Abstract
A structural profile-based computational screen was used to identify neuropoietin (NP), a new cytokine. The np gene is localized in tandem with the cardiotrophin-1 gene on mouse chromosome 7. NP shares structural and functional features with ciliary neurotrophic factor (CNTF), cardiotrophin-1, and cardiotrophin-like cytokine. It acts through a membrane receptor complex comprising CNTF receptor-α component (CNTFRα), gp130, and leukemia inhibitory factor receptor to activate signal transducer and activator of transcription 3 signaling pathway. NP is highly expressed in embryonic neuroepithelia. Strikingly, CNTFRα, but not its alternate ligands, CNTF and cardiotrophin-like cytokine, is expressed at the same developmental stages. NP is also observed in retina and to a lesser extent in skeletal muscle. Moreover, NP could sustain the in vitro survival of embryonic motor neurons and could increase the proliferation of neural precursors when associated to epidermal growth factor and fibroblast growth factor 2. Thus, NP is a new ligand for CNTFRα, with important implications for murine nervous system development.
Aparticular group of mediators referred to as the IL-6 family of cytokines activates the signal transducing receptor protein, gp130. IL-6, IL-11, leukemia inhibitory factor (LIF), oncostatin M, cardiotrophin-1 (CT-1), ciliary neurotrophic factor (CNTF), and cardiotrophin-like cytokine (CLC) belong to this family of cytokines (1, 2). These cytokines, which display a four-helix bundle structure (3, 4), utilize one or both of the shared receptor signal transducing subunits, gp130 and LIF receptor (LIFR), in their respective receptors (5, 6). In addition to these shared signaling receptor subunits, the CNTF receptor complex implicates a third binding component, CNTFRα (1, 7, 8). Downstream signaling involves activation of the Janus kinase 1/signal transducer and activator of transcription (STAT) 3 pathway (9). CNTF was characterized through its ability to sustain the survival of parasympathetic and motor neurons in vitro and in vivo (10–12). CNTF can also attenuate the motor deficits found in different strains of mice with neuromuscular deficiencies (13–16). Reductions in CNTF have been associated with increased severity of familial forms of amyotrophic lateral sclerosis and with early onset of multiple sclerosis (17, 18). Beside its activities on the nervous system, CNTF, which also displays trophic effects on denervated skeletal muscle, is a regulator of muscular strength in aging and can reduce body fat (19–21). CNTF-deficient mice display only a mild phenotype involving motor neuron degeneration, which is most significant in older mice (22). In contrast, newborn mice deficient in CNTFRα fail to initiate the feeding process, die shortly after birth, and show a dramatic loss of motor neurons (23). These contrasting phenotypes point to the existence of alternative and developmentally important ligand(s) for CNTFRα. This is further supported by the observation that, whereas CNTFRα is strongly expressed in embryo, CNTF expression is barely detectable during development (24, 25). Additionally, CNTF lacks a leader sequence and appears to be released from cells only during trauma (12). We previously established that CLC, in association with either cytokine-like factor, or with a soluble form of CNTFRα, generates soluble composite cytokines that also bind and activate the CNTF receptor complex (26–28). In the present study, we report the characterization of a family member of the long-chain four-helix bundle cytokines we named neuropoietin (NP). We show that NP is only expressed during embryonic life when CNTF and CLC are absent, or barely detectable, postulating for an important role of NP in nervous system development. We demonstrate that this protein, closely related to CNTF, CT-1, and CLC, also binds to the tripartite CNTF receptor complex and can sustain motor neuron survival and could increase neurosphere proliferation in vitro.
Materials and Methods
Computational Analysis. A structural alignment of helical IL-6 cytokine family was profiled with t-coffee (29) and was refined by manual adjustments. A position-specific scoring matrix for the B, C, and D helices was created by using psi-blast (30) to screen public mouse databases of genscan-predicted genes (31). Human and rat sequences were identified by a blastn of the nucleotide sequence of mouse np on public genome databases. The chimpanzee genome fragments (ftp://ftp.infobiogen.fr/db/Trace/pan_troglodytes) were concatened to form a database. Secondary structure prediction of NP was computed with phd (32). A distance matrix was created with protdist, and used to build the phylogenic tree, by using neighbor with the upgma algorithm (Phylip package, which can be accessed at http://evolution.genetics.washington.edu/phylip.html). Graphical representation of the tree was obtained with treeview 1.6.5, which can be accessed at (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
cDNA Cloning, Protein Expression, and Purification. mRNAs were isolated from embryonic day (E)15 mouse brain. Full-length NP cDNA was amplified by using the 5′-CACCATGTACTGCCTGCTGGCA-3′ and 5′-TGCTGAGTACTTAGCCTTGAGGAG-3′ primers, before being inserted in pcDNA3.1D/V5-His-Topo vector (Invitrogen). Resulting cDNA was fused to a sequence encoding histidine hexamer and V5 epitopes. Cos-7 cells were transfected by DEAE dextran method. After a 72-h culture period, NP was affinity purified by using Ni+ beads. Eluted fractions were next submitted to SDS/PAGE silver staining and Western blot analyses. For some of the experiments, NP was expressed in HEK 293 cells and similarly purified.
PCR and Real-Time PCR. For tissue distribution analyses, cDNA were amplified by 35 cycles of PCR by using the following primers: 5′-CTCTCTACATGCAGAAGAACACGTC-3′ and 5′-GACCATATTCTCGGGTCACTATGTA-3′ for NP. Quantitative PCR was performed on capillary real-time LightCycler (Roche Diagnostics) by using the same NP primers. The following primers were designed to amplify mouse CNTF: 5′-TGTCGACAAGTAAATCCACA-3′ and 5′-GGAGCACCACATTAAATACC-3′; mouse CLC: 5′-GGTGACTGTACGCCTCATAG-3′ and 5′-GCTACCTGGAGCATCAACT-3′; and mouse CNTFRα: 5′-GTGAATTCGTCAAAGGTGAT-3′ and CTACATCCCCAATACCTACA-3′. Human and chimpanzee genomic DNA fragments were amplified by 30 cycles of PCR using the following primers: exon 2, 5′-CCCGCCTCTGCCTGCTGACC-3′ and 5′-ATAAGTCCGCAGCAGCGCTG-3′; and exon 3, 5′-CTCCAGTACCAGGGCAGCCC-3′ and 5′-TGCTGAGTACTTAGCCTTGA-3′. DNA fragments then were cloned in Topo TA vector (Invitrogen).
