Abstract
Soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) catalyze compartment-specific membrane fusion. Whereas most SNAREs are bona fide type II membrane proteins, Ykt6 lacks a proteinaceous membrane anchor but contains a prenylation consensus motif (CAAX box) and exists in an inactive cytosolic and an active membrane-bound form. We demonstrate that both forms are farnesylated at the carboxyl-terminal cysteine of the CCAIM sequence. Farnesylation is the prerequisite for subsequent palmitoylation of the upstream cysteine, which permits stable membrane association of Ykt6. The double-lipid modification and membrane association is crucial for intra-Golgi transport in vitro and cell homeostasis/survival in vivo. The membrane recruitment and palmitoylation is controlled by the N-terminal domain of Ykt6, which interacts with the SNARE motif, keeping it in an inactive closed conformation. Together, these results suggest that conformational changes control the lipid modification and function of Ykt6. Considering the essential and central role of Ykt6 in the secretory pathway, this spatial and functional cycle might provide a mechanism to regulate the rate of intracellular membrane flow.
The dynamic and specific trafficking of proteins and lipids along the secretory and endocytic pathways relies on precisely choreographed membrane fusion events, in turn relying on the faithful pairing of cognate soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs) between membranes (1, 2). In vitro membrane fusion assays employing purified SNAREs reconstituted into liposomes, as well as in vivo cell–cell fusion experiments using flipped SNAREs [cognate-vesicle (v) and target-membrane (t) SNAREs expressed on the extracellular surface of two cell populations, respectively], provide compelling evidence that SNAREs are the driving force for fusion and, in addition, encode targeting specificity (3–7). Furthermore, as reported for intra-Golgi transport, some SNAREs function also as inhibitory SNAREs to fine-tune specific fusion events (8). Thus, the intracellular distribution of cognate SNAREs outlines the fusion potential of distinct compartments and provides a road map for membrane trafficking (9, 10).
The rate at which compartments containing cognate SNAREs fuse is at least in part determined by the conformational state of SNAREs. Most SNAREs contain regulatory domains in addition to their SNARE motifs, which form the four-helix bundle at the core of the SNARE complex (11). These regulatory domains control the assembly of the three-helix t-SNARE (one helix derived from a syntaxin heavy chain and two from t-SNARE light chains) and/or the subsequent v-/t-SNARE complex formation. Additional components can either bind or posttranslationally modify SNAREs, increasing or decreasing SNARE activity.
A unique and therefore interesting SNARE is Ykt6, an essential protein that is highly conserved from yeast to man (12). Ykt6 is special in that it is lipid-anchored to membranes, lacking the usual hydrophobic anchor sequence. Ykt6 is localized to both the cytosol and membranes (12, 13). In mammalian cells, the majority of membrane-bound pool of Ykt6 is associated with the Golgi (13); however, a broader perinuclear distribution has also been observed (14). In some specialized cells, like neurons, Ykt6 antibodies show a punctuated labeling pattern of compartments of unknown origin (15). Consistent with such a broad cellular distribution, Ykt6 has been implicated in multiple transport steps in the secretory pathway of yeast, including retrograde transport from the Golgi to the endoplasmic reticulum, intra-Golgi transport, and trafficking to the vacuole (12, 15–18).
This functional diversity and the dual localization of Ykt6 suggest tight regulation. Indeed, Ykt6 contains an N-terminal regulatory domain, which adopts a profilin-like structure (also found in the SNARE Sec22) with a hydrophobic surface that folds back on the SNARE motif resulting in a closed conformation (19). Destabilization of the closed conformation enhances SNARE complex formation (19). However, it remains to be shown whether the regulatory domain has additional functions, e.g., in membrane recruitment of Ykt6. Furthermore, the exact nature of the lipid anchor of Ykt6 and its role in the interconversion of the cytosolic and membrane-bound pools need to be determined.
In this study, we refined the cycle of Ykt6 in the Golgi, showing that farnesylation, a previously unanticipated palmitoylation, and a conformational switch in Ykt6 control membrane association and SNARE activity.
Materials and Methods
Materials. Lovastatin, 2-Br-palmitate, Triton X-114, and saponin were purchased from Sigma. FTI-277, a farnesyltransferase-specific inhibitor, and GGTI-298, a geranylgeranyltransferase-specific inhibitor, were purchased from Calbiochem. Effective concentrations of FTI-277 (30 μM) and GGTI-298 (20 μM) in HeLa cells were determined by using N-ras and Rap1 as a control, respectively.
Antibodies. mAb against p115 was purchased from BD Transduction Laboratories. Anti-GFP and anti-myc mAbs were purchased from Roche Biochemicals. Goat anti-Ykt6 polyclonal antibodies were prepared and affinity-purified as described (14).
Ykt6 Plasmid Construction. GFP fusion proteins were constructed by using monomeric L221/K GFP (20) in pEGFP-C1 vector (pmEGFP-C1) (Clontech). pmGFP-ykt6 (full-length Ykt6) and pmGFP-ykt6-ΔN (encoding amino acids 134–198 of Ykt6) were generated by inserting PCR fragments into the HindIII–PstI site of pmEGFP-C1. myc-Ykt6 was generated by inserting PCR fragment of the full-length human Ykt6 into the BamHI–MluI site of pTRE-2 (Clontech) containing the 3× cMyc epitope at the SacII–BamHI site. F42/E (Ykt6-F42E), C194C195/SS (Ykt6-SS), C194/S (Ykt6-SC), and C195/S (Ykt6-CS) mutants of human Ykt6 constructs were created by PCR-based site-directed mutagenesis. pJM128-SC containing the C196/S mutation was generated by PCR-based site-directed mutagenesis of pJM128 coding GST-ykt6 (21). Yeast ykt6 expression vectors p416–ykt6 were generated by inserting PCR fragments of full-length yeast ykt6 into the BamHI–EcoRI site of p416 GAL1 (American Type Culture Collection). p416-ykt6-SC containing C196/S mutation were created by PCR-based site-directed mutagenesis.
