Abstract
Steroid receptors bind as dimers to a degenerate set of response elements containing inverted repeats of a hexameric half-site separated by 3 bp of spacer (IR3). Naturally occurring selective androgen response elements have recently been identified that resemble direct repeats of the hexameric half-site (ADR3). The 3D crystal structure of the androgen receptor (AR) DNA-binding domain bound to a selective ADR3 reveals an unexpected head-to-head arrangement of the two protomers rather than the expected head-to-tail arrangement seen in nuclear receptors bound to response elements of similar geometry. Compared with the glucocorticoid receptor, the DNA-binding domain dimer interface of the AR has additional interactions that stabilize the AR dimer and increase the affinity for nonconsensus response elements. This increased interfacial stability compared with the other steroid receptors may account for the selective binding of AR to ADR3 response elements.
The androgen receptor (AR) is a ligand-activated transcription factor that plays a central role in male sexual development and in the etiology of prostate cancer (1, 2). It is a member of the steroid and nuclear hormone receptor superfamily, which also includes receptors for glucocorticoids (GR), mineralocorticoids (MR), progesterone (PR), estrogens (ER), and vitamin D (VDR) (3). Members of this family contain conserved, discrete, DNA-binding domains (DBDs) and ligand-binding domains. The amino-terminal domain and the hinge region connecting the central DBD to the C-terminal ligand-binding domain diverge among family members.
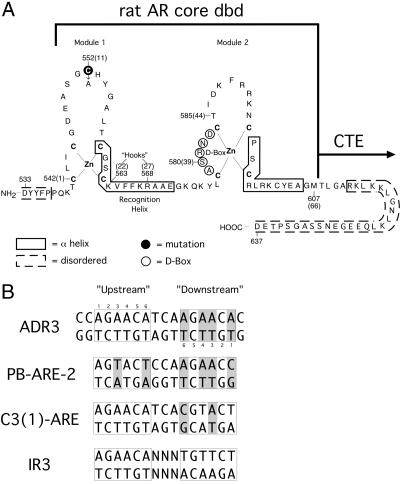
The hormone receptor DBD consists of a highly conserved 66-residue core made up of two zinc-nucleated modules, shown schematically in Fig. 1 A (4, 5). With VDR as the only reported exception (6), the isolated DBD and associated C-terminal extension are necessary and sufficient to generate the same pattern of DNA response element selectivity, partner selection, and dimerization as the full-length receptor from which it is derived (6–11).
Fig. 1.
Protein and DNA constructs. (A) The rat AR DBD. Sequence numbers in parentheses refer to the common receptor DBD-numbering scheme. Residues in dashed boxes are disordered in both protomers of the homodimeric complex. (B) The DNA used in cocrystallization, labeled ADR3, two naturally occurring AR response elements, PB-ARE-2 and C3 (1)-ARE, and a canonical IR3 steroid response element. Differences from the IR3 sequence are shaded gray.
Although ligand binding elicits distinct hormone-specific responses, all classical steroid receptors (AR, PR, MR, and GR) recognize identical DNA response elements, which consist of two hexameric half-sites (5′-AGAACA-3′) arranged as inverted repeats with 3 bp of separating DNA, producing the 2-fold IR3 sequence pattern (Fig. 1B) (12). A question that continues to engage the steroid receptor field is how these transcription factors achieve DNA target specificity despite this degeneracy. As seen in the structures of the GR and ER DBDs bound to IR3 elements (4, 13), the receptors bind as “head-to-head” homodimers whose symmetric displacement across the DNA pseudodyad reflects the underlying half-site arrangement. Differences in steroid metabolism, receptor expression, local chromatin structure, and the availability of cofactors all contribute to steroid-specific responses (14–17). However, recent work has now also identified selective androgen response elements (AREs). The AREs consist of two hexameric half-sites arranged as an androgen direct repeat separated by 3 bp of spacer (ADR3) (18–21), with the half-site repeating on the same strand (Fig. 1B). The expanded binding repertoire of AR, including both the common IR3 and specific ADR3 elements, breaks the degeneracy of the steroid response elements, allowing specific AR activation from certain response elements but disfavoring interaction with PR, MR, or GR. This finding could further account for steroid-specific actions in vivo.
The crystal structures of nuclear receptors bound to direct-repeat elements, including the VDR DBD bound to a similar DR3 element, reveal a “head-to-tail” protein dimer bound to the DNA (6, 22–24). For AR to bind to ADR3-type elements in a head-to-tail orientation, the DBD would require a second dimerization interface that is distinct from the canonical D box region used to dimerize on IR3 elements (25). To visualize this unusual homodimeric assembly, we have solved the crystal structure of an AR DBD homodimer bound to an ADR3 response element. The structure we report here reveals that the proteins do not adopt the expected head-to-tail orientation on the DNA, but, instead, they retain the symmetric mode of dimerization observed previously for the GR DBD bound to an IR3 DNA element. We describe the protein–protein and protein–DNA interactions that allow for this unexpected arrangement, and we propose that AR-specific dimerization contacts account for the AR specificity of ADR3 elements.
