SUMMARY
Microvascular endothelial cells (ECs) within different tissues are endowed with distinct but as yet unrecognized structural, phenotypic, and functional attributes. We devised EC purification, cultivation, profiling, and transplantation models that establish tissue-specific molecular libraries of ECs devoid of lymphatic ECs or parenchymal cells. These libraries identify attributes that confer ECs with their organotypic features. We show that clusters of transcription factors, angiocrine growth factors, adhesion molecules, and chemokines are expressed in unique combinations by ECs of each organ. Furthermore, ECs respond distinctly in tissue regeneration models, hepatectomy, and myeloablation. To test the data set, we developed a transplantation model that employs generic ECs differentiated from embryonic stem cells. Transplanted generic ECs engraft into regenerating tissues and acquire features of organotypic ECs. Collectively, we demonstrate the utility of informational databases of ECs toward uncovering the extravascular and intrinsic signals that define EC heterogeneity. These factors could be exploited therapeutically to engineer tissue-specific ECs for regeneration.
INTRODUCTION
Endothelial cells (ECs) are a heterogeneous population of cells, not only with respect to the macrovasculature, including arterial, venous, and lymphatic systems (Aird, 2007), but also among microvascular capillary beds of different organs. The unique properties of ECs in the brain and kidney glomeruli have long been appreciated. Capillary ECs of the blood brain barrier (BBB) form a restrictive environment for passage between the brain tissue and the circulating blood. Many of the trafficking processes that are passive in other vascular beds are tightly controlled in the brain (Rubin and Staddon, 1999). As opposed to the BBB, the capillary ECs of the kidney glomeruli are fenestrated for the filtration of the blood (Churg and Grishman, 1975). Although the structural differences between these representative organs are well described, the molecular signatures of the microvascular ECs and the extravascular and intrinsic signals that dictate their unique tissue-specific properties are poorly understood.
In vitro studies have advanced the concept that tissue-specific ECs respond uniquely to stimuli (Molema, 2010; Müller et al., 2002). During inflammatory responses, TNF-α stimulation elicits discrete responses from the ECs of various organs. Although the interpretations of these in vitro studies are appropriately limited (Børsum et al., 1982), they suggest that EC heterogeneity in vivo is partially determined by intrinsic signals and maintained after ECs are removed from their microenvironment. ECs are exposed to a large and dynamic cadre of stimuli, including blood-borne cytokines, extracellular matrix proteins, and biophysical signals. Thus, reductive in vitro studies cannot address EC heterogeneity sufficiently because without an in vivo reference, the results will remain ambiguous.
It is now evident that the endothelium is more than an inert conduit for blood flow. Tissue-specific ECs, by expression of unique repertoires of trophic growth factors, known as angiocrine factors, support the homeostasis and regeneration of stem and progenitor cells after tissue injury. Notably, sinusoidal ECs in the bone marrow (BM), by expression of Notch-ligands (Butler et al., 2010), epidermal growth factor (EGF) (Doan et al., 2013a), pleiotrophin (Himburg et al., 2012), and stem cell factor (SCF, Kit-ligand) support hematopoiesis (Butler et al., 2010; Ding et al., 2012; Hooper et al., 2009). Moreover, sinusoidal ECs in the liver express Wnt2 and hepatocyte growth factor (HGF) to orchestrate liver regeneration after 70% partial hepatectomy (Ding et al., 2010). Furthermore lung, but not liver ECs, supply MMP14 and EGF-like ligands that support alveolar regeneration (Ding et al., 2011). Thus, the microvascular ECs within each organ are unique and may be programmed to satisfy the angiocrine function and metabolic demands of that particular organ.
Nonetheless, the signatures of organ-specific ECs and microenvironmental cues that sustain those signatures remain poorly understood. Transcriptional profiling has been employed to identify druggable targets on tumor ECs (Peters et al., 2007), whereas others have focused on arterial-venous distinctions (Swift and Weinstein, 2009). However, these studies did not achieve a global view of the vascular state. Furthermore, existing approaches for the isolation of tissue-specific microvasculature result in contamination with various perivascular cells and lymphatic ECs. As such, sample purity is paramount for the meaningful identification of the molecular signatures that determine the heterogeneity of microvascular ECs. To this end, we have developed an approach to purify capillary ECs devoid of any contaminating lymphatic ECs or parenchymal cells. Employing microarray profiling, we have developed informational databases of steady-state and regenerating capillary ECs, which serve as platforms to unravel the molecular determinants of vascular heterogeneity. We demonstrate that the microvascular bed of each organ is composed of specialized ECs, endowed with unique modules of angiocrine factors, adhesion molecules, chemokines, transcription factors (TFs), and metabolic profiles. Mining of these databases will enable identification of unique factors deployed by the tissue-specific microvascular ECs that sustain tissue homeostasis at steady state and regeneration during organ repair.
RESULTS
Intravital Staining Establishes Multiparameter Definitions for Tissue-Specific Capillary ECs
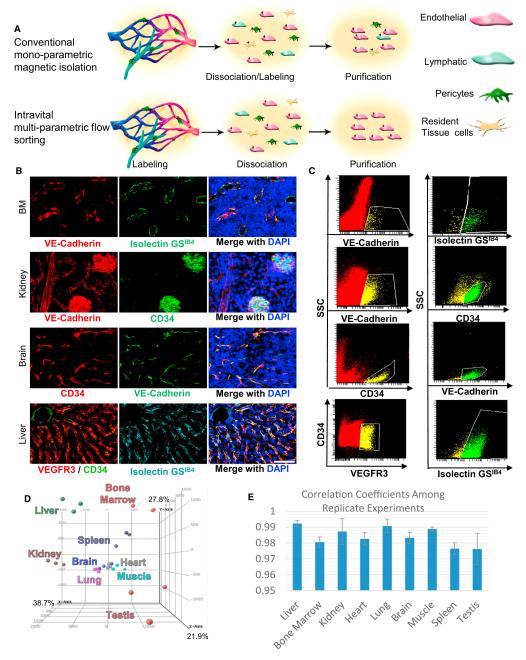
Conventional monoparametric labeling with magnetic particles for isolation of tissue-specific capillaries is incapable of distinguishing lymphatic ECs, clusters of two or more contaminating cells, and hematopoietic and parenchymal cells sharing markers with ECs (Figure 1A). In order to profile tissue-specific microvascular ECs devoid of lymphatic ECs and perivascular and parenchymal cells, we established a high fidelity approach to purify and immediately profile ECs from an in vivo source. Numerous antibodies to EC markers were assayed for their ability to transit through circulation and mark ECs, a process termed intravital labeling. Candidate antibodies were only considered if they yielded a high signal-to-noise ratio, stained the target population entirely and exhibited a high degree of specificity. Conjugated antibodies, such as VE-Cadherin Alexa Fluor 647 and CD34 Alexa Fluor 488, that bound surface antigens shared among all vascular beds were used for consistency. The technique of intravital labeling resulted in superior purities compared to magnetic isolation technologies (Figure 1A; Figures S1A and S1B available online).
Figure 1. Determination of Tissue-Specific Vascular Signature of ECs Purified by Intravital Staining.
(A) Schematic model of conventional EC isolations utilizing magnetic beads after tissue dissociation compared to intravital labeling with multiple fluorescent markers in vivo, which results in enhanced purities.
(B) Wild-type (WT) animals were coinjected with fluorescently labeled antibodies and IsolectinGSIB4 8 min prior to sacrifice. Primary channels (left) provided for the clearest resolution of the ECs, secondary channels (middle) confirmed the cell as EC via microscopy. Sections were counterstained with DAPI (right).
(C) The identical markers from (B) were applied to flow cytometric analysis. Tissues from intravitally labeled animals were enzymatically dissociated. Red cells noted in the scatterplots only include live single cells without highly autofluorescent, nonspecific IgG binding cells, or aggregates of two or more cells. Cells highlighted in yellow are positive for the primary-specific EC marker and then interrogated in a secondary channel. Double positive cells are shown in green.
(D) Genome-wide principal component analysis (PCA) of the nine tissues profiled showing the individual replicates from each tissue. Tissues are color-coded corresponding to their label.
(E) Correlation coefficients are presented for the transcriptional profiling among the biological triplicates demonstrating high fidelity. Scale bars represent 100 μm, error bars represent SD.
See also Figures S1 and S2.
