Abstract
Cajal bodies (CBs) are complex organelles found in the nuclei of a wide variety of organisms, including vertebrates, invertebrates, plants, and yeast. In most cell types CBs are <1 μm in diameter, severely limiting the range of experimental observations that can be made on them. By contrast, CBs in the amphibian oocyte nucleus (also called the germinal vesicle) are 2–10 μm in diameter. We have taken advantage of this large size to carry out kinetic studies on coilin, a protein that is specifically enriched in CBs. We labeled coilin with photoactivatable green fluorescent protein and analyzed the movement of the protein by confocal microscopy in unfixed germinal vesicles isolated in oil. We showed that coilin leaves the CB relatively slowly (minutes rather than seconds) with kinetics similar to earlier measurements on its entrance. We also showed that coilin diffuses very slowly within the CB, consistent with its being in a large macromolecular complex. Finally, we found that the movement of coilin is not directly affected by the transcriptional state of the nucleus or ongoing nucleocytoplasmic exchange. These data on the kinetics of coilin reinforce the conclusion that CB components are in a constant state of flux, consistent with models that postulate an active role for CBs in nuclear physiology.
In 1903, the Spanish neurobiologist Santiago Ramón y Cajal described small, silver-staining structures in the nuclei of vertebrate neurons (1), which he named accessory bodies. Only in the past decade, with the discovery of useful molecular markers, was it realized that homologous structures occur in a wide variety of animals and plants, including the yeast, Saccharomyces cerevisiae (2–4). These structures are now called Cajal bodies (CBs) in honor of their discoverer. One of the most commonly used markers for CBs is the protein p80-coilin. Coilin is highly enriched in CBs (5, 6) and thus can be used to identify CBs by immunofluorescence. Earlier studies suggested that coilin is involved in some step in the transport of small nuclear ribonucleoproteins (snRNPs) to the CBs in the nucleus (7, 8). More recent data from coilin knockout mice support this view (9, 10), as does biochemical evidence that coilin can associate with the survival of motor neurons (SMN) protein (11, 12), which is part of the machinery for assembly of snRNPs (13, 14).
In an earlier study, we used fluorescence recovery after photobleaching (FRAP) to show that coilin in the CB is in dynamic equilibrium with coilin in the nucleoplasm. Analysis of the FRAP curves revealed three kinetic components with residence times in the CB from several seconds to >30 min. FRAP data give direct information about entry of components into a structure, but exit kinetics must be inferred on the assumption that the system is at equilibrium. To learn more about the exit of coilin from the CB, we have carried out experiments with coilin labeled with photoactivatable green fluorescent protein (PA-GFP) (15). By activating PA-GFP fluorescence inside the CB, we could monitor the loss of coilin from the CB. Furthermore, by examining the distribution of fluorescence as a function of time after photoactivation, we showed that coilin diffuses very slowly within the CB. Finally, we showed that the flux of coilin in and out of the CB is independent of ongoing transcription or nucleocytoplasmic exchange.
Materials and Methods
Plasmids and Transcripts. The ORF of the Xenopus coilin gene (16) was cloned downstream of PA-GFP in the pPA-GFP-C1 vector (15). A 9-aa hemagglutinin (HA) tag was included at the C terminus of the coilin sequence. To generate a template for sense-strand transcripts with a poly(A) tail, we made a PCR product from the plasmid by using primers CM163 (or ZW33) and SD5. Finally, the PCR product was transcribed with T3 or T7 RNA polymerase.
Plasmids were as follows: CM163, 5′-GCAATTAACCCTCACTAAAGGGAAGATCCGCTAGCGCTACCGG-3′; ZW33, 5′-AGTAATACGACTCACTATAGGGAGAGAGCTGGTTTAGTGAACC-3′; SD5, 5′(T)30-GCAGTGAAAAAAATGCTTTATTTG-3′.