Cell Fractionation. All manipulations were performed at 4°Coron ice. After being washed with PBS, HeLa cells were harvested by scraping and precipitated by centrifugation at 300 × g for 5 min. The precipitated cells were resuspended in 10 mM Tris·HCl (pH 7.35), containing 150 mM NaCl and a protease inhibitor mixture (Roche Biochemicals) and lysed by sonication. After centrifugation of the lysate at 2,500 × g for 5 min, cytosol and membranes were separated from the postnuclear supernatant by centrifugation at 100,000 × g for 30 min. The precipitated fraction (membranes) was resuspended in the same buffer and repurified twice by centrifugation at 100,000 × g for 30 min. Protein concentrations of preparations were determined with a protein assay kit (Bio-Rad).
Fluorescence Microscopy. HeLa cells expressing GFP constructs were fixed with 4% paraformaldehyde/PBS and observed with a TSC SP2 laser-scanning confocal microscope (Leica, Deerfield, IL).
For immunofluorescent microscopy, fixed cells were permeabilized with PBS containing 0.1% (wt/vol) saponin or 0.2% (vol/vol) Triton X-100 for 20 min or 10 min, respectively, and they were incubated with affinity-purified goat anti-ykt6 polyclonal antibody (1:200) and/or mouse anti-p115 mAb (1:400) overnight at 4°C. Cells were incubated with secondary antibody (Alexa 488 donkey anti-goat IgG and/or Alexa 633 rabbit anti-mouse IgG, Molecular Probes) for 1 h and observed with the confocal microscope.
Metabolic Lipid Labeling of Ykt6. [3H]Mevalonolactone and [3H]palmitate metabolic labelings were performed essentially as described (22). Cells grown on six-well dishes were labeled with 50 μCi/ml (1 Ci = 37 GBq) [3H]mevalonolactone (American Radiolabeled Chemicals, St. Louis, 40 Ci/mmol) or 100 μCi/ml [3H]palmitate (Perkin–Elmer, 50 Ci/mmol) for 15 h and 5 h, respectively. In some experiments, labeled cells were fractionated into 100,000 × g supernatant (cytosolic) and 100,000 × g membrane fractions. Cells were lysed in detergent buffer (50 mM Tris·HCl, pH 7.35/150 mM NaCl/1% Nonidet P-40/0.5% deoxycholate and a protease inhibitor mixture), and insoluble material was removed by centrifugation at 100,000 × g for 30 min. Epitope-tagged Ykt6 was immunoprecipitated by using anti-GFP or anti-myc antibodies and Protein G-agarose (Roche Biochemicals) according to the manufacturer's instructions. The immunoprecipitates were fractionated by electrophoresis as described below. The gels were dried and subjected to fluorography by using EN3HANCE (NEN) according to the manufacturer's instructions.
Triton X-114 Partitioning. Cells grown in a 10-cm dish were washed with PBS and extracted for 1 h in ice-cold 2% (vol/vol) Triton X-114 in TBS (10 mM Tris·HCl, pH 7.35/150 mM NaCl). Detergent-insoluble material was removed by a 100,000 × g centrifugation for 10 min at 4°C. The clarified supernatant was transferred to a new microcentrifuge tube and clouded for 10 min at 37°C, followed by centrifugation at 100,000 × g for 10 min at room temperature. The aqueous phase was transferred to a new tube, and the aqueous and detergent phases were back-extracted three times according to the method of Brusca and Radolf (23). The volume of samples was adjusted by addition of TBS or Triton X-114, followed by electrophoresis and immunoblotting.
Electrophoresis and Immunoblotting. Electrophoresis was performed in precast NuPAGE 10% or 12% [bis(2-hydroxyethyl) amino]tris(hydroxymethyl)methane or 2-[bis(2-hydroxyethyl) amino]-2-(hydroxymethyl)-1,3-propanediol (Bis-Tris) gels (Invitrogen). For immunoblot analysis, the proteins were transferred onto poly(vinylidene difluoride) membrane, and the blotted membrane was incubated with primary antibody at a 1:2,000 dilution for 90 min. Peroxidase-conjugated secondary antibody was incubated with the blot for 90 min at a 1:2,000 dilution. Detection was performed by using ECL (Amersham Biosciences) and fluorography.
Expression, Purification, and Lipid Modification of Recombinant Yeast Ykt6 Proteins. Recombinant GST-ykt6 (pJM128) and GST-ykt6-SC (pJM128-SC) proteins were expressed and purified as described (21, 24).
Lipid modification of recombinant Ykt6 proteins with maleimidopropionic acid farnesylester (C15) or maleimidopropionic acid geranylgeranylester (C20) was performed as described (3, 6). After dialysis against 25 mM Tris·HCl, pH 7.35/25 mM KCl, recombinant proteins were used for the in vitro intra-Golgi transport assay.
In Vitro Intra-Golgi Transport Assay. Intra-Golgi transport assay was performed according to the standard protocol (25). Cytosol was prepared from HeLa cells as described (26). For each experiment, the assay was conducted at least three times.