Cell Cultures. Cos-7, HEK 293, neuroblastoma, glioblastoma, and hepatoma cell lines were cultured in RPMI medium 1640 supplemented with 10% FCS. Factor-dependent cell lines were grown in the same culture medium supplemented with appropriate cytokines as indicated in Fig. 5 (7, 26, 27). For proliferation assays, Ba/F3 and TF1 cell lines were seeded in 96-well plates at 5 × 103 cells per well in RPMI medium 1640 containing 5% FCS. Serial dilutions of tested cytokines were performed in triplicate. After a 72-h incubation, 0.5 μCi of 3H thymidine (1 Ci = 37 GBq) was added to each well for the last 4 h of the culture and the incorporated radioactivity determined. Motor neurons were isolated from E14.5 Sprague–Dawley rat ventral spinal cords as described (33). Purified motor neurons were plated at a density of 1,000 neurons per well. Motor neuron survival was quantified by counting the number of large phase-bright neurons with long axonal processes in the total area of triplicate wells. Neurospheres were kindly provided by M. Mitjavila-Garcia and L. Coulombel (Institut National de la Santé et de la Recherche Médicale U421, Créteil, France) and were generated from E15 murine cortex and striatum by using a protocol adapted from Gritti et al. (34). For proliferation assays, neurospheres were seeded in 96-well plates at 2 × 103 cells per well in DMEM/F12 culture medium supplemented with 20 ng/ml epidermal growth factor (EGF) and fibroblast growth factor (FGF) 2. Serial dilutions of tested cytokines were performed in triplicate. After a 7-day incubation, 0.5 μCi of 3H thymidine was added and the incorporated radioactivity determined.
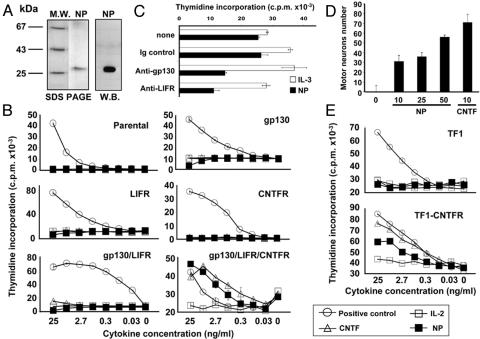
Fig. 5.
Tripartite functional receptor for NP. (A) Silver staining and Western blot analyses of NP purified from a Ni+ agarose column. (B) Proliferation of transfected Ba/F3 cells to NP. Murine IL-3 was used as positive control for parental-, LIFR-, and CNTFRα-transfected Ba/F3 cells, whereas IL-6/sol.IL-6R, LIF, and CNTF were used as controls for BAF gp130, Ba/F3 cells expressing only gp130 and LIFR, and BAF GLC, respectively (most of the SE deviation values were inferior to the symbol size). (C) Inhibition of NP-induced BAF GLC cell proliferation by using neutralizing anti-gp130 and LIFR Abs. Cells were incubated with 5 ng/ml NP (black bars) or murine IL-3 as control (open bars), alone (none) or with 15 μg/ml of an anti-gp130 (AN-HH1), anti-LIFR (12D3), or an IgG-matched control mAb, as indicated. (D) NP sustains motor neuron survival. Survival effects were measured on motor neurons from E14.5 rat embryos maintained in culture for 3 days in the presence of indicated cytokine concentrations, in ng/ml. (E) Proliferation of human TF1 and CNTFRα-transfected TF1 cells to NP. Human LIF was used as a positive control for both studied cell lines (most of the SE deviation values were inferior to the symbol size).
Reagents. Recombinant human IL-2, IL-6, granulocyte/macrophage colony-stimulating factor, CNTF, soluble IL-6R, and soluble CNTFRα were from R & D Systems. Human LIF, murine IL-3, soluble LIFR-Fc, soluble CNTFRα-Fc, soluble gp130-Fc, AN-HH1 (anti-gp130), AN-G30 (anti-gp130), AN-E1 (anti-LIFR), AN-D3 (anti-CNTFRα) mAbs, and biotinylated polyclonal anti-CNTF Ab were produced in the laboratory, as described (26, 27). The 12D3 anti-LIFR-neutralizing Ab was obtained from Becton Dickinson. Anti-STAT3 Abs were bought from New England Biolabs. Anti-phosphotyrosine (4G10) and anti-V5 epitope Abs, coupled or not to peroxidase, were from Upstate Biotechnology (Lake Placid, NY) and Invitrogen, respectively.
Tyrosine Phosphorylation, Immunoprecipitation, and Western Blot Analyses. Cells were incubated overnight in serum-free medium before being stimulated for 10 min with the indicated cytokines. Cells were lysed with 1% Brij 96 buffer (50 mM Tris·HCl, pH 7.5/150 mM NaCl/1% Brij 96), and protein tyrosine phosphorylations or protein contacts were subsequently analyzed as described (27). For in vitro coprecipitation experiments, proteins were purified from the reaction medium by using either AN-D3 anti-CNTFRα Ab or protein A beads as described (27). Tagged NP and CNTF were then visualized by Western blots by using an anti-V5 epitope Ab labeled with peroxidase and a biotinylated polyclonal anti-CNTF Ab, respectively.