Materials and Methods
Protein and DNA Purification. The rat AR DBD (residues 533–637, C552A) was expressed in Escherichia coli BL21/DE3 cells as a GST fusion and purified with a glutathione-Sepharose column (Sigma). The GST was cleaved with thrombin at 4°C overnight. Further purification was performed with SP Sepharose FastFlow (pH 7.4) and Source 15S (pH 6.9) columns. Protein concentration and purity was determined by UV absorbance and SDS/PAGE.
Synthetic oligonucleotides (W. M. Keck Facility, Yale University) were detritylated and purified by reversed-phase HPLC (Rainin Dynamax-300). Concentrated, purified strands were annealed by heating to 95°C and slowly cooling to room temperature.
Crystallization and Data Collection. Samples for cocrystallization contained DNA and protein concentrations of 0.15 and 0.30 mM, respectively, in 5 mM Tris (pH 7.6)/150 mM LiCl/10 mM DTT. Crystals were grown by hanging drop vapor diffusion at 18°C with the addition of 2 μl of the complex to an equal volume of reservoir solution (50 mM Mes, pH 5.6/0–20 mM MgCl2/0–2% polyethylene glycol 400). Diffraction quality crystals (0.15 × 0.15 × 0.4 mm) grew in 2–6 weeks.
Crystals were equilibrated into reservoir solution supplemented with 35% glycerol before being flash-cooled in liquid nitrogen. Diffraction data were collected at –180°C on beamline 22ID at the Advanced Photon Source with a CCD detector (Marresearch, Norderstedt, Germany). Data were indexed and reduced by using hkl2000 (26).
Structure Determination and Refinement. Four zinc sites were found by using solve (27) and data from the peak anomalous wavelength. Experimental phases were generated with these sites; and, in the anomalous difference Fourier maps, the four zinc sites had peaks of >30 σ, whereas the next highest peak was 3 σ, indicating one AR dimer was in the asymmetric unit. Only one of the two possible enantiomeric space group choices yielded zinc sites that corresponded to possible AR dimers. Visual inspection of the zinc sites revealed that the proteins were arranged in a palindromic orientation. This finding led to construction of a molecular replacement model by using the ER DBD-IR3 structure (13) (PDB ID code 1HCQ). Because of its higher sequence homology to AR, the ER DBD was replaced with the core GR DBD (4) (PDB ID code 1GLU) by using least-squares fitting. A molecular replacement solution was obtained by using molrep (28).
Multiwavelength anomalous dispersion phases were calculated by using the remote and peak wavelength data to 3.4 Å and also used in refinement, which was done in cns (29) by using the maximum likelihood Hendrickson–Lattman target. Model building was done by using O (30). Even at 3.1 Å, the number of unique reflections used was eight times the number of modeled atoms because of the very large (>80%) solvent content of the crystal, allowing for restrained individual B factor refinement in later rounds. Visualization of hydrogen bonds, van der Waals interactions, and clashes was aided by use of all atom contacts in king and probe (31). Graphics used ribbons (32) and pymol (DeLano Scientific, San Carlos, CA).
Results
Crystallization and Structure Solution. Initial crystals of AR DBD–ADR3 complexes grew as thin needles from complexes containing AR DBD (residues 533–619) and diffracted to 4 Å with synchrotron radiation. These crystals were resistant to dissolution, suggesting crosslinking within the lattice. The AR DBD contains a nonconserved cysteine at position 552[11] (common receptor DBD numbering is given in brackets), which was predicted to be solvent-exposed based on modeling from the GR DBD structure. When Cys-552[11] in the AR DBD was changed to alanine, complexes containing this mutant yielded bar-shaped crystals that were isomorphous with the initial crystal form. These crystals were used to determine the structure of the AR DBD–DNA complex (PDB ID code 1R4I).
The structure of AR DBD(533–637)Cys552Ala in complex with ADR3 DNA (Fig. 1) was determined at 3.1 Å by a combined MAD and molecular replacement approach with diffraction data collected at the zinc anomalous edge. The arrangement of the proteins on the ADR3 DNA was determined from zinc anomalous data that revealed the location of the four zinc atoms in the complex. Data collection and refinement statistics are presented in Table 1, and representative electron density maps are shown Fig. 7, which is published as supporting information on the PNAS web site.
