Abstract
By monitoring the end-to-end extension of a mechanically stretched, supercoiled, single DNA molecule, we have been able directly to observe the change in extension associated with unwinding of approximately one turn of promoter DNA by RNA polymerase (RNAP). By performing parallel experiments with negatively and positively supercoiled DNA, we have been able to deconvolute the change in extension caused by RNAP-dependent DNA unwinding (with ≈1-bp resolution) and the change in extension caused by RNAP-dependent DNA compaction (with ≈5-nm resolution). We have used this approach to quantify the extent of unwinding and compaction, the kinetics of unwinding and compaction, and effects of supercoiling, sequence, ppGpp, and nucleotides. We also have used this approach to detect promoter clearance and promoter recycling by successive RNAP molecules. We find that the rate of formation and the stability of the unwound complex depend profoundly on supercoiling and that supercoiling exerts its effects mechanically (through torque), and not structurally (through the number and position of supercoils). The approach should permit analysis of other nucleic-acid-processing factors that cause changes in DNA twist and/or DNA compaction.
Transcription initiation involves a series of reactions (1–2): (i) RNA polymerase holoenzyme (RNAP) binds to promoter DNA to form an RNAP–promoter closed complex; (ii) RNAP unwinds approximately one turn of the promoter DNA to form an RNAP–promoter open complex (in a process referred to as “promoter unwinding”); and (iii) RNAP escapes the promoter and enters into productive synthesis of RNA as an RNAP–DNA elongation complex (in a process referred to as “promoter clearance”).
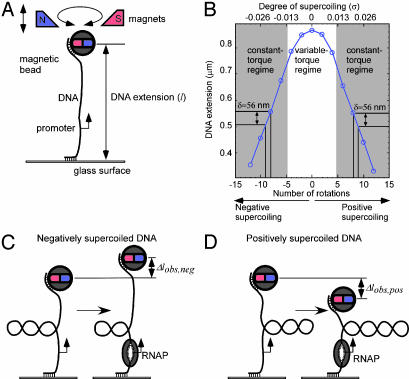
We have developed a single-molecule DNA-nanomanipulation approach that enables us to detect and characterize promoter unwinding and promoter clearance by RNAP. Our approach uses an experimental setup originally developed for analysis of DNA polymer physics (Fig. 1A and refs. 3–5). In this experimental setup, a double-stranded DNA molecule containing a single promoter site is attached at one end, through multiple linkages, to a paramagnetic bead, and at the other end, through multiple linkages, to a glass surface; the DNA is torsionally constrained and mechanically stretched between the bead and the glass surface by application of a pair of magnets above the DNA helix axis; and the distance between the bead and the glass surface (which reflects the DNA end-to-end extension, l) is monitored in real time by using videomicroscopy. Upon rotation of the pair of magnets, the bead is rotated in lock-step register, superhelical turns are introduced into the torsionally constrained DNA molecule in lock-step register, supercoils are formed, and, correspondingly, l is changed. With this experimental setup, it readily is possible to construct an experimental calibration curve relating l to the number of clockwise or counterclockwise rotations of the pair of magnets, and thus to the number of negative or positive superhelical turns (Fig. 1B; refs. 3–5). Over a broad range of negative or positive supercoiling, l changes linearly with the number of negative or positive superhelical turns, with, under our conditions, a change in l of 56 ± 5 nm per superhelical turn (Fig. 1B).
Fig. 1.
Experimental approach. (A) Experimental setup. A double-stranded 4-kb DNA molecule containing a single promoter is tethered at one end, through multiple linkages, to a paramagnetic bead, and at the other end, through multiple linkages, to a glass surface. The DNA is torsionally constrained and mechanically stretched between the bead and the glass surface by application of a pair of magnets above the DNA helix axis. The distance between the bead and the surface, which reflects the DNA end-to-end extension (l), is monitored in real time by videomicroscopy. Upon rotation of the pair of magnets, the bead is rotated in lock-step register, superhelical turns are introduced into the DNA in lock-step register, plectonemic supercoils are formed, and, correspondingly, l is changed. (B) Calibration of l vs. number of superhelical turns. Over a broad range of positive and negative supercoiling, there is a linear relationship between l and the number of superhelical turns, with a change in l (δ) of 56 ± 5 nm per superhelical turn. (C and D) Detection of promoter unwinding. According to the relationship Lk = Tw + Wr (6), in a torsionally constrained DNA molecule with constant linking number (Lk), a change in twist (Tw; unwinding), must be compensated by an equal, but opposite, change in writhe (Wr; number of supercoils). With negatively supercoiled DNA, unwinding of approximately one turn of promoter DNA by RNAP must result in a compensatory loss of approximately one negative supercoil and, correspondingly, an increase in l (Δlobs,neg). With positively supercoiled DNA, unwinding of approximately one turn of promoter DNA by RNAP must result in a compensatory gain of approximately one positive supercoil and, correspondingly, a decrease in l (Δlobs,pos).