The resulting protocol utilized intravital labeling adapting to multiparametric definitions via flow sorting. Tissue-specific ECs, which are predominantly composed of capillary ECs, were labeled intravitally with two markers (e.g., VEGFR3 and Isolectin GSIB4) at the lowest workable concentration and then validated by microscopy (Figures 1B and S1C) and flow cytometry (Figures 1C and S1D). Liver sinusoidal ECs were defined as VEGFR3+IsolectinGSIB4+CD34dim/−IgG−. Bone marrow, heart, lung, and spleen ECs were defined as VE-Cadherin+ Isolectin+ IgG−. Kidney ECs were specifically selected for the specialized glomeruli ECs with a definition of VE-Cadherin+CD34brightIgG−. Testis and brain ECs were defined as CD34+VE-Cadherin+IgG−. Muscle ECs were defined as CD34+Isolectin+IgG− (Figures 1B, 1C, S1C, and S1D).
Exclusion of nonspecific binding was achieved by the addition of fluorescently labeled Rat IgG (similar results were obtained with CD45 and TER119 antibodies). An additional channel lacking any fluorescent label was also acquired to detect and exclude auto-fluorescence (Figures S2A and S2B). Most importantly, VECadherin+ lymphatic ECs (Alva et al., 2006) remained unstained when using intravital staining (Figure S2C). All cells were interrogated by examining FSC-H and FSC-W in order to discern single cells from two or more cells in close proximity to each other. This was repeated by comparing SSC-H to SSC-W, to ensure that only ECs were collected without perivascular, lymphatic, and stromal cell contamination. These parameters resulted in consistently pure EC population (>95% ECs) that stained negative for CD45+ (hematopoietic) and PDGFRβ+ perivascular contaminants, such as smooth muscle cells (data not shown). This approach, which enables procurement of authentic microvascular ECs devoid of lymphatic and perivascular ECs, is superior to conventional magnetic isolation techniques that result in suboptimal purification of the ECs (Figures S1A and S1B).
Global Transcriptional Profiling of Tissue-Specific ECs Reveals Vascular Heterogeneity
Tissue-specific EC harvests yielded high quality RNA for gene expression analysis (data not shown). Each tissue was analyzed in biological triplicates. A principal component analysis (PCA) shows the relationships of the EC transcriptomes derived from each organ (Figure 1D). The ECs of the brain, heart, lung, and muscle clustered tightly with each other. BM, liver, and spleen ECs clustered apart from the aforementioned group. The kidney and testis ECs were the most dissimilar clusters among those tested. The correlation coefficients of the replicates were found to be high (Figure 1E). The reproducibility among biological triplicates and the tight clustering among distinct tissues demonstrate the fidelity of the identification, isolation, and profiling of tissue-specific ECs. Pairwise comparisons of the tissues were performed by determining the correlational coefficients of the pairs. The most closely related ECs were from the heart and muscle (R2 = 0.9761), whereas the least similar pair was the lung and bone marrow ECs (R2 = 0.79551) (Table 1). Collectively, these data demonstrate that ECs derived from different organs possess significant differences in their transcriptomes and have unique relationships to each other. The entirety of the data set has been supplied to the Gene Expression Omnibus public database (Series GSE47067).
Table 1.
Correlational Coefficients between Pairs of Tissue-Specific ECs
| Tissue | Most Similar | Least Similar | Difference from Mean |
|---|---|---|---|
| liver | spleen (0.89418) | lung (0.82104) | 0.87188 |
| bone marrow | spleen (0.94495) | lung (0.79551) | 0.87267 |
| kidney | heart (0.94132) | bone marrow (0.84675) |
0.91029 |
| heart | muscle (0.97609) | bone marrow (0.86225) |
0.91852 |
| lung | brain (0.91577) | bone marrow (0.79551) |
0.88435 |
| brain | heart (0.9253) | bone marrow (0.93719) |
0.90341 |
| muscle | heart (0.9761) | liver (0.85624) | 0.91744 |
| spleen | bone marrow (0.944952) |
lung (0.8563) | 0.9109 |
| testis | brain (0.89475) | liver (0.82186) | 0.88177 |
Genome-wide expression levels were used for the generation of correlational coefficients between pairs of ECs. The most and least similar results are presented. Additionally, all nine tissues were pooled to create a mean sample with the correlational coefficient of each individual sample also provided.
Regulation of EC Fate and Heterogeneity
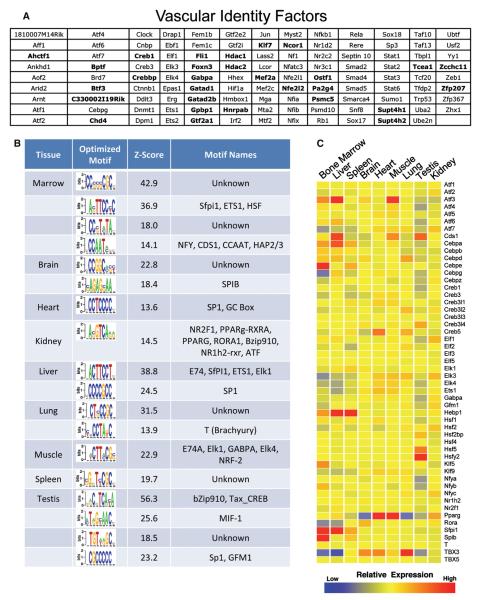
To uncover the factors that contribute to ECs, the profiling data were mined for Gene Ontology annotated transcription factors (TFs) that are expressed in most EC beds. These factors may represent a group of genes needed for the identity of the tissue-specific ECs at homeostatic conditions. A total of 116 genes annotated as TFs were identified that were highly expressed (in the top 20% of transcript intensities) in at least seven of the nine profiled tissues in each of the three replicates (Figure 2A). When the stringency of examined TFs was increased to only the top 10% of transcript intensities in nine of nine tissues, the number of differentially expressed TFs was reduced to 29 genes (Figure 2A, bold text). The normalized intensities of the genes listed in Figure 2A demonstrated highly consistent expression, with only five genes (Septin10, Nfib, Sox17, Epas1, and Ebf1) out of 116 deviating 2-fold or greater from the mean in any tissue (Figure S3).
Figure 2. Transcriptional Regulation of EC Fate and Tissue-Specific Heterogeneity.
(A) A list of Gene Ontology-annotated transcription factors is presented, selected for consistent expression in seven of nine of the profiled ECs and in the top 20th percentile of transcripts of each of the biological triplicates. Further emphasis was placed on transcription factors present in all nine tissues in triplicate in the top tenth percentile with bold lettering
(B) High-scoring DNA motifs uncovered by de novo motif analysis in the promoters of genes with high expression in tissue-specific ECs are presented. The Z score indicates the statistical strength of motif overrepresentation in this tissue. Motif names are shown when a match to a transcription factor-binding site in JASPAR or TRANSFAC could be found. Motif matching was performed using the CompareACE approach, using 0.8 as threshold.
(C) Corresponding transcript levels of transcription factor candidates represented in (B) are shown.
See also Figure S3.
The TFs that dictate organ-specific vascular identity are not known. The data set was interrogated to find factors that might contribute to EC heterogeneity. A discriminative motif discovery approach (Elemento et al., 2007) was used to identify DNA motifs that were overrepresented in the promoters of genes that were differentially expressed among the various organotypic ECs (Figure 2B). When coupled with the transcriptional profiling data of the TFs themselves, vascular heterogeneity among expression of TFs was found that corresponded with the candidate motif partners (Figure 2C). These analyses resulted in identification of numerous known and several unrecognized, yet repeated, motifs in the promoters of upregulated genes.
The ETS family of TFs emerged as a potential regulator of EC diversity. This family of transcription factors is known to play essential roles in EC development and homeostasis (Meadows et al., 2011). However, the tissue-specific expression of ETS family members has not been thoroughly studied, raising the possibility that EC diversity is regulated by the expression of specific members of the ETS family among vascular beds. We found that different vascular beds did indeed express different levels of numerous ETS TFs (Figure 2C). For example, bone marrow and liver ECs expressed much higher levels of SFPI1 compared to other EC populations. Importantly, many target DNA motifs discovered with known binding proteins are either part of the ETS family of transcription factors or known to be cofactors in ETS signaling, either enhancing (SP1, CREB) (Gory et al., 1998; Papoutsopoulou and Janknecht, 2000), or suppressing (PPARG) (Kitamura et al., 1999) gene expression. This finding demonstrates the ability of the tissue-specific EC TF profiling established here to unravel specific transcriptional networks that may dictate vascular heterogeneity.
Tissue-Specific Clustering of Angiocrine Factors
Capillary ECs play important roles in tissue growth and regeneration through the expression of angiocrine factors that govern resident stem and progenitor cell proliferation and differentiation (Butler et al., 2010, 2012; Ding et al., 2010, 2011, 2012; Ding and Morrison, 2013; Himburg et al., 2012). However, the diversity of angiocrine factor signatures among the different vascular beds is unknown. This concept prompted us to determine whether organotypic ECs express tissue-specific combinations of angiocrine factors.