Xenopus U7 small nuclear RNA (snRNA) construct 401 (17) was linearized with PvuII and transcribed with T7 RNA polymerase to generate sense-strand RNA for injection. Alexa 546-UTP (Molecular Probes) was added to the reaction mix to provide a fluorescent label. Transcripts of Xenopus GFP-coilin were synthesized as described (16).
Microinjections and Germinal Vesicle (GV) Spreads. Methods for microinjection of oocytes, isolation of GVs, and preparation of GV spreads were as described (18). All photoactivation experiments were carried out on CBs in GVs that had been isolated and squashed in mineral oil. PA-GFP-coilin transcripts were injected along with Alexa 546-U7 snRNA at an ≈10:1 ratio to visualize CBs before photoactivtion.
Photoactivation of PA-GFP. A suitable CB was found in the microscope field by the red fluorescence of Alexa 546-U7 snRNA. Imaging, photoactivation, and bleaching were then conducted with a laser scanning confocal microscope (Leica TCS SP2, Leica Microsystems, Exton, PA), using a ×63, 1.4 numerical aperture oil immersion objective. Images were taken with the 488-nm laser at a single focal plane through the middle of a CB. Whole CB photoactivation was performed by scanning six times with the full intensity of a 405-nm laser. For spot activation within a CB, the 405-nm laser beam was parked for 25 ms without scanning. A bleached spot within a photoactivated CB was obtained by applying full intensity of the 405-nm laser for 1 s without scanning. After photoactivation, images were collected with the 488-nm laser every 15 s for 1 min, then every minute for 15 min, and finally every 10 min for a total of 1 h. Controls for minor loss of PA-GFP fluorescence during imaging were performed on GV spreads mounted in PBS. Data were analyzed with iplab spectrum (Scanalytics, Fairfax, VA) and kaleida-graph (Synergy Software, Reading, PA).
Results
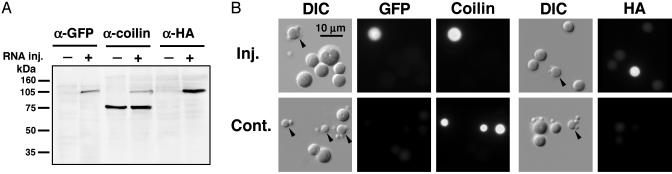
Targeting of PA-GFP-Coilin to CBs. In Xenopus oocytes, newly translated coilin is rapidly imported into the GV and specifically targeted to CBs (19). To verify that the same is true for coilin labeled with PA-GFP, we injected in vitro-synthesized transcripts of PA-GFP-coilin into the cytoplasm of Xenopus oocytes. The PA-GFP tag is at the N terminus of coilin and an HA tag is at the C terminus. After overnight incubation, we isolated GVs from the oocytes and extracted their proteins for analysis by Western blotting (Fig. 1A). Antibodies against GFP and the HA tag reacted with a single band at ≈105 kDa, as expected for full-length PA-GFP-coilin. An antibody against coilin reacted with this same band and also with endogenous coilin at ≈75 kDa. We made spread preparations of the GV contents, fixed with paraformaldehyde and stained with the same antibodies (Fig. 1B). Staining of CBs with all three antibodies confirmed that full-length coilin is synthesized from the construct and is properly targeted to CBs.
Fig. 1.
PA-GFP-coilin is correctly translated in the oocyte cytoplasm, imported into the GV, and targeted to CBs. (A) Western blot of GV proteins from oocytes that had been injected with transcripts of PA-GFP-coilin. The newly translated protein is detectable as a band at ≈105 kDa with antibodies against GFP, coilin, or the HA tag. Endogenous coilin in both control and injected oocytes appears as a band at ≈75 kDa with anti-coilin antibody. (B) Nuclear organelles from control oocytes and oocytes injected with transcripts of PA-GFP-coilin. In the injected oocytes, CBs are labeled with antibodies against GFP, coilin, or the HA tag, demonstrating that PA-GFP-coilin is targeted to CBs. In control oocytes, endogenous coilin is detectable in CBs only with the antibody against coilin. Arrowheads point to CBs.