Growth of Yeast Strains Having Temperature-Sensitive ykt6. Wild-type ykt6 vector (p416-ykt6) and unpalmitoylated mutant ykt6 vectors having C196/S mutation (p416-ykt6-SC) were transfected into a yeast strain SARY166 [MATα ykt6Δ::LEU2 (CEN6, TRP1, ykt6–1) leu2–1, 112 ura3–52 his3-Δ200 trp1-Δ901 suc2-Δ9] (27), carrying temperature-sensitive ykt6 (gift from David K. Banfield, Hong Kong University of Science and Technology, Kowloon, Hong Kong). Transfectants were selected on the SC (–URA, –TRP) plates at 25°C. Growth of yeasts was observed at 25°C or 37°C (restrictive temperature) on the SC (–URA, –TRP) containing 2% galactose.
Results
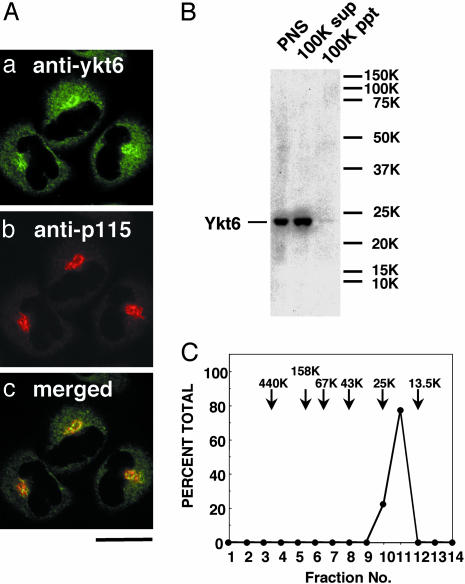
Endogenous Ykt6 Is Localized to both the Cytosol and the Golgi in HeLa Cells. We first tested the intracellular localization of endogenous Ykt6 in human HeLa cells. Immunofluorescence labeling of endogenous Ykt6 in permeabilized cells revealed a cytosolic and perinuclear distribution pattern, with predominant Golgi staining as shown by the colocalization with the Golgi marker p115 (Fig. 1A). Consistent with this result, cell fractionation demonstrated that >90% of the total Ykt6 fractionated with the cytosol (100,000 × g supernatant) and <10% with membranes (100,000 × g pellet) (Fig. 1B). Gel-filtration analysis showed that the majority of cytosolic Ykt6 exists in a 20-kDa monomeric form (Fig. 1C). A similar intracellular localization of Ykt6 was detected in other cell lines such as HEK293 cells and neuroteratocarcinoma NTera2 cells (data not shown).
Fig. 1.
Localization of the endogenous Ykt6 in HeLa cells. (A) Cells were fixed, permeabilized with 0.1% saponin, and coimmunostained with anti-ykt6 antibodies (a) and an antibody against the Golgi marker p115 (b). (Bar = 10 μm.) (B) Postnuclear supernatant (PNS, 60 μg; lane 1), 100,000 × g supernatant (cytosol; lane 2), and 100,000 × g pellet (membranes; lane 3) fractions of HeLa cells were analyzed by immunoblotting with anti-ykt6 antibodies. (C) HeLa cytosol was fractionated by Superdex 200 gel filtration, and Ykt6 in each fraction was analyzed by immunoblotting.
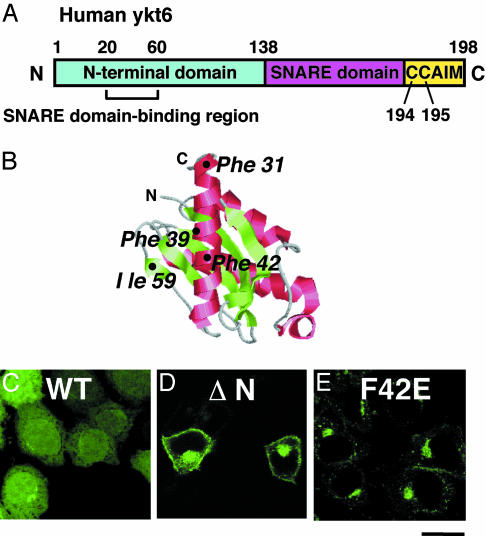
Interaction of the N-Terminal Domain of Ykt6 with Its SNARE Domain Is Required for the Cytosolic Localization of Ykt6. Ykt6 has three conserved domains: an N-terminal regulatory domain, a SNARE motif, and a carboxyl-terminal CAAX box, which contains double cysteines (Fig. 2A). To determine the role of regulatory N-terminal domain in the localization of Ykt6, we constructed a deletion mutant of a GFP-tagged human Ykt6 (GFP-ykt6-ΔN), and we examined its intracellular distribution in unpermeabilized HeLa cells. The full-length GFP-ykt6 showed some Golgi labeling but was mainly localized in the cytosol and the nucleus (Fig, 2C). The partial nuclear localization is likely a result of the GFP tag because GFP itself is targeted to the nucleus, as has been reported (28). In contrast, GFP-ykt6-ΔN showed very little cytosolic staining and no nuclear labeling, but it showed a predominant perinuclear and plasma membrane distribution (Fig. 2D). These results indicate that the N-terminal domain of Ykt6 masks a membrane localization signal. The regulatory domain seems not to be essential for the Golgi localization but may retain Ykt6 in the Golgi area, preventing its transfer to the plasma membrane.
Fig. 2.