Table 1. Summary of data collection and refinement.
| Diffraction data | ||
| Space group,*† Å | P3221 137.89, 85.71 | |
| Data set | Native/remote | Zn peak |
| Wavelength, Å | 1.0000 | 1.2831 |
| Resolution, Å | 50-3.1 | 50-3.4 |
| Last shell, Å | 3.21-3.1 | 3.52-3.4 |
| Unique reflections | 17,313 | 25,060 |
| Completeness, % (last shell) | 99.7 (99.1) | 99.5 (98.9) |
| Average I/σσ (last shell) | 20.4 (2.5) | 19.6 (2.0) |
| Rmerge, % (last shell) | 9.6 (62) | 7.8 (58) |
| FOM (after DM)‡ | 0.41 (0.96) | |
| Crystallographic refinement | ||
| Resolution range, Å | 50-3.1 | |
| Reflections (F > 2σF) | 14,839 (12,418) | |
| Atoms | 1,813 | |
| rms bond lengths, Å | 0.0076 | |
| rms bond angles, ° | 1.29 | |
| R value (F > 2σF)† | 24.6 (22.7) | |
| Rfree (F > 2σF) | 26.4 (24.9) | |
Rmerge = ΣhklΣi|Ii(hkl) - 〈I(hkl)〉| /ΣhklΣiI(hkl).
R = Σ|Fo - Fc|/ΣFo. 5% of the reflections were used for Rfree.
Figure of merit = 〈|ΣP(α)eiα / ΣP(α)|〉, where α is the phase and P(α) is the phase-probability distribution.
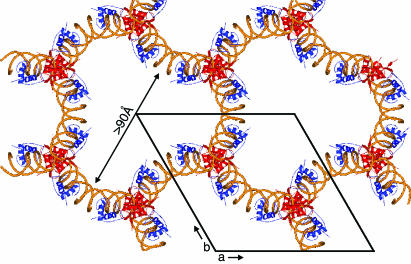
Anomalous difference Fourier maps confirmed that the asymmetric unit consists of just one AR DBD homodimer–DNA complex, yielding a Matthews number of 6.9 and a solvent content of 82%. The main crystal-packing interactions are made by the junction near protomer A, which contains neither a pseudocontinuous DNA interaction nor a biologically plausible alternative protein dimer interface. The downstream AR DBD (protomer B) makes only two crystal contacts by residues Phe-589[48] and Arg-590[49] and, except for the interaction with protomer A and the DNA, it is otherwise completely exposed to the large solvent channels (Fig. 2).
Fig. 2.
Crystal packing of the AR DBD–ADR3 complex. Red and blue ribbons are the upstream and downstream subunits, respectively, with the DNA backbone shown in gold. The view is parallel to the c axis of the crystal, and the unit cell is shown.
Examination of the crystal-packing interactions can explain the refractory effect of C552[11] on crystallization. Residue 552[11] from protomer A is in position to crosslink with Cys-578[37] of protomer A in the adjacent symmetry-related complex. Cys-578[37] coordinates a zinc atom in the first Zn module. Formation of a C552[11]-C578[37] disulfide link is likely to disrupt the native AR DBD conformation and adversely affect crystal order.
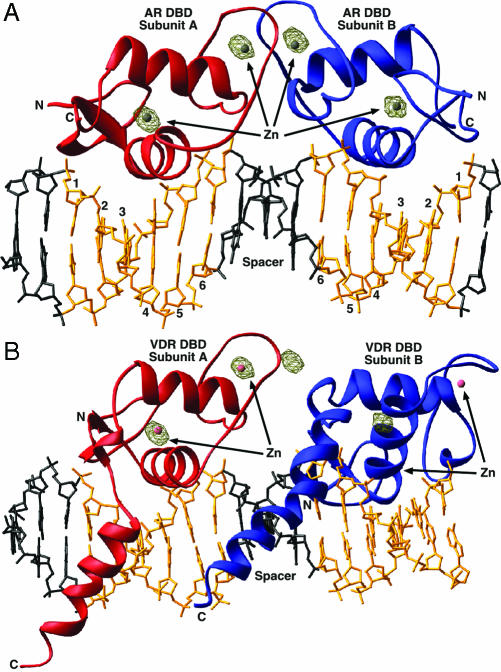
The AR DBDs Are Arranged as an Inverted Repeat on a Direct-Repeat DNA Target. In all the dimeric hormone receptor DBD–DNA complexes determined to date, the two DBDs adopt the same relative orientation as that of the underlying DNA target. Surprisingly, however, in the structure of AR DBD bound to ADR3 DNA, the two AR DBD protomers are not arranged as a head-to-tail dimer, as would be expected of receptors bound to a direct-repeat DNA element. Instead, the proteins form a symmetric, head-to-head dimer that is nearly identical with the dimer seen in the ER DBD–DNA and GR DBD–DNA structures (rms deviation for α-carbons of 1.09 and 0.89 Å, respectively) (4, 13). This finding was confirmed unambiguously by inspection of the positions of the four zinc sites determined from anomalous difference maps calculated from single wavelength anomalous dispersion phases (Fig. 3). The arrangement of the AR dimer is unlikely to be an artifact of crystal packing, because there are only two small crystal contacts between the downstream DBD (protomer B) and the neighboring molecules in the crystal lattice (Fig. 2).
Fig. 3.