Photoactivation of PA-GFP-Coilin in the CB. We simultaneously injected two in vitro-synthesized transcripts into the cytoplasm of Xenopus oocytes: PA-GFP-coilin mRNA and Alexa 546-labeled U7 snRNA. The coilin transcripts are translated in the cytoplasm into coilin protein, whereas the U7 snRNA is assembled into the U7 snRNP (20). Both then enter the nucleus and accumulate in the CBs. The U7 snRNA, which fluoresces red, allows us to identify CBs before photoactivation of the PA-GFP and serves as a control for the integrity of the CBs throughout the experiment. The kinetics of PA-GFP-coilin were examined in GVs isolated in oil. Earlier studies showed that such GVs retain physiological activity for hours, including RNA transcription (21, 22). Because of its large size (400-μm diameter), the GV must be squashed under a coverslip to visualize the CBs and other nuclear components (18).
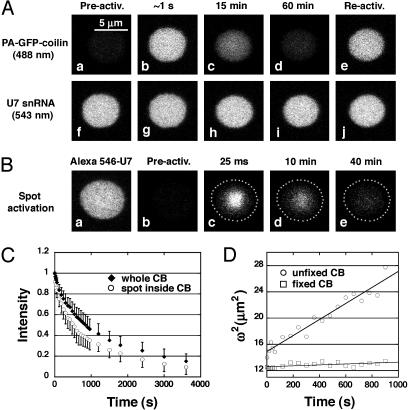
A suitable CB was located by means of the red fluorescence of its U7 snRNA. PA-GFP-coilin was then photoactivated in the whole CB with light from a 405-nm laser. The fluorescence of the CB was monitored simultaneously at 488 nm (green PA-GFP-coilin) and 543 nm (red U7 snRNA). Typical results are shown in Fig. 2A. Over a period of ≈1 h, the green fluorescence of PA-GFP-coilin disappears (Fig. 2 A a–d), whereas the red fluorescence of U7 snRNA remains invariant (Fig. 2 A f–i). If the same CB is again photoactivated with the 405-nm laser, the green fluorescence returns almost to its initial value (Fig. 2 A e), whereas the red fluorescence remains constant (Fig. 2 A j). The interpretation of these results is relatively straightforward, based partly on what we already know from FRAP experiments with GFP-coilin and U7 snRNA (18). Both coilin and U7 snRNA are constantly cycling in and out of the CB. At any given time, there is a small amount of each component in the CB at relatively high concentration and a much greater amount in the nucleoplasm at low concentration. PA-GFP-coilin that is activated at the beginning of the experiment leaves the CB over a period of 1 h, resulting in loss of fluorescence. It is replaced by an equal amount of unactivated PA-GFP-coilin, which is invisible until it is photoactivated by another exposure of the CB to 405-nm light. Cycling of the U7 snRNA cannot be inferred from this experiment, but is known from previous FRAP studies and from the FRAP experiment illustrated in Fig. 3.
Fig. 2.