Intracellular localization of GFP-tagged ykt6 constructs in HeLa cells. (A) Domain organization of human Ykt6. (B) The structure of the N-terminal domain of yeast Ykt6 (19). The image was generated from the data (Protein Data Bank ID 1H8M) by using rasmac and photoshop (Adobe Systems, Mountain View, CA) software. (C–E) Localization of GFP-ykt6 in HeLa cells. Cells were transfected with GFP-ykt6 (C), GFP-ykt6-ΔN (amino acids 134–198) (D), and GFP-ykt6-F42E (E) and observed with a confocal microscope. (Bar = 10 μm.)
To evaluate the role of the N-terminal domain further, a ykt6 mutant locked in an open conformation was constructed by following the work of Tochio et al. (19), by changing the phenylalanine in position 42 to a glutamate (position 42 is located at the hydrophobic surface of the regulatory domain) (ref. 19 and Fig. 2B). This F42E mutation destabilizes the interaction between the N-terminal domain and SNARE motif (19). Similar to GFP-ykt6-ΔN, GFP-ykt6-F42E localizes mainly to the perinuclear region (Fig. 2E). The observed perinuclear localizations of GFP-ykt6-F42E, myc-tagged ykt6-F42E (data not shown), and endogenous Ykt6 are brefeldin A-sensitive, suggesting a Golgi distribution for both forms of Ykt6 (Fig. 1 and ref. 14). These results demonstrate that the interaction of the N-terminal domain with the SNARE domain prevents membrane recruitment or, alternatively, keeps a normally membrane-attached Ykt6 in a soluble conformation.
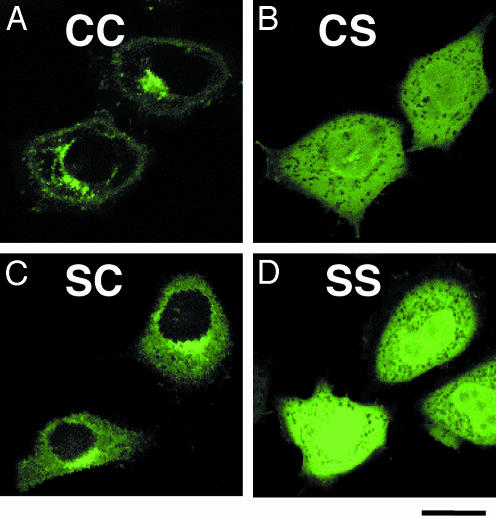
Cys-195 of Ykt6 Is Farnesylated and Essential for the Golgi Localization of Ykt6. To examine the function of the double cysteine residues at the carboxyl-terminal domain of Ykt6, we mutated these cysteines of the Golgi-associated GFP-ykt6-F42E. In contrast to the Golgi-localized C194C195 Ykt6 (Fig. 3A), both the S194S195 (Ykt6-SS) and C194S195 (Ykt6-CS) mutants were distributed to the cytosol and the nucleus (Fig. 3 B and D). The S194C195 (Ykt6-SC) mutant showed both cytosolic and Golgi localization (Fig. 4C). These results suggest that Cys-195 is essential for (and Cys-194 enhances) the membrane (Golgi) association of Ykt6.
Fig. 3.
The carboxyl-terminal Cys-194 and Cys-195 of Ykt6 are required for the Golgi association of GFP-ykt6-F42E. HeLa cells were transfected with GFP-ykt6-F42E containing double cysteines at the carboxyl terminus (C194C195, wild type) (A), C194C195/CS (B), C194C195/SC (C), and C194C195/SS (D) and visualized with a confocal microscope. (Bar = 10 μm.)
Fig. 4.
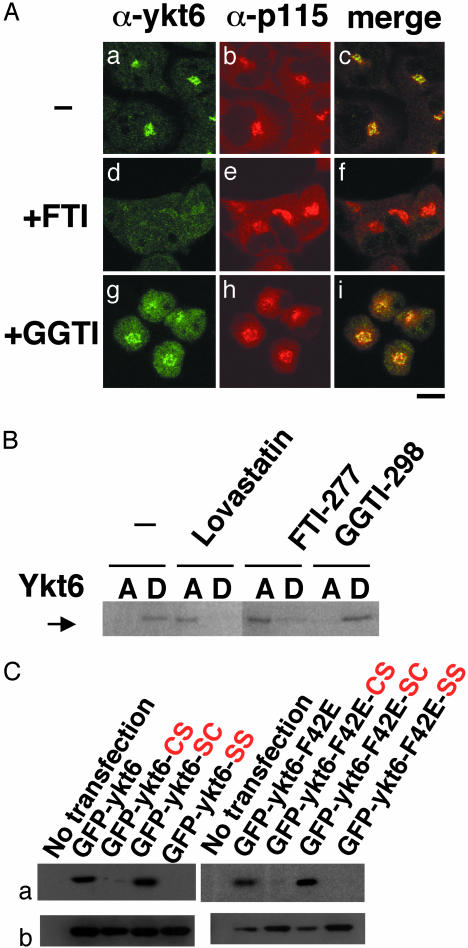
Cys-195 of Ykt6 is farnesylated in vivo.(A) HeLa cells were treated for 2 days with 30 μM FTI-277 or 20 μM GGTI-298, fixed, and coimmunostained with anti-ykt6 and anti-p115 antibodies. (Bar = 10 μm.) (B) HeLa cells were treated for 1 day with 30 μM lovastatin or for 2 days with 30 μM FTI-277 or 20 μM GGTI-298. Cells were solubilized with 2% Triton X-114 on ice and partitioned into aqueous (A) and detergent (D) phases at 37°C. Ykt6 derived from both phases was analyzed by immunoblot with anti-ykt6 antibodies. (C) HeLa cells were transfected with GFP constructs fused with the wild-type Ykt6 or Ykt6-F42E mutant. After a 15-h incubation with [3H]mevalonolactone, both forms of GFP-ykt6 containing the single or double substitutions of the carboxyl-terminal cysteines were immunoprecipitated from cell lysates with antibodies to GFP. Each immunoprecipitate was resolved by SDS/PAGE and subjected to fluorography to visualize prenylated proteins (a) or to immunoblot analysis using anti-GFP antibody (b).