In Situ Hybridization. Fixed embryos were cryopreserved in 30% sucrose and cut at 16 μm. In situ hybridization was performed as described (35). In brief, sections were hybridized overnight with a DIG-labeled NP riboprobe. After washes and blocking, slides were incubated with an anti-DIG-alkaline-phosphatase conjugated Ab (Roche Diagnostics) and revealed with NBT/BCIP reagent. To confirm that muscle fibers, per se, express np double in situ hybridization/immunohistochemistry was carried out as described (35) on sections from E14.5 MLCnlacZ mice, which express the nlacZ-reporter gene under the control of a muscle-specific myosin light chain promoter (36). After in situ hybridization, slides were processed for staining by using diaminobenzidine.
Results
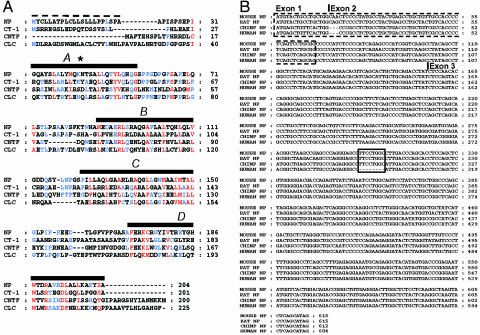
NP, a New Member of the IL-6 Cytokine Family. We describe another helical cytokine identified by analyzing mouse genome. We searched sequence databases of predicted genes (31) with a computationally derived profile of IL-6 cytokine family members. This search led to the identification of a new cytokine we named NP, according to its structural features and CNTF similarity. The murine cDNA sequence isolated from E15 brain embryo encodes a 204-aa protein with a potential hydrophobic signal peptide of 24 residues and a single predicted N-glycosylation site at position 44 (Fig. 1A). A pairwise comparison between the deduced amino acid sequence of NP and other members of the neuropoietic cytokine family demonstrates a 16% identity with CNTF and an 11–27% identity with the other IL-6 family members. It also includes prediction of four helices, a hallmark of cytokines belonging to hematopoietic/neuropoietic superfamily (1, 4). We also identified orthologs of NP in rat, chimpanzee, and human genomes (Fig. 1B). To confirm computational data, the exon regions of chimpanzee and human np genes were PCR amplified and cloned from genomic DNA. Obtained sequences were identical to those present in the databases. An 83% identity was observed between human and mouse np genes, and 98% between human and chimpanzee ones. Importantly, an 8-nt deletion was observed in the third putative exon of human np gene leading to a shift and a loose of the reading frame (Fig. 1B). Genomic DNA surrounding this deletion area was analyzed by pyrosequencing in 93 individuals (data not shown), and all of the samples gave the same 8-nt deletion, suggesting that in human, np has evolved toward a pseudogene. In contrast, chimpanzee np gene is intact giving the possibility for a functional protein in ape.
Fig. 1.
NP amino acid sequence and its comparison with related IL-6 family members. (A) Sequence alignment of mouse NP with mouse CT-1, CNTF, and CLC. Strictly conserved and 80% conserved amino acids are in red and blue, respectively. Black bars indicate the four predicted α-helices. Predicted signal peptide is highlighted with a dashed line. An asterisk represents a potential N-glycosylation site on NP. (B) Alignment of concatened exons of mouse, rat, chimpanzee, and human NP orthologs. Indicative exon positions were based on mouse np sequence. A dashed box indicates the predicted signal peptide-coding regions. A plain box highlights the deletion in the putative third exon of human NP.
The phylogenetic tree demonstrates that murine NP is most closely related to CT-1 and contributes to form a subgroup with CLC and CNTF into the IL-6 family of cytokines (Fig. 2A). The np and ct-1 genes were both localized on mouse chromosome 7F3 and were only 1.7 kb distant from each other with opposite transcriptional orientations (Fig. 2B). Analysis of exon organization revealed that np gene contained 3 exons, a mostly conserved gene structure for cytokines that bind LIFR (Fig. 2C and ref. 4). General intron/exon organization of np and ct-1 genes, their chromosomal proximity and similarity level, suggest that they arose from a gene duplication event.
Fig. 2.
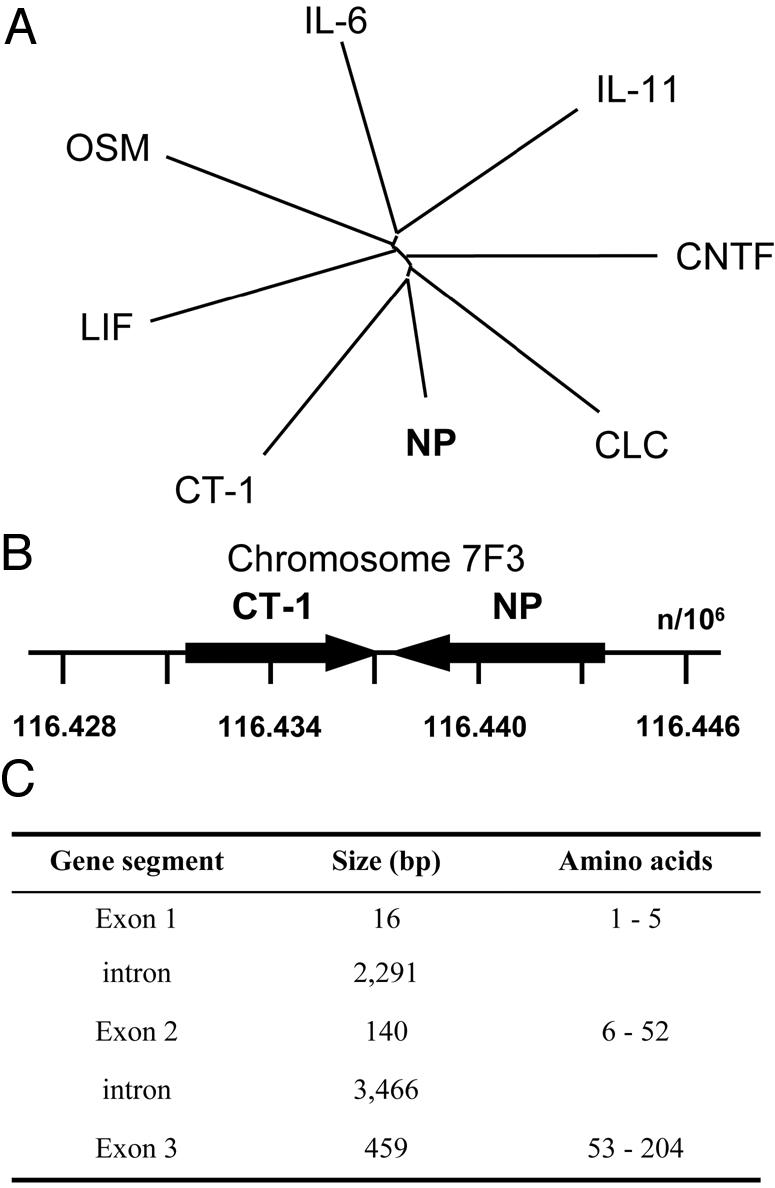
Evolution of the np gene. (A) Evolutionary dendrogram for members of the IL-6 family cytokines. (B) Gene localization of mouse np on chromosome 7F3 located in tandem with ct-1. Positions are expressed in kilobytes. (C) Exon/intron organization of np gene.