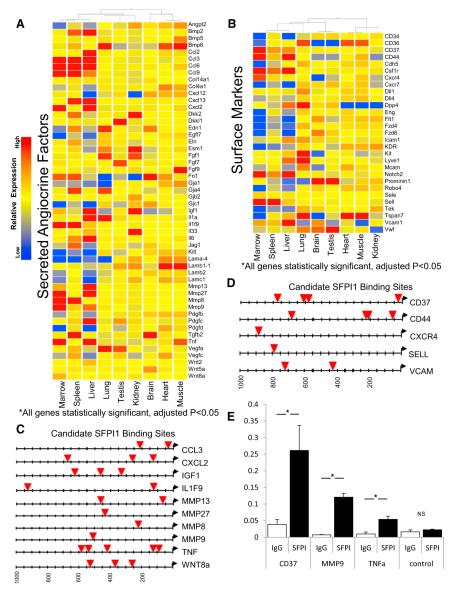
A group of angiocrine factors was selected for hierarchical clustering that significantly differed from mean expression (adjusted p < 0.05) in at least one tissue (Figure 3A). Specifically, genes were selected for 2-fold or greater expression either above or below the mean. We found the hierarchical clustering among various tissue-ECs were similar to the genome-wide PCA (Figure 1D), i.e., the bone marrow, liver, and spleen were closely related and the heart and muscle were closely related. We also observed high expression levels in limited numbers of tissues of certain angiocrine factors. Interleukin 33 (IL33) expression was only found in the kidney, Wnt5a in the brain, FGF1 in the kidney and lung, and BMP5 in the muscle. Conversely, certain factors manifested reduced expression, such as CXCL12 (SDF1) in the liver and kidney and PDGF-D in the bone marrow and liver (Figure 3A). The angiocrine signature that defines the vascular niche in each organ attains its specificity through combinatorial expression of numerous angiocrine factors rather than any one specific factor.
Figure 3. Hierarchical Clustering of Heterogeneous Factors with ETS-Factor Promoter Binding.
(A) Hierarchical clustering of selected organ-specific angiocrine factors deviating 2-fold or greater from mean expression with statistical significance (Benjamini-Hochberg adjusted p < 0.05), including growth factors, cytokines, and ECM are presented. Red denotes higher than average expression, blue denotes lower than average expression.
(B) Selected cell surface receptors are also depicted in a hierarchical cluster, all genes listed are statistically significant (Benjamini-Hochberg adjusted p < 0.05).
(C and D) The first 1,000 bases upstream of the start codon of angiocrine factors and surface markers were upregulated in the bone marrow (BM). ECs were analyzed for potential SFPI1 binding sites and are marked by red triangles. SFPI1 binding in the promoters of CD37, MMP9, and TNF promoter was analyzed by ChIP targeting potential SFPI1 binding sites and a control region without SFPI binding.
(E) Antibodies and genomic regions are indicated on the x axis and the amount of recovered DNA as indicated by percent of the input DNA is indicated on the y axis. Error bars represent SD. Asterisk (*) denotes statistical significance p < 0.05.
See also Figure S4.
Analysis of histone modifiers, cell death modifiers, and metabolic genes revealed divergence among the organs tested (Figure S4). Similarly, a group of differentially expressed surface markers was analyzed (Figure 3B). A large diversity of known EC markers was found among various vascular beds, notably vWF, Tek (Tie-2), CD36, and KDR (VEGFR2). For example, Cdh5 (VE-Cadherin) transcript was lower in bone marrow than in the other tissues, yet it was still in the top 10% of all transcripts in bone marrow-derived ECs (data not shown). Several receptors had preferential expression in just one or few organs, such as CD37 in bone marrow, liver and spleen; Kit (CD117) in the lung, CD36 in the heart, muscle, and lung, and Prominin1 (CD133) in the brain and testis. Taken together, these data indicate that angiocrine factors and many other specialized genes are differentially expressed among tissue-specific ECs, supporting the notion that capillary EC heterogeneity is based on the differential expression of key EC genes.
To demonstrate the utility of the libraries of tissue-EC expression data, we tested whether a TF associated with an enriched motif and expressed in a specific vascular bed did indeed directly bind tissue-EC angiocrine and marker genes. We identified ETS binding sites within the promoter regions of angiocrine factors that were highly expressed in BM (Figure 3C). Similarly, all of the highly expressed surface receptors found on bone marrow-ECs had promoters with at least one SFPI1 binding site (Figure 3D). We analyzed candidate genes for sequence conservation with their human homologs in the first 1 kb upstream of the start codon. Among the genes listed in Figures 3C and 3D, we identified conserved candidate binding sites for SFPI1 in the promoter regions of CD37, MMP9, and TNFα between mouse and human. To test whether SFPI1 could bind these regions, human umbilical vein endothelial cells (HUVECs) overexpressing SFPI1 were used for chromatin immunoprecipitation (ChIP). Indeed, SFPI1 binding was enriched at the promoter regions of CD37, MMP9, and TNFα. Specific SFPI1 binding was not observed at a control genomic region located 3.6 kb away and outside of the TNF-α promoter (Figure 3E). This example of SFPI1 binding illustrates the predictive power of our database and demonstrates that organ EC signatures are governed, at least in part, by inherent transcriptional programs.
Phenotypic Validation of the Genome-wide Signatures of Tissue-Specific ECs
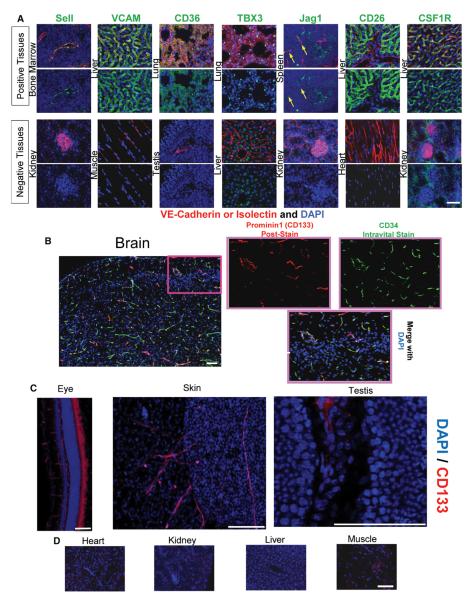
Differences in the phenotypic signatures among EC sources (Figure 3B) can be attributable to different levels among subpopulations of ECs, a binary present-and-absent scenario, or uniform levels within a tissue differing from the mean. Figure 4A presents examples of the binary expression of markers; L-Selectin (Sell) was found on bone marrow ECs, but not kidney glomeruli ECs; VCAM was found on liver ECs, but not muscle ECs; CD36 was abundant on lung EC, but not testis ECs; and CSF1R was well-expressed in liver ECs, but not kidney glomeruli ECs. The resolution of cells during flow sorting was capable of subfractionating ECs within a tissue, as demonstrated by the ability to discern CSF1R− glomeruli ECs from the remaining CSF1R+ ECs of the kidney. In contrast to these binary examples, Jag1 was found only on a subset of spleen ECs (yellow arrows), whereas no significant expression could be detected in kidney ECs. The TF TBX3 was found to be widely present to varying degrees in the lung ECs, yet absent in the liver ECs despite most hepatocytes expressing the protein.
Figure 4. Validations of Differentially Expressed Targets among Vascular Beds.
(A) Representative images of various markers confirmed to have higher expression among organotypic ECs. ECs with positive intravital labeling in some or all vessels are shown as the top pair of images; tissues with no discernible staining are shown as the lower pair of images. Each pair of images represent merged images with VE-Cadherin or Isolectin (red) and the targeted stain alone (green). Scale bar represents 50 μm.
(B) Representative image of animal tissues intravitally labeled with fluorescent CD34 antibody and poststained for Prominin1 (CD133) after cryopreservation and sectioning in both a low-magnification and high-magnification region (highlighted in pink) showing costaining by both markers.
(C) Intravital injections of Prominin1 antibody confirm protein expression on the ECs of the eye, skin, and dimly on the testis. Scale bars represent 100 μm.
(D) An example of tissues with no detectable Prominin1 expression after intravital injections is shown. Scale bars represent 100 μm.
Examination of trancripts of cell surface markers among ECs revealed the expression of CD133 by brain ECs (Figure 3B). Validation of CD133 protein was scrutinized by intravital injection of a labeled CD34 antibody followed by traditional postsectioning staining with CD133 and subsequent microscopic interrogation (Figure 4B). CD133 was specifically expressed in the brain ECs with no discernible perivascular staining. The ECs of the eye, skin, and testis were also partially positive for CD133 expression (Figure 4C). Other than these tissues, CD133 expression on other vascular beds was not found, even on a minority of cells (Figure 4D). Although the intensity and percentage varied, CD133 on ECs appears to be restricted to the testis, eye, skin and brain.