To test whether Ykt6 is prenylated in vivo, we examined the effect of prenylation inhibitors on the intracellular localization of Ykt6. When HeLa cells were treated with a farnesyltransferase-specific inhibitor FTI-277 (29), perinuclear staining of endogenous Ykt6 disappeared (Fig. 4A a and d). In contrast, a geranylgeranyltransferase-specific inhibitor GGTI-298 (30) showed no effect on the Ykt6 localization (Fig. 4A g–i). Neither inhibitor affected p115 localization, suggesting that the Golgi is intact (Fig. 4Ae and h). These results are consistent with the idea that protein farnesylation is required for the Golgi localization of Ykt6.
To confirm the biochemical effect of prenylation inhibitors on Ykt6, we performed Triton X-114 phase partitioning analysis. Fig. 4B demonstrates that Ykt6 fractionates with the detergent phase, indicating that Ykt6 is lipid-modified. When cells were treated either with the HMG-CoA reductase inhibitor lovastatin, which inhibits both farnesylation and geranylgeranylation, or with FTI-277, most of endogenous Ykt6 have partitioned into the aqueous phase. In contrast, GGTI-298 had no effect. These results further substantiate that endogenous human Ykt6 is farnesylated.
Next, we confirmed farnesylation of Ykt6 by metabolic radio-labeling of cysteine mutants of GFP-tagged Ykt6 with [3H]mevalonolactone. As shown in Fig. 4C, both Ykt6 and Ykt6-F42E containing C194C195 and S194C195 were radiolabeled, whereas C194S195 and S194S195 mutants were not. Thus, it is the carboxyl-terminal cysteine of Ykt6, Cys-195, that is farnesylated.
Membrane-Associated Ykt6 Is Palmitoylated. The observation that the S194C195 mutant and the C194C195 wild type showed distinct intracellular localization patterns (Fig. 3) suggests the Cys-194 could also be lipid-modified. The similar prenyl/protein ratio of the C194C195 (wild type) and S194C195 mutant (Fig. 4C) would exclude a second site (Cys-194) prenylation.
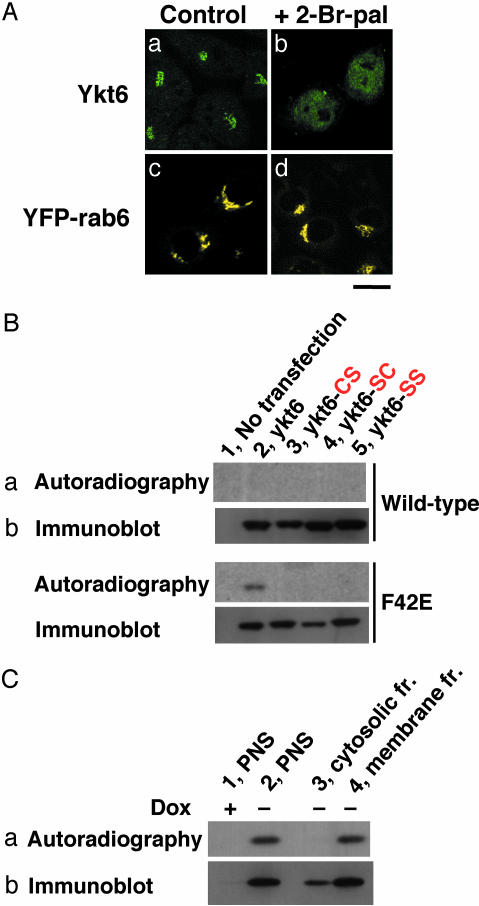
Thus, we tested the possibility of other lipid modifications of Ykt6. When cells were treated with the protein palmitoylation inhibitor 2-Br-palmitate, the perinuclear staining of endogenous Ykt6 but not that of YFP-rab6 disappeared (Fig. 5A). These results suggest that protein palmitoylation is required for the Golgi localization of Ykt6.
Fig. 5.
Palmitoylation of Ykt6 in vivo.(A) Wild-type HeLa cells (a and b) and HeLa cells stably expressing YFP-tagged human rab6 localized to the Golgi (c and d) were treated for 1 day with 150 μM 2-Br-palmitate (b and d). The cells were immunostained with anti-ykt6 antibodies and observed with a confocal microscope. (Bar = 10 μm.) (B) HeLa cells were transfected with GFP-tagged wild-type or F42E mutant forms of Ykt6. The following mutations were introduced into both constructs: C195/S (CS; lane 3), C194/S (SC; lane 4), and C194C195/SS (SS; lane 5). After a 5-h incubation with [3H]palmitate, GFP-tagged proteins were immunoprecipitated from cell lysates. (C) HeLa (tet-off) cells stably expressing myc-tagged Ykt6-F42E were metabolically labeled with [3H]palmitate as described above in the presence (lane 1) or absence (lanes 2–4) of 500 ng/ml doxycyclin (Dox). myc-tagged proteins from the postnuclear supernatants (PNS; lanes 1 and 2); 100,000 × g supernatant (cytosolic) fraction (lane 3); and 100,000 × g precipitated (membrane) fraction (lane 4) were immunoprecipitated and analyzed by fluorography (a) and immunoblotting (b) using anti-GFP or anti-myc antibody.