Photoactivated PA-GFP-coilin disappears from CBs over a time period of ≈1h. (A) (a) Before activation, the CB is not detectable at 488 nm. (b) After photoactivation of PA-GFP-coilin with light of 405 nm, the CB fluoresces strongly at 488 nm. (c and d) Over time, the fluorescence disappears. (e) Fluorescence in the CB can be reactivated at the end of the observations, demonstrating that unactivated PA-GFP-coilin from the nucleoplasm has replaced activated PA-GFP-coilin in the CB. (f–j) The fluorescence of Alexa 546-labeled U7 snRNA in the same CB remains unchanged during the experiment. (B) An experiment similar to that in A, except that only a small spot inside the CB was photoactivated. (a and b) Before photoactivation Alexa-546-U7 snRNA in the CB fluoresces at 543 nm (a), but PA-GFP-coilin in the same CB is not detectable at 488 nm (b). (c–e) After photoactivation, PA-GFP-coilin disappears from the CB over a period of ≈1 h. (C) Quantitative data showing loss of PA-GFP-coilin from CBs as a function of time after photoactivation. The kinetics are similar for activation of the whole CB (filled diamonds) and of a spot within the CB (open circles). (D) Quantitative data showing very slow diffusion of PA-GFP-coilin within a CB. The increasing diameter (ω) of a photoactivated spot within a CB was determined for a period of 15 min after photoactivation. From the plot of ω2 versus t, one can calculate a diffusion coefficient (D) from the relationship D = ω2/8t (23). A spot within a fixed CB showed no increase in diameter.
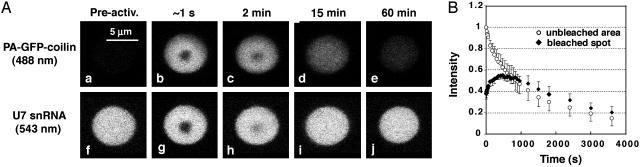
Fig. 3.
Behavior of a photobleached spot within a photoactivated CB. (A) Before photoactivation, the CB is not detectable at 488 nm (PA-GFP-coilin) (a), but fluoresces strongly at 543 nm (Alexa 546-U7 snRNA) (f). When illuminated with the full intensity of the 405-nm laser for 1 s, PA-GFP-coilin is photoactivated throughout the CB and a spot is photobleached where the beam is most intense (b). A spot is also bleached in the Alexa-546-U7 snRNA (g). Over time, the PA-GFP-coilin disappears, whereas the Alexa-546-U7 snRNA undergoes conventional FRAP (c–e and h–j). (B) Quantitative data showing typical loss of intensity for PA-GFP-coilin in the photoactivated (unbleached) area of the CB (open circles). Within the bleached spot, the intensity increases for a few minutes and then decreases with kinetics similar to the unbleached area (filled diamonds).
To control for photobleaching during imaging, we isolated GVs in a saline solution and centrifuged the nuclear contents onto a microscope slide. In such preparations the nucleoplasm is washed away and all organelles are in direct contact with the saline solution. When such isolated CBs are illuminated with 405-nm light, the PA-GFP becomes activated. However, when the CB is imaged over a period of 1 h at 488 nm, there is only minimal loss of fluorescence intensity. We used data from such control CBs for minor correction of PA-GFP intensities obtained in the experiments with oil-isolated GVs (Fig. 2C).
A major advantage of CBs in the oocyte is their large size, which permits detailed analysis of macromolecules within individual CBs. We photoactivated a small spot of PA-GFP-coilin within a single CB by using the 405-nm laser, and then followed the loss of fluorescence intensity from this spot (Fig. 2B). Two features of this experiment were immediately obvious. First, the loss of intensity was slow, as in the experiments where the entire CB was photoactivated. Second, the photoactivated area did not expand to fill the entire CB, as it would have, if the contents of the CB were in rapid motion.
A detailed analysis showed that three kinetic components were necessary to fit the loss of fluorescence, and these components had roughly the same relative sizes and rate constants for whole CBs and a spot within a CB (Fig. 2C and supporting information, which is published on the PNAS web site). Moreover, the rates and relative sizes of the components were similar to those obtained in our earlier FRAP experiments (18).