Tissue Regeneration Induces Expression of Unique Angiocrine Profiles
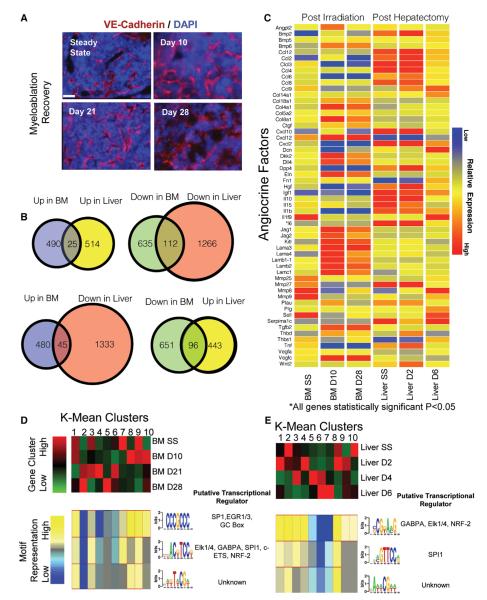
Our laboratory and others have recently shown that sinusoidal ECs in the liver and bone marrow guide tissue regeneration after partial hepatectomy and myeloablation, respectively (Butler et al., 2010; Ding et al., 2010; Ding and Morrison, 2013; Doan et al., 2013b; Himburg et al., 2012; Wang et al., 2012). The same profiling protocol was used to study the distinct responses of ECs to defined physiological stresses. Bone marrow-ECs were harvested at 10, 21, and 28 days after exposure to a sublethal irradiation dose (650 Rads). This approach resulted in a profound decrease in the hematopoietic cells, followed by EC-driven hematopoietic recovery by day 28 postsublethal irradiation. Another cohort of mice underwent the surgical removal of 70% of the three liver lobes (partial hepatectomy), which leads to compensatory liver growth in the remaining intact lobes of the liver without transplantation of any exogenous cells or introduction of growth factors.
Despite vascular remodeling within the BM compartment after myeloablation, the sinusoidal ECs maintain blood flow (Figure 5A). Likewise, the vasculature within the regenerating liver also remained functional without any compromise in the perfusion capacity of sinusoidal ECs (Ding et al., 2010). Thus, ECs from regenerating BM and liver could be intravitally labeled and purified in the exact manner as their steady-state counterparts. Transcriptional profiling of the regenerating ECs purified from liver and BM manifested profound tissue-specific alterations in the angiocrine profiles. Despite the structural similarities between the sinusoidal ECs of the BM and liver, these reparative responses were distinct from each other. The sinusoidal ECs from both tissues were analyzed for genes whose expression was 2-fold up- or downregulated to a statistical significance of p < 0.05. Compared to steady state conditions, 1,262 genes in total were found at days 10, 21, and 28 after sublethal irradiation to be either up- or downregulated. Similarly, the liver yielded 1,917 genes at days 2, 4, and 6 posthepatectomy compared to the control conditions (Figure 5B).
Figure 5. Tissue-Specific Angiocrine Responses Are Dynamic and Modular during Organ Regeneration.
(A) Representative images of intravital labeling for VE-Cadherin (red) and counterstained with DAPI (blue) demonstrating that the bone marrow (BM) vasculature maintains functional blood flow throughout myeloablation and recovery. Scale bar represents 50 μm.
(B) Venn diagrams depicting statistically significant (p < 0.05), 2-fold differentially regulated genes between recovering BM ECs and liver ECs after myeloablation and hepatectomy across all time points, respectively (B).
(C) Selective groups of angiocrine factors between steady state ECs in the BM (BM SS) and 10 and 28 days postsublethal 650 Rad radiation along with steady state liver ECs (Liver SS) and ECs 2 and 6 days after partial hepatectomy (p < 0.05 for all genes shown.
(D and E) The upper red and green heat map represents K-Means clusters of genes, which are specific to steady state, the early phases of recovery, the late phase of recovery, and combinations of these phases. Each column in the heat map represents a different group of clustered genes, numbered 1–10. The lower heat map depicts the results of de novo motif discovery in either BM (D) or liver (E) on the upregulated genes of the corresponding columns. Identified motifs found via de novo motif analysis in each cluster and their corresponding potential binding factors are listed. In each row, the color indicates whether the motif is overrepresented in a group (yellow) or underrepresented (blue).
Only a fraction of genes were commonly regulated between the regenerating liver and BM (Figure 5B). Our studies identified a number of genes that were differentially regulated during recovery. For example, various members of the Notch family were altered as the BM ECs supported hematopoietic recovery, yet these levels in the liver ECs were unchanged during the recovery from hepatectomy. Conversely, HGF was significantly upregulated at day 2 posthepatectomy, yet these levels remained low in the BM ECs (Figure 5C). Importantly, many genes manifested stage-specific expression during either the earlier or later phases of regeneration, such as Angpt2 in the BM ECs and BMP2 and MMP8 in the liver. These results demonstrate that ECs from different tissues mount distinct reparative angiocrine responses, presumably due to the distinct metabolic and physiological demands of recovering tissue.
The global tissue-specific vascular responses to the regenerative challenges were interrogated to identify the transcriptional machinery that might underlie organ-specific angiocrine factor deployment. Unsupervised clustering was performed (using the k-means approach) to organize genes into ten clusters based on their correlated expression and timing of expression across the four time points in both BM and liver (Figures 5D and 5E, upper panels). These clusters recapitulate distinct expression patterns at steady state, early, middle, and late stage recoveries. We then identified overrepresented DNA motifs in the promoter regions of genes in these clusters (Figures 5D and 5E, lower panels). Referring to Figure 5D cluster 10 as an example, genes that are preferentially upregulated at day 10 postmyeloablation were associated with the CCCGCCC DNA motif. This motif was underrepresented in clusters 2, 4, and 5, which included genes mostly present at days 21 and 28. De novo sequence analysis of these discrete clusters again highlighted the importance of the ETS family of transcription factors in EC biology. Unexpectedly, although our results found a potential Elk1/4 and GABPA binding site in genes upregulated in regenerating liver and BM at days 2 and 10, respectively, we found only modest alterations in TF expression, and there was only ~5% overlap in the gene expression pattern. This finding points to the complexity of the transcriptional regulation of angiocrine heterogeneity and suggests that other complementory mechanism(s), such as extravascular cues, dictate tissue-specific expression of angiocrine factors.
Human Embryonic Stem Cell-Derived ECs to Model Developmental Specification of Organotypic ECs
Endothelial cells from different organs in normal and stressed conditions revealed extensive differences in gene expression patterns, transcriptional programs and angiocrine responses. It is probable that EC heterogeneity is established in response to intrinsic and extravascular signals that tailor ECs to adapt to their microenvironment. The molecular mechanism(s) that dictates such heterogeneity is unexplored. Therefore, we employed a model of human embryonic stem cell (hESC) differentiation into ECs to investigate early phenotypes shortly after the specification of EC identity.
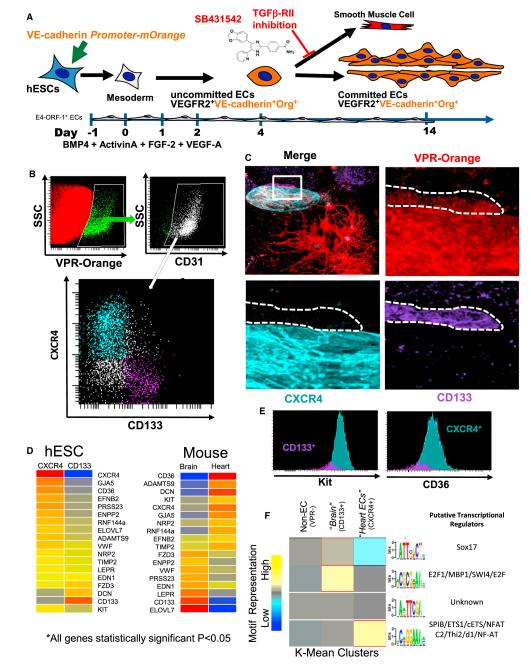
The VE-Cadherin promoter (VPR)-reporter cassette allowed tracking the temporal and spatial emergence of embryonic ECs via the expression of the mOrange fluorescent protein. VPR+ cells are VEGFR2+ VE-Cadherin (protein)+ ECs differentiating from mesodermal precursors of the hESCs (Figure 6A) (James et al., 2010). As the cardiopulmonary system and neural system specify early in development (Gasser, 1975), hESC-ECs were surveyed for some of the most divergent markers predicted in the database between the heart and brain in the adult mouse.