Next, we conducted metabolic labeling studies of GFP-tagged Ykt6 and Ykt6-F42E by using [3H]palmitate. GFP-ykt6-F42E was labeled with [3H]palmitate (Fig. 5B), but GFP-ykt6 showed only very faint labeling even after prolonged exposure (data not shown), consistent with the relatively low amounts of membrane-associated wild-type ykt6 compared with the ykt6-F42E mutant (Fig. 2 C and E). Palmitoylation of myc-tagged Ykt6-F42E was detected only in ykt6 in the membrane fraction and not in the cytosolic fraction (Fig. 5C). These results indicate that only the membrane-associated form of Ykt6 is palmitoylated and, therefore, that posttranslational palmitoylation is a key step in the efficient targeting of Ykt6 to the Golgi apparatus.
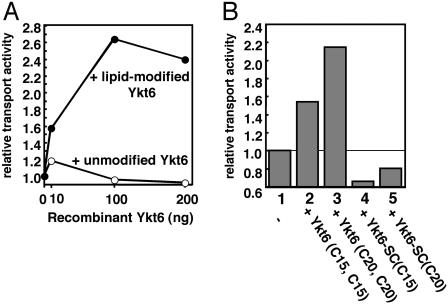
Double-Lipid-Modified but Not Single-Lipid-Modified Ykt6 Enhances Intra-Golgi Transport Activity. We examined the effect of the recombinant Ykt6 containing various lipid modifications (see Materials and Methods for details) on intra-Golgi transport by using in vitro intra-Golgi transport assay (25). Because it was difficult to introduce prenyl and acyl groups simultaneously into isolated recombinant Ykt6 proteins, we chemically coupled either one or two prenyl groups to Ykt6. The recombinant Ykt6 proteins were added to the standard assay, and the transport activity was measured. Unmodified Ykt6 had no effect on the intra-Golgi transport activity, whereas lipid-modified Ykt6 with two prenyl groups attached to the double-cysteine sequence-enhanced transport in a dose-dependent manner (Fig. 6A). Whereas double-lipid-modified Ykt6s containing farnesyl (C15) or geranylgeranyl (C20) groups both enhanced intra-Golgi transport, single-lipid-modified Ykt6 reduced transport activity (Fig. 6B). These results indicate that double-lipid-modified Ykt6, but not a single-lipid-modified Ykt6, is active in intra-Golgi transport. Furthermore, functionally active, double-lipid-anchored Ykt6 is a rate-limiting factor in the transport assay, indicating that Ykt6 could regulate membrane flow.
Fig. 6.
Effect of the recombinant Ykt6 on intra-Golgi transport. (A) The in vitro intra-Golgi transport assay was performed as described in Materials and Methods. The increasing amounts of unmodified recombinant Ykt6 (○) or a double-lipid-modified recombinant form of Ykt6 containing C20 (•) were added to the reaction. (B) The in vitro intra-Golgi transport activity was measured in the absence (column 1) or presence of 200 ng of the double-lipid-modified Ykt6 (columns 2 and 3) and the single-lipid-modified Ykt6 (columns 4 and 5) containing C15 (columns 2 and 4) or C20 (columns 3 and 5).
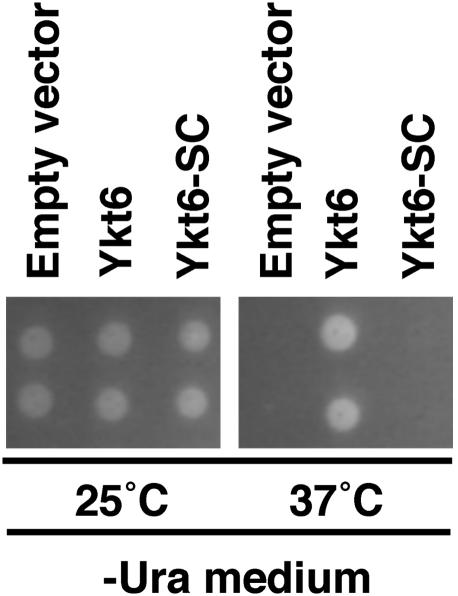
Deletion of the Palmitoylation Site in Ykt6 Is Lethal. To establish the role of Ykt6 palmitoylation in vivo, an S196C197 mutant or wild-type yeast Ykt6 was introduced into a yeast strain (SARY166) carrying a temperature-sensitive Ykt6 mutant. In contrast to the Ykt6 wild type, the S196C197 mutant did not rescue cell growth at the restrictive temperature (37°C), demonstrating that Cys-196 and bilipidation are essential for Ykt6 function and cell survival (Fig. 7).
Fig. 7.
Palmitoylation site in Ykt6 is essential for yeast growth. Yeast strain SARY166 carrying temperature-sensitive mutant ykt6 transfected with empty vectors, wild-type Ykt6 vectors, and S196C197 mutant of Ykt6 vectors were plated on the selection media as described in Materials and Methods and incubated at 25°C or 37°C for 5 and 3 days, respectively. Two independent clones were shown in each transfectant. We confirmed that the expression levels of ykt6 at 25°C were comparable between wild-type and SC mutant transfectants in immunoblot analysis (data not shown).