Overall architecture of the AR DBD–ADR3 and VDR DBD–DR3 complexes. (A) The AR DBD–ADR3 complex. The two protomers are in red and blue, the hexameric half-site DNA is gold, and the spacer and flanking base pairs are black. In brown is a 20-σ contour of the experimental anomalous Fourier difference map. (B) The VDR DBD–DR3 complex. VDR DBD protomer A is shown in the same orientation as the AR DBD subunit A in A. The zincs of subunit B fail to occupy the peaks in the anomalous difference Fourier map in this dimeric arrangement, indicating the AR DBD does not form a head-to-tail dimer.
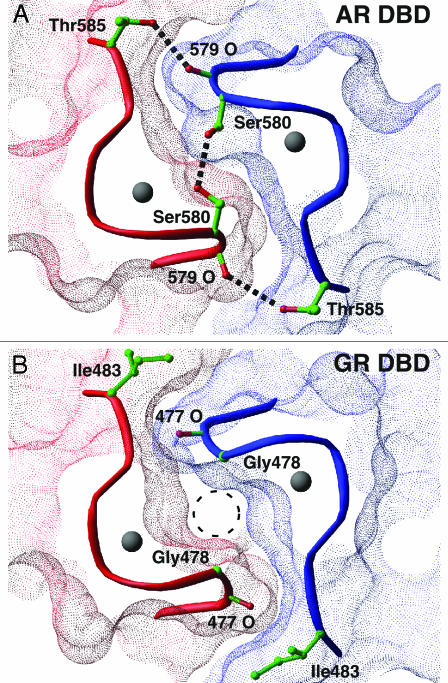
The AR DBD Homodimer Interface. The subunit interface of the AR DBD homodimer is symmetric and closely resembles that seen in the GR DBD–DNA complex (4). As in the GR DBD– and ER DBD–DNA complexes, the majority of the cross-subunit contacts are made in the D box region of the second zinc module. In the GR homodimer, the subunit interface is stabilized both by a network of hydrogen bonds between D box residues and by an extensive complementary surface. As seen in Fig. 4B, however, the GR interface contains a void formed where the Gly-478[39] from the opposing subunits face each other. This “glycine hole” is also a feature of the MR and PR. In the AR DBD, however, glycine is replaced by Ser-580[39]. This serine packs into the glycine hole of the dimer interface, filling the void and making van der Waals contact with its counterpart in the other subunit. In addition, the arrangement of the two serines is optimal for the formation of a hydrogen bond across the molecular pseudodyad. The substitution of serine for glycine in the AR D box is likely to increase the relative strength of the dimer interface of the AR DBD.
Fig. 4.
(A) The AR DBD dimer interface. The molecular surfaces of the AR subunits are shown in red and blue. Dashed black lines are hydrogen bonds. (B) A similar view of the GR DBD dimer interface. The “glycine hole” is noted by the dashed circle.
The AR DBD also makes an additional pair of symmetrical contacts between Thr-585[44] and the carbonyl oxygen of Ala-579[38] in the opposing protomer. In the GR DBD the residue at this position is an isoleucine, and replacement with a threonine as seen in the AR is likely to increase the stability of the dimer because of the enthalpic contribution of the additional two hydrogen bonds. In addition, the change from Ile in GR to Thr in AR removes a nonpolar residue from the solvent-exposed surface of the DBD, thus entropically stabilizing the AR as well.
The AR DBD (P.L.S. and D.T.G., unpublished work) and GR DBD (33) are monomers in solution. Because cooperative dimerization greatly increases the affinity of receptors for their bipartite response elements, these two changes should also increase the relative affinity of the AR for a given response element compared with GR. In support of this hypothesis, GR DBD mutants containing a serine in place of Gly-478[39] in the D box or a threonine in place of GR Ile-483[44] show increased affinity for both palindromic and direct-repeat response elements compared with wild type (34), confirming the importance of these interactions for dimer stability.
Protein–DNA Interactions. The DNA used for cocrystallization has a DR3 arrangement of hexameric half-sites, with the sense strand sequence 5′-CC AGAACA TCA AGAACA G-3′. However, the AR proteins were observed to bind in a symmetric, head-to-head arrangement, as was seen with steroid receptors bound to an IR3 response element (symmetrized consensus sequence of 5′-AGAACA NNN TGTTCT-3′). One half-site, bound by protomer A and shown here as upstream, is common to both DR3 and IR3 elements and is a high-affinity, consensus-binding site for steroid DBDs. Protomer B, on the other hand, binds to the downstream half-site that contains the consensus IR3-type bases at only the second and fifth positions. Experimentally phased electron density maps were used to identify the length of the asymmetric flanking sequences and unambiguously assign the orientation of the DNA. Within the limitations imposed by the diffraction resolution, the DNA does not exhibit significant deviations from B form.
