094) | 0.496 (0.119) | H23–H28 |
| SCA | 6 | 0.00608 (0.00155) | 0.733 (0.155) | H09, H23, H24 |
| NSA | 6 | 0.00104 (0.00067) | 0.333 (0.215) | H16, H20 |
| CNESA | 97 | 0.00315 (0.00027) | 0.828 (0.027) | H01–H04, H06, H07, H09–H11, H13–H17, H21, H22 |
| ESA | 6 | 0.00000 (0.00000) | 0.000 (0.000) | H03 |
| CSSA | 15 | 0.00246 (0.00047) | 0.810 (0.078) | H05–H09, H12 |
| SWSA | 20 | 0.00334 (0.00032) | 0.595 (0.073) | H07, H18, H19 |
| Total | 174 | 0.00553 (0.00030) | 0.919 (0.010) | |
Geographic acronyms are as in Table 1.
N = number of individuals sampled for each continent/region. All samples (n = 186) were used for the sub-continental analysis, while 12 individuals lacking detailed geographic information were excluded from the regional analysis, resulting in a sample of 174 animals.
Evolutionary relationships among haplotypes were inferred using the median-joining network approach (Figure 2A), which showed a clear separation between lineages from South America (H01–H08, H10-H22) and those from North America and most of the haplotypes from Central America (H23–H28). The main exception was a haplotype (H09) found in southern Central America (Costa Rica), which is also common in central South America and is equidistant (five mutational steps) from the two closest haplotypes found in Central and North America (H23 and H24).
Among the 22 South American haplotypes, two (H16 and H20) were found north of the Amazon River (Brazil and Venezuela), with H16 also being sampled in one individual from southern Brazil. We observed very high levels of genetic diversity in central South America (including Bolivia, Paraguay, Uruguay, northern Argentina and the central-western, southeastern and southern regions of Brazil), where 15 distinct haplotypes were sampled (H01–H12, H14, H15 and H21). Among these, some haplotypes exhibited a localized distribution, such as H05, H08 and H12, which were found only in the Pantanal biome. A peculiar case was that of haplotype H03, sampled east and southeast of the São Francisco River (see Figure 1), and also in one individual from the Brazilian central-western region. This haplotype is five mutational steps away from its closest relatives, a distance which is equal to or larger than that observed between South American lineages and most of those sampled in North and Central America (4 mutational steps: H23 and H24; 5 mutational steps: H25, H26, H27 and H28). Individuals sampled along the Andes harbored two different haplotypes (H18 and H19), with H18 being more broadly distributed across the central and southern Cordillera, all the way to its extreme south in Patagonia.
In contrast, a single main haplotype (H23) was sampled in all surveyed countries from North and Central Americas (see Figure 2A). Four other haplotypes were connected to this common sequence by a single mutational step each, leading to a star-like appearance of this portion of the network. Among these haplotypes related to H23, H25 was found in the Florida peninsula (USA), H27 in Nicaragua, H28 in Guatemala, and H26 was sampled in both of the latter countries. Another haplotype (H24) found in Central and North America (Panama, Costa Rica and Florida peninsula [USA]) was quite distinct, differing by 8 mutational steps from H23, and by 4 steps from its closest relative, the common South American haplotype H02.
When the P. yagouaroundi sequences were used as outgroups in the network (Figure 2A) they were connected by 85 mutational steps to puma haplotype H01, supporting this position for the puma mtDNA root. Haplotype H01 is found in central South America (Paraguay, as well as the Brazilian states MS, SP and MG), differing by at least six mutational steps from the main North and Central American haplotypes. When the M. trumani sequence was included in the network (Figure 2B), the sequence alignment length was reduced to 286 bp to match the segment available for this outgroup. Due to this reduction in the number of sites, the network became less resolved, but the results still suggested that the outgroup was connected more closely to South American and southern Central American pumas than to northern Central and North American haplotypes.
To investigate the population genetic structure in more quantitative detail we performed several rounds of AMOVA, employing different geographic clustering strategies. In the first stage, when only South American pumas were analyzed, Φst values were quite high and statistically significant, with strategy 1c yielding the highest overall Φst (Table 3). In the second round of AMOVA, using the full sample set, we found even higher Φst values. The highest overall value was obtained with strategy 2d, employing a two-level AMOVA (Table 3). Central America was best represented as two distinct groups, northern and southern, with the former being merged with North America. In the best two-level scenario (see Table 3), we also noted that the Φsc and Φct were 0.41904 (p = 0.000±0.000) and 0.43693 (p = 0.01871+−0.00135), respectively. Pairwise estimates of Φst were carried out at the various spatial scales tested in these scenarios, and helped assess the distinctiveness of large-scale as well as regional population units (Tables S4 and S5).