Fig. 1 C and D shows how we use this experimental setup to detect promoter unwinding by RNAP. Conservation of linking number (Lk), according to the established relationship Lk = Tw + Wr (6), implies that in a torsionally constrained DNA molecule a change in twist (Tw; unwinding) must be compensated by an equal, but opposite, change in writhe (Wr; number of supercoils). With a negatively supercoiled DNA molecule, unwinding of approximately one turn of promoter DNA by RNAP must result in a compensatory loss of approximately one negative supercoil and, correspondingly, an increase in l of ≈56 nm (Δlobs,neg; Fig. 1C). With a positively supercoiled DNA molecule, unwinding of approximately one turn of promoter DNA by RNAP must result in a compensatory gain of approximately one positive supercoil and, correspondingly, a decrease in l of ≈56 nm (Δlobs,pos; Fig. 1D). Thus, our approach couples RNAP-dependent promoter unwinding to movement of the bead. This results in an immense amplification of signal: converting a subnanometer scale, effectively undetectable, change in local DNA geometry, to a tens-of-nanometer scale, readily detectable, change in the position of the bead.
Methods
DNA Fragments. lacCONS-4kb and rrnBP1–4kb are 4-kb G/C-rich (67% G/C) DNA fragments that contain centrally located single copies of, respectively, the lacCONS promoter (7, 8) and the rrnB P1 promoter (23). lacCONS-4kb and rrnBP1–4kb were prepared, attached to beads, attached to glass surfaces, and calibrated as described for rrnBP1–4kb in ref. 5.
Data Collection. Unless noted otherwise, reaction mixtures contained 0.5 nM RNAP (Epicentre) and negatively supercoiled, mechanically stretched, DNA molecule lacCONS-4kb or rrnBP1–4kb (ref. 5; σ = –0.018; extending force = 0.3 pN), or 5 nM RNAP (Epicentre) and positively supercoiled, mechanically stretched, DNA molecule lacCONS-4kb or rrnBP1–4kb (ref. 5; σ = 0.018; extending force = 0.3 pN), in transcription buffer (25 mM Hepes·NaOH, pH 7.9/100 mM NaCl/10 mM MgCl2/0.1% Tween 20/0.1 mg/ml BSA/3 mM 2-mercaptoethanol) at 34°C. Reactions were initiated by introduction of transcription buffer containing RNAP by flow (1 ml; 1 min), followed by cease of flow and data collection. Reactions were terminated, and the DNA molecule was recycled for further experiments, by application of an extending force and introduction of at least five positive supercoils, followed by introduction of transcription buffer (3 ml; 15 min).
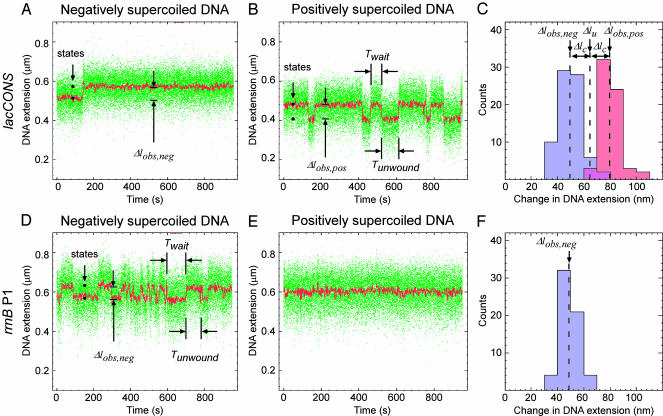
Data Analysis: DNA Unwinding and DNA Compaction. DNA unwinding has opposite effects on l in experiments with negatively and positively supercoiled DNA (increases l with negatively supercoiled DNA; decreases l with positively supercoiled DNA), whereas DNA compaction has equivalent effects on l in experiments with negatively and positively supercoiled DNA (decreases l with both negatively and positively supercoiled DNA). Therefore, effects of unwinding and compaction can be deconvoluted from data with negatively and positively supercoiled DNA by use of simple algebra (Fig. 2C and Fig. 5, which is published as supporting information on the PNAS web site; see ref. 5). From experimental time traces with data averaged by using a 1-s window (red points in Fig. 2), extents of DNA unwinding can be determined to within ±3 bp, and extents of DNA compaction can be determined to within ±20 nm; from experimental time traces with data averaged by using a 20-s window, extents of unwinding can be determined to within ± 1 bp, and extents of compaction can be determined to within ±5 nm.