Figure 6. ECs Derived from hESCs Phenocopy Adult Mouse Tissue-Specific Capillaries.
(A) Schema of in vitro conditions to support the differentiation and identification of hESC-derived vasculature. hESCs are grown on an E4-ORF1 EC feeder layer and transduced with a VE-Cadherin-Orange reporter gene. VE-Cadherin-Orange+ vascular networks are readily identifiable by day 10.
(B) Flow cytometry data depicting the expression of VPR-Orange on hESC-derived CD31+ ECs. These VPR+ ECs have distinct populations based on the expression of either CXCR4 (teal) or CD133 (purple).
(C) VPR+CXCR4+CD133− and VPR+CD133+CXCR4− ECs are capable of forming distinct clusters of ECs in hESC cultures.
(D) Heat maps of the genes, which were common in their statistically significant differential expression (Benjamini-Hochberg adjusted p < 0.05) between hESC-derived vasculature and adult mouse heart and brain tissues.
(E) VPR+CXCR4+CD133− and VPR+CD133+CXCR4− ECs were analyzed for cKit and CD36 levels via flow cytometry. Validation of the higher expression of CD36 and Kit in the CXCR4+ ECs is shown.
(F) Heat map of K-Mean clusters depicting the results of de novo motif discovery among non-ECs, CXCR4+VPR+ ECs, and CD133+VPR+ ECs. Candidate binding partners to the motifs are listed.
The expression of CXCR4 and CD133 was mostly mutually exclusive on hESC-derived vasculature (Figure 6B). The ECs defined by either the expression of CD133 or CXCR4 also formed cohesive regions within the hESC cultures, creating a specific niche of hESC-derived ECs within culture (Figure 6C). To define the signatures of VPR+CD31+CD133+CXCR4− and VPR+CD31+ CD133−CXCR4+ ECs, cells were sorted and profiled. The CD133+ and CXCR4+ hESC-ECs were compared to adult mouse brain and heart ECs, respectively. Of the genes with statistically significant deviations in each pairwise comparison (Benjamini-Hochberg adjusted p < 0.05), 18 genes were found in common. Twelve of 18 (67%, CXCR4, GJA5, CD36, EFNB2, NRP2, CD133, Kit, ADAMTS9, TIMP2, EDN1, FZD3) genes followed the same trends in regulation, i.e., the genes upregulated in CD133+ hESC-ECs were also upregulated in adult mouse brain ECs, when compared to CXCR4+ hESC-derived ECs and adult mouse heart ECs, respectively (Figure 6D). A striking finding was that seven of these 18 genes were capable of directly modifying their local microenvironment as angiocrine factors (ADAMTS9, TIMP2, EDN1, FZD3, PRSS23, ENPP2, DCN). Four of seven angiocrine genes (57%, ADAMTS9, TIMP2, EDN1, FZD3) maintained the trend from adult mouse to hESC-derived ECs. Of the remaining 11 nonangiocrine genes, nine are present on the cell surface and capable of sensitizing the EC to environmental cues. Seven of nine cell surface proteins maintained the trend (78%, CXCR4, GJA5, CD36, EFNB2, NRP2, CD133, KIT), with the levels of KIT and CD36 protein levels validated by flow cytometry to have an ~4-fold difference in each case, in agreement with the profiling data in both the mouse and hESC-EC profiling (Figure 6E). Thus, ECs generated in vitro from ESCs exhibit heterogeneity and the EC subtypes that we observed had strong in vivo correlates with their respective adult counterparts.
Next, to identify TFs that are differentially expressed in distinctly marked hESC-ECs, we employed de novo DNA motif discovery in the promoters of genes with transcriptional differences between CD133+, CXCR4+, and VPR− cells. The promoters of upregulated genes within CXCR4+ hESC ECs had an abundance of potential ETS1 binding sites, along with strong levels of ETS1 transcript (Figure 6F and data not shown). Of note, 42% of all upregulated genes in this group had this consensus sequence. As for the CD133+ hESC-ECs, which phenocopy adult mouse brain ECs, a SWI consensus-binding site was discovered as a potential binding candidate. As with several examples in steady-state adult organs, SWI does not belong to the ETS family, yet is documented to directly interact with ETS members (Ahn et al., 2005). Thus, adult tissue-specific vascular heterogeneity may be determined early in specification process and refined during progression through the specification process, yet the identity of intrinsic and extrinsic cues that establish this heterogeneity, are unknown. The entirety of the human data set has also been supplied to the Gene Expression Omnibus public database (Series GSE47067).
Murine ECs Derived from ESCs Engraft in Regenerating Tissue and Undergo In Vivo Tissue-Specific Education
Beyond the EC-astrocyte published coculture experiments (Janzer and Raff, 1987), the effects of the tissue-specific extravascular signals on ECs are unknown. To address the influence of microenvironmental cues on determining vascular heterogeneity, an EC transplantation model was developed. To this end, we adapted a murine ESC (mESC) model by combining previously discovered aspects of mESC-derived cells (McDevitt et al., 2005) and EC differentiation and expansion (James et al., 2010; Kobayashi et al., 2010). To this end, mESCs were differentiated into ECs (mESC-ECs) with stepwise stimulation with BMP4, Activin-A, VEGF-A, and FGF2. Next, VE-Cadherin protein expression was used to identify and purify a uniform population of ECs, followed by transduction with myrAkt1 to generate stable and proliferative mESC-ECs.
The purified cultures of mESC-ECs manifest a stable “generic EC.” By employing this differentiation protocol, the purified cultures of mESC-ECs manifast a stable population that was distinct from any definition found in the adult tissues tested. Prominin1 (CD133), which marks brain-like ECs (Figures 5B and 6) and stem cells of various lineages (Shmelkov et al., 2005), was absent on any substantial population of mESC-ECs (data not shown). CD44 and VCAM expression was minimal, although CD34 and c-Kit were universally present on all cultured mESC-ECs (Figure S5A). Purified mESC-ECs maintained 99.3% VE-Cadherin and CD31 positivity for at least 4 weeks after purification (Figure 7A). Cultured without any instructive cues from surrounding embryonic-derived cells, the mESC-ECs did not drift toward other lineages and thus represent generic ECs that could undergo microenvironmental education and adopt tissue-specific gene expression patterns. The vascular heterogeneity database established here provided the means to demonstrate the extent of these effects and the plasticity of the mESC-ECs upon engraftment into various tissues.
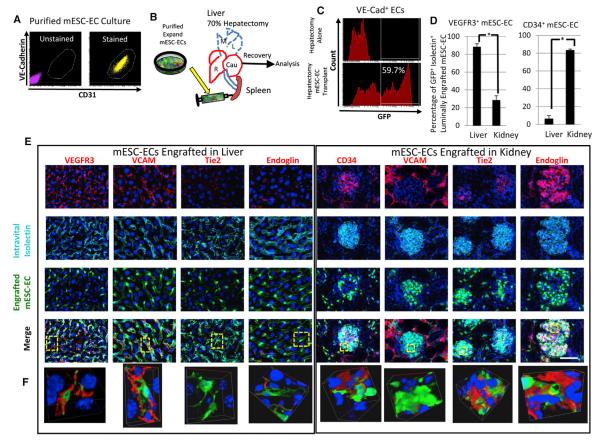
Figure 7. Transplantation and In Vivo Education of Engrafted mESC-ECs in Multiple Tissues.
(A) C57BL/6 mESC cultures were induced toward the EC fate. Two weeks after plating mouse embryoid bodies (EBs), VE-Cadherin+ cells were purified and expanded independently of other cell types. Purified mESC-ECs maintain vascular identity as evident by sustained VE-Cadherin and CD31 expression.
(B) C57BL/6 animals underwent 70% partial hepatectomy by the removal of the three most anterior lobes (dashed blue line) and simultaneously were injected intrasplenically with 500,000 syngeneic GFP-labeled mESC-ECs.
(C) Transplanted animals were intravitally labeled with VE-Cadherin and IgG antibodies to identify ECs. GFP+ mESC-ECs were found to consist of ~60% of the vasculature in the regenerating liver by flow cytometric analysis.
(D) Microscopic quantification of engrafted mESC-ECs in the kidney and liver is presented depicting the percentage within the liver and kidney expressing VEGFR3 and CD34. Results are statistically significant (p < 0.05, t test).