Diffusion of PA-GFP-Coilin Within the CB. Although it was clear that a photoactivated spot did not expand rapidly to fill the whole CB, visual inspection suggested that the spot diameter did increase slowly with time. To gain further insight into possible diffusion of PA-GFP-coilin within the CB, we measured the average fluorescence intensity in a small circle at the center of the photoactivated spot and in nine concentric annuli around it. A plot of intensity against distance from the spot center was fitted to a Gaussian curve. Curve parameters were determined for the initial image at time 0, and for each successive image up to 15 min after photoactivation. The width of the Gaussian curve (ω) increased slowly with time. A plot of ω2 against time (Fig. 2D) yielded a straight line, consistent with a spatially random transport process. Thus, from the slope one can calculate a diffusion coefficient for PA-GFP-coilin within the CB (23) (for details, see supporting information). Diffusion coefficients were determined for 12 CBs, with a range of 0.5–3.7 × 10–3 μm2·sec–1. As a control, similar measurements were made on CBs from GVs that had been centrifuged onto a microscope slide and fixed in paraformaldehyde. The diameter of the photoactivated spot remained constant in these CBs (Fig. 2D), and there was only a slight decrease in intensity due to photobleaching.
FRAP Within a Region of Photoactivated PA-GFP-Coilin. To obtain additional evidence concerning movement of coilin molecules within the CB, we illuminated a CB in a single spot with the full intensity of the 405-nm laser for 1 s. After switching to the 488-nm laser, one could see photoactivation of PA-GFP over the whole of the CB, plus a central bleached spot, where the laser intensity was highest (Fig. 3Ab). A similar image was seen when the Alexa 546-U7 snRNA was viewed: a uniformly stained CB with a central bleached spot. In the case of U7, however, the CB was fluorescent from the beginning, the new feature being the bleached area (Fig. 3Ag). During the next hour, the behavior of the bleached spots was different for coilin and U7 snRNA. The U7 snRNA behaved as in a typical FRAP experiment (Fig. 3A h–j). That is, the bleached spot recovered its full intensity due to import and binding of fluorescent U7 snRNA. The kinetics were similar to those observed in earlier FRAP experiments (18). The situation for PA-GFP-coilin was different. Because of exchange with nonfluorescent PA-GFP-coilin in the nucleoplasm, the CB gradually lost its fluorescence (Fig. 3B “unbleached area”). A similar decrease in intensity should occur in the central spot, if there is no bulk movement of photoactivated PA-GFP-coilin within the CB itself. However, analysis of the intensity data for the central bleached spot revealed a transient increase during the first few minutes (Fig. 3B “bleached spot”). After this time, the central spot lost intensity with kinetics essentially identical to the rest of the CB. We conclude, therefore, that a certain amount of bulk movement of coilin occurs within the CB on a time scale of minutes. This observation confirms the more quantitative analysis made in the spot-photoactivation experiment.
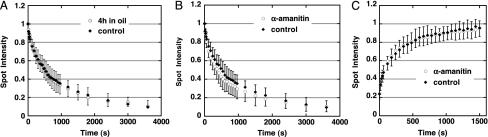
Coilin Kinetics in the CB Are Independent of Ongoing Nucleocytoplasmic Exchange. In our typical FRAP or photoactivation experiments, we isolate a GV in oil, squash it under a coverslip, and carry out observations as quickly as feasible. Under these conditions, the GV is no longer in contact with the cytoplasm and there is no ongoing nucleocytoplasmic exchange. To determine whether the movement of coilin in and out of the CB subsides after the GV is removed from the oocyte, we isolated GVs in oil and left them for 4 h before squashing. The kinetics of fluorescence loss of PA-GFP-coilin (Fig. 4A) or FRAP kinetics of GFP-coilin (data not shown) were essentially identical to measurements made on freshly isolated GVs. These results suggest that the flux of coilin in and out of CBs is independent of ongoing nucleocytoplasmic exchange, at least over a time period of 4 h.
Fig. 4.
The kinetics of coilin in the CB are independent of ongoing nucleocytoplasmic exchange or transcription. (A) PA-GFP-coilin leaves a photoactivated spot in a CB at the same rate in freshly isolated GVs (filled diamonds) as in isolated GVs held in oil for 4 h before observation (open circles). (B) PA-GFP-coilin leaves a photoactivated spot in a CB at the same rate in control GVs (filled diamonds) as in GVs from transcriptionally inactive oocytes (α-amanitin injected) (open circles). (C) FRAP kinetics of GFP-coilin are the same in control (filled diamonds) and transcriptionally inactive oocytes (α-amanitin injected) (open circles).