Fig. 2.
Detection of promoter unwinding. (A and B) Single-molecule traces of DNA extension vs. time for interaction of RNAP with a consensus promoter, as assessed with negatively supercoiled DNA (A; σ =–0.018; stable, effectively irreversible, promoter unwinding) and with positively supercoiled DNA (B; σ = 0.018; unstable, reversible promoter unwinding). Green points, raw data obtained at video rate (30 frames per s); red points, averaged data (1-s window); Δlobs,neg, transition amplitude with negatively supercoiled DNA; Δlobs,pos, transition amplitude with positively supercoiled DNA; Twait, time interval between a rewinding event and the next unwinding event; Tunwound, time interval between an unwinding event and the next rewinding event. (C) Histograms of Δlobs,neg (blue) and Δlobs,pos (red). The change in l attributable to DNA unwinding (Δlu) and the change in l attributable to DNA compaction arising from wrapping and/or bending (Δlc) are calculated as Δlu = (Δlobs,neg +Δlobs,pos)/2 and Δlc = (Δlobs,pos–Δlobs,neg)/2. The extent of unwinding (ΔTw) is calculated as ΔTw =Δlu/δ. (See Fig. 5; see also discussion in ref. 5.) (D–F) As in A–C, but with the rrnB P1 promoter (unstable, reversible promoter unwinding with negatively supercoiled DNA; no promoter unwinding with positively supercoiled DNA).
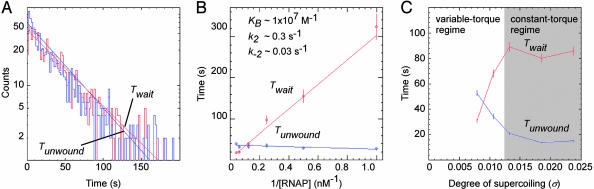
Data Analysis: Twait and Tunwound. Histograms of observed values of Twait and Tunwound were prepared from ≥100 individual unwinding/rewinding events in raw time traces. Histograms displayed single-exponential distributions, with mean equal to standard deviation, allowing determination of mean values of Twait and Tunwound as half-lives of distributions (Fig. 3A).
Fig. 3.
Kinetics and supercoiling-dependence of promoter unwinding. (A) Histograms of Twait and Tunwound (consensus promoter, positively supercoiled DNA; σ = 0.018; n = 1,127 events). (B) Tau plot (13) of values of Twait and Tunwound measured at a series of RNAP concentrations (consensus promoter, positively supercoiled DNA; n ≥ 100 events per RNAP concentration). KB, k2, and k–2, are, respectively, the equilibrium constant for formation of closed complex, the rate constant for unwinding, and the rate constant for rewinding in the standard kinetic scheme (refs. 1, 2, and 13; see Eq. 1). The slope and y intercept of the tau plot of Twait yields (KBk2)–1 and (k2)–1; the y intercept of the tau plot of Tunwound yields (k–2)–1. (C) Supercoiling-dependence of Twait and Tunwound (consensus promoter, positively supercoiled DNA; σ = 0.008 to 0.024; n ≥ 100 events per σ value). The variable-torque and constant-torque regimes are defined based on the nonlinear and linear regimes of the experimental calibration curve in Fig. 1B (see ref. 18). Torque, |Γ|, is estimated to be 4.2, 4.4, 4.6, 5.0, and 5.0 pN nm for superhelical densities of 0.008, 0.011, 0.013, 0.019, and 0.024, respectively (refs. 4, 18, and 21 and T.R.S., unpublished results). The magnitude of the supercoiling dependence in the variable-torque regime is consistent with a simple Arrhenius-law model in which torque biases the free energy of promoter unwinding/rewinding (see ref. 26).