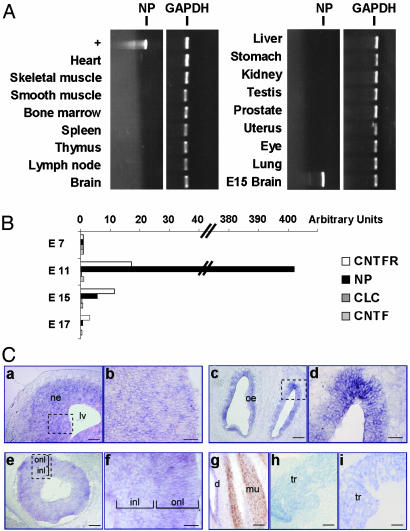
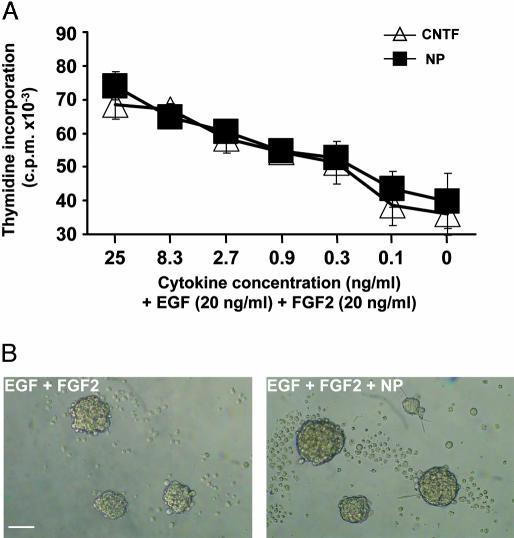
NP Is Expressed During Embryonic Life and Triggers the Proliferation of Neuronal Precursors. Distribution of NP was analyzed by RT-PCR and real-time PCR by using a panel of different poly(A)+ mRNAs obtained from embryo or adult mice tissues (Fig. 3 A and B). No transcripts could be detected in any tested adult tissues. However, NP was expressed in embryonic life, with a dramatic peak at day 11 of gestation. Importantly, a similar kinetic of expression was observed for CNTFRα component (Fig. 3B), in agreement with previous observations (24, 25). In contrast, CNTF and CLC transcripts were barely detectable at this developmental stage. To further determine the biological function of the cytokine, in situ hybridization was performed to localize the NP-expressing cells (Fig. 3C). By E10, NP is prominently expressed in cells scattered throughout all neuroepithelia: those lining of the brain vesicles, the olfactory and retina neuroepithelia (Fig. 3 Ca–f), suggesting an important role for NP in neuronal precursor development and maturation. NP was also readily detectable in cranial and dorsal root sensory ganglia, as well as in spinal cord (Fig. 3Ch and data not shown). At E14, outside the nervous system, NP is detectable in vibrissae, dermis, and to a lesser extent in skeletal muscle, lung, and kidney (Fig. 3Cg and data not shown); all these tissues are known to express the CNTFRα (Fig. 3Ci and ref. 24). Neurospheres are undifferentiated multipotent neural progenitors that can be amplified in vitro in the presence of EGF plus FGF2 (34, 37). We studied the possibility for NP to directly trigger the proliferation of these neural stem cells (Fig. 4). When NP was tested alone, no modification in the cultures could be evidenced (data not shown). In contrast, when associated to FGF2 and EGF, NP induced a strong increase in thymidine incorporation and on the neurosphere volume, as observed for CNTF (38). This evidences a direct trophic effect of NP on dividing neuronal precursors.
Fig. 3.
Expression profiles of NP. (A) NP expression was studied by RT-PCR in adult mice tissues and in E15 brain. NP plasmid DNA was used as positive control. (B) Expression of NP, CLC, CNTF, and CNTFRα were measured in developing embryo by quantitative PCR, as described in Materials and Methods. (C) In situ localization of NP mRNA in mouse embryonic tissues. (a) Transverse section of the lateral ventricle of E12.5 embryo. (c) Sagittal section through E16 olfactory neuroepithelium. NP signal was detected throughout neuroepithelia in scattered cells as shown in higher magnifications of squares in a (b) and c (d). (e) Sagittal section through E14.5 retina. (f) Higher magnification of the square in e showing that NP expression is dispersed throughout the retina in both inner nuclear layer and outer nuclear layer where neural retina progenitors are actively differentiating. (g) Transverse section of forelimb muscles from E14.5 MLCnlacZ mice, which express the nlacZ-reporter gene in muscles under the control of myosin light-chain promoter, double stained by in situ hybridization for NP and by immunohistochemical staining for β-galactosidase. β-galactosidase-positive cells (brown nuclear staining) are also weakly NP positive (blue staining). (h and i) Sagittal sections through trigeminal ganglion hybridized for NP (h) or CNTFRα (i). ne, neuroepithelium; lv, lateral ventricle; oe, olfactory epithelium; inl, inner nuclear layer; onl, outer nuclear layer; d, dermis; mu, muscle; tr, trigeminal ganglion. (Scale bars: 100 μm for a, c, e, and g–i;30 μm for b, d, and f.)