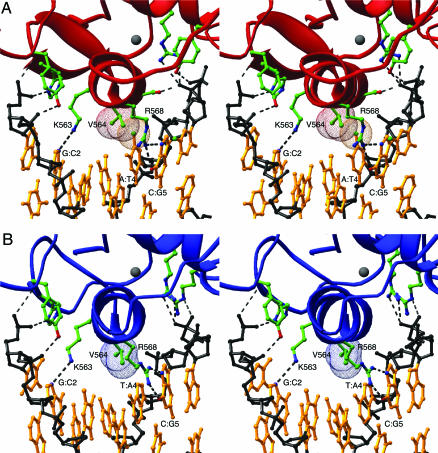
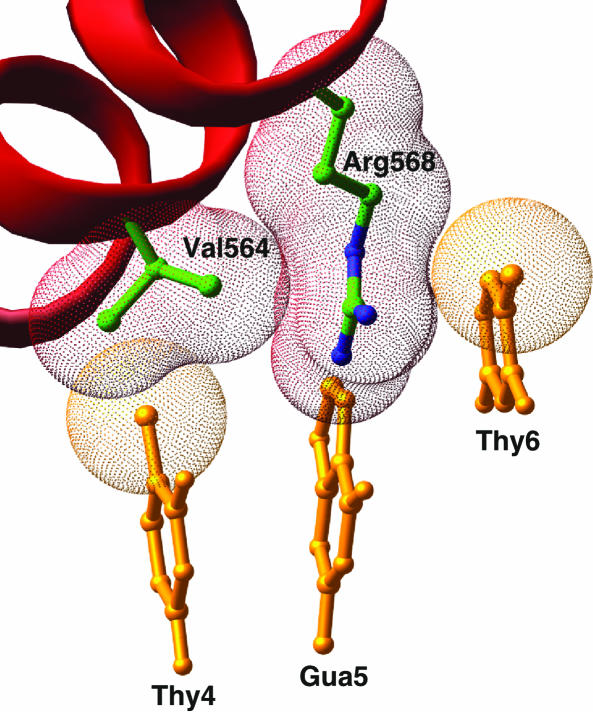
Backbone DNA contacts are similar for both AR protomers (Fig. 5) and show the pattern seen previously in structures of steroid receptor–DNA complexes (4, 35). The base-specific contacts between the AR DBD and the consensus half-site are also nearly identical with those of the GR DBD to its cognate half-site and are shown in Fig. 5A. In addition to these previously described interactions, we also note that the aliphatic portion of the Arg-568[27] side chain makes additional van der Waals contacts with Val-564[23] and the C5 methyl group of the thymine at the sixth position of the consensus half-site. Thymine is the only base that can form the second half of this van der Waals “sandwich,” and this specific contact likely explains why an A:T base pair is commonly observed at the sixth position of AR-specific half-sites (Fig. 6). Because the interaction between the conserved arginine and thymine is also present in consensus half-sites in the GR, ER, 9-cis-retinoic acid receptor, and other steroid and nuclear hormone receptor DBD structures, this can explain the preference for the A:T base pair at the sixth position in these protein–DNA complexes as well.
Fig. 5.
Stereoview of the AR DBD–DNA interfaces. (A) The upstream, cognate, protein–DNA interface. (B) The downstream, noncognate interface. The protein is shown in the same orientation as in A.
Fig. 6.
The arginine “sandwich.” Val-564 and Arg-568 of the AR DBD subunit along with bases T4, G5, and T6 of the antisense strand of the upstream, cognate half-site are shown. The C5 methyl group of T6 forms van der Waals interactions with one face of Arg-568, whereas the other side packs against Val-564.
The nonconsensus half-site interaction seen in the AR DBD–ADR3 structure contains the top strand sequence 5′-AGAACA-3′, with the two bases that match the consensus for a downstream IR3 half-site underlined. These two bases lie at the correct IR3 positions because they are symmetric within the hexameric half-site. This serendipitous match to the consensus IR3 half-site allows Lys-563[22] and Arg-568[27] of protomer B to recapitulate the hydrogen bonds to the GC base pairs at positions 2 and 5 of the hexameric half-site, as seen in the upstream element. These two “hooks” are common elements that position the recognition helix within the major groove of the hexameric half-site (36).
In the cognate AR DBD half-complex, the side chain of Val-564[23] makes van der Waals contact with the 5-methyl group of the T4 of the antisense strand. This interaction between the two nonpolar substituents is the discriminating feature of specific steroid receptor–DNA interfaces, and the resulting dehydration of the protein–DNA interface contributes entropic stabilization to the binding (35, 37). In the nonconsensus AR half-complex, A replaces the T at position 4 of the sense strand, resulting in the loss of the Val-564[23]-T4 contact. Although this replacement reduces the number of specific, stabilizing, interactions with the DNA half-site, the substitution of an A base for the consensus T does not cause a steric clash that might disfavor binding to this element. As befits the reduced complementarity between the AR DBD and the nonconsensus half-site, the cognate half-complex buries slightly more surface area from solvent (1,230 Å2) than the noncognate one (960 Å2).