Results and Discussion
We have performed experiments using Escherichia coli RNAP and the consensus promoter lacCONS (7, 8). Fig. 2 A and B document detection of formation of the unwound complex in experiments with negatively and positively supercoiled DNA, respectively. With negatively supercoiled DNA, under conditions where, based on conventional assays, the RNAP–promoter open complex is expected to be stable and effectively irreversible, experimental single-molecule traces of l vs. time reveal single, abrupt, effectively irreversible, increases in DNA extension (Δlobs,neg = 50 ± 5 nm; Fig. 2 A; see also Fig. 6A, which is published as supporting information on the PNAS web site). With positively supercoiled DNA, under conditions where, based on conventional assays, the RNAP–promoter open complex is expected to be unstable and reversible, experimental single-molecule traces of l vs. time reveal cycles of abrupt decreases in l (Δlobs,neg = 80 ± 5 nm) followed by abrupt increases in l, returning to the initial state (Δlobs,neg = 80 ± 5 nm; Fig. 2 A; see also Fig. 6B). Extensive control experiments establish that the observed transitions correspond to single promoter-unwinding and promoter-rewinding events. Thus: (i) no events are observed in the absence of RNAP; (ii) no events are observed in the absence of the initiation factor σ; (iii) no events are observed in the absence of a promoter; (iv) one, two, or three levels of events are observed with DNA molecules containing, respectively, one, two, or three promoters; (v) no events are observed at low temperatures (which are known to prevent formation of open complexes refs. 1 and 2); (vi) events are prevented by prior addition of heparin (which is known to prevent formation of open complexes; ref. 9); and (vii) events are not affected by subsequent addition of heparin (which is known not to affect open complexes once formed; ref. 9) (Figs. 7–11, which are published as supporting information on the PNAS web site, and additional data not shown).
The experimental time traces yield four observables. The first observable is the number of states, which reflects the number of unwinding intermediates in promoter unwinding (Fig. 2 A and B). The experimental time traces show only two states: an initial state and a final state, with abrupt, sharp transitions between them. Thus, there are no unwinding intermediates in promoter unwinding, at least none with lifetimes equal to or greater than the temporal resolution of the analysis (≈1 s).
The second observable is the amplitude of the transition between the initial and final states (Δlobs,neg with negatively supercoiled DNA; Δlobs,pos with positively supercoiled DNA; Fig. 2 A and B). The amplitude of the transition is a function of the extent of DNA unwinding, and of the extent of DNA compaction (arising from DNA wrapping and/or DNA bending). From the experimental time traces obtained with negatively supercoiled DNA and with positively supercoiled DNA, we are able to deconvolute the extent of DNA unwinding and the extent of DNA compaction (Figs. 2C and 5). The inferred extent of unwinding is 1.2 ± 0.1 turns, which is equivalent to 13 ± 1 bp, which corresponds within experimental error to the extent of unwinding defined by DNA footprinting (1, 2, 10). The inferred extent of compaction is 15 ± 5 nm, which corresponds within experimental error to the extent of compaction defined by scanning probe microscopy (refs. 11 and 12 and C. Rivetti, N. Naryshkin, E. Kortkhonjia, and R.H.E., unpublished data).
The third observable is the time interval between unwinding events (Twait; Figs. 2 A and B and 3A). Twait is a function of the rate of formation of the unwound complex. From experimental time traces determined at a series of RNAP concentrations (1–30 nM), using tau-plot analysis (13), we are able to deconvolute Twait into the equilibrium-binding constant for formation of closed complex (KB) and the rate constant for isomerization of closed complex to yield unwound complex (k2) in the standard kinetic scheme (Fig. 3B):
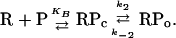 |
[1] |
The inferred values of KB (1 × 107 M–1) and k2 (0.3 s–1) are consistent with expectation based on conventional experiments (14–17).
The fourth observable is the time interval between an unwinding event and the subsequent rewinding event (Tunwound; Figs. 2 A and B and 3A). Tunwound directly yields the lifetime of the unwound state and the rate constant for rewinding (k–2). As expected, Tunwound is independent, within experimental error, of RNAP concentration (Fig. 3B). The inferred value of k–2 is 0.03 s–1, consistent with expectation based on conventional experiments (14–17).