Fig. 4.
Proliferation of neurospheres to NP. (A) Proliferation assay of neurospheres by using serial dilution of CNTF (▵) or NP(▪) in presence of 20 ng/ml EGF and FGF2 (some of the SE deviation values were inferior to the symbol size). In the absence of EGF and FGF2, or by using NP alone, the obtained values were 305 and 2,153 cpm, respectively. (B) Phase-contrast photomicrograph of neurosphere cultures supplemented with 20 ng/ml EGF and FGF2 (Left) or with 20 ng/ml EGF and FGF2 plus 25 ng/ml NP (Right) during 7 days. (Scale bar, 50 μm.)
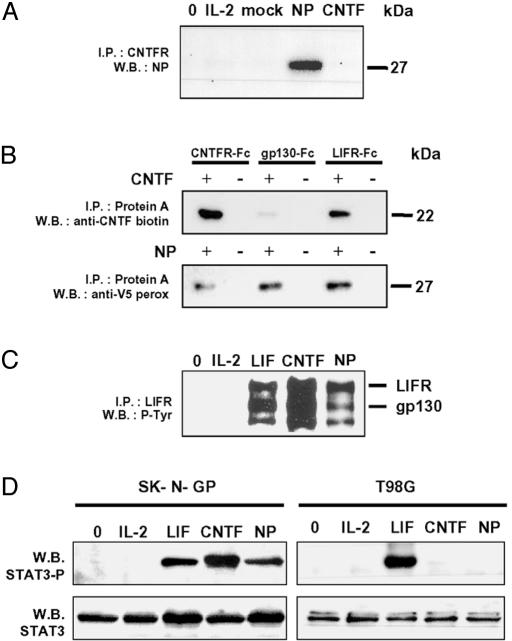
NP Binds to and Signals Through the Tripartite CNTF Receptor. NP was expressed as a tagged protein and purified from culture supernatants of Cos-7- or HEK 293-transfected cells. SDS/PAGE gels and Western blot analyses show that the polypeptide had an Mr of 27 kDa, corresponding, after subtracting tag molecular weight, to a mature protein of 22 kDa (Fig. 5A). To investigate the functional activity of NP, we used derivatives of IL-3-dependent Ba/F3 cell line rendered responsive to cytokines of the IL-6 family by transfection with the appropriate receptor chains (26). NP specifically induced proliferation of Ba/F3 cells expressing the functional receptor for CNTF [gp130, LIFR, and CNTFRα (BAF GLC)] on their surface (Fig. 5B). Those cells expressing only gp130 and LIFR failed to proliferate in the presence of the cytokine, implicating CNTFRα in NP receptor complex formation. Furthermore, NP did not provide significant signals on parental Ba/F3 cells or on Ba/F3 cells expressing gp130, LIFR, or CNTFRα alone. The observation was further reinforced by using the human TF1 erythroleukemia cell line spontaneously expressing gp130 and LIFR components (7). Parental TF1 cells failed to respond to NP, but a proliferative response to the cytokine could be induced by expressing CNTFRα to reconstitute a functional tripartite receptor for NP (Fig. 5E). The involvement of LIFR and gp130 signaling subunits in NP receptor complex was confirmed by the inhibition of proliferation after addition of anti-gp130- and anti-LIFR-neutralizing Abs to BAF GLC cell cultures (Fig. 5C). These results demonstrate that NP and CNTF require the same cell surface receptor components to induce functional responses. CNTF is known to sustain the in vitro survival of embryonic motor neurons (14). We then demonstrated that CNTF and NP had related biological properties on this tissue. Embryonic motor neurons were isolated (33), and a 72-h survival assay was performed (Fig. 5D). As with CNTF, NP was found to keep motor neurons alive.
We also demonstrated a direct effect of NP by studying the contact between the cytokine and CNTFRα. In coprecipitation experiments, BAF GLC cells were incubated with NP. The cell lysates were then analyzed after a CNTFRα immunoprecipitation, and associated NP was evidenced by Western blotting (Fig. 6A). In the in vitro experiments, CNTF or NP was added to soluble Fc forms of CNTFRα, gp130, and LIFR before being precipitated (Fig. 6B). Direct contacts of CNTF to CNTFRα and LIFR, but not to gp130, were detected as reported before (39, 40). On its side, NP was found to directly bind each of the components involved in its tripartite receptor formation.
Fig. 6.
Physical interaction and recruitment of CNTF receptor complex by NP. (A) Coimmunoprecipitation of NP and CNTFRα from the surface of BAF GLC cells. BAF GLC were incubated with IL-2 (50 ng/ml)-, CNTF (50 ng/ml)-, mock-, or NP-containing culture media for 10 min at 37°C. Cells were lysed with Brij 96 buffer, and proteins were immunoprecipitated with AN-D3 anti-CNTFRα mAb and protein A beads. NP was detected by using an anti-V5 epitope mAb. (B) Coimmunoprecipitation of CNTF (Upper) or NP(Lower) with CNTFRα-Fc, gp130-Fc, or LIFR-Fc fusion proteins. Receptor subunits were incubated at a concentration of 4 nM for 16hat4°C in the presence of 4 nM NP or CNTF before being immunoprecipitated with protein A beads. NP and CNTF were detected by using an anti-V5 epitope mAb and a polyclonal biotinylated anti-CNTF Ab, respectively. (C) Tyrosine phosphorylation of gp130 and LIFR in SK-N-GP cells in response to NP. After an exposure to 50 ng/ml of indicated cytokines, SK-N-GP cells were lysed with Brij 96 buffer. Then, LIFR and gp130 were coprecipitated by using the AN-E1 anti-LIFR mAb. Tyrosine phosphorylation content was determined with the 4G10 anti-phosphotyrosine mAb. (D) Induction of STAT3 tyrosine phosphorylation by NP in SK-N-GP neuroblastoma (gp130+, LIFR+, and CNTFRα+) and T98G glioblastoma cells (gp130+, LIFR+, and CNTFRα–). After exposure to 50 ng/ml of indicated cytokines, cells were lysed with 1% Nonidet P-40 buffer and analyzed by Western blotting by using an anti-phospho-STAT3 Ab.