Table 3.
Best Φst values for different population structure scenarios in Puma concolor, as assessed with an AMOVA approach.
| Group | Φst |
|---|---|
| 1. South America only (144 individuals) | |
| a) 4 SA subspecies proposed by Culver et al. (2000) | 0.22001* |
| b) Grouped biomes from SAa | 0.25871* |
| c) Five groups from SA (NSA, CNESA, ESA, CSSA and CWSA)b | 0.35993* |
| 2. Total sample (174 individuals) | |
| a) 6 subspecies proposed by Culver et al. (2000) | 0.42353* |
| b) 7 groups [(NA+NCA), SCA, NSA, CNESA, ESA, CSSA and CWSA]b | 0.57314* |
| c) NA+NCA x SCA x SA subcontinentsc | 0.55740* |
| d) 2-level [(NA+NCA) x SCA x (NSA + CNESA + ESA + CSSA + CWSA)]d | 0.67290* |
| 3. Barriers | |
| Amazon River acting as a barriere | 0.34739* |
| Paraná River acting as a barriere | 0.09041* |
Significant Φst (p < 0.01).
3 groups: 1st-northern Amazon Rainforest (Tropical Moist Broadleaf Forests), Tropical Dry Broadleaf Forests and Tropical Grasslands, Savannas and Shrublands in Venezuela; 2nd-southern Amazon Rainforest and Atlantic Forests (Tropical and Subtropical Moist Broadleaf Forests), Pantanal (Flooded Grasslands and Shrublands), Caatinga (Xeric Shrublands), Cerrado, Pampas, Gran Chaco and Beni Savanna (Tropical and Subtropical Grasslands, Savannas and Shrublands), Patagonian Pampas (Temperate Grasslands, Savannas and Shrublands) and Tropical Dry Broadleaf Forests in Bolivia; 3rd-Andean biomes: Temperate Broadleaf and Mixed Forests, Montane Grasslands and Shrublands, Desert and Xeric Shrublands and Mediterranean Forests, Woodlands and Scrubs (Olson et al., 2001; IBGE 2010).
Central America was divided into two separate groups, northern (NCA) and southern (SCA).
1st level: 3 groups: NA+NCA (2 populations), SCA (1 population), SA (5 populations); 2nd level: 7 populations ([NA + NCA] x [SCA] x [NSA + CNESA + ESA + CSSA + CWSA]);
Considering only samples collected near the river (see text for details).
Finally, we performed a third round of AMOVA to test two large rivers as possible barriers to historical gene flow in this species. We observed high Φst values when the Amazon River was assessed (see Table 3). The Φst value found for the Parana River was rather low (0.09), although still statistically significant.
Mantel Tests (Figure S1) revealed a significant correlation between genetic and geographic distances for the total geographic sample (r = 0.48; p < 0.001), indicating that isolation by distance plays a relevant role in the genetic structuring of this species. As for the analysis of South American samples alone, the resulting value was 0.085 (p = 0.09347), which suggests that within this subcontinent there is no significant influence of isolation by distance.
The spatial autocorrelation analysis (performed only for the full data set) yielded results that were consistent with those of the Mantel test. There was a trend for an increase in the statistic Ay (representing the average pairwise genetic distance within geographic distance class ‘y’) as the geographic distance increased, and the overall relationship was statistically significant (p = 0.0048), supporting the inference that isolation by distance affected the patterns observed in our global data set.