To assess effects of supercoiling, we performed parallel experiments at superhelical densities of 0.008, 0.011, 0.013, 0.019, and 0.024 [where superhelical densities of 0.008, 0.011, and 0.013 are in the variable-torque regime (where torque increases with increasing superhelical density), and superhelical densities of 0.019 and 0.024 are in the constant-torque regime (where the torque is constant with increasing superhelical density) (ref. 18; Figs. 1B and 3C)]. In the variable-torque regime, Twait increases with increasing superhelical density (4-fold increase for an increase in superhelical density of 0.005), and Tunwound decreases with increasing superhelical density (4-fold decrease for an increase in superhelical density of 0.005). Thus, in the variable-torque regime, the ratio Twait/Tunwound changes by fully a factor of 16 for a change in superhelical density of 0.005. We conclude that, in the variable-torque regime, supercoiling affects the rate of formation of the unwound complex, the stability of the unwound complex, and, profoundly, the ratio of times in the unwound and wound states (see refs. 14–17). In contrast, in the constant-torque regime, Twait and Tunwound are constant. Based on the supercoiling dependence of Twait and Tunwound in the variable-torque regime and supercoiling independence of Twait and Tunwound in the constant-torque regime, we conclude that effects of supercoiling on promoter unwinding are manifest mechanically (i.e., through changes in torque), and not structurally (i.e., through changes in number and positions of supercoils; hypothesis in refs. 19 and 20). [In this work, where extending force is constant and l is variable, the transition between variable-torque and constant-torque regimes occurs at relatively low torque, achieved at relatively low negative or positive superhelical densities (|Γ| ≈ 5 pN nm; |σ| ≈ 0.014; Figs. 1B and 3C) (4, 18, 21). In vivo, where extending force is variable and l is constant, the transition between variable- and constant-torque regimes is expected to occur only at high torque, achieved only at high negative or positive superhelical densities (|Γ| ≥ 15 pN nm; |σ| ≥ 0.06) (4, 17, 18, 21, 22). Therefore, in vivo, Twait and Tunwound are expected to vary with supercoiling across essentially the full physiological range of supercoiling.]
To assess effects of promoter sequence, we performed parallel experiments with the rrnB P1 promoter, which, based on the literature, forms exceptionally unstable, reversible open complexes (23). Fig. 2D documents that, with negatively supercoiled DNA, the rrnB P1 promoter exhibits unstable, reversible unwinding (Tunwound = 16 s; compare Tunwound > 10,000 s for the consensus promoter). Fig. 2E documents that, with positively supercoiled DNA, the rrnB P1 promoter exhibits no detectable unwinding (Tunwound = 0 s; compare Tunwound = 34 s for the consensus promoter). The transition amplitude for promoter unwinding at the rrnB P1 promoter is indistinguishable from that at the consensus promoter (Δlobs,neg = 50 ± 5 nm vs. Δlobs,neg = 50 ± 5 nm; Fig. 2 A, B, and D), suggesting that the extents of unwinding and the extents of compaction at the two promoters are identical.
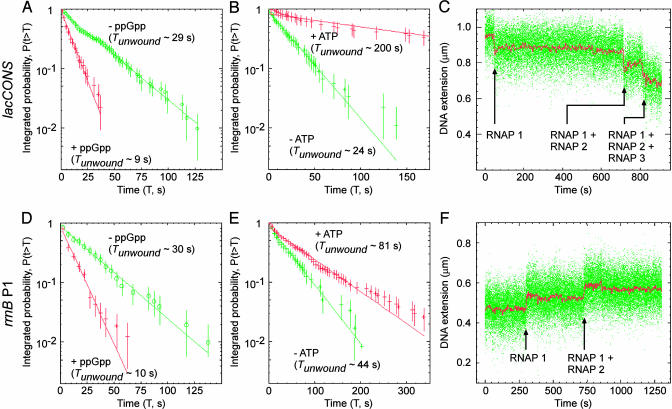
The effector ppGpp and initiating nucleotides have been reported to have significant effects on the stability of the open complex (24, 25). Fig. 4 A and B document effects of ppGpp on the stability of the unwound complex at, respectively, the consensus promoter and the rrnB P1 promoter. At both promoters, addition of ppGpp results in a large (≈3-fold) decrease in the lifetime of the unwound complex, in quantitative agreement with expectation based on the literature (24). Fig. 4 C and D documents effects of initiating nucleotides on the stability of the unwound complex at, respectively, the consensus promoter and the rrnB P1 promoter. At the consensus promoter, where ATP is able to serve as both the initiating nucleotide and the second nucleotide (permitting synthesis of the dinucleotide pppApA), addition of ATP results in a large (≈10-fold) increase in Tunwound. At the rrnB P1 promoter, where ATP is able to serve as the initiating nucleotide but not as the second nucleotide, addition of ATP results in a moderate, but reproducible (≈2-fold), increase in Tunwound. Control experiments show that the effects are nucleotide-specific. Thus, addition of the noncomplementary nucleotide GTP results in no increase in Tunwound, neither at the consensus promoter nor at the rrnB P1 promoter (data not shown). We conclude that initiating nucleotides stabilize the unwound complex, consistent with expectation based on the literature (25).