(E) Tissue sections of the regenerated liver (left) and kidney (right) were postfixation stained for VEGFR3, CD34, VCAM, Endoglin, and Tie2. Luminal incorporated mESC-ECs acquire structural and phenotypic attributes of native ECs. Scale bar represents 50 μm, error bars represent SD.
(F) Regions from (E) highlighted in yellow are presented in 3D to highlight the expression of the markers directly on the functionally engrafted mESC-ECs.
See also Figure S5.
To determine whether mESC-ECs could undergo in vivo vascular education, we designed an approach to facilitate engraftment into regenerating adult liver sinusoidal vessels and compare the acquired phenotypic signature of engrafted mESC-ECs to the signature of the ECs described in the database. Toward this end, 5 × 105 syngeneic mESC-ECs were transplanted intrasplenically in mice subsequent to 70% partial hepatectomy (Figure 7B). Animals were intravitally labeled with Isolectin GSIB4 to identify perfused blood vessels. The regenerated livers were normal and lacked teratomas. GFP+ mESC-ECs had functionally incorporated into vasculature forming mosaic vessels with native liver sinusoidal ECs (LSECs). This finding was reminiscent of a previous study demonstrating engraftment of xeno-transplanted human reprogrammed amniotic cell-derived vascular endothelial cells (rAC-VECs) into the regenerating liver of immunocompromised mice without provoking malformations that were structurally indistinguishable from host vasculature (Ginsberg et al., 2012). However, injection of species-specific, syngeneic cells resulted in drastically higher engraftment rates than the human rAC-VECs into immunocompromised mice. Functionally engrafted mESC-ECs consistently demonstrated robust contributions to the sinusoidal ECs within the liver (Figure 7C). Unexpectedly, engrafted mESC-ECs were also detected in other noninjured tissues. Although engrafted tissues varied, we found that the kidney was reproducibly engrafted in numerous transplants, yet only after significant tissue damage (data not shown). Thus, surgical trauma creates a distal permissive environment for mESC-EC engraftment.
Engrafted mESC-ECs acquired expression of VEGFR3 to comparable levels of intensity as neighboring native sinusoidal liver ECs (LSECs). To compare these cells to the intravital labeling (Figure 1B), microscopic quantification of three independently engrafted livers identified 88% as VEGFR3+. Conversely, only 28% of mESC-ECs were found to be dimly VEGFR3+ in kidney glomeruli, which is comparable to endogenous host vasculature. The mESC-ECs presented a negligible 6% positivity for CD34 compared to 84% positivity for CD34 of engrafted cells in the glomeruli (Figures 7D and S5B). In comparison to in-vitro-cultured CD34+ c-Kit+ mESC-ECs, few engrafted cells expressed CD34 in the liver and no detectable expression of c-Kit was found in those transplanted mESC-ECs that had incorporated into the regenerated liver (Figure S5B).
The reference vascular heterogeneity database regarding expression levels of various targets (Figures 1, 2, 3, and 4) was interrogated to predict expression levels of tissue-specific markers on transplanted mESC-ECs, which engrafted and had undergone in vivo education into LSECs and glomeruli type ECs. VCAM expression was robust in engrafted mESC-ECs in the liver; however, cells within the kidney glomeruli were incapable of matching such intensity. Both Tie2 and Endoglin were expressed in the endogenous and engrafted mESC-ECs within the glomeruli. Yet these markers were barely detectable within engrafted cells within the liver sinusoids (Figure 7E). Three-dimensional images of regions from the incorporated GFP+ ECs (Figure 7E) confirmed that the markers are indeed from the functionally engrafted mESC-ECs (Figure 7F). Therefore, engrafted mESC-ECs were influenced by the liver and kidney microenvironments and underwent in vivo education. The EC transplantation-engraftment model described here provides an instructive platform to identify the microenvironmental cues that confer generic ECs tissue-specific vascular signatures and demonstrates the utility of the database for studying EC fate and heterogeneity.
DISCUSSION
Although much is known about the unique structural attributes of ECs, the molecular signatures defining tissue-specific EC heterogeneity have remained unclear. Previous studies focusing on tumor-specific markers (Seaman et al., 2007) and in vitro responses to stimuli (Müller et al., 2002) suggested that EC heterogeneity exists, but these studies did not analyze the molecular repertoire of tissue-specific endothelium. Furthermore, lack of strategies to isolate, cultivate, and transplant mouse ECs posed major obstacles for the functional identification of intrinsic TFs and microenvironmental cues that govern heterogeneity. Here, we have developed EC purification, cultivation, profiling, differentiation, and transplantation models to establish a reference microvascular heterogeneity database and study their operational attributes. The microvascular ECs purified by this intravital labeling strategy are devoid of lymphatic ECs or parenchymal cells. This allowed for the interrogation of the molecular signatures of authentic microvascular ECs. This informational database identified distinct clusters of TFs, angiocrine factors, adhesion molecules, metabolic profiles, and surface receptors expressed on the microvascular ECs of each organ at steady state or during regeneration. Human and mouse embryonic stem cell-EC derivation models partially recapitulated vascular heterogeneity observed in the adults, and subsequent transplantation showed that ECs are phenotypically and functionally educated by their microenvironment. The data presented here supports the notion that one single factor or marker is not sufficient to define tissue-specific ECs, but rather unique combinations of factors need to be considered as organotypic-determining factors.
Crosstalk between tissue-specific ECs and corresponding stem cells orchestrate organ regeneration. Indeed, we show that modules of tissue-specific angiocrine signals balance self-renewal and differentiation of organotypic stem cells and correlate well with the findings of our profiling approach. BMP signaling modulates the differentiation of the muscle satellite cells (Dimitriou et al., 2005), with BMP5 and BMP6 expression detected predominantly in muscle ECs. FGF-7 was only found to be expressed in the testis ECs and has been demonstrated to have similarities with testosterone in male reproduction (Sugimura et al., 1996). Also, the Wnt family of genes has demonstrated the ability to stimulate neuronal cells (Castelo-Branco et al., 2003), with the brain ECs specifically expressing Wnt-5a. None of these factors were previously appreciated to be of EC in origin.
Notably, the induction of these tissue-specific angiocrine factors is dynamic, as physiological stress stimulates the expression of distinct combinations of angiocrine factors throughout the regenerative process. The differences between the liver and bone marrow angiocrine profiles in response to insult were profound, establishing the notion that vascular-derived signals are unique among tissues during regeneration. The data presented here is in agreement with previous studies that found Notch ligands are critical for hematopoietic recovery after irradiation (Butler et al., 2010) and that Wnt2 and HGF supplied by sinusoidal ECs are indispensable for hepatic recovery after partial hepatectomy (Ding et al., 2010; Ober et al., 2006). Further examination of the genes expressed in regenerating ECs reveals a temporal and modular expression of distinct angiocrine factors during the proliferation and resolution phases of liver and BM regeneration. In the liver, for example, HGF is markedly upregulated in ECs in the early phase of tissue challenge at day 2 posthepatectomy (Figure 5C). The mRNA levels are lowered by day 6 as the regeneration reaches completion. Also at day 6, Decorin (Dcn) is strongly expressed in liver ECs and is capable of antagonizing HGF’s ability to stimulate the c-Met receptor (Goldoni et al., 2009). Similar patterns of initiation and resolution are seen in the BM ECs. There are numerous genes whose higher expression at day 10 postirradiation was reduced by day 28, such as Dll1, Jag1, Jag2, and Angpt2. Moreover, the expression of TNF, which is a factor that maintains a quiescent pool of stem and progenitor cells (Dybedal et al., 2001; Zhang et al., 1995), is markedly reduced during hematopoietic recovery (Figure 5C).
Our bioinformatic analysis of the transcriptional profiles repeatedly pointed to the ETS family as being one of the most predominant groups of TFs governing EC biology, expanding on the known roles in the development of ECs (Dejana et al., 2007) and other cell types (Maroulakou and Bowe, 2000). Notably, the ETS TF, FLI1 was highly expressed in all of the tissue-specific ECs examined, whereas other ETS factors, such as SFPI1 and ELF4 were found only in specific tissue-ECs. This suggests modular hierarchy of ETS transcriptional regulation with some factors being critical for specifying general EC identity and function from nascent embryonic ECs, whereas other TFs endow generic ECs with specialized functions of the individual vascular beds. The final tier of ETS-mediated regulation, likely along with various cofactors, guides EC-mediated tissue repair, such as with Elk4 in the BM and liver. Validation of such complex networks would benefit greatly from using the EC transplantation-engraftment model developed here (Figure 7) to piece together the progression of vascular fate from specification into generic ECs to committed tissue-specific capillaries.