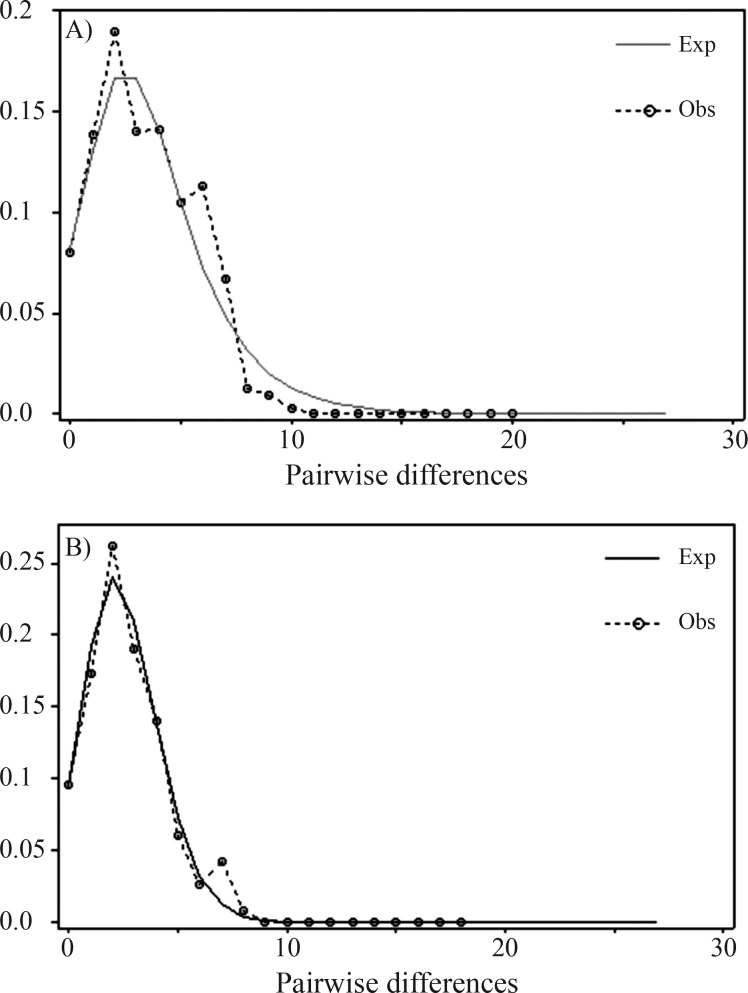
Another front of analyses pertained to demographic history assessments. When we conducted a Mismatch Distribution analysis, the results indicated a unimodal pattern for both data sets (full sample and South American samples only) (Figure 3) compatible with a scenario of recent demographic expansion. In addition, the neutrality tests yielded negative values for all indices, with those of Fu’s Fs being significantly negative with respect to neutral expectations for both data sets (Table 4).
Figure 3.
Graphs depicting the results of the Mismatch Distribution analysis for the total sample (A) and South American samples alone (B). The analysis was performed with 669 bp of P. concolor ND5 sequences (excluding all sites with missing information or gaps). The dashed line represents the observed pattern, while the continuous line depicts the pattern expected under a model of sudden demographic expansion.
Table 4.
Neutrality tests for Puma concolor ND5 sequences, considering both the ‘Total’ and ‘South American’ (SA) sample sets.
| Total | SA | |
|---|---|---|
| Tajima’s D | −1.16291 | −0.98375 |
| Fu and Li’s D* | −0.82174 | −0.39340 |
| Fu and Li’s F* | −1.15989 | −0.75225 |
| Fu’s Fs | −9.322a | −7.283a |
Significant values (p < 0.02).
The Bayesian analyses performed with Beast further helped shed light onto the historical demography pumas as revealed by the mtDNA. In the first round of analyses, using five individuals with the most divergent haplotypes and two jaguarundis, we estimated the substitution rate of the analyzed ND5 segment to be 2.054% per million years (MY). Applying this substitution rate in the second set of analyses, we inferred the mean tMRCA of pumas to be 0.237 MYA (95% HPD: 0.105–0.391 MYA). For South American pumas, the mean tMRCA was estimated to be 0.211 (0.091–0.353) MYA. When North American (NA) and Central American (CA) samples were analyzed separately, their mean estimated tMRCA was 0.231 (0.102–0.387) MYA. Finally, we estimated the tMRCA for a subset of 24 NA+CA samples that formed an endemic monophyletic cluster (see Figure 2A), yielding a value of 0.037 (0.006–0.077) MYA.