AR Mutations. Mutations in the AR DBD associated with partial or complete androgen insensitivity (see ww2.mcgill.ca/androgendb) can be understood mechanistically in light of the structure determined here. Many of these were correctly analyzed earlier based on the structure of the GR DBD (38). More recently, within the D box, Ala579Thr (39–41) and Ser580Thr (42) mutations have been reported to lead to loss of AR dimerization. Modeling the Ser580Thr mutation on the AR DBD dimer leads to bad steric clashes in any possible Thr conformation, forcing backbone shifts that presumably disfavor dimerization. Modeling of the Ala579Thr substitution is more problematic, because the Thr side chains can each be accommodated with modest steric overlaps of 0.3–0.4 Å. However, that may be enough to force structural changes in the interface, and the imprecision of low resolution may underestimate the problem. The Ala579Thr mutation can be relieved by a compensatory change in Thr-585 to Ala (43), close to residue 579 across the dimer interface. This further change may relieve strains in the dimer interface or in the Zn ligand geometry caused by the Ala579Thr mutation.
Discussion
We have determined the structure of the AR DBD bound to an idealized steroid DR3 response element. Based on studies of the VDR DBD (6), which also binds to a DR3-type response element, we expected the tandem arrangement of half-sites to direct head-to-tail binding of the AR DBD to the DNA. Surprisingly, however, the AR DBDs bind to the direct-repeat response element as head-to-head symmetrical dimers. This mismatch between receptor dimer- and response element-arrangement results in one AR DBD bound to a high-affinity cognate half-site, and the partner DBD bound to a lower-affinity half-site. This finding indicates that the energetic penalty incurred by binding to a less favored half-site sequence is more than offset by maintaining the preferred IR3-type dimer interface. This finding is analogous to an earlier observation that the GR DBD maintains the IR3 dimer interface and spacing even when challenged with an IR4 response element (4).
Both the AR and the GR exhibit similar interactions with steroid response elements, yet the AR exhibits consistently stronger binding to direct repeat-type response elements than does the GR. Some of this difference in affinity may be attributable to differences in the C-terminal extension of each DBD, although in both GR and AR these regions were disordered in the crystal structure and may contribute only general electrostatic interactions without affecting selectivity or discrimination. Within the core of the DBD, however, the protein–DNA interactions are nearly identical for both receptor DBDs, and much of the difference in response element affinity is therefore likely to reside in the ability of each receptor to cooperatively form head-to-head dimers on bipartite response elements where the interaction with one or both hexameric half-sites is nonoptimal.
The second zinc module has been shown to be necessary for AR to bind cooperatively to ADR3s (44). The steroid receptor DBD dimerization interface is contained within this module, and between AR and GR it differs at just four positions. The increased AR dimer affinity can be explained by two of these four substitutions, one in the D box, and the other two residues beyond. In the D box, AR is the only steroid receptor that has a Ser residue at the second position, Ser-580[39], and this serine packs into the core of the dimer interface, making both van der Waals interactions and a cross-subunit hydrogen bond. All other steroid receptors have a Gly at this position, which lacks this additional hydrogen bond and leaves a void in the interface. Two residues beyond the D box, an Ile-to-Thr substitution in AR allows both a favorable cross-subunit side chain-to-backbone hydrogen bond and removes the nonpolar Ile side chain from exposure to solvent. Together these two substitutions appear to account for the stronger AR dimer interface. These substitutions in turn allow the receptor to bind to a more diverse set of response elements with higher affinity and cooperativity than the GR.
Biochemical evidence for the increased cooperativity of the AR DBD dimer correlates with these structural observations. All the steroid receptors (MR, PR, GR, and AR) show a 5- to 10-fold lower affinity for the naturally occurring PB-ARE-2 DR3-type element than the C3 (1) IR3-type element (34). However, the AR DBD binds 3- to 10-fold better to both elements relative to the other steroid receptors. Thus, the binding constant for AR on an apparent DR3 target (23 ± 5 nM) is the same as that of the other receptors for the more optimal IR3 element (the average of the other three is 23 ± 9 nM) (44). Because the concentration of individual steroid receptors in the cell is approximately nanomolar, differences in binding constants of this order are likely to be significant. AR substitutions in the GR dimerization interface, including Gly483Ser and Ile483Thr, show higher affinity binding to both DR3 and IR3 response elements (34), thus mimicking the behavior of the AR. Together with the structural data, these observations suggest a model where, because of the increased strength of the AR dimer interface, AR-selective gene activation arises from the ability of the AR to bind to IR3 response elements that have a greater deviation from the consensus half-site sequence. The reverse cross-activation of GR-responsive genes by the AR would likely be disfavored by the highly tissue-specific expression pattern of the AR compared with the GR.