Discussion
Among the various SNARE proteins, Ykt6 has exceptional properties linked to its unique cellular distribution. At steady state, only a small fraction of Ykt6 is targeted to Golgi membranes, whereas the majority of it is found in the cytoplasm (Fig. 1), consistent with studies in NRK cells (13). Thus, it is likely that cytoplasmic Ykt6 provides a reserve pool for the Golgi-associated Ykt6 needed for its SNARE function in intra-Golgi transport.
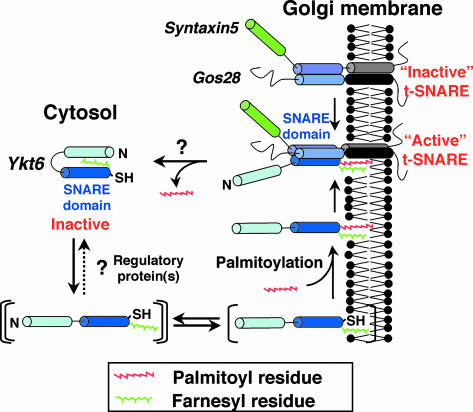
A model by which cyclical palmitoylation-dependent targeting of ykt6 to the Golgi could regulate membrane flow is outlined in Fig. 8. The following lines of evidence suggest that the cytosolic Ykt6 exists as an inactive “ready-to-go” pool of molecules. (i) Soluble Ykt6 is monomeric (Fig. 1C) even though it is already farnesylated (Fig. 4) but not palmitoylated (Fig. 5), and (ii) soluble Ykt6 has a closed conformation with its N-terminal domain (19), and disruption of this interaction by mutation (Fig. 2E) or deletion (Fig. 2D) results in complete translocation of Ykt6 to the membranes. We deduce that a farnesyl residue of Ykt6 is masked by the N-terminal domain, which in turn prevents binding of Ykt6 to the Golgi membranes where it is palmitoylated and locked in place. That the cytosolic pool of Ykt6 is inactive is also supported by the fact that a double-cysteine mutant of Ykt6 shows the growth defect in yeast (12).
Fig. 8.
Schematic representation of the association of Ykt6 with the membranes.
When the interaction of the N terminus with the SNARE domain of Ykt6 is altered by mutation, Ykt6 can now associate with the membranes by means of its farnesyl residue (Fig. 2E).
Presumably, this mutation mimics the effect of (still unknown) regulatory proteins in the cells. The membrane association efficiency of the single-prenylated Ykt6 is low, as the SC mutant of Ykt6-F42E shows both cytosolic and membrane localization (Fig. 3C). Additional palmitoylation of Ykt6 is required for its efficient retention in the Golgi (Fig. 5) and also for its full fusogenic activity as a SNARE molecule (Fig. 6). Consistent with this, the SC unpalmitoylated mutant form of Ykt6 does not complement the growth defect of yeast carrying the temperature-sensitive Ykt6 (Fig. 7).
Palmitoylation is a frequent posttranslational modification involved in membrane association and protein sorting (31). As an example, SNAP-25 and the calcium sensor synaptotagmin I are palmitoylated (32, 33); palmitoylation stimulates SNARE complex formation in vivo (34) probably by stabilizing them in the membranes (35). Recently, two new endoplasmic reticulum/Golgi-localized palmitoyltransferases specific for Ras2p and Yck2p have been identified in yeast and have been shown to control their localization and function (36, 37). The identification of the Ykt6 palmitoyltransferase could be helpful information establishing the link between membrane fusion at the Golgi and the palmitoyl modification, which was reported in early studies from our laboratory (38) but is still not well understood.
Recently, Dietrich et al. (39) reported that the N-terminal domain of yeast Ykt6 binds palmitoyl CoA and can mediate palmitoylation of Vac8 involved in homotypic fusion of vacuoles. It will be important to establish whether this results from true function as an enzyme, as distinct from chemical acylation by an activated fatty acid.
Acknowledgments
We thank Dr. David K. Banfield for providing yeast strain SARY166. This work was supported by a fellowship from the Naito Foundation (to M.F.) and Grant 5 R01 DK27044 from the National Institutes of Health (to J.E.R.).
Abbreviation: SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor.