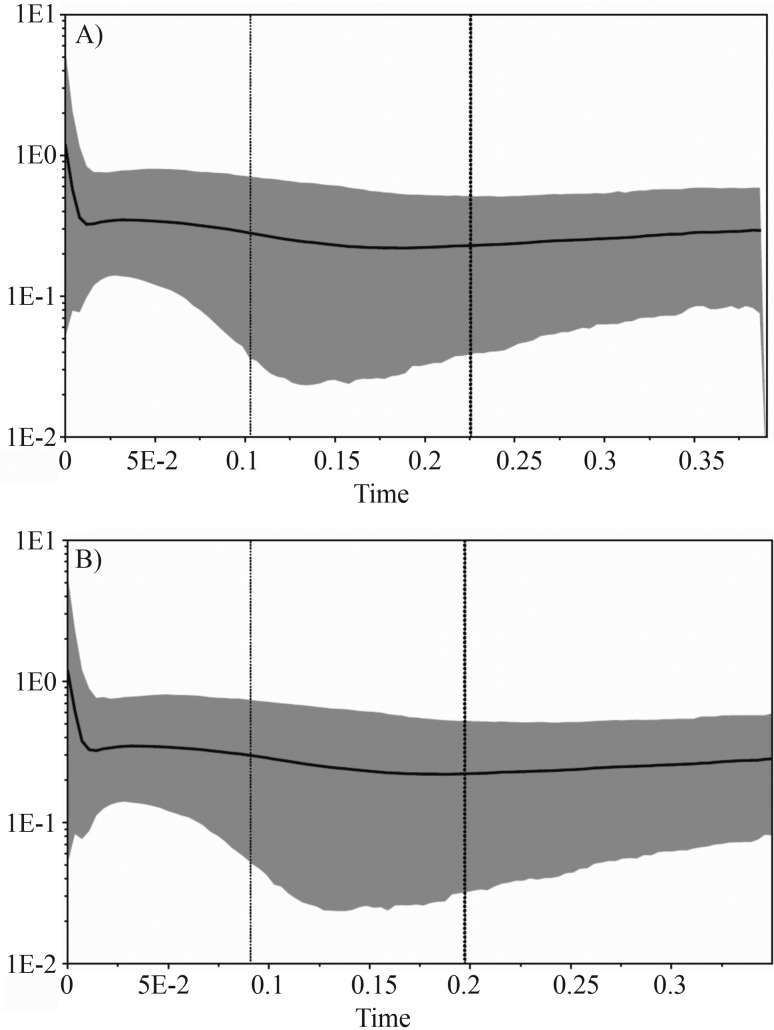
The Bayesian skyline plot analysis revealed evidence of a very recent (ca. 8,000 years ago) demographic expansion in pumas (Figure 4). The signal for this expansion was apparent with both data sets (full sample and South American pumas only), with only minor differences in the estimated parameters.
Figure 4.
Bayesian Skyline plots derived from an alignment of puma ND5 sequences. In (A) the analysis was run for all puma samples, and in (B) for South American pumas only. The thick solid line represents the mean, while the 95% HPD (highest posterior density) limits are shown by the blue areas. The vertical dashed lines refer to the tMRCA, with the thin line indicating the lower 95% HPD limit, and the thick line representing the mean. The x-axis is depicted on a scale of millions of years (myr). The y-axis represents the effective population size (Ne) multiplied by the generation time (also in myr). Assuming a generation time of 5 years, the numbers on this axis should be multiplied by 200,000 to yield interpretable values.
Discussion
mtDNA diversity and genealogical history
The mtDNA data set analyzed in this study allowed a deeper understanding of the matrilineal history of pumas, refining, extending and in some cases adjusting the inferences previously put forth by Culver et al. (2000). Our initial observation was that ND5 is indeed a very informative coding mtDNA segment for phylogeographic studies of felids, and that the longer segment employed here (relative to the one reported by Culver et al., 2000) does provide useful additional information regarding this species’ diversity and evolutionary history. For example, the ND5 data set used here contained 33 variable sites and allowed the identification of 28 different puma haplotypes, while the full data set employed by Culver et al. (2000), including ND5, ATP8 and 16S segments, contained 15 variable sites and defined 14 different haplotypes.
Pumas exhibited moderately high levels of diversity, which could be directly compared to only two other felid species that have so far been sequenced for the same mtDNA segment and whose sequences for available for assessment (Trigo et al., 2008). Both of these Neotropical cats, Leopardus tigrinus and L. geoffroyi, were sequenced for a slightly shorter fragment, so we adjusted our alignment to include only the matching 567 bp and thus allow an exact comparison (Table S3). This assessment revealed that pumas possess higher gene diversity than either of these other species, as well as slightly higher nucleotide diversity. Indeed, the observed levels of haplotype (gene) diversity in pumas were quite high, while nucleotide diversity was more modest, suggestive of rather recent mtDNA coalescence in this species.
When subcontinental or regional groups of pumas were compared, we observed that haplotype and nucleotide diversity indices were consistently higher for South and Central America, relative to those estimated for North America (Table 2). Gene diversity was highest for South America, while nucleotide diversity reached a maximum in Central America. North American pumas presented low values for both indices, which were at least two-fold lower than those of the other sub-continental groups. When the South American regional groups defined by the AMOVA (see Figure 1 and Table 3 for their definition) were assessed, we observed that the CNESA and CSSA groups exhibited the highest haplotype diversity, with the SWSA and NSA groups also presenting high values (Table 2).