Coilin Kinetics in the CB Are Independent of RNA Polymerase (Pol) II Transcription. Because coilin may play a role in the transport of snRNPs to the CB, one might expect the kinetics of coilin in the CB to be influenced by the transcriptional state of the cell. With this possibility in mind, we examined the behavior of GFP-coilin and PA-GFP-coilin after inhibition of transcription with α-amanitin. Oocytes were injected with in vitro synthesized transcripts of GFP-coilin or PA-GFP-coilin as usual. After overnight incubation for GFP-coilin or >24 h incubation for PA-GFP-coilin, 10–20 ng of α-amanitin was injected into the GV, yielding a final concentration of ≈10–20 μg/ml, on the assumption that the inhibitor diffuses throughout the oocyte volume. To assess the effect of the inhibitor, GVs were isolated in saline after ≈20 min and examined by immunostaining. Such preparations exhibited two features diagnostic of pol II inhibition: the lampbrush chromosomes were devoid of lateral loops, and they failed to stain with mAb H14, which recognizes the C-terminal domain of pol II when serine-5 is phosphorylated (24). In keeping with earlier α-amanitin experiments, CBs in such preparations still stained with mAb H14, showing that they contained phosphorylated pol II (25). GVs were also isolated in oil and squashed for observation in the confocal microscope. The CBs in these oil-isolated GVs were subjected to FRAP for GFP-coilin or photoactivation for PA-GFP-coilin. In both cases the kinetics of fluorescence recovery or loss were indistinguishable from the kinetics in control GVs (Fig. 4 B and C). These experiments demonstrate that ongoing transcription is not necessary for the exchange of coilin between CBs and the nucleoplasm.
Discussion
Earlier studies of GFP-coilin in the nuclei of cultured mammalian cells (26–28) and in the Xenopus GV (18) showed that coilin in the CB continuously exchanges with coilin in the nucleoplasm. In the GV, there are three kinetic components with half times of ≈14 s, 7.2 min, and 33 min (18). FRAP studies give information primarily about the entrance of a component from the nucleoplasm to the CB. Exit of components can be determined by fluorescence loss in photobleaching (FLIP) experiments, also called inverse FRAP or iFRAP (29). In a FLIP experiment, the area outside the fluorescent object of interest is repeatedly bleached until all fluorescence has left the object (and the nucleus or cell, as the case may be). Because of the enormous volume of the GV, a FLIP experiment to visualize loss of fluorescence from CBs is not possible. Here, photoactivation of PA-GFP-coilin provides a direct measure of exit. Our data show that coilin leaves the CB with kinetics very similar to those of its entrance. Thus, the data for coilin leaving the CB (Figs. 2C, 3B, and 4 A and B) cannot be fitted to a single exponential curve, but require at least three components. The half times of these components are 53 s, 7.3 min, and 33.5 min (see supporting information for details). In an earlier study of coilin, we suggested that the relatively slow kinetics of CB components are consistent with the assembly of large macromolecular complexes inside the CB (18).
The fine structure of CBs from the Xenopus GV has been examined by electron microscopy (30). The entire interior of the body consists of (at least) two kinds of interspersed granules. One granule has a diameter of 20–30 nm, is heavily contrasted by osmium and uranium, and closely resembles the interchromatin granules that make up the speckles or B snurposomes. The other granule is 30–50 nm in diameter and is of lower contrast. Like other nuclear organelles, CBs are devoid of a detectable limiting membrane or other type of investing material. Furthermore, there is no evident internal skeleton or fibrous network that could supply rigidity to the body. Granules with diameters of 20–50 nm, if free in a dilute aqueous solution, would have diffusion coefficients (D) ranging from ≈10 to 20 μm2·s–1 (31), some four orders of magnitude greater than the measured D for PA-GFP-coilin inside the CB. If we assume that coilin is in one of the granules, the measured D implies that there are severe constraints on the free movement of the granules.