We also studied NP-induced signal transduction. The role of gp130 and LIFR as signaling components for NP was further reinforced when analyzing their tyrosine phosphorylation level after activation by NP. In response to LIF and CNTF, a strong induction of tyrosine phosphorylation was detected for gp130 and LIFR in SK-N-GP neuroblastoma cells (gp130+, LIFR+, and CNTFRα+; Fig. 6C). Similarly, NP induced a clear tyrosine phosphorylation of gp130 and LIFR in the same cells. Downstream signaling events were investigated by measuring induction of STAT3 tyrosine phosphorylation in response to the cytokine. NP induced the phosphorylation of STAT3 in SK-N-GP neuroblastoma cells but failed to recruit this protein in T98G glioblastoma cells, which lack expression of CNTFRα (Fig. 6D). Additionally, NP was tested in a series of cells expressing the tripartite CNTF receptor (SK-N-FL, SY5Y neuroblastoma, and EW-1 Ewing sarcoma cells), or specifically lacking expression of CNTFRα component (Go-G-UVM, Go-G-CCM glioblastoma, and HepG2 hepatoma cells) (27). In all tested cell lines, an expression of CNTFRα was strictly required to allow NP signaling.
Discussion
The present study reports the identification of a new four-helix bundle cytokine, we named NP and which shares features with the IL-6 family members. We demonstrate, that mouse NP acts through the tripartite CNTF receptor complex to generate similar signaling cascades and biological activities in vitro. The np gene is localized on mouse chromosome 7 in tandem with ct-1 gene, a structurally related cytokine. Similarly, cntf and clc are also twin genes localized on chromosome 19, suggesting that this functionally related cytokine subgroup arose from successive duplication events.
The existence of an alternate developmentally regulated ligand for CNTFRα was initially postulated based on the CNTF–/– and CNTFRα–/– mice phenotype comparison (22, 23). Inactivation of CNTF led to a mild loss of motor neurons in adult. In contrast CNTFRα subunit is essential for survival of nearly one-half of motor neurons during development, and mice harboring a homozygous CNTFRα–/– mutation die shortly after birth. Whereas CNTF is normally expressed at very low levels in embryo, it is abundantly stored inside of adult glial cells and released by injury (24, 25). Similarly, CLC transcripts are highly expressed in adult tissues, mainly in ovary, heart, liver, colon, kidney, and spleen, suggesting a not yet known function for CLC in these tissues (41, 42). A recent study demonstrates that CLC mRNA was also found in skeletal muscle fibers of embryonic mice during the motor neuron cell death period (43). This data indicates that CLC can play an important role on developing motor neurons. In contrast, NP was not present in adult tissues, and a specific and high expression of its transcript was detected in neuroepithelium by E10 and later in olfactory neuroepithelium and retina. To a lesser extent, NP is also detectable in skeletal muscle. LIFR, gp130, and CNTFRα are all present at high levels as early E11 (24). Importantly, we show that NP can strongly trigger the in vitro proliferation of neural precursors when associated to EGF plus FGF2. These suggest a crucial function for NP in nervous system development, mirroring CNTFRα expression in mitotically active neuronal precursors of neuroepithelium (24). A small proportion of the human population (2.3%) is homozygous for a mutation inactivating the cntf gene without any sign of neurological defects (44, 45). We previously showed that CLC could substitute to CNTF to activate the tripartite receptor, suggesting the functional redundancy of both ligands (26–28).
Regarding human np gene, we failed to detect any transcript in all tested tissues (data not shown). Importantly, an 8-nt deletion in the area corresponding to a putative exon 3 and leading to a frameshift was evidenced, whereas the corresponding portion of DNA was preserved in chimpanzee. This data strongly suggests that np has evolved toward a pseudogene in human. A recent study (46) reported a rapid acceleration of pseudogene formation in several functional classes of genes in human lineage compared to chimpanzee and mouse lineages. This is particularly true for olfactory receptors in which 70% of the genes become nonfunctional in human (47, 48). Interestingly, a protein pheromone affecting female receptivity and displaying structural features and a similarity of 45% with NP was described in salamander (49). The possibility that NP may also act on the sense of smell, and has been lost in human, is open.
Perhaps the most important aspects of the current study are the high expression level of NP during mouse development in neuroepithelium, a region known to be essential for neural precursor expansion (50) and the ability of NP to trigger the self-renewal of neural stem cells. Therefore, it will be important to compare np gene inactivation phenotype with the deletion of cntf and clc genes coding for alternate CNTFRα ligands (22, 51). Because genetic deletion of cntf fails to perturb neuronal development before birth, we can hypothesize some in vivo functional redundancies that will require an analysis of double or triple knockout mice for CNTFRα ligands to clarify their respective involvement in mouse neural development.