The high gene diversity estimated for pumas could be examined in more detail with the use of haplotype networks (Figure 2A), which depicted intricate relationships among sequences, especially in South America. Such a pattern is indicative of a complex evolutionary history, with some suggestion of geographic structuring but no clear-cut separation of well-defined clades. Some portions of the network are suggestive of a recent population expansion involving South American lineages, with central haplotypes giving rise to multiple derived sequences differing by a single mutational step each. Central America was also observed to bear a complex mixture of haplotypic lineages, with more than one connection to basal South American sequences and evidence of internal structure (see Figure 2A and Table 3). In contrast, the lineages sampled in North America exhibited a much simpler pattern, dominated by a common haplotype (H23) shared with Central America, from which four other lineages have recently evolved. This shape is indicative of a recent population expansion involving these haplotypes, whose sampling locales indicate a present distribution across North America and also in Central America, especially in the northern portion of the latter (Figures 1 and 2).
In North America, an unusual pattern was observed in an individual from Florida (USA) that carried haplotype H24, which is shared with animals from southern Central America (Panama and Costa Rica) and is genetically distant from the other lineages found throughout North America. Given the record of human-induced introgression of puma mtDNA lineages from Central America to Florida in the 1960s (O’Brien et al., 1990), it is possible that this individual descends from a female translocated during that period, implying that such haplotype sharing may be a human-induced artifact. If this were indeed the case, our results would in fact be an underestimate of the differentiation between SCA and NA+NCA, since haplotype H24 may only naturally occur in the former.
Central America and northern South America (NSA) did not share haplotypes, implying no evidence of historical matrilineal gene flow between these regions. Possible barriers include the northernmost Andean Cordillera or the Darien Straits in Panama, as suggested for jaguars by Eizirik et al. (2001) (but see Ruiz-Garcia et al., 2013 for a different view). Detailed sampling in that region would be required to distinguish between these two hypotheses. Although no haplotype was shared between Central America and northern South America, we did observe a shared haplotype (H09) between southern Central America (Costa Rica) and the central region of South America. This suggests some continued matrilineal connectivity between these areas (necessarily going through northern South America), or the retention of an ancestral polymorphism that predates the implicated phylogeographic subdivision. Since the available sampling for northern South America is still modest (see Figure 1), both hypotheses remain viable and should be the focus of additional scrutiny. Moreover, northwestern South America has been reported to harbor high levels of genetic diversity and a complex evolutionary history for several mammalian taxa (e.g. Eizirik et al., 1998; Ruiz-García et al., 2009, 2010, 2012; Thoisy et al., 2010), highlighting the need to further investigate pumas from this portion of their range to assess their role in the overall phylogeographic structure of the species.
The network root based on the outgroup Puma yagouaroundi indicates that the most ancestral lineage of current pumas is located primarily in central South America. When we considered M. trumani as the outgroup, once again the rooting was placed primarily in South America, reasserting the area of occurrence of the earliest ancestor of the current lineages and helping reveal the direction of evolution of pumas throughout their distribution.
Population structure
The AMOVA results revealed extremely high Φst values in many of the assessed scenarios (see Table 3). The highest Φst value, used to define the scenario that best explains the current partitioning of mtDNA diversity, was found when a 2-level AMOVA was conducted. The first level separated “subcontinents” as major units (NA+NCA, SCA and SA), while the second level identified subdivisions within the “subcontinents”. Several analyses, following the steps outlined in the AMOVA strategy 2b (data not shown), demonstrated that other possible groupings were almost as good, indicating that there is no clear-cut definition of the structure in central South America, but demonstrating that this region is a genetic hotspot for this species, with considerable structure but complex historical relationships.
Our next step was to assess the role of major rivers as historical barriers to female gene flow. Our sampling allowed a test of two riverine barriers proposed by Culver et al. (2000), which postulated that the Amazon and Paraná rivers might have induced the differentiation of historical population units. Our results supported an important role for the Amazon River, corroborating the hypothesis previously raised for pumas (Culver et al., 2000), and showing a pattern consistent with inferences made for other Neotropical felids such as Leopardus pardalis, L. wiedii and Panthera onca (Eizirik et al., 1998, 2001). On the other hand, the influence of the Paraná River appears to be much lower than previously hypothesized, although we still observed significant Φst values when we compared populations separated by this watercourse. Although they were significant, the magnitude of the differentiation was considerably lower than that estimated for the Amazon River (see Table 3), and several AMOVA assessments comparing regional populations on the same side of the Paraná River yielded higher Φst values than those obtained in the comparison between the two sides. These findings suggest that, although this river may play a role in limiting regional maternal gene flow in pumas, it does not induce major differentiation between clear-cut historical units. Such an observation has implications for the definition and delimitation of puma subspecies, two of which (P. c. cabrerae and P. c. capricornensis) have been suggested to be separated by this river (Culver et al., 2000).