The structure of the AR DBD bound as an inverted repeat to a direct-repeat response element highlights the fact that DNA target recognition by hormone receptors is strongly governed by the dimerization behavior of the two interacting protomers, even at the cost of losing specific interactions with the target DNA. With the exception of the Ecdysone receptor, which binds to IR1 rather than IR3 targets consisting of AGGTCA rather than AGAACA half-sites (45), no physiologically relevant dimerization interface within the classical steroid receptor DBDs, other than the primary one, has been observed to date in structural studies. Moreover, attempts to capture such potential alternative interfaces, as described in this report, and previously for GR (4), have been unfruitful. This in turn implies that selective hormone response elements that appear to have alternative arrangements of their hexameric half-sites, such as the pemARE with a proposed 5-bp spacer between half-sites (46), may instead simply be further examples of the ability of these receptors to exploit the strength of their DBD dimerization interfaces to accommodate suboptimal protein–half-site interactions. This ability is likely to be not only a mechanism of response element discrimination, but also an effective way of modulating transcription from different hormone-responsive genes.
Supplementary Material
Acknowledgments
We thank Nikki Fetter and Jenna Vanliere for help with crystallization and Karen Soldano for preparative assistance. This work was supported by U.S. Army Prostate Cancer Research Program grants (to D.T.G. and F.C.).
Abbreviations: AR, androgen receptor; GR, glucocorticoid receptor; MR, mineralocorticoid receptor; PR, progesterone receptor; ER, estrogen receptor; VDR, vitamin D receptor; DBD, DNA-binding domain; ARE, androgen response element.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1R4I).
References
- 1.McPhaul, M. J. (1999) J. Steroid Biochem. Mol. Biol. 69, 315–322. [DOI] [PubMed] [Google Scholar]
- 2.Gottlieb, B., Lehvaslaiho, H., Beitel, L. K., Lumbroso, R., Pinsky, L. & Trifiro, M. (1998) Nucleic Acids Res. 26, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf, D. J., Thummel, C., Beato, M., Herrlich, P., Schutz, G., Umesono, K., Blumberg, B., Kastner, P., Mark, M., Chambon, P., et al. (1995) Cell 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luisi, B. F., Xu, W. X., Otwinowski, Z., Freedman, L. P., Yamamoto, K. R. & Sigler, P. B. (1991) Nature 352, 497–505. [DOI] [PubMed] [Google Scholar]
- 5.Khorasanizadeh, S. & Rastinejad, F. (2001) Trends Biochem. Sci. 26, 384–390. [DOI] [PubMed] [Google Scholar]
- 6.Shaffer, P. L. & Gewirth, D. T. (2002) EMBO J. 21, 2242–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mader, S., Chen, J. Y., Chen, Z., White, J., Chambon, P. & Gronemeyer, H. (1993) EMBO J. 12, 5029–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perlmann, T., Rangarajan, P. N., Umesono, K. & Evans, R. M. (1993) Genes Dev. 7, 1411–1422. [DOI] [PubMed] [Google Scholar]
- 9.Towers, T. L., Luisi, B. F., Asianov, A. & Freedman, L. P. (1993) Proc. Natl. Acad. Sci. USA 90, 6310–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zechel, C., Shen, X. Q., Chambon, P. & Gronemeyer, H. (1994) EMBO J. 13, 1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zechel, C., Shen, X. Q., Chen, J. Y., Chen, Z. P., Chambon, P. & Gronemeyer, H. (1994) EMBO J. 13, 1425–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beato, M., Herrlich, P. & Schutz, G. (1995) Cell 83, 851–857. [DOI] [PubMed] [Google Scholar]
- 13.Schwabe, J. W., Chapman, L., Finch, J. T. & Rhodes, D. (1993) Cell 75, 567–578. [DOI] [PubMed] [Google Scholar]
- 14.Strahle, U., Boshart, M., Klock, G., Stewart, F. & Schutz, G. (1989) Nature 339, 629–632. [DOI] [PubMed] [Google Scholar]
- 15.Funder, J. W. (1993) Science 259, 1132–1133. [DOI] [PubMed] [Google Scholar]
- 16.Muller, J. M., Isele, U., Metzger, E., Rempel, A., Moser, M., Pscherer, A., Breyer, T., Holubarsch, C., Buettner, R. & Schule, R. (2000) EMBO J. 19, 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.List, H. J., Lozano, C., Lu, J., Danielsen, M., Wellstein, A. & Riegel, A. T. (1999) Exp. Cell Res. 250, 414–422. [DOI] [PubMed] [Google Scholar]