High viscosity and/or high physical density (molecular crowding) could contribute to reduced diffusion. In an earlier study, we presented qualitative FRAP data suggesting that the viscosity of CBs is not a significant factor (18). In addition, we have recently completed an interferometric study of nuclear organelles and found that the protein concentration inside CBs is only slightly higher than that of the nucleoplasm in which they are suspended (150 mg/ml for CBs versus 110 mg/ml for the nucleoplasm). Because neither high viscosity nor high physical density is likely to be a significant factor impeding the movement of granules inside CBs, there must be other constraints. Attachment of the granules to a scaffold or matrix could provide such a constraint, but until now there is no morphological evidence for this type of structure within the CB. In the absence of other factors, it may be that CB granules have limited mobility because of strong cohesive forces. Such forces might also help to explain why CBs so often have a perfectly spherical shape (4, 32). Lacking both an external limiting membrane and a rigid skeleton, CBs should assume a spherical shape, if their constituent granules are constrained primarily by high cohesive forces.
In summary, our study of PA-GFP-coilin confirms earlier FRAP analyses showing that CBs are dynamic structures. At least some of their macromolecular constituents, including coilin, are in dynamic equilibrium with the same constituents in the nucleoplasm. Our study also reveals the unanticipated fact that coilin within the CB exhibits extremely slow diffusion. One possibility is that coilin resides in some of the 25- to 50-nm granules detected by electron microscopy within the CB, and that these granules exhibit strong cohesive forces.
Supplementary Material
Acknowledgments
We thank George Patterson and Jennifer Lippincott-Schwartz for kindly providing the pPA-GFP-C1 vector. This work was supported by National Institute of General Medical Sciences/National Institutes of Health Grant GM 33397.
Abbreviations: CB, Cajal body; FRAP, fluorescence recovery after photobleaching; GV, germinal vesicle; PA, photoactivatable; snRNP, small nuclear ribonucleoprotein; HA, hemagglutinin; snRNA, small nuclear RNA.
References
- 1.Ramón y Cajal, S. (1903) Trab. Lab. Invest Biol. (Madrid) 2, 129–221. [Google Scholar]
- 2.Ogg, S. C. & Lamond, A. I. (2002) J. Cell Biol. 159, 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gall, J. G. (2000) Annu. Rev. Cell Dev. Biol. 16, 273–300. [DOI] [PubMed] [Google Scholar]
- 4.Gall, J. G. (2003) Nat. Rev. Mol. Cell. Biol. 4, 975–980. [DOI] [PubMed] [Google Scholar]
- 5.Andrade, L. E. C., Chan, E. K. L., Raška, I., Peebles, C. L., Roos, G. & Tan, E. M. (1991) J. Exp. Med. 173, 1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raška, I., Andrade, L. E. C., Ochs, R. L., Chan, E. K. L., Chang, C.-M., Roos, G. & Tan, E. M. (1991) Exp. Cell Res. 195, 27–37. [DOI] [PubMed] [Google Scholar]
