Abstract
The signaling activities of multiple developmental ligands require sulfated heparan sulfate (HS) proteoglycans as coreceptors. QSulf1 and its mammalian orthologs are cell surface HS 6-O-endosulfatases that are expressed in embryonic mesodermal and neural progenitors and promote Wnt signal transduction. In this study, we have investigated the function of QSulf1 in fibroblast growth factor (FGF) signaling, which requires 6-O-sulfated HS for FGF receptor (FGFR) dimerization and tyrosine kinase activation. Here, we report that QSulf1 inhibits FGF2- and FGF4-induced mesoderm formation in the Xenopus embryo and FGF-dependent angiogenesis in the chicken embryo through 6-O-desulfation of cell surface HS. QSulf1 regulates FGF signaling through inhibition of HS-mediated FGFR1 activation by interfering with FGF–HS–FGFR1 ternary complex formation. Furthermore, QSulf1 can produce enzymatically modified soluble heparin that acts as a potent inhibitor of FGF2-induced angiogenesis in the chicken embryo. QSulf1, therefore, has dual regulatory functions as a negative regulator of FGF signaling and a positive regulator of Wnt signaling. Therefore, QSulf1 provides another reagent to produce enzymatically modified heparin compounds, in vivo and in vitro, to modulate cellular signaling in stem cell-based therapies to promote tissue regeneration and in cancer therapies to control cell growth and block angiogenesis.
Heparan sulfate (HS) proteoglycans regulate cell surface signaling during embryogenesis and contribute to the pathophysiology of numerous diseases (1, 2). HS proteoglycans include a protein core with O-linked HS chains composed of 50–200 disaccharide repeats of uronic acid and glucosamine residues. HS chains are selectively sulfated at the 2-O position of uronic acid and the 6-O, 3-O, and N positions of glucosamine residues for molecular interactions with signaling ligands and matrix components (1–3). HS chains have highly sulfated and undersulfated domains along their lengths, creating structural heterogeneity likely related to their complex biological functions (4).
Sulfation of HS chains is required for their functions in developmental signaling. Loss of HS sulfation in Drosophilia sulfateless (5, 6) and slalom (7) mutants leads to defects in Wingless (Wg), fibroblast growth factor, and Hedgehog (Hh