Fig. 4.
Effects of ppGpp, effects of initiating nucleotide, and observation of promoter clearance. (A) Effects of ppGpp (0 or 100 μM) on stability of the unwound complex (consensus promoter, positively supercoiled DNA; σ = 0.018; n = 200 events). (B) Effects of ATP (0 or 2 mM) on stability of the unwound complex (consensus promoter, positively supercoiled DNA; σ = 0.018; n = 100 events). (C) Observation of promoter clearance by detection of successive, cumulative unwinding events, each corresponding to promoter clearance by RNAP molecule n, followed by promoter binding and unwinding by RNAP molecule n + 1 (consensus promoter, positively supercoiled DNA; 2 mM each NTP). (D–F) As in A–C, but for the rrnB P1 promoter, with negatively supercoiled DNA.
To demonstrate that our experimental approach also can detect promoter clearance, we performed experiments in the presence of all four nucleoside triphosphates (NTPs) (ATP, CTP, GTP, and UTP). The rationale is as follows: in the presence of all four NTPs, RNAP molecule 1 will bind and unwind the promoter (just as in the absence of NTPs); after an interval of time (Tclear), RNAP molecule 1 will clear the promoter, rendering the promoter accessible, and permitting RNAP molecule 2 to bind and unwind, yielding a second, cumulative unwinding event; and so forth. Thus, by this logic, experiments in the presence of all four NTPs should yield successive, cumulative unwinding events, with each successive, cumulative unwinding event reporting promoter clearance by RNAP molecule n, followed by promoter binding and unwinding by RNAP molecule n + 1. Fig. 4 E and F shows that precisely such patterns of successive, cumulative unwinding events are observed, with both the consensus promoter (with positively supercoiled DNA) and the rrnB P1 promoter (with negatively supercoiled DNA). Extensive control experiments establish that the observed successive, cumulative unwinding events in fact reflect promoter clearances by RNA polymerase n and binding and unwinding by RNA polymerase molecule n + 1 (data not shown). In principle, the experimental time traces for each successive, cumulative unwinding event yields three observables relevant to the mechanism and kinetics of promoter clearance: (i) the number of states in the event (which reflects the number of unwinding intermediates in promoter clearance); (ii) the transition amplitude of the event (which reflects the extent of DNA unwinding and extent of DNA compaction in promoter clearance and permits determination of the extent of unwinding and extent of compaction in the transcription elongation complex); and (iii) the time interval between events (which reflects Tclear + Twait and permits determination of the kinetics of promoter clearance). Further experiments will be essential to realize the full potential of the approach for analysis of promoter clearance.
Our approach and results provide a single-molecule study of transcription initiation.
We note that our approach should be generalizable to analysis of other nucleic-acid-processing factors that affect DNA twist and/or DNA compaction. Integration of our approach (which permits control and readout of DNA torsional state) with single-molecule fluorescence (which permits readout of binding, translocation, and conformational states) should provide unprecedented opportunities for structural, mechanistic, and kinetic analysis of nucleic-acid-processing factors.
Supplementary Material
Acknowledgments
We thank D. Bensimon, V. Croquette, T. Record, R. Saecker, and K. Severinov for discussion. This work was supported by National Institutes of Health Grant GM41376 and a Howard Hughes Medical Investigatorship (to R.H.E.) and by a Cold Spring Harbor Laboratory Fellowship (to T.R.S.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RNAP, RNA polymerase holoenzyme; NTP, nucleoside triphosphate.