- 7.Bauer, D. W. & Gall, J. G. (1997) Mol. Biol. Cell 8, 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellini, M. & Gall, J. G. (1998) Mol. Biol. Cell 9, 2987–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucker, K. E., Berciano, M. T., Jacobs, E. Y., LePage, D. F., Shpargel, K. B., Rossire, J. J., Chan, E. K. L., Lafarga, M., Conlon, R. A. & Matera, A. G. (2001) J. Cell Biol. 154, 293–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jády, B. E., Darzacq, X., Tucker, K. E., Matera, A. G., Bertrand, E. & Kiss, T. (2003) EMBO J. 22, 1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert, M. D., Szymczyk, P. W., Shpargel, K. B. & Matera, A. G. (2001) Genes Dev. 15, 2720–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hebert, M. D., Pillai, R., Shpargel, K. B., Ospina, J. K., Schümperli, D. & Matera, A. G. (2002) Dev. Cell 3, 329–337. [DOI] [PubMed] [Google Scholar]
- 13.Meister, G., Eggert, C. & Fischer, U. (2002) Trends Cell Biol. 12, 472–478. [DOI] [PubMed] [Google Scholar]
- 14.Fischer, U., Liu, Q. & Dreyfuss, G. (1997) Cell 90, 1023–1029. [DOI] [PubMed] [Google Scholar]
- 15.Patterson, G. H. & Lippincott-Schwartz, J. (2002) Science 297, 1873–1877. [DOI] [PubMed] [Google Scholar]
- 16.Handwerger, K. E., Wu, Z., Murphy, C. & Gall, J. G. (2002) J. Cell Sci. 115, 2011–2020. [DOI] [PubMed] [Google Scholar]
- 17.Wu, C.-H. H., Murphy, C. & Gall, J. G. (1996) RNA 2, 811–823. [PMC free article] [PubMed] [Google Scholar]
- 18.Handwerger, K. E., Murphy, C. & Gall, J. G. (2003) J. Cell Biol. 160, 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu, Z., Murphy, C. & Gall, J. G. (1994) Mol. Biol. Cell 5, 1119–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefanovic, B., Hackl, W., Lührmann, R. & Schümperli, D. (1995) Nucleic Acids Res. 23, 3141–3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paine, P. L., Johnson, M. E., Lau, Y.-T., Tluczek, L. J. M. & Miller, D. S. (1992) BioTechniques 13, 238–245. [PubMed] [Google Scholar]
- 22.Lund, E. & Paine, P. (1990) Methods Enzymol. 181, 36–43. [DOI] [PubMed] [Google Scholar]
- 23.Cardullo, R. A., Mungovan, R. M. & Wolf, D. E. (1991) in Biophysical and Biochemical Aspects of Fluorescence Spectroscopy, ed. Dewey, T. G. (Plenum, New York), pp. 231–260.
- 24.Patturajan, M., Schulte, R. J., Sefton, B. M., Berezney, R., Vincent, M., Bensaude, O., Warren, S. L. & Corden, J. L. (1998) J. Biol. Chem. 273, 4689–4694. [DOI] [PubMed] [Google Scholar]
- 25.Doyle, O., Corden, J. L., Murphy, C. & Gall, J. G. (2002) J. Struct. Biol. 140, 154–166. [DOI] [PubMed] [Google Scholar]
- 26.Snaar, S., Wiesmeijer, K., Jochemsen, A., Tanke, H. & Dirks, R. (2000) J. Cell Biol. 151, 653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sleeman, J. E., Trinkle-Mulcahy, L., Prescott, A. R., Ogg, S. C. & Lamond, A. I. (2003) J. Cell Sci. 116, 2039–2050. [DOI] [PubMed] [Google Scholar]
- 28.Dundr, M., Hebert, M. D., Karpova, T. S., Stanek, D., Xu, H., Shpargel, K. B., Meier, U. T., Neugebauer, K. M., Matera, A. G. & Misteli, T. (2004) J. Cell Biol. 164. [DOI] [PMC free article] [PubMed]
- 29.Pederson, T. (2001) Cell 104, 635–638. [DOI] [PubMed] [Google Scholar]
- 30.Gall, J. G., Bellini, M., Wu, Z. & Murphy, C. (1999) Mol. Biol. Cell 10, 4385–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saltzman, W. M., Radomsky, M. L., Whaley, K. J. & Cone, R. A. (1994) Biophys. J. 66, 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callan, H. G. & Lloyd, L. (1960) Philos. Trans. R. Soc. London Ser. B 243, 135–219. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.