The discovery of phenotypically similar ECs with unique overlapping signatures between adult mice and hESCs cultures speaks to the developmentally conserved pathways common to tissue-specific ECs. Notably, the expression of endothelin1 (Edn1) is conserved from human hESC-EC to adult mouse. Edn1 has long been appreciated for its capacity to effect sympathetic nerve activity in various structures within the brain (Kuwaki et al., 1997) and is upregulated in the brain/CD133+ ECs. Timp2 is critical for cardiac development (Brauer and Cai, 2002), after adult heart failure (Peterson et al., 2000), and is upregulated in the heart’s CXCR4+ ECs (Figure 6D). These results point to the ECs as early sources for angiocrine growth factors essential for tissue development. Thus, during mouse development, the formation of vasculature precedes the development of tissues, such as the pancreas (Lammert et al., 2001), liver (Matsumoto et al., 2001), kidney (Serluca et al., 2002), and testis (Cool et al., 2011). The paradigm of EC angiocrine function may therefore have conserved properties from vasculogenesis and throughout adulthood, representing as yet untapped target for therapies and tissue engineering.
The mESC-EC transplantation and engraftment model developed here offers an approach to study the specification of vascular heterogeneity in vivo. This naive EC population is not tainted by any epigenetic memory from a tissue of origin, as the cell never existed in a fetal or adult tissue. This platform has proven the capacity for the liver and kidney microenvironments to dictate the operational attributes of LSECs and glomeruli ECs. Finally, the conclusion that the generic ECs did indeed successfully undergo in vivo education is grounded in the informational vascular heterogeneity database described here. The comprehensive vascular heterogeneity reference library from organotypic ECs provides the means to identify various vascular-niche-dependent angiocrine pathways involved in safeguarding the integrity of tissue-specific stem and progenitor cells at steady states and during organ regeneration. Unraveling the molecular determinants of vascular heterogeneity brings us closer to develop strategies to capitalize on the instructive potential of tissue-specific ECs to promote functional organ regeneration.
EXPERIMENTAL PROCEDURES
Intravital Staining and Tissue Harvest
Antibodies were conjugated to Pacific Blue, Alexa Fluor 488, Alexa Fluor 594, or Alexa Fluor 647 (Invitrogen/Molecular Probes). The degree of labeling (DOL) was calculated by using a Nanodrop. Rat IgG Pacific Blue was maintained at a DOL of 15–20. All remaining Alexa Fluor Dyes were kept at a DOL of 8–12. Each protocol was reviewed and approved by Institutional Animal Care and Use Committee. Twenty-five micrograms of each antibody and 100 mg of Isolectin GSIB4 488 (Invitrogen/Molecular Probes) was injected retro-orbitally under anasthesia 8 min prior to sacrifice and organ harvest. The EC-specific labels used were CD34 (RAM34, BD PharMingen), VE-Cadherin (BV13, BioLegend), and VEGFR3 (31C1, ImClone). Nonendothelial antibodies used were rat and mouse IgG (Jackson Laboratories), CD45 (30-F11, BD PharMingen), CD11b (M1/70, BD PharMingen), and TER119 (TER119, BD PharMingen). For flow cytometry, organs were minced and incubated with Collagenase A (25 mg/ml), Dispase II (25 mg/ml), and DNase (250 μg/ml) (Roche) at 37°C for 20–30 min to create a single cell suspension. Hematopoietic and erythroid cells were removed via CD45 and TER119 microbeads (Miltenyi Biotech). Cells were filtered through a 40 μm filter immediately prior to analysis. For microscopy, the organs were fixed in paraformaldehyde and cryopreserved in 30% sucrose.
RNA Isolation, Amplification, and Microarray Analysis
RNA was isolated using the PicoPure Isolation kit (Arcturus). Cells were sorted into chilled serum-free medium, pelleted, and resuspended in RNA extraction buffer. All samples were subjected to on-column DNase (QIAGEN) treatments according to the Arcturus protocol. Total harvest time from antibody injection to resuspension in RNA buffer was 70–90 min, depending on tissue. Quality of the RNA was assessed using a Bioanalyzer (Agilent). Satisfactory RNA was amplified using the WT-Ovation RNA amplification system. Fragmentation and labeling was done using the WT-Ovation Exon and Encore Biotin modules (NuGEN). Samples were then hybridized to GeneChip 1.0 ST arrays (Affymetrix). RMA normalized data were analyzed by Genespring 11.0 software, which also performed all statistical analysis. Specifically, ANOVA was utilized with Benjamini-Hochberg adjusted p values to include multiple test correction. The false discovery rate was set to 5% (adjusted p < 0.05).
Additional procedures are included in the Supplemental Experimental Procedures, including descriptions of flow cytometry, ChIP, human and mouse embryonic stem cell culture, mice, de novo motif analysis, and microscopy.
Supplementary Material
ACKNOWLEDGMENTS
S.R. is supported by the Ansary Stem Cell Institute, the Howard Hughes Medical Institute, the Empire State Stem Cell Board, the New York State Department of Health (NYSTEM C024180, C026438, and C026878), NHLBI (R01s HL097797 and DK095039), the Qatar National Priorities Research Foundation (NPRP08-663-3-140), and the Qatar Foundation BioMedical Research Program (BMRP). D.J.N. is supported by the Tri-Institutional Weill Cornell Starr Stem Cell Scholar program. A.R. is supported by the Qatar National Priorities Research Foundation (NPRP09-1087-3-274).
Footnotes
ACCESSION NUMBERS All microarray analysis is available through the Gene Expression Omnibus Series GSE47067.
SUPPLEMENTAL INFORMATION Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.devcel.2013.06.017.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahn J, Ko M, Lee K, Oh J, Jeon SH, Seong RH. Expression of SRG3, a core component of mouse SWI/SNF chromatin-remodeling complex, is regulated by cooperative interactions between Sp1/Sp3 and Ets transcription factors. Biochem. Biophys. Res. Commun. 2005;338:1435–1446. doi: 10.1016/j.bbrc.2005.10.107. [DOI] [PubMed] [Google Scholar]
- Aird WC. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ. Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- Alva JA, Zovein AC, Monvoisin A, Murphy T, Salazar A, Harvey NL, Carmeliet P, Iruela-Arispe ML. VE-Cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev. Dyn. 2006;235:759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- Børsum T, Hagen I, Henriksen T, Carlander B. Alterations in the protein composition and surface structure of human endothelial cells during growth in primary culture. Atherosclerosis. 1982;44:367–378. doi: 10.1016/0021-9150(82)90011-9. [DOI] [PubMed] [Google Scholar]
- Brauer PR, Cai DH. Expression of tissue inhibitor of metalloproteinases (TIMPs) during early cardiac development. Mech. Dev. 2002;113:175–179. doi: 10.1016/s0925-4773(02)00016-3. [DOI] [PubMed] [Google Scholar]
- Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, Seandel M, Shido K, White IA, Kobayashi M, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JM, Gars EJ, James DJ, Nolan DJ, Scandura JM, Rafii S. Development of a vascular niche platform for expansion of repopulating human cord blood stem and progenitor cells. Blood. 2012;120:1344–1347. doi: 10.1182/blood-2011-12-398115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc. Natl. Acad. Sci. USA. 2003;100:12747–12752. doi: 10.1073/pnas.1534900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churg J, Grishman E. Ultrastructure of glomerular disease: a review. Kidney Int. 1975;7:254–261. doi: 10.1038/ki.1975.37. [DOI] [PubMed] [Google Scholar]