References
- 1.Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P. & Rothman, J. E. (1993) Nature 362, 318–324. [DOI] [PubMed] [Google Scholar]
- 2.Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., Söllner, T. H. & Rothman, J. E. (1998) Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- 3.McNew, J. A., Weber, T., Parlati, F., Johnston, R. J., Melia, T. J., Söllner, T. H. & Rothman, J. E. (2000) J. Cell Biol. 150, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parlati, F., McNew, J. A., Fukuda, R., Miller, R., Söllner, T. H. & Rothman, J. E. (2000) Nature 407, 194–198. [DOI] [PubMed] [Google Scholar]
- 5.Paumet, F., Brugger, B., Parlati, F., McNew, J. A., Söllner, T. H. & Rothman, J. E. (2001) J. Cell Biol. 155, 961–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parlati, F., Varlamov, O., Paz, K., McNew, J. A., Hurtado, D., Söllner, T. H. & Rothman, J. E. (2002) Proc. Natl. Acad. Sci. USA 99, 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu, C., Ahmed, M., Melia, T. J., Söllner, T. H., Mayer, T. & Rothman, J. E. (2003) Science 300, 1745–1749. [DOI] [PubMed] [Google Scholar]
- 8.Varlamov, O., Volchuk, A., Rahimian, V., Doege, C. A., Paumet, F., Eng, W. S., Arango, N., Parlati, F., Ravazzola, M., Orci, L., et al. (2004) J. Cell Biol. 164, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pelham, H. R. (2001) Trends Cell Biol. 11, 99–101. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Y. A. & Scheller, R. H. (2001) Nat. Rev. Mol. Cell. Biol. 2, 98–106. [DOI] [PubMed] [Google Scholar]
- 11.Sutton, R. B., Fasshauer, D., Jahn, R. & Brunger, A. T. (1998) Nature 395, 347–353. [DOI] [PubMed] [Google Scholar]
- 12.McNew, J. A., Søgaard, M., Lampen, N. M., Machida, S., Ye, R. R., Lacomis, L., Tempst, P., Rothman, J. E. & Söllner, T. H. (1997) J. Biol. Chem. 272, 17776–17783. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, T. & Hong, W. (2001) J. Biol. Chem. 276, 27480–27487. [DOI] [PubMed] [Google Scholar]
- 14.Volchuk, A., Ravazzola, M., Perrelet, A., Eng, W. S., Di Liberto, M., Varlamov, O., Fukasawa, M., Engel, T., Söllner, T. H., Rothman, J. E. & Orci, L. (2004) Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 15.Hasegawa, H., Zinsser, S., Rhee, Y., Vik-Mo, E. O., Davanger, S. & Hay, J. C. (2003) Mol. Biol. Cell 14, 698–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kweon, Y., Rothe, A., Conibear, E. & Stevens, T. H. (2003) Mol. Biol. Cell 14, 1868–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dilcher, M., Köhler, B. & von Mollard, G. F. (2001) J. Biol. Chem. 276, 34537–34544. [DOI] [PubMed] [Google Scholar]
- 18.Liu, Y. & Barlowe, C. (2002) Mol. Biol. Cell 13, 3314–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tochio, H., Tsui, M. M., Banfield, D. K. & Zhang, M. (2001) Science 293, 698–702. [DOI] [PubMed] [Google Scholar]
- 20.Zacharias, D. A., Violin, J. D., Newton, A. C. & Tsien, R. Y. (2002) Science 296, 913–916. [DOI] [PubMed] [Google Scholar]
- 21.McNew, J. A., Parlati, F., Fukuda, R., Johnston, R. J., Paz, K., Paumet, F., Söllner, T. H. & Rothman, J. E. (2000) Nature 407, 153–159. [DOI] [PubMed] [Google Scholar]
- 22.Hancock, J. F., Magee, A. I., Childs, J. E. & Marshall, C. J. (1989) Cell 57, 1167–1177. [DOI] [PubMed] [Google Scholar]
- 23.Brusca, J. S. & Radolf, J. D. (1994) Methods Enzymol. 228, 182–193. [DOI] [PubMed] [Google Scholar]
- 24.McNew, J. A., Coe, J. G., Søgaard, M., Zemelman, B. V., Wimmer, C., Hong, W. & Söllner, T. H. (1998) FEBS Lett. 435, 89–95. [DOI] [PubMed] [Google Scholar]
- 25.Balch, W. E., Dunphy, W. G., Braell, W. A. & Rothman, J. E. (1984) Cell 39, 405–416. [DOI] [PubMed] [Google Scholar]
- 26.Malhotra, V., Serafini, T., Orci, L., Shepherd, J. C. & Rothman, J. E. (1989) Cell 58, 329–336. [DOI] [PubMed] [Google Scholar]
- 27.Tsui, M. M. & Banfield, D. K. (2000) J. Cell Sci. 113, 145–152. [DOI] [PubMed] [Google Scholar]
- 28.Gerdes, H. H. & Kaether, C. (1996) FEBS Lett. 389, 44–47. [DOI] [PubMed] [Google Scholar]
- 29.Lerner, E. C., Qian, Y., Hamilton, A. D. & Sebti, S. M. (1995) J. Biol. Chem. 270, 26770–26773. [DOI] [PubMed] [Google Scholar]
- 30.McGuire, T. F., Qian, Y., Vogt, A., Hamilton, A. D. & Sebti, S. M. (1996) J. Biol. Chem. 271, 27402–27407. [DOI] [PubMed] [Google Scholar]
- 31.Bijlmakers, M. J. & Marsh, M. (2003) Trends Cell Biol. 13, 32–42. [DOI] [PubMed] [Google Scholar]
- 32.Hess, D. T., Slater, T. M., Wilson, M. C. & Skene, J. H. (1992) J. Neurosci. 12, 4634–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Veit, M., Söllner, T. H. & Rothman, J. E. (1996) FEBS Lett. 385, 119–123. [DOI] [PubMed] [Google Scholar]
- 34.Vogel, K. & Roche, P. A. (1999) Biochem. Biophys. Res. Commun. 258, 407–410. [DOI] [PubMed] [Google Scholar]
- 35.Kammer, B., Schmidt, M. F. & Veit, M. (2003) Mol. Cell. Neurosci. 23, 333–340. [DOI] [PubMed] [Google Scholar]
- 36.Lobo, S., Greentree, W. K., Linder, M. E. & Deschenes, R. J. (2002) J. Biol. Chem. 277, 41268–41273. [DOI] [PubMed] [Google Scholar]
- 37.Roth, A. F., Feng, Y., Chen, L. & Davis, N. G. (2002) J. Cell Biol. 159, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pfanner, N., Glick, B. S., Arden, S. R. & Rothman, J. E. (1990) J. Cell Biol. 110, 955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietrich, L. E., Gurezka, R., Veit, M. & Ungermann, C. (2004) EMBO J. 23, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]