Acknowledgments
We thank Drs. M. Mitjavila-Garcia and L. Coulombel for providing murine neurospheres; the Association Française des Vétérinaires de Parcs Zoologiques; Dr. P. Rodien for providing chimpanzee genomic DNA; and Dr. C. E. Henderson for correcting the English. This work was supported by Post Genome Program from Contrat de Plan Etat-Région and by Association Française Contre les Myopathies Grants 9212 and 9300.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: En, embryonic day n; CLC, cardiotrophin-like cytokine; CNTF, ciliary neurotrophic factor; CNTFRα, CNTF receptor α; CT-1, cardiotrophin-1; LIF, leukemia inhibitory factor; LIFR, LIF receptor; NP, neuropoietin; STAT, signal transducer and activator of transcription; EGF, epidermal growth factor; FGF, fibroblast growth factor; BAF GLC, Ba/F3 cells expressing gp130, LIFR, and CNTFRα.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. NP_031821 (mouse CT-1), NP_740756 (mouse CNTF), NP_063336 (mouse CLC), AY363390 (mouse NP), AY518205 (rat NP), AY518206 (human NP), and AY518207 (chimpanzee NP)].
References
- 1.Grotzinger, J. (2002) Biochim. Biophys. Acta 1592, 215–223. [DOI] [PubMed] [Google Scholar]
- 2.Bravo, J. & Heath, J. K. (2000) EMBO J. 19, 2399–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grotzinger, J., Kurapkat, G., Wollmer, A., Kalai, M. & Rose-John, S. (1997) Proteins 27, 96–109. [PubMed] [Google Scholar]
- 4.Bazan, J. F. (1991) Neuron 7, 197–208. [DOI] [PubMed] [Google Scholar]
- 5.Hibi, M., Murakami, M., Saito, M., Hirano, T., Taga, T. & Kishimoto, T. (1990) Cell 63, 1149–1157. [DOI] [PubMed] [Google Scholar]
- 6.Gearing, D. P., Comeau, M. R., Friend, D. J., Gimpel, S. D., Thut, C. J., McGourty, J., Brasher, K. K., King, J. A., Gillis, S., Mosley, B., et al. (1992) Science 255, 1434–1437. [DOI] [PubMed] [Google Scholar]
- 7.Davis, S., Aldrich, T. H., Ip, N. Y., Stahl, N., Scherer, S., Farruggella, T., DiStefano, P. S., Curtis, R., Panayotatos, N., Gascan, H., et al. (1993) Science 259, 1736–1739. [DOI] [PubMed] [Google Scholar]
- 8.Davis, S., Aldrich, T. H., Stahl, N., Pan, L., Taga, T., Kishimoto, T., Ip, N. Y. & Yancopoulos, G. D. (1993) Science 260, 1805–1808. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich, P. C., Behrmann, I., Muller-Newen, G., Schaper, F. & Graeve, L. (1998) Biochem. J. 334, 297–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stockli, K. A., Lottspeich, F., Sendtner, M., Masiakowski, P., Carroll, P., Gotz, R., Lindholm, D. & Thoenen, H. (1989) Nature 342, 920–923. [DOI] [PubMed] [Google Scholar]
- 11.Arakawa, Y., Sendtner, M. & Thoenen, H. (1990) J. Neurosci. 10, 3507–3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oppenheim, R. W., Prevette, D., Yin, Q. W., Collins, F. & MacDonald, J. (1991) Science 251, 1616–1618. [DOI] [PubMed] [Google Scholar]
- 13.Curtis, R., Adryan, K. M., Zhu, Y., Harkness, P. J., Lindsay, R. M. & DiStefano, P. S. (1993) Nature 365, 253–255. [DOI] [PubMed] [Google Scholar]
- 14.Sendtner, M., Schmalbruch, H., Stockli, K. A., Carroll, P., Kreutzberg, G. W. & Thoenen, H. (1992) Nature 358, 502–504. [DOI] [PubMed] [Google Scholar]
- 15.Mitsumoto, H., Ikeda, K., Klinkosz, B., Cedarbaum, J. M., Wong, V. & Lindsay, R. M. (1994) Science 265, 1107–1110. [DOI] [PubMed] [Google Scholar]
- 16.Linker, R. A., Maurer, M., Gaupp, S., Martini, R., Holtmann, B., Giess, R., Rieckmann, P., Lassmann, H., Toyka, K. V., Sendtner, M. & Gold, R. (2002) Nat. Med. 8, 620–624. [DOI] [PubMed] [Google Scholar]
- 17.Giess, R., Holtmann, B., Braga, M., Grimm, T., Muller-Myhsok, B., Toyka, K. V. & Sendtner, M. (2002) Am. J. Hum. Genet. 70, 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giess, R., Maurer, M., Linker, R., Gold, R., Warmuth-Metz, M., Toyka, K. V., Sendtner, M. & Rieckmann, P. (2002) Arch. Neurol. 59, 407–409. [DOI] [PubMed] [Google Scholar]
- 19.Helgren, M. E., Squinto, S. P., Davis, H. L., Parry, D. J., Boulton, T. G., Heck, C. S., Zhu, Y., Yancopoulos, G. D., Lindsay, R. M. & DiStefano, P. S. (1994) Cell 76, 493–504. [DOI] [PubMed] [Google Scholar]
- 20.Guillet, C., Auguste, P., Mayo, W., Kreher, P. & Gascan, H. (1999) J. Neurosci. 19, 1257–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lambert, P. D., Anderson, K. D., Sleeman, M. W., Wong, V., Tan, J., Hijarunguru, A., Corcoran, T. L., Murray, J. D., Thabet, K. E., Yancopoulos, G. D., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4652–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masu, Y., Wolf, E., Holtmann, B., Sendtner, M., Brem, G. & Thoenen, H. (1993) Nature 365, 27–32. [DOI] [PubMed] [Google Scholar]