- Cool J, DeFalco TJ, Capel B. Vascular-mesenchymal crosstalk through VEGF and PDGF drives organ patterning. Proc. Natl. Acad. Sci. USA. 2011;108:167–172. doi: 10.1073/pnas.1010299108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejana E, Taddei A, Randi AM. Foxs and Ets in the transcriptional regulation of endothelial cell differentiation and angiogenesis. Biochim. Biophys. Acta. 2007;1775:298–312. doi: 10.1016/j.bbcan.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Dimitriou R, Tsiridis E, Giannoudis PV. Current concepts of molecular aspects of bone healing. Injury. 2005;36:1392–1404. doi: 10.1016/j.injury.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding BS, Nolan DJ, Guo P, Babazadeh AO, Cao Z, Rosenwaks Z, Crystal RG, Simons M, Sato TN, Worgall S, et al. Endothelial-derived angiocrine signals induce and sustain regenerative lung alveolarization. Cell. 2011;147:539–553. doi: 10.1016/j.cell.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan PL, Himburg HA, Helms K, Russell JL, Fixsen E, Quarmyne M, Harris JR, Deoliviera D, Sullivan JM, Chao NJ, et al. Epidermal growth factor regulates hematopoietic regeneration after radiation injury. Nat. Med. 2013a;19:295–304. doi: 10.1038/nm.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan PL, Russell JL, Himburg HA, Helms K, Harris JR, Lucas J, Holshausen KC, Meadows SK, Daher P, Jeffords LB, et al. Tie2(+) bone marrow endothelial cells regulate hematopoietic stem cell regeneration following radiation injury. Stem Cells. 2013b;31:327–337. doi: 10.1002/stem.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dybedal I, Bryder D, Fossum A, Rusten LS, Jacobsen SE. Tumor necrosis factor (TNF)-mediated activation of the p55 TNF receptor negatively regulates maintenance of cycling reconstituting human hematopoietic stem cells. Blood. 2001;98:1782–1791. doi: 10.1182/blood.v98.6.1782. [DOI] [PubMed] [Google Scholar]
- Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol. Cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser RF. Atlas of Human Embryos. Harper & Row; Hagerstown, MD: 1975. [Google Scholar]
- Ginsberg M, James D, Ding BS, Nolan D, Geng F, Butler JM, Schachterle W, Pulijaal VR, Mathew S, Chasen ST, et al. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ETS factors and TGFβ suppression. Cell. 2012;151:559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldoni S, Humphries A, Nyström A, Sattar S, Owens RT, McQuillan DJ, Ireton K, Iozzo RV. Decorin is a novel antagonistic ligand of the Met receptor. J. Cell Biol. 2009;185:743–754. doi: 10.1083/jcb.200901129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gory S, Dalmon J, Prandini MH, Kortulewski T, de Launoit Y, Huber P. Requirement of a GT box (Sp1 site) and two Ets binding sites for vascular endothelial cadherin gene transcription. J. Biol. Chem. 1998;273:6750–6755. doi: 10.1074/jbc.273.12.6750. [DOI] [PubMed] [Google Scholar]
- Himburg HA, Harris JR, Ito T, Daher P, Russell JL, Quarmyne M, Doan PL, Helms K, Nakamura M, Fixsen E, et al. Pleiotrophin regulates the retention and self-renewal of hematopoietic stem cells in the bone marrow vascular niche. Cell Rep. 2012;2:964–975. doi: 10.1016/j.celrep.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper AT, Butler JM, Nolan DJ, Kranz A, Iida K, Kobayashi M, Kopp HG, Shido K, Petit I, Yanger K, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Nam HS, Seandel M, Nolan D, Janovitz T, Tomishima M, Studer L, Lee G, Lyden D, Benezra R, et al. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFbeta inhibition is Id1 dependent. Nat. Biotechnol. 2010;28:161–166. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzer RC, Raff MC. Astrocytes induce blood-brain barrier properties in endothelial cells. Nature. 1987;325:253–257. doi: 10.1038/325253a0. [DOI] [PubMed] [Google Scholar]
- Kitamura S, Miyazaki Y, Hiraoka S, Toyota M, Nagasawa Y, Kondo S, Kiyohara T, Shinomura Y, Matsuzawa Y. PPARgamma inhibits the expression of c-MET in human gastric cancer cells through the suppression of Ets. Biochem. Biophys. Res. Commun. 1999;265:453–456. doi: 10.1006/bbrc.1999.1715. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Butler JM, O’Donnell R, Kobayashi M, Ding BS, Bonner B, Chiu VK, Nolan DJ, Shido K, Benjamin L, Rafii S. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat. Cell Biol. 2010;12:1046–1056. doi: 10.1038/ncb2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwaki T, Kurihara H, Cao WH, Kurihara Y, Unekawa M, Yazaki Y, Kumada M. Physiological role of brain endothelin in the central autonomic control: from neuron to knockout mouse. Prog. Neurobiol. 1997;51:545–579. doi: 10.1016/s0301-0082(96)00063-9. [DOI] [PubMed] [Google Scholar]
- Lammert E, Cleaver O, Melton D. Induction of pancreatic differentiation by signals from blood vessels. Science. 2001;294:564–567. doi: 10.1126/science.1064344. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Bowe DB. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene. 2000;19:6432–6442. doi: 10.1038/sj.onc.1204039. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Yoshitomi H, Rossant J, Zaret KS. Liver organogenesis promoted by endothelial cells prior to vascular function. Science. 2001;294:559–563. doi: 10.1126/science.1063889. [DOI] [PubMed] [Google Scholar]
- McDevitt TC, Laflamme MA, Murry CE. Proliferation of cardiomyocytes derived from human embryonic stem cells is mediated via the IGF/PI 3-kinase/Akt signaling pathway. J. Mol. Cell. Cardiol. 2005;39:865–873. doi: 10.1016/j.yjmcc.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows SM, Myers CT, Krieg PA. Regulation of endothelial cell development by ETS transcription factors. Semin. Cell Dev. Biol. 2011;22:976–984. doi: 10.1016/j.semcdb.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molema G. Heterogeneity in endothelial responsiveness to cytokines, molecular causes, and pharmacological consequences. Semin. Thromb. Hemost. 2010;36:246–264. doi: 10.1055/s-0030-1253448. [DOI] [PubMed] [Google Scholar]
- Müller AM, Hermanns MI, Cronen C, Kirkpatrick CJ. Comparative study of adhesion molecule expression in cultured human macro- and microvascular endothelial cells. Exp. Mol. Pathol. 2002;73:171–180. doi: 10.1006/exmp.2002.2446. [DOI] [PubMed] [Google Scholar]
- Ober EA, Verkade H, Field HA, Stainier DY. Mesodermal Wnt2b signalling positively regulates liver specification. Nature. 2006;442:688–691. doi: 10.1038/nature04888. [DOI] [PubMed] [Google Scholar]
- Papoutsopoulou S, Janknecht R. Phosphorylation of ETS transcription factor ER81 in a complex with its coactivators CREB-binding protein and p300. Mol. Cell. Biol. 2000;20:7300–7310. doi: 10.1128/mcb.20.19.7300-7310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BA, St Croix B, Sjöblom T, Cummins JM, Silliman N, Ptak J, Saha S, Kinzler KW, Hatzis C, Velculescu VE. Large-scale identification of novel transcripts in the human genome. Genome Res. 2007;17:287–292. doi: 10.1101/gr.5486607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson JT, Li H, Dillon L, Bryant JW. Evolution of matrix metalloprotease and tissue inhibitor expression during heart failure progression in the infarcted rat. Cardiovasc. Res. 2000;46:307–315. doi: 10.1016/s0008-6363(00)00029-8. [DOI] [PubMed] [Google Scholar]
- Rubin LL, Staddon JM. The cell biology of the blood-brain barrier. Annu. Rev. Neurosci. 1999;22:11–28. doi: 10.1146/annurev.neuro.22.1.11. [DOI] [PubMed] [Google Scholar]
- Seaman S, Stevens J, Yang MY, Logsdon D, Graff-Cherry C, St Croix B. Genes that distinguish physiological and pathological angiogenesis. Cancer Cell. 2007;11:539–554. doi: 10.1016/j.ccr.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serluca FC, Drummond IA, Fishman MC. Endothelial signaling in kidney morphogenesis: a role for hemodynamic forces. Curr. Biol. 2002;12:492–497. doi: 10.1016/s0960-9822(02)00694-2. [DOI] [PubMed] [Google Scholar]
- Shmelkov SV, St Clair R, Lyden D, Rafii S. AC133/CD133/Prominin-1. Int. J. Biochem. Cell Biol. 2005;37:715–719. doi: 10.1016/j.biocel.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Sugimura Y, Foster BA, Hom YK, Lipschutz JH, Rubin JS, Finch PW, Aaronson SA, Hayashi N, Kawamura J, Cunha GR. Keratinocyte growth factor (KGF) can replace testosterone in the ductal branching morphogenesis of the rat ventral prostate. Int. J. Dev. Biol. 1996;40:941–951. [PubMed] [Google Scholar]
- Swift MR, Weinstein BM. Arterial-venous specification during development. Circ. Res. 2009;104:576–588. doi: 10.1161/CIRCRESAHA.108.188805. [DOI] [PubMed] [Google Scholar]
- Wang L, Wang X, Xie G, Wang L, Hill CK, DeLeve LD. Liver sinusoidal endothelial cell progenitor cells promote liver regeneration in rats. J. Clin. Invest. 2012;122:1567–1573. doi: 10.1172/JCI58789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Harada A, Bluethmann H, Wang JB, Nakao S, Mukaida N, Matsushima K. Tumor necrosis factor (TNF) is a physiologic regulator of hematopoietic progenitor cells: increase of early hematopoietic progenitor cells in TNF receptor p55-deficient mice in vivo and potent inhibition of progenitor cell proliferation by TNF alpha in vitro. Blood. 1995;86:2930–2937. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.