Demographic history
The tMRCA estimates derived from both data sets were very similar, indicating that the age of the common ancestor for all samples was essentially the same as that for the South American subset. Taken together with the other results (e.g. network, diversity indices), this finding supports the inference that most of the extant diversity of the puma mtDNA resides in South America, and that the deepest history of coalescence of its lineages is fully represented in this subcontinent.
The signal of demographic expansion on both subcontinents suggests that this process seems to have occurred simultaneously, after the last glacial maximum. This inference is consistent with the hypothesis raised by Culver et al. (2000), of an extinction of North American pumas in the late Pleistocene, followed by re-colonization from South America. Our results corroborate and extend this hypothesis, showing evidence for a substantial and very recent demographic expansion in South America, which likely preceded or included the re-colonization of North America. Such a pattern was not clearly observed in previous studies and sheds light onto the historical demography of present-day puma lineages. Our findings indicate that pumas were substantially affected by faunal changes induced by the Late Pleistocene extinction of large mammals, likely disappearing from North America, as previously postulated, with subsequent recolonization from the south. What is new in our results is the inference of a substantial demographic expansion in South America ca. 8,000 years ago, possibly induced by release from competitive interactions with other large carnivores that went extinct during the Late Pleistocene event. We see no evidence of a secondary expansion in North America, as the full data set mirrors the results observed with South America alone. We thus conclude that pumas likely re-colonized North America spurred by this Holocene demographic expansion stemming from South America. If corroborated by additional genetic markers and more refined sampling, especially in northern South America, this hypothesis would provide a useful demographic framework for pumas against which local patterns of genetic structure, genomic variation and population connectivity can be assessed.
Acknowledgments
We thank C.R. Sarturi, A. Schnorr, A.C.G. Escobar, C.B. Pires, A.T. Hahn, F. Britto, F.G. Grazziotin, T. Haag, T.C. Trigo and T.L.L. Simão for laboratory assistance and helpful discussions regarding data analysis and interpretation, as well as J. Schiefelbain and M. G. da Silva for additional support. This work was funded by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil, and Companhia Energética de São Paulo (CESP), Brazil. Part of this research was also supported by funds from the Intramural Research Program of the National Institutes of Health, National Cancer Institute, USA awarded to WEJ and SJO.
Footnotes
Associate Editor: Fabrício Rodrigues dos Santos
Supplementary Material
The following online material is available for this article:
Table S1-Geographic locales included in this study.
Table S2 - Puma concolor samples analyzed in this study.
Table S3 - Nucleotide and gene diversity levels observed in Puma concolor mtDNA ND5 sequences.
Table S4 - Pairwise Φst values among subcontinental groups of Puma concolor.
Table S5 - Pairwise Φst values among seven major groups of Puma concolor.
Figure S1 - Graph depicting the correlation between genetic and geographic distances calculated for P. concolor ND5 mtDNA sequences.
This material is available as part of the online article from http://www.scielo.br/gmb
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Autom Contr. 1974;19:716–723. [Google Scholar]
- Avise JC. The history and purview of phylogeography: A personal reflection. Mol Ecol. 1998;7:371–379. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-Joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barnett R, Barnes I, Phillips MJ, Martin LD, Harington CR, Leonard JA, Cooper A. Evolution of the extinct Sabre-tooths and the American cheetah-like cat. Curr Biol. 2005;15:R589–R590. doi: 10.1016/j.cub.2005.07.052. [DOI] [PubMed] [Google Scholar]
- Beier P, Vaughan MR, Conroy MJ, Quigley H. A review of literature related to the Florida panther. Bureau of Wildlife Diversity Conservation - Florida Fish and Wildlife Conservation Commission, Tallahassee. 2003:203. [Google Scholar]
- Castilho CS, Marins-Sá LG, Benedet RC, Freitas TO. Landscape genetics of mountain lions (Puma concolor) in southern Brazil. Mammal Biol. 2011;76:476–483. [Google Scholar]
- Culver M, Johnson WE, Pecon-Slattery J, O’Brien SJ. Genomic ancestry of the American Puma (Puma concolor) J Hered. 2000;91:186–197. doi: 10.1093/jhered/91.3.186. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:e214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Eizirik E. A molecular view on the evolutionary history and biogeography of Neotropical carnivores (Mammalia, Carnivora) In: Patterson BD, Costa LP, editors. Bones, Clones, and Biomes: An Extended History of Recent Neotropical Mammals. University of Chicago Press; Chicago: 2012. pp. 123–142. [Google Scholar]
- Eizirik E, Bonatto SL, Johnson WE, Crawshaw PG, Jr, Vie JC, Brousset DM, O’Brien SJ, Salzano FM. Phylogeographic patterns and evolution of the mitochondrial DNA control region in two Neotropical cats (Mammalia, Felidae) J Mol Evol. 1998;47:613–624. doi: 10.1007/pl00006418. [DOI] [PubMed] [Google Scholar]
- Eizirik E, Kim J-H, Menotti-Raymond MA, Crawshaw PG, Jr, O’Brien SJ, Johnson WE. Phylogeography population history and conservation genetics of jaguars (Panthera onca, Mammalia, Felidae) Mol Ecol. 2001;10:65–79. doi: 10.1046/j.1365-294x.2001.01144.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer H. Arlequin suite ver. 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resourc. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Higgings DG, Thompson JD, Gibson TJ. Using CLUS-TAL for multiple sequence alignments. Meth Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Culver M, Iriarte JA, Eizirik E, Seymour KL, O’Brien SJ. Tracking the evolution of the elusive Andean mountain cat (Oreailurus jacobita) from mitochondrial DNA. J Hered. 1998;89:227–232. doi: 10.1093/jhered/89.3.227. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Pecon-Slattery J, Eizirik E, Kim JH, Menotti-Raymond M, Bonacic C, Cambre R, Crawshaw P, Nunes A, O’Brien SJ, et al. Disparate phylogeography patterns of molecular genetic variation in four closely related South American small cat species. Mol Ecol. 1999;8:S79–S92. doi: 10.1046/j.1365-294x.1999.00796.x. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O’Brien SJ. The late Miocene radiation of modern Felidae: A genetic assessment. Science. 2006;311:73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- Kurtén B, Anderson E. Pleistocene Mammals of North America. Columbia University Press; New York: 1980. p. 442. [Google Scholar]
- Librado P, Rozas J. DnaSP ver. 5: A software for comprehensive analysis of DNA polymorphism data. Bioinform Appl Note. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Logan KA, Sweanor LL. Desert puma: evolutionary ecology and conservation of an enduring carnivore. Island Press; Washington, D.C: 2001. p. 470. [Google Scholar]
- Maehr DS, Land ED, Shindle DB, Bass OL, Hoctor TS. Florida panther dispersal and conservation. Biol Conserv. 2002;106:187–197. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Miller MP. Alleles in Space (AIS): Computer software for the joint analysis of interindividual spatial and genetic information. J Hered. 2005;96:722–724. doi: 10.1093/jhered/esi119. [DOI] [PubMed] [Google Scholar]
- Miotto RA, Cervini M, Figueiredo MG, Begotti RA, Galetti PM., Jr Genetic diversity and population structure of pumas (Puma concolor) in southeastern Brazil: Implications for conservation in a human-dominated landscape. Conserv Genet. 2011;12:1447–1455. [Google Scholar]
- Miotto RA, Cervini M, Begotti RA, Galetti PM., Jr Monitoring a puma (Puma concolor) population in a fragmented landscape in southeast Brazil. Biotropica. 2012;44:98–104. [Google Scholar]
- Nowak RM. Walker’s Mammals of the World. 6th ed. Vol. 2. The Johns Hopkins University Press; Baltimore: 1999. pp. 818–820. [Google Scholar]
- O’Brien SJ, Roelke M, Yuhki N, Richards KW, Johnson WE, Franklin WL, Anderson AE, Bass OL, Jr, Belden RC, Martenson JS. Genetic introgression within the Florida Panther Felis concolor coryi. Natl Geogr Res. 1990;6:485–494. [Google Scholar]
- Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D’Amico JA, Itoua I, Strand HE, Kassem KR, et al. Terrestrial ecoregions of the World: A new map of life on earth. BioScience. 2001;51:933–938. [Google Scholar]
- Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pusey AE, Packer C. The evolution of sex-biased dispersal in lions. Behaviour. 1987;101:275–310. [Google Scholar]
- Redford KH, Eisenberg JF. Mammals of the Neotropics. Vol. 2, The Southern Cone. University of Chicago Press; Chicago: 1992. p. 460. [Google Scholar]
- Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- Ruiz-García M, Pacheco LF, Álverez D. Genetic characterization of the Bolivian Andean puma (Puma concolor) at the Sajama National Park (SNP) and relationships with other north-western South American puma populations. Rev Chil Hist Nat. 2009;82:97–117. [Google Scholar]
- Ruiz-García M, Pinedo-Castro MO. Molecular systematics and phylogeography of the genus Lagothrix (Atelidae, Primates) by means of the mitochondrial COII gene. Folia Primatol. 2010;81:109–128. doi: 10.1159/000315070. [DOI] [PubMed] [Google Scholar]
- Ruiz-García M, Vásquez C, Pinedo-Castro MO, Sandoval S, Castellanos A, Kaston F, Thoisy B, Shostell J. Phylogeography of the Mountain Tapir (Tapirus pinchaque) and the Central American Tapir (Tapirus bairdii) and the origins of the three Latin-American tapirs by means of mtCyt-B sequences. In: Anamthawat-Jónsson K, editor. Current Topics in Phylogenetics and Phylogeography of Terrestrial and Aquatic Systems. InTech; Rijeka: 2012. pp. 83–116. [Google Scholar]
- Ruiz-Garcia M, Vásquez C, Murillo A, Pinedo-Castro M, Alvarez D. Population genetics and phylogeography of the largest wild cat in the Americas: An analysis of the jaguar by means of microsatellites and mitochondrial gene sequences. In: Ruiz-García M, Shostell J, editors. Molecular Population Genetics, Evolutionary Biology and Biological Conservation of the Neotropical Carnivores. Nova Science Publishers Inc; New York: 2013. pp. 413–464. [Google Scholar]
- Saiki RK, Scharf S, Fallona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of ß-globin genomic sequences and restriction site analysis for diagnosis of sickle-cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sambrook E, Fritsch F, Maniatis T. Molecular Cloning. 2nd edition. Cold Spring Harbor Press; Cold Spring Harbor, New York: 1989. [Google Scholar]
- Schneider S, Excoffier L. Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: Application to human mitochondrial DNA. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JLD. The role of dispersal in structuring the Chitwan tiger population. Behaviour. 1993;124:165–195. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software ver. 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tchaicka L, Eizirik E, Oliveira TG, Candido F, Jr, Freitas TRO. Phylogeography and population history of the crab-eating fox (Cerdocyon thous) Mol Ecol. 2007;16:819–838. doi: 10.1111/j.1365-294X.2006.03185.x. [DOI] [PubMed] [Google Scholar]
- Thoisy B, Silva AG, Ruiz-García M, Tapia A, Ramirez O, Arana M, Quse V, Paz-y-Miño C, Tobler M, Pedraza C, et al. Population history, phylogeography, and conservation genetics of the last Neotropical mega-herbivore, the lowland tapir (Tapirus terrestris) BMC Evol Biol. 2010;10:e278. doi: 10.1186/1471-2148-10-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigo TC, Freitas TRO, Kunzler G, Cardoso L, Silva JCR, Johnson WE, O’Brien SJ, Bonatto SL, Eizirik E. Inter-species hybridization among Neotropical cats of the genus Leopardus, and evidence for an introgressive hybrid zone between L. geoffroyi and L. tigrinus in southern Brazil. Mol Ecol. 2008;17:4317–4333. doi: 10.1111/j.1365-294X.2008.03919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinca CS, De Thoisy B, Rosas FCW, Waldemarin HF, Koepfli KP, Vianna JA, Eizirik E. Phylogeography and demographic history of the Neotropical otter (Lontra longicaudis). J. Hered. 2012;103:479–492. doi: 10.1093/jhered/ess001. [DOI] [PubMed] [Google Scholar]
- Woodburne MO. The great american biotic interchange: Dispersals, tectonics, climate, sea level and holding pens. J Mammal Evol. 2010;17:245–264. doi: 10.1007/s10914-010-9144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- IBGE, Instituto Brasileiro de Geografia e Estatística http://www.ibge.gov.br (April 30, 2012).
- Chromas Lite ver. 2.01 Program ( http://www.technelysium.com.au) (March 12, 2011).
- FinchTV ver. 1.4.0 Program ( http://www.geospiza.com) (March 12, 2011).
- Network ver. 4.5.1.6 Program ( www.fluxus-engineering.com) (April 3, 2011).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1-Geographic locales included in this study.
Table S2 - Puma concolor samples analyzed in this study.
Table S3 - Nucleotide and gene diversity levels observed in Puma concolor mtDNA ND5 sequences.
Table S4 - Pairwise Φst values among subcontinental groups of Puma concolor.
Table S5 - Pairwise Φst values among seven major groups of Puma concolor.
Figure S1 - Graph depicting the correlation between genetic and geographic distances calculated for P. concolor ND5 mtDNA sequences.