References
- 1.Record, M. T. J., Reznikoff, W., Craig, M., McQuade, K. & Schlax, P. (1996) in Escherichia coli and Salmonella, ed. Neidhart, F. C. (Am. Soc. Microbiol. Press, Washington, DC), Vol. 1, pp. 792–820. [Google Scholar]
- 2.deHaseth, P., Zupancic, M. & Record, M. T. J. (1998) J. Bacteriol. 180, 3019–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strick, T., Allemand, J., Bensimon, D., Bensimon, A. & Croquette, V. (1996) Science 271, 1835–1837. [DOI] [PubMed] [Google Scholar]
- 4.Strick, T., Allemand, J.-F., Bensimon, D. & Croquette, V. (1998) Biophys. J. 74, 2016–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Revyakin, A., Allemand, J.-F., Croquette, V., Ebright, R. & Strick, T. (2003) Methods Enzymol. 370, 577–598. [DOI] [PubMed] [Google Scholar]
- 6.White, J. (1969) J. Math. 91, 693–728. [Google Scholar]
- 7.Mukhopadhyay, J., Kapanidis, A., Mekler, V., Kortkhonjia, E., Ebright, Y. & Ebright, R. (2001) Cell 106, 453–463. [DOI] [PubMed] [Google Scholar]
- 8.Mekler, V., Kortkhonjia, E., Mukhopadhyay, J., Knight, J., Revyakin, A., Kapanidis, A., Niu, W., Ebright, Y., Levy, R. & Ebright, R. (2002) Cell 108, 599–614. [DOI] [PubMed] [Google Scholar]
- 9.Cech, C. & McClure, W. (1980) Biochemistry 19, 2440–2447. [DOI] [PubMed] [Google Scholar]
- 10.Zaychikov, E., Denissova, L., Meier, T., Gotte, M. & Heumann, H. (1997) J. Biol. Chem. 272, 2259–2267. [DOI] [PubMed] [Google Scholar]
- 11.Rivetti, C., Guthold, M. & Bustamante, C. (1999) EMBO J. 18, 4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naryshkin, N., Revyakin, A., Kim, Y., Mekler, V. & Ebright, R. (2000) Cell 101, 601–611. [DOI] [PubMed] [Google Scholar]
- 13.McClure, W. (1980) Proc. Natl. Acad. Sci. USA 77, 5634–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malan, T. P., Kolb, A., Buc, H. & McClure, W. (1984) J. Mol. Biol. 180, 881–909. [DOI] [PubMed] [Google Scholar]
- 15.Amouyal, M. & Buc, H. (1987) J. Mol. Biol. 195, 795–808. [DOI] [PubMed] [Google Scholar]
- 16.Borowiec, J. & Gralla, J. D. (1987) J. Mol. Biol. 195, 89–97. [DOI] [PubMed] [Google Scholar]
- 17.Su, T. T. & McClure, W. R. (1994) J. Biol. Chem. 269, 13511–13521. [PubMed] [Google Scholar]
- 18.Strick, T., Allemand, J.-F., Bensimon, D. & Croquette, V. (2000) Annu. Rev. Biophys. Biomol. Struct. 29, 523–543. [DOI] [PubMed] [Google Scholar]
- 19.ten Heggeler-Bordier, B., Wahli, W., Adrian, M., Stasiak, A. & Dubochet, J. (1992) EMBO J. 11, 667–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavigne, M., Kolb, A., Yeramian, E. & Buc, H. (1994) EMBO J. 13, 4983–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strick, T., Bensimon, D. & Croquette, V. (1999) Genetica 106, 57–62. [DOI] [PubMed] [Google Scholar]
- 22.Kanaar, R. & Cozzarelli, N. (1992) Curr. Opin. Struct. Biol. 2, 369–379. [Google Scholar]
- 23.Gourse, R. (1988) Nucleic Acids Res. 16, 9789–9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barker, M., Gaal, T., Josaitis, C. & Gourse, R. (2001) J. Mol. Biol. 305, 673–688. [DOI] [PubMed] [Google Scholar]
- 25.Gaal, T., Bartlett, M., Ross, W., Turnbough, C. & Gourse, R. (1997) Science 278, 2092–2097. [DOI] [PubMed] [Google Scholar]
- 26.Rief, M., Fernandez, J. & Gaub, H. (1998) Phys. Rev. Lett. 81, 4765–4767. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.