- 23.DeChiara, T. M., Vejsada, R., Poueymirou, W. T., Acheson, A., Suri, C., Conover, J. C., Friedman, B., McClain, J., Pan, L., Stahl, N., et al. (1995) Cell 83, 313–322. [DOI] [PubMed] [Google Scholar]
- 24.Ip, N. Y., McClain, J., Barrezueta, N. X., Aldrich, T. H., Pan, L., Li, Y., Wiegand, S. J., Friedman, B., Davis, S. & Yancopoulos, G. D. (1993) Neuron 10, 89–102. [DOI] [PubMed] [Google Scholar]
- 25.Sendtner, M., Carroll, P., Holtmann, B., Hughes, R. A. & Thoenen, H. (1994) J. Neurobiol. 25, 1436–1453. [DOI] [PubMed] [Google Scholar]
- 26.Elson, G. C., Lelievre, E., Guillet, C., Chevalier, S., Plun-Favreau, H., Froger, J., Suard, I., de Coignac, A. B., Delneste, Y., Bonnefoy, J. Y., et al. (2000) Nat. Neurosci. 3, 867–872. [DOI] [PubMed] [Google Scholar]
- 27.Plun-Favreau, H., Elson, G., Chabbert, M., Froger, J., deLapeyriere, O., Lelievre, E., Guillet, C., Hermann, J., Gauchat, J. F., Gascan, H., et al. (2001) EMBO J. 20, 1692–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lelievre, E., Plun-Favreau, H., Chevalier, S., Froger, J., Guillet, C., Elson, G. C., Gauchat, J. F. & Gascan, H. (2001) J. Biol. Chem. 276, 22476–22484. [DOI] [PubMed] [Google Scholar]
- 29.Notredame, C., Higgins, D. G. & Heringa, J. (2000) J. Mol. Biol. 302, 205–217. [DOI] [PubMed] [Google Scholar]
- 30.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burge, C. & Karlin, S. (1997) J. Mol. Biol. 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 32.Rost, B. & Sander, C. (1993) J. Mol. Biol. 232, 584–599. [DOI] [PubMed] [Google Scholar]
- 33.Arce, V., Garces, A., de Bovis, B., Filippi, P., Henderson, C., Pettmann, B. & deLapeyriere, O. (1999) J. Neurosci. Res. 55, 119–126. [DOI] [PubMed] [Google Scholar]
- 34.Gritti, A., Frolichsthal-Schoeller, P., Galli, R., Parati, E. A., Cova, L., Pagano, S. F., Bjornson, C. R. & Vescovi, A. L. (1999) J. Neurosci. 19, 3287–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carroll, P., Gayet, O., Feuillet, C., Kallenbach, S., de Bovis, B., Dudley, K. & Alonso, S. (2001) Mol. Cell. Neurosci. 17, 611–623. [DOI] [PubMed] [Google Scholar]
- 36.Kelly, R. G., Zammit, P. S., Schneider, A., Alonso, S., Biben, C. & Buckingham, M. E. (1997) Dev. Biol. 187, 183–199. [DOI] [PubMed] [Google Scholar]
- 37.Reynolds, B. A. & Weiss, S. (1996) Dev. Biol. 175, 1–13. [DOI] [PubMed] [Google Scholar]
- 38.Shimazaki, T., Shingo, T. & Weiss, S. (2001) J. Neurosci. 21, 7642–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gearing, D. P., Ziegler, S. F., Comeau, M. R., Friend, D., Thoma, B., Cosman, D., Park, L. & Mosley, B. (1994) Proc. Natl. Acad. Sci. USA 91, 1119–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Serio, A., Graziani, R., Laufer, R., Ciliberto, G. & Paonessa, G. (1995) J. Mol. Biol. 254, 795–800. [DOI] [PubMed] [Google Scholar]
- 41.Shi, Y., Wang, W., Yourey, P. A., Gohari, S., Zukauskas, D., Zhang, J., Ruben, S. & Alderson, R. F. (1999) Biochem. Biophys. Res. Commun. 262, 132–138. [DOI] [PubMed] [Google Scholar]
- 42.Senaldi, G., Varnum, B. C., Sarmiento, U., Starnes, C., Lile, J., Scully, S., Guo, J., Elliott, G., McNinch, J., Shaklee, C. L., et al. (1999) Proc. Natl. Acad. Sci. USA 96, 11458–11463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Forger, N. G., Prevette, D., deLapeyriere, O., de Bovis, B., Wang, S., Bartlett, P. & Oppenheim, R. W. (2003) J. Neurosci. 23, 8854–8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi, R., Yokoji, H., Misawa, H., Hayashi, M., Hu, J. & Deguchi, T. (1994) Nat. Genet. 7, 79–84. [DOI] [PubMed] [Google Scholar]
- 45.Giess, R., Goetz, R., Schrank, B., Ochs, G., Sendtner, M. & Toyka, K. (1998) Muscle Nerve 21, 236–238. [DOI] [PubMed] [Google Scholar]
- 46.Clark, A. G., Glanowski, S., Nielsen, R., Thomas, P. D., Kejariwal, A., Todd, M. A., Tanenbaum, D. M., Civello, D., Lu, F., Murphy, B., et al. (2003) Science 302, 1960–1963. [DOI] [PubMed] [Google Scholar]
- 47.Rouquier, S., Blancher, A. & Giorgi, D. (2000) Proc. Natl. Acad. Sci. USA 97, 2870–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gilad, Y., Man, O., Paabo, S. & Lancet, D. (2003) Proc. Natl. Acad. Sci. USA 100, 3324–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rollmann, S. M., Houck, L. D. & Feldhoff, R. C. (1999) Science 285, 1907–1909. [DOI] [PubMed] [Google Scholar]
- 50.McKay, R. (1997) Science 276, 66–71. [DOI] [PubMed] [Google Scholar]
- 51.Valenzuela, D. M., Murphy, A. J., Frendewey, D., Gale, N. W., Economides, A. N., Auerbach, W., Poueymirou, W. T., Adams, N. C., Rojas, J., Yasenchak, J., et al. (2003) Nat. Biotechnol. 21, 652–659. [DOI] [PubMed] [Google Scholar]