- 18.Claessens, F., Alen, P., Devos, A., Peeters, B., Verhoeven, G. & Rombauts, W. (1996) J. Biol. Chem. 271, 19013–19016. [DOI] [PubMed] [Google Scholar]
- 19.Rennie, P. S., Bruchovsky, N., Leco, K. J., Sheppard, P. C., McQueen, S. A., Cheng, H., Snoek, R., Hamel, A., Bock, M. E. & MacDonald, B. S. (1993) Mol. Endocrinol. 7, 23–36. [DOI] [PubMed] [Google Scholar]
- 20.Verrijdt, G., Schoenmakers, E., Alen, P., Haelens, A., Peeters, B., Rombauts, W. & Claessens, F. (1999) Mol. Endocrinol. 13, 1558–1570. [DOI] [PubMed] [Google Scholar]
- 21.Verrijdt, G., Schoenmakers, E., Haelens, A., Peeters, B., Verhoeven, G., Rombauts, W. & Claessens, F. (2000) J. Biol. Chem. 275, 12298–12305. [DOI] [PubMed] [Google Scholar]
- 22.Rastinejad, F., Perlmann, T., Evans, R. M. & Sigler, P. B. (1995) Nature 375, 203–211. [DOI] [PubMed] [Google Scholar]
- 23.Rastinejad, F., Wagner, T., Zhao, Q. & Khorasanizadeh, S. (2000) EMBO J. 19, 1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao, Q., Chasse, S. A., Devarakonda, S., Sierk, M. L., Ahvazi, B. & Rastinejad, F. (2000) J. Mol. Biol. 296, 509–520. [DOI] [PubMed] [Google Scholar]
- 25.Verrijdt, G., Haelens, A. & Claessens, F. (2003) Mol. Genet. Metab. 78, 175–185. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 27.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vagin, A. A. & Isupov, M. N. (2001) Acta Crystallogr. D. 57, 1451–1456. [DOI] [PubMed] [Google Scholar]
- 29.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 30.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 31.Word, J. M., Lovell, S. C., LaBean, T. H., Taylor, H. C., Zalis, M. E., Presley, B. K., Richardson, J. S. & Richardson, D. C. (1999) J. Mol. Biol. 285, 1711–1733. [DOI] [PubMed] [Google Scholar]
- 32.Carson, M. (1991) J. Appl. Crystallogr. 24, 958–961. [Google Scholar]
- 33.Freedman, L. P., Yamamoto, K. R., Luisi, B. F. & Sigler, P. B. (1988) Cell 54, 444 (lett.). [DOI] [PubMed] [Google Scholar]
- 34.Schoenmakers, E., Verrijdt, G., Peeters, B., Verhoeven, G., Rombauts, W. & Claessens, F. (2000) J. Biol. Chem. 275, 12290–12297. [DOI] [PubMed] [Google Scholar]
- 35.Gewirth, D. T. & Sigler, P. B. (1995) Nat. Struct. Biol. 2, 386–394. [DOI] [PubMed] [Google Scholar]
- 36.Nelson, C. C., Hendy, S. C., Shukin, R. J., Cheng, H., Bruchovsky, N., Koop, B. F. & Rennie, P. S. (1999) Mol. Endocrinol. 13, 2090–2107. [DOI] [PubMed] [Google Scholar]
- 37.Lundback, T., Cairns, C., Gustafsson, J. A., Carlstedt-Duke, J. & Hard, T. (1993) Biochemistry 32, 5074–5082. [DOI] [PubMed] [Google Scholar]
- 38.Luisi, B. F., Schwabe, J. W. & Freedman, L. P. (1994) in Vitamins and Hormones, ed. Litwack, G. (Academic, New York), Vol. 49, pp. 1–47. [DOI] [PubMed] [Google Scholar]
- 39.Gast, A., Neuschmid-Kaspar, F., Klocker, H. & Cato, A. C. (1995) Mol. Cell Endocrinol. 111, 93–98. [DOI] [PubMed] [Google Scholar]
- 40.Holterhus, P. M., Wiebel, J., Sinnecker, G. H., Bruggenwirth, H. T., Sippell, W. G., Brinkmann, A. O., Kruse, K. & Hiort, O. (1999) Pediatr. Res. 46, 684–690. [DOI] [PubMed] [Google Scholar]
- 41.Lundberg Giwercman, Y., Nikoshkov, A., Lindsten, K., Bystrom, B., Pousette, A., Knudtzon, J., Alm, J. & Wedell, A. (2000) Horm. Res. 53, 83–88. [DOI] [PubMed] [Google Scholar]
- 42.Nordenskjold, A., Friedman, E., Tapper-Persson, M., Soderhall, C., Leviav, A., Svensson, J. & Anvret, M. (1999) Urol. Res. 27, 49–55. [DOI] [PubMed] [Google Scholar]
- 43.Kaspar, F., Klocker, H., Denninger, A. & Cato, A. C. (1993) Mol. Cell. Biol. 13, 7850–7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenmakers, E., Alen, P., Verrijdt, G., Peeters, B., Verhoeven, G., Rombauts, W. & Claessens, F. (1999) Biochem. J. 341, 515–521. [PMC free article] [PubMed] [Google Scholar]
- 45.Devarakonda, S., Harp, J. M., Kim, Y., Ozyhar, A. & Rastinejad, F. (2003) EMBO J. 22, 5827–5840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Geserick, C., Meyer, H. A., Barbulescu, K. & Haendler, B. (2003) Mol. Endocrinol. 17, 1738–1750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.