Abstract
Anfinsen showed that a protein's fold is specified by its sequence. Although it is clear why mutant proteins form amyloid, it is harder to rationalize why a wild-type protein adopts a native conformation in most individuals, but it misfolds in a minority of others, in what should be a common extracellular environment. This discrepancy suggests that another event likely triggers misfolding in sporadic amyloid disease. One possibility is that an abnormal metabolite, generated only in some individuals, covalently modifies the protein or peptide and causes it to misfold, but evidence for this is sparse. Candidate metabolites are suggested by the recently appreciated links between Alzheimer's disease (AD) and atherosclerosis, known chronic inflammatory metabolites, and the newly discovered generation of ozone during inflammation. Here we report detection of cholesterol ozonolysis products in human brains. These products and a related, lipid-derived aldehyde covalently modify Aβ, dramatically accelerating its amyloidogenesis in vitro, providing a possible chemical link between hypercholesterolemia, inflammation, atherosclerosis, and sporadic AD.
Anfinsen's classic experiments demonstrated that a protein's amino acid sequence specifies its conformation (1). These ideas were extended to explain the misfolding susceptibility of mutant proteins associated with a growing number of familial amyloid diseases (2–5). Although it is thus clear why mutant proteins might be more susceptible to misfolding, it is harder to understand why a wild-type protein or peptide adopts a native conformation in some individuals but it misfolds in others in what should be a common extracellular environment, leading to sporadic amyloid diseases. This discrepancy suggests that other events likely trigger misfolding in sporadic amyloid disease, but their nature remains elusive.
The misfolding of secreted amyloid β peptides (Aβ) 39–43 residues in length is linked by a plethora of evidence to the pathology of Alzheimer's disease (AD) (6, 7). Aβ misfolding occurs when the soluble, monomeric, extracellular ensemble of extended conformations and low Mr oligomers is transformed first into spherical assemblies, then into a number of intermediates, and lastly into fibrillar cross β-sheet quaternary structures known as amyloid (8–12). Amyloid fibrils and related structures recruit soluble Aβ to the aggregate by a seeded polymerization mechanism (10). The direct neurotoxicity of Aβ aggregates (8, 13) combined with their role in mediating chronic inflammation by microglia (14) and complement cascade activation (15) suggests that aggregation then mediates inflammation (16), which in turn promotes aggregation, in a vicious cycle of AD pathology.
It is known that atherosclerosis and AD share many risk factors, including hypercholesterolemia and inflammation. The apoE-ε4 allele, which exacerbates hypercholesterolemia, has been linked to AD by data from both epidemiological and transgenic mouse studies (17–20). It has also recently been shown that antibodies catalyze ozone production during inflammation (21) and that metabolites resulting from cholesterol ozonolysis (1 and 2; see Fig. 1) are generated during the inflammatory component of atherosclerosis (22). From a chemical perspective, these ozonolysis products are special in that they contain an aldehyde group attached to a large hydrocarbon. The aldehyde group could covalently modify Aβ by condensing with amines present in the peptide (the side chains of Lys-16 and Lys-28 and the N terminus). This modification would increase the hydrophobicity of the peptide, possibly increasing its propensity to misfold and assemble into the quaternary structures associated with AD. We have therefore evaluated whether cholesterol metabolites 1 and 2, along with an aldehyde-containing lipid oxidation product (8) known to be associated with AD (23–27), could initiate Aβ misfolding.
Fig. 1.
(A) Aβ aggregation (100 μM, pH 7.5, 37°C) as detected by TfT fluorescence at several concentrations of 1. Fluorescence is measured in arbitrary units. (B) As in A, except with 2.(C and D) AFM images of Aβ (100 μM) samples in the presence (C) and absence (D) of 1 (50 μM). The images shown here are representative of four experiments.
Methods
Preparation of Seed-Free Aβ. Aβ1–40 and 1–42 (SynPep, Dublin, CA) were characterized by sequencing and liquid chromatography (LC)-MS. The pretreatment used was adapted from the method of Fezoui et al. (28). In brief, lyophilized Aβ powder was dissolved in aqueous NaOH (2 mM). The pH was adjusted to 10.5 with aqueous NaOH (100 mM). The solution was sonicated (20 min, 25°C), then filtered sequentially through 0.2 μm and 10-kDa cutoff filters. The concentration of protein was determined by UV absorption at 280 nm (ε = 1,280 M–1·cm–1). The peptide in the filtrate was diluted to 200 μM by using aqueous NaOH (pH 10.5) and used immediately. Note that ≈50% of the Aβ sample was retained on the 10-kDa filter. After pretreatment, the final peptide solution was seed-free according to atomic force microscopy (AFM) analysis and the reproducibility of experiments with these preparations described below. This pretreatment method yielded the same seed-free preparations with Aβ1–42.
Time Courses of Aβ Aggregation with Test Compounds. The test compounds (1-8 and 10) were added as stock solutions (0.2–20 mM in isopropyl alcohol) to phosphate buffer (100 mM sodium phosphate/600 mM NaCl, pH 7.2) to achieve the desired concentration of test compound (for compounds 1-8) or precursor (compound 10). The isopropyl alcohol concentration was 1% throughout. Aggregation experiments were initiated by adding an equal volume of seed-free Aβ1–40 (200 μM) in aqueous NaOH (pH 10.5) to the aldehyde solution described above at 4°C. Thus, the assay solution comprised Aβ (100 μM) and test compound (1–100 μM) in phosphate buffer (50 mM sodium phosphate/300 mM NaCl, pH 7.5/0.5% vol/vol isopropyl alcohol). Each assay solution was divided into several aliquots (100 μl) and incubated at 37°C without shaking. Aliquots (20 μl) were removed from the assay solutions at desired time points, vortexed briefly, and added to a thioflavin T (TfT) solution (480 μl, 20 μM) in sodium phosphate (50 mM, pH 7.5). The fluorescence of the solution was measured by using an Aviv ATF-105 fluorometer (excitation, 440 nm; emission, 485 nm). The fluorescence of TfT is reported to be proportional to the amount of aggregate in solution (29). The Aβ aggregation experiments were reproducible in that the time required for the reactions to reach 50% completion (where the amplitude of the reaction was 50% of its value at its plateau) varied by <20% between experiments. In some cases, the reactions had very small amplitudes and did not reach completion. For these samples, we note that their small amplitudes were reproducible.
AFM. Aliquots (20 μl) were removed from the assay solutions described in the aggregation experiments above and placed on freshly cleaved mica (1 × 1 cm). After 1 min, the solvent was absorbed into filter paper. Water (30 μl) was then placed on the mica surface and immediately absorbed into filter paper. This process was repeated twice to remove salts from the sample. Tapping-mode images were obtained on a MultiMode scanning probe microscope with a Nanoscope IIIa controller (Veeco, Woodbury, NY).
Sodium Borohydride Reduction of Aβ-Aldehyde Adducts. A solution of Aβ (100 μM) in phosphate buffer (prepared as described above, pH 7.5) and aldehyde 1 (100 μM) containing 0.5% vol/vol isopropyl alcohol was incubated for 3 h at 37°C without shaking. A solution of sodium borohydride (NaBH4, 280 mM) in water was then added to the assay solution for 30 min. Under these conditions (final NaBH4 concentration, 8.2 mM), we surmise that Aβ-1(2) adducts and any remaining 1 and 2 in solution would be reduced. The NaBH4-treated sample was centrifuged (2 h at 100,000 × g), the supernatant was decanted and saved, and the pellet was washed three times with phosphate buffer. It should be noted that centrifuging before separately reducing the pellet and supernatant (NaBH4) afforded the same HPLC results (see below).
HPLC Quantification of Aβ and Reduced Aβ-1(2) Adducts. Both the supernatant and the pellet from the reduction experiment discussed above were analyzed by HPLC. The HPLC system comprised a Waters 600 controller and pump and a Waters 2487 detector measuring absorbance at 214 nm. The column was an Agilent Zorbax C3 reverse-phase HPLC column (5 μm, 4.6 × 150 mm; Agilent Technologies, Palo Alto, CA). The mobile phase involved a gradient elution [flow rate, 1 ml/min; gradient, 1–51% B for 25 min, then to 100% B for 15 min (A, 0.05% trifluoroacetic acid in H2O; B, 0.05% trifluoroacetic acid in acetonitrile)]. The supernatant was injected directly onto the HPLC system, whereas the pellet was taken up in phosphate buffer (50 mM sodium phosphate/300 mM NaCl, pH 7.5, 100 μl) and sonicated for 20 min before injection. Peaks were assigned as either Aβ or Aβ-1(2) (monoadduct) by collection of the eluted peaks and analysis by matrix-assisted laser desorption ionization MS conducted by the Center for Mass Spectrometry at the Scripps Research Institute. The retention times (RT) were ≈22.5 min for Aβ and ≈28.5 min for Aβ-1(2). The concentration of the bisadduct, Aβ-1(2)2 was not high enough to be identified in this way. Two peaks eluting at ≈31 and 33 min have tentatively been assigned as the bisadducts, pending further experiments to identify them.
Estimating the Critical Concentrations for Aggregation of the Aβ-1(2) Adducts. AsolutionofAβ (100 μM) and 1 (100 μM) in phosphate buffer (prepared as described above, pH 7.5) was incubated for 16 h at 37°C without shaking. A solution of NaBH4 (280 mM) in water was then added to the assay solution for 30 min. The NaBH4-treated sample was centrifuged (2 h, 100,000 × g), and the supernatant was decanted. A solution of fluorescamine (30, 31) in acetone (50 μl, 3 mg/ml) was added to this supernatant (150 μl) and the resulting solution was vortexed (1 min) and analyzed by HPLC by using a Hitachi D-7000 system equipped with a pump, and a fluorescence detector (excitation, 400 nm; emission, 480 nm). The column and gradient elution were as described above. The peak corresponding to fluorescamine-modified Aβ (RT ≈ 27 min) was identified by comparing its elution time with the elution time of an authentic sample derivatized with fluorescamine. The monoadduct Aβ-1(2) was identified by collection of the eluted peak (RT = 30.5 min) and analysis by matrix-assisted laser desorption ionization MS as described above. The concentration of the fluorescamine-derivatized Aβ adduct in the supernatant was determined by using a calibration curve, assuming that fluorescamine-labeled Aβ and its adducts equally (if not, an error of at most 33% would be introduced, because Aβ has three reactive sites and an Aβ monoadduct has two).
Metabolite-Initiated Aβ Aggregation in the Presence of Albumin and Transthyretin. Solutions of seed-free Aβ (100 μM) in aqueous NaOH (pH 10.5) or aqueous NaOH alone (pH 10.5) were each added to equal volumes of solutions of human serum albumin (30 μM) or transthyretin (TTR, 7.2 μM) in phosphate buffer (100 mM sodium phosphate/600 mM NaCl, pH 7.2), resulting in four solutions. To these solutions was added either a solution of 1 (10 mM) in isopropyl alcohol or pure isopropyl alcohol, such that the final concentration of 1 was 50 μM and/or the final concentration of isopropyl alcohol was 0.5%. This procedure resulted in the preparation of three pairs of solutions: one with albumin (15 μM) or TTR (3.6 μM) and 1 (50 μM), one with albumin (15 μM) or TTR (3.6 μM) and Aβ (50 μM), and one with albumin (15 μM) or TTR (3.6 μM) and both Aβ (50 μM) and 1 (50 μM). The solutions were divided into several aliquots (100 μl), and aggregation at 37°C was followed by TfT fluorescence as described above.
Clinical Brain Samples. Frontal cortex brain specimens (four from Alzheimer's brains and seven from control brains) were obtained from the Institute for Brain Aging and Dementia Tissue Repository at the University of California, Irvine. The AD diagnosis was based on clinical cognitive testing according to the Braak dementia scale and was confirmed by postmortem examination based on plaque and neurofibrillary tangle criteria and the absence of other types of pathology.
Quantification of the Concentrations of 1 and 2 in Brain Tissue. Brain tissue specimens (400–600 mg) from above were homogenized, diluted with water (1 ml), and extracted with hexane (3 × 5 ml). The combined organic fractions were evaporated in vacuo. The residue was resuspended in a suspension of zinc powder in glacial acetic acid (1 ml, 1 mg/ml) and stirred for 2 h. The resulting suspension was diluted with water (1 ml) and extracted with dichloromethane (3 × 5 ml). The combined organic fractions were evaporated in vacuo. The residue was dissolved in methanol (1 ml), and a solution of 2,4-dinitrophenylhydrazine (0.01 M, 200 μl) in acidic ethanol (0.8% HCl) was added. This solution was allowed to stand overnight at 37°C and then analyzed by inline HPLC-MS on a Hitachi D-7000 system equipped with a Hitachi L-7100 pump, a Hitachi L-7455 diode array detector measuring absorbance at 360 nm, and an inline Hitachi M-8000 ion trap mass spectrometer. The column was a reverse-phase Vydac C18 MS column (Vydac, Hesperia, CA), and the mobile phase was isocratic (75% acetonitrile/5% water/20% methanol) with a flow rate of 1 ml/min. Data were recorded with hitachi d-7000 software on a Dell PC. Peak areas measured at least in duplicate were converted to concentrations by comparison with authentic synthetic material (22). Mass analysis was performed with an Hitachi M-8000 ESI mass analyzer and 3dq/ms station software.
Results and Discussion
Incubation of either ketoaldehyde 1 or its aldol product 2 with seed-free Aβ1–40 (pH 7.5, 37°C, no stirring) leads to Aβ amyloidogenesis (Fig. 1 A and B). In contrast, no Aβ amyloidogenesis is observed within 72 h in the absence of 1 and 2. The extent and rate of amyloidogenesis depended on the concentration of 1 or 2. Amyloidogenesis was followed by TfT fluorescence, which is proportional to the amount of amyloid formed (29). AFM images (Fig. 1C) show that Aβ in the presence of 1 (50 μM) forms abundant, roughly spherical assemblies 5–8 nm in diameter within 1 h. These assemblies are similar to those thought to be the principal neurotoxic Aβ aggregates in AD (8, 13, 32, 33). Comparable aggregates are barely detectable without 1 (Fig. 1D). AFM time courses of Aβ aggregation with and without 1 are shown in Fig. 6, which is published as supporting information on the PNAS web site. They show that aggregates began forming within 10 min of initiating the aggregation reaction of Aβ (100 μM) with 1 (50 μM). The number of aggregates adsorbed to the substrate peaked at 1 h, after which fewer, but larger, aggregates were observed. Few aggregates were observed in the sample of Aβ in the absence of 1, even after 64 h. It has been shown that 1 is converted to 2 in the presence of whole blood, plasma, serum, and several amino acids (22). Preliminary experiments indicate that Aβ also catalyzes the conversion of 1 to 2 by Schiff base formation with Aβ (data not shown). We therefore make no distinction between Aβ-1 and Aβ-2 and represent monoadducts of Aβ with cholesterol ozonolysis products as Aβ-1(2).
The formation of fibrillar aggregates is a hallmark of AD (34). To determine whether the spherical aggregates formed by incubation of Aβ with 1 were competent to form fibrils, fibrillar seeds (1% wt/wt) were added to the aggregation reaction after a 48 h incubation of Aβ (100 μM) with 1 (50 μM). On seed addition, the spherical aggregates rapidly converted into fibrils, as indicated both by the jump in TfT fluorescence (Fig. 2A) and by AFM images obtained before seeding (Fig. 2B, which shows a few large aggregates, consistent with the time course in Fig. 6) and 1 h after seeding (Fig. 2C). This process is much slower in the absence of 1, as revealed by AFM time courses of Aβ aggregation with and without 1 seeded with fibrils after 48 h (see Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
(A) Aβ aggregation at several concentrations of 1 was monitored for 48 h, as described in Fig. 1A. After 48 h, 1% wt/wt of Aβ seeds was added, and aggregation was monitored for another week. (B and C) AFM images obtained just before (B) and 1 h after (C) seed addition. The images shown here are representative of two experiments. (D) Aggregation of Aβ1–40 (50 μM, black lines), 1–42 (50 μM, red lines), Aβ1–40:1–42 in a 3:1 ratio (50 μM, green lines) were monitored as described in Fig. 1A (but note the lower Aβ concentrations).
Aβ exists as a mixture of 40 and 42 residue peptides in vivo (35). As with Aβ1–40 alone, incubation of 1 with a mixture of Aβ1–40 and Aβ1–42 (3:1 ratio, 50 μM) initiates aggregation (Fig. 2D). The extent of aggregation in the presence of 1 with this peptide composition is slightly (10–20%) but consistently higher than that of Aβ1–40 alone (Fig. 2D). A comparison of Aβ1–40 with Aβ1–42 aggregation versus the concentration of 1 reveals that the rate of Aβ1–40 amyloidogenesis becomes similar to that of Aβ1–42 when the concentration of 1 is ≥50 μM (Fig. 2D). These data support the hypothesis that metabolite-modified Aβ1–40 could be as efficient at initiating AD pathology as familial mutations that lead to increased concentrations of Aβ1–42 relative to Aβ1–40 (35).
To assess to what extent covalent modification of Aβ occurs during incubation with 1, aggregation reactions of Aβ1–40 (100 μM) in the presence of 1 (100 μM) that had been incubated at 37°C for 3 h were centrifuged (100,000 × g), and the supernatant and pellet were separated. NaBH4 was added to the supernatant and pellet fractions to reduce the presumed Schiff base adducts of Aβ with 1(2) and any unreacted 1(2). HPLC analysis of the NaBH4-treated supernatant and pellet fractions revealed that 6% of the initial quantity of Aβ was present in the pellet as unmodified Aβ, whereas 8% was present in the pellet as Aβ-1(2) (Fig. 3A). Another 1% could be the bisadduct Aβ-1(2)2 (eluting at 31 and 33 min in Fig. 3A), but the small quantity precluded its identification. Proteolysis of the Aβ adducts followed by preliminary mass spectrometry analysis (see Supporting Text, which is published as supporting information on the PNAS web site) indicate modification at Lys-16, Lys-28, and the N terminus of the Aβ in the pellet, but it is not yet possible to know which adducts were the most amyloidogenic. The distribution of adducts among the various sites at which Aβ is susceptible to modification requires further investigation.
Fig. 3.
(A) HPLC traces from the NaBH4-reduced supernatant and pellet of Aβ (100 μM) incubated with 1 (100 μM) for 3 h.(B) Fluorescence-detection HPLC trace from the NaBH4-reduced supernatant of Aβ (100 μM) incubated with 1 (100 μM) for 16 h, after fluorescamine derivatization. Fluorescence is measured in arbitrary units.
Given that Aβ misfolding is generally described by a nucleation-dependent polymerization model, the concentration of soluble Aβ remaining after an aggregation reaction has reached completion is equivalent to the critical concentration for aggregation [the concentration below which aggregation is negligible (10)]. No Aβ-1(2) adducts were found in the NaBH4-treated supernatant samples analyzed by HPLC with UV detection. This finding implies that the critical concentration for Aβ-1(2) adducts was below the limit of detection of this method (≈500 nM). The limit of detection can be decreased to ≈50 nM by derivatizing Aβ with fluorescamine and by using fluorescence detection for the HPLC analysis. To determine whether the critical concentration of Aβ-1(2) could be determined by using fluorescamine derivatization, Aβ (100 μM) was incubated in the presence of 1 (100 μM) for 16 h and centrifuged at 100,000 × g. The supernatant and pellet were separated, treated with NaBH4 as described above, and derivatized with fluorescamine. HPLC analysis with fluorescence detection (Fig. 3B) showed that the concentration of Aβ-1(2) was still near the limit of detection; a small shoulder on the main fluorescamine-derivatized Aβ peak, corresponding to a concentration of at most 90 nM, was the only peak that eluted at the retention time corresponding to fluorescamine-derivatized Aβ-1(2)(RT ≈ 30.5 min). The critical concentration for Aβ-1(2) was therefore at most 90 nM. This value is much lower than the ≈15 μM critical concentration reported for unmodified Aβ (10, 36). This dramatic lowering of the Aβ critical concentration on metabolite modification may explain how physiological concentrations of Aβ (typically in the nanomolar range) could form amyloid in individuals lacking predisposing mutations.
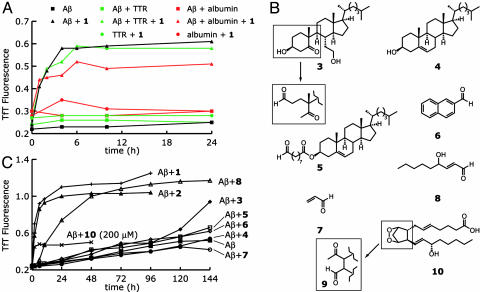
In the brain, Aβ has to compete with other, much more abundant proteins to be modified by metabolites. To determine whether the presence of other proteins could affect Aβ adduct formation, Aβ was incubated with 1 in the presence of human serum albumin (15 μM, 885 μM in reactive Lys residues) and TTR (3.6 μM, 115 μM in reactive Lys residues). TTR had a negligible effect on Aβ amyloidogenesis in the presence of 1, whereas albumin reduced the extent of amyloidogenesis by ≈15% (Fig. 4A). It is reasonable to surmise that the reactions of albumin and TTR with 1 are essentially reversible, leaving the concentration of 1 available for reaction with Aβ roughly unchanged. We also note that 1 does not induce albumin or TTR to form amyloid, perhaps because their critical concentrations for aggregation are not lowered substantially. The origin of the selective effect of 1 and 2 on Aβ aggregation requires further investigation.
Fig. 4.
(A) Influence of albumin (15 μM, 885 μM in Lys residues) or TTR (3.6 μM, 115μM in Lys residues) on Aβ (50 μM, pH 7.5, 37°C) amyloidogenesis with 1 (50 μM). (B) Compounds related to 1 and 2 or otherwise of interest for their potential to accelerate Aβ amyloidogenesis (see text). (C) Aβ1–40 (100 μM) aggregation in the presence of 50μM concentrations of 3-8 was monitored as in Fig. 1 A. Compound 10, the precursor of 9, was used at 200 μM, which should yield an ≈40 μM concentration of 9.
To determine whether 1 and 2 are unique in their ability to accelerate Aβ amyloidogenesis, we evaluated a panel of other small molecules for their ability to enhance Aβ misfolding (Fig. 4B). Of compounds 3-9, only 8, 4-hydroxynonenal, accelerated Aβ aggregation to an extent similar to 1 and 2 (Fig. 4C). It is plausible that the reactivity of 8 with Aβ may be enhanced, because in addition to the 1,2 addition between the Lys ε-amine and the aldehyde group, it can undergo a Michael-type alkylation of Aβ through both His and Lys side chains (25). Seco-prostaglandin 9 is a γ-ketoaldehyde that is a minor byproduct arising from cyclooxygenase I- and II-mediated oxidation of arachidonic acid. This molecule has previously been shown to enhance Aβ1–42 oligomer formation (37). Aldehyde 9 is not commercially available, but its endoperoxide precursor 10 is (Fig. 4B). Salomon and coworkers (38) have shown that the prostaglandin endoperoxide 10 undergoes a solvent-induced fragmentation in phosphate buffer (0.2 M sodium phosphate, pH 7.9) at 37°Ctogive 9 in 22% yield. Fig. 4C shows that the extent of Aβ amyloidogenesis in the presence of 10 (200 μM), which, according to the data above, should yield 9 at ≈40 μM, is ≈25% of that seen with 1, 2, and 8. (Note that the concentration of 9 was not directly measured.) Alcohol 3 did not affect Aβ amyloidogenicity during the first 120 h of incubation, demonstrating that the aldehyde is a critical structural feature for metabolite-initiated Aβ amyloidogenesis on this timescale. However, the slight effect of 3 on the course of the aggregation reaction observed after 120 h becomes much more pronounced after 168 h (the TfT fluorescence increases to 1.1 at 168 h; data not shown). Although we have not yet sought the cause of this phenomenon, we speculate that it may be due to the formation of an aldehyde by a retro-aldol reaction in the A ring of 3 (Fig. 4B). The 9-oxononanoyl ester of cholesterol (5), derived from low-density lipoprotein oxidation (39), also had a negligible effect on Aβ amyloidogenicity. Preliminary experiments indicate that this may be due to its not reacting efficiently with Aβ (data not shown). Aldehyde 6, which also did not accelerate Aβ amyloidogenesis, was investigated because Mihara and Takahashi (40) had shown that increasing the hydrophobicity of the termini of coiled-coil peptides with similar structures accelerated their conversion to amyloid. Cholesterol (4), which lacks an aldehyde, was unable to accelerate Aβ aggregation. These data suggest that an aldehyde is necessary but not sufficient to trigger amyloidogenesis. A hydrophobic structural component in the correct orientation with respect to the aldehyde is also required.
Given that 1 and 2 greatly enhance Aβ amyloidogenesis, we turned to an analysis of brain tissue to see whether these molecules could be detected. Eleven age-matched, frozen, and unfixed brain specimens (four from AD brains and seven from brains without any pathology) were analyzed as described (22). In brief, the aldehydes were extracted from brain tissue, derivatized with 2,4-dinitrophenylhydrazine, and analyzed by LC-MS. Peaks corresponding to the hydrazones of 1 and 2 were present in the chromatograms of all but two of the samples, with combined concentrations as high as 1.38 pmol/mg (Fig. 5A). In one sample, the concentrations of 1 and 2 were high enough to be unambiguously detected by MS (Fig. 5B). The average combined concentrations of 1 and 2 were approximately the same in the AD (0.44 pmol/mg) and control (0.35 pmol/mg) brains. This similarity in the concentrations of cholesterol ozonolysis products is not inconsistent with a model for metabolite-initiated misfolding whereby the levels of 1 and 2 transiently increase, initiating the nucleation of Aβ amyloidogenesis. Once nucleated, the propagation of Aβ amyloidogenesis is fast, making the initiating event traceless (see below).
Fig. 5.
(A) Quantification of the concentrations of 1 and 2 in four human AD and seven age-matched control brains by HPLC analysis after derivatization with 2,4-dinitrophenylhydrazine. The combined concentrations of 1 and 2 in each brain are indicated by black circles. The averages and standard deviations are indicated by the dashes and the vertical lines, respectively. (B) LC-MS chromatogram detecting the total ion count at mass/charge = 597, which corresponds to the 2,4-dinitrophenylhydrazones of 1 and 2.
The results above demonstrate that metabolite modification of a peptide or protein must now be considered in addition to sequence and environment as factors capable of influencing the conformational energy landscape and aggregation propensity of a given polypeptide chain. Metabolite modification of physiological proteins has precedent in the case of the hedgehog tissue-patterning family of proteins, which must be esterified by cholesterol for their function (41). Although other modifications of Aβ have been implicated in amyloidogenesis [e.g., Met oxidation, glycation, proteolysis of the hydrophilic N terminus, deamination, and pyroGlu formation (42)], none of these match the enhancement of amyloidogenicity seen with 1, 2, or 8 [although they can be significant for other amyloidogenic peptides (43)]. Schiff base formation by 1(2) converts either or both of the Lys side chains in Aβ to the most hydrophobic of all of the α-amino acid side chains, rationalizing the lowered critical concentration and accelerated amyloidogenicity. Covalent modifications of Aβ and α-synuclein that inhibit amyloidogenesis are also known (44, 45).
The convergence of risk factors for AD and atherosclerosis (18), including the apoE ε4 genotype, hypercholesterolemia, and inflammation, suggest overlapping causes. Abnormal metabolites 1(2) have already been associated with atherosclerosis (22), and the results described above suggest a scenario where inflammation (not necessarily initiated by AD) leads to a transient or sustained increase in the concentration of aldehydes 1, 2, and 8 that initiates or exacerbates the process of Aβ amyloidogenesis, leading ultimately to the neurodegeneration associated with AD (14, 15). The ozonolysis of cholesterol may be the chemical link between atherosclerosis and AD.
The concepts presented here differ from the usual view of how changes in the chemistry of the organism lead to disease. Typically, proteins and nucleic acids change their function by mutation and/or up- or down-regulation, leading to diseases like the hemoglobinopathies or cancer. There are also several diseases that result from fluctuations in the concentration of one or more chemically unaltered metabolites, e.g., glycolipids in lysosomal storage diseases (46). Here we suggest a new paradigm where a ubiquitous metabolite (e.g., cholesterol) is transformed into an abnormal metabolite with unusual reactivity [e.g., 1(2)], whereas the protein remains unaltered in sequence and concentration. Pathology is initiated only when the new functional group(s) on the reactive metabolite forms a covalent bond with a protein or related macromolecule. This hypothesis is supported by the ability of 1(2) and 8 to enhance Aβ amyloidogenicity in vitro and by the detection of 1(2) in human brains reported herein. In some instances, metabolite initiation of misfolding may be traceless because the covalent modification may accelerate the slow (nucleation) step but not the fast propagation step. Propagation involves the energetically favorable addition of the unmodified amyloidogenic peptide to the metabolite-modified seed, leading to substantial dilution of Aβ-1(2) or Aβ-8 by Aβ, making metabolite-modified Aβ difficult or impossible to detect. In other words, once the seed is formed in a transient event there is no further need for 1(2) or 8. The detection of metabolite-modified Aβ adducts, e.g., Aβ-1(2), is rendered even more challenging by their hydrolytic instability.
In summary, we have shown that cholesterol ozonolysis products 1 and 2 exist in human brains. These and related metabolites (e.g., 8) modify Aβ, lowering its critical concentration and accelerating its amyloidogenesis. These modifications perturb the conformational energy landscape of Aβ to the extent that Aβ1–40 becomes as aggregation-prone as the Aβ1–42-rich peptide distribution found in familial Alzheimer's patients. Taken together, these results indicate that metabolite-initiated Aβ amyloidogenesis may explain the genesis of sporadic AD and related amyloidoses.
Supplementary Material
Acknowledgments
We thank P. Herrling (Novartis), A. R. Hurshman and D. M. Fowler (Scripps) for helpful discussions, M. R. Ghadiri for use of his AFM, and the Skaggs Institute of Chemical Biology and the Lita Annenberg Hazen Foundation for financial support.
Abbreviations: AD, Alzheimer's disease; Aβ, amyloid β peptide; AFM, atomic force microscopy; RT, retention time; LC, liquid chromatography; TTR, transthyretin; TfT, thioflavin T.
References
- 1.Anfinsen, C. B. (1973) Science 181, 223–230. [DOI] [PubMed] [Google Scholar]
- 2.Colon, W. & Kelly, J. W. (1992) Biochemistry 31, 8654–8660. [DOI] [PubMed] [Google Scholar]
- 3.Kelly, J. W. (1996) Curr. Opin. Struct. Biol. 6, 11–17. [DOI] [PubMed] [Google Scholar]
- 4.Hammarstrom, P., Jiang, X. & Kelly, J. W. (2002) Proc. Natl. Acad. Sci. USA 99, 16427–16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, D. R., Sunde, M., Bellotti, V., Robinson, C. V., Hutchinson, W. L., Fraser, P. E., Hawkins, P. N., Dobson, C. M., Radford, S. E., Blake, C. C. F. & Pepys, M. B. (1997) Nature 385, 787–793. [DOI] [PubMed] [Google Scholar]
- 6.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 7.Selkoe, D. J. (1997) Science 275, 630–631. [DOI] [PubMed] [Google Scholar]
- 8.Lamber, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 6448–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitan, G., Kirkitadze, M. D., Lomakin, A., Vollers, S. S., Benedek, G. B. & Teplow, D. B. (2003) Proc. Natl. Acad. Sci. USA 100, 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harper, J. D. & Lansbury, P. T., Jr. (1997) Annu. Rev. Biochem. 66, 385–407. [DOI] [PubMed] [Google Scholar]
- 11.Lashuel, H. A., Hartley, D., Petre, B. M., Walz, T. & Lansbury, P. T. (2002) Nature 418, 291. [DOI] [PubMed] [Google Scholar]
- 12.Soreghan, B., Joseph, K. & Glabe, C. (1994) J. Biol. Chem. 269, 28551–28554. [PubMed] [Google Scholar]
- 13.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J. & Selkoe, D. J. (2002) Nature 416, 535–539. [DOI] [PubMed] [Google Scholar]
- 14.Meda, L., Cassatella, M. A., Szendrei, G. I., Otvos, L. J., Baron, P., Villalba, M., Ferrari, D. & Rossi, F. (1995) Nature 374, 647–650. [DOI] [PubMed] [Google Scholar]
- 15.Bradt, B. M., Kolb, W. P. & Cooper, N. R. (1998) J. Exp. Med. 188, 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eikelenboom, P., Zhan, S.-S., van Gool, W. A. & Allsop, D. (1994) Trends Pharmacol. Sci. 15, 447–450. [DOI] [PubMed] [Google Scholar]
- 17.Jarvik, G. P., Wijsman, E. M., Kukull, W. A., Schellenberg, G. D., Yu, C. & Larson, E. (1995) Neurology 45, 1092–1096. [DOI] [PubMed] [Google Scholar]
- 18.Hartmann, T. (2001) Trends Neurosci. 24, S45–S48. [DOI] [PubMed] [Google Scholar]
- 19.Refolo, L. M., Pappolla, M. A., Malester, B., LaFrancois, J., Bryant-Thomas, T., Wang, R., Tint, G. S., Sambamurti, K. & Duff, K. (2000) Neurobiol. Dis. 7, 321–331. [DOI] [PubMed] [Google Scholar]
- 20.Simons, M., Keller, P., De Strooper, B., Beyreuther, K., Dotti, C. G. & Simons, K. (1998) Proc. Natl. Acad. Sci. USA 95, 6460–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wentworth, P., Jr., McDunn, J. E., Wentworth, A. D., Takeuchi, C., Nieva, J., Jones, T., Bautista, C., Ruedi, J. M., Gutierrez, A., Janda, K. D., et al. (2002) Science 298, 2195–2199. [DOI] [PubMed] [Google Scholar]
- 22.Wentworth, P., Jr., Nieva, J., Galve, R., Takeuchi, C., Wenworth, A. D., Dilley, R. B., DeLaria, G. A., Saven, A., Babior, B. M., Janda, K. D., et al. (2003) Science 302, 1053–1056. [DOI] [PubMed] [Google Scholar]
- 23.Sayre, L. M., Zelasko, D. A., Harris, P. L. R., Perry, G., Salomon, R. G. & Smith, M. A. (1997) J. Neurochem. 68, 2092–2097. [DOI] [PubMed] [Google Scholar]
- 24.Ando, Y., Brannstrom, T., Uchida, K., Nyhlin, N., Nasman, B., Suhr, O., Yamashita, T., Olsson, T., El Salhy, M., Uchino, M. & Ando, M. (1998) J. Neurol. Sci. 156, 172–176. [DOI] [PubMed] [Google Scholar]
- 25.Magni, F., Galbusera, C., Tremolada, L., Ferrarese, C. & Kienle, M. G. (2002) Rapid Commun. Mass Spec. 16, 1485–1493. [DOI] [PubMed] [Google Scholar]
- 26.Markesbery, W. R. & Lovell, M. A. (1998) Neurobiol. Aging 19, 33–36. [DOI] [PubMed] [Google Scholar]
- 27.Zarkovic, K. (2003) Mol. Aspects Med. 24, 293–303. [DOI] [PubMed] [Google Scholar]
- 28.Fezoui, Y., Hartley, D. M., Harper, J. D., Khurana, R., Walsh, D. M., Condron, M. M., Selkoe, D. J., Lansbury, P. T., Jr., Fink, A. L. & Teplow, D. B. (2000) Amyloid 7, 166–178. [DOI] [PubMed] [Google Scholar]
- 29.LeVine, H., III (1999) Methods Enzymol. 309, 274–284. [DOI] [PubMed] [Google Scholar]
- 30.Lorenzan, A. & Kennedy, S. W. (1993) Anal. Biochem. 214, 346–348. [DOI] [PubMed] [Google Scholar]
- 31.Bantan-Polak, T., Kassai, M. & Grant, K. B. (2001) Anal. Biochem. 297, 128–136. [DOI] [PubMed] [Google Scholar]
- 32.Wang, H.-W., Pasternak, J. F., Kuo, H., Ristic, H., Lambert, M. P., Chromy, B., Viola, K. L., Klein, W. L., Stine, W. B., Krafft, G. A. & Trommer, B. L. (2002) Brain Res. 924, 133–140. [DOI] [PubMed] [Google Scholar]
- 33.Caughey, B. & Lansbury, P. T., Jr. (2003) Annu. Rev. Neurosci. 26, 267–298. [DOI] [PubMed] [Google Scholar]
- 34.Sipe, J. D. (1994) Crit. Rev. Clin. Lab. Sci. 31, 325–354. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki, N., Cheung, T. T., Cai, X. D., Odaka, A., Otvos, L. J., Eckman, C., Golde, T. E. & Younkin, S. G. (1994) Science 264, 1336–1340. [DOI] [PubMed] [Google Scholar]
- 36.Sengupta, P., Garai, K., Sahoo, B., Shi, Y., Callaway, D. J. E. & Maiti, S. (2003) Biochemistry 42, 10506–10513. [DOI] [PubMed] [Google Scholar]
- 37.Boutaud, O., Ou, J. J., Chaurand, P., Caprioli, R. M., Montine, T. J. & Oates, J. A. (2002) J. Neurochem. 82, 1003–1006. [DOI] [PubMed] [Google Scholar]
- 38.Salomon, R. G., Miller, D. B., Zagorski, M. G. & Coughlin, D. J. (1984) J. Am. Chem. Soc. 106, 6049–6060. [Google Scholar]
- 39.Kawai, Y., Saito, A., Shibata, N., Kobayashi, M., Yamada, S., Osawa, T. & Uchida, K. (2003) J. Biol. Chem. 278, 21040–21049. [DOI] [PubMed] [Google Scholar]
- 40.Mihara, H. & Takahashi, Y. (1997) Curr. Opin. Struct. Biol. 7, 501–508. [DOI] [PubMed] [Google Scholar]
- 41.Porter, J. A., Young, K. E. & Beachy, P. A. (1996) Science 274, 255–259. [DOI] [PubMed] [Google Scholar]
- 42.Head, E., Garzon-Rodriguez, W., Johnson, J. K., Lott, I. T., Cotman, C. W. & Glabe, C. (2001) Neurobiol. Dis. 8, 792–806. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson, M. R., Driscoll, M. & Raleigh, D. P. (2002) Protein Sci. 11, 342–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conway, K. A., Rachet, J.-C., Bieganski, R. M. & Lansbury, P. T., Jr. (2001) Science 294, 1346–1349. [DOI] [PubMed] [Google Scholar]
- 45.Dickerson, T. J. & Janda, K. D. (2003) Proc. Natl. Acad. Sci. USA 100, 8182–8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawkar, A., Cheng, W.-C., Beutler, E., Wong, C. H., Balch, W. E. & Kelly, J. W. (2002) Proc. Natl. Acad. Sci. USA 99, 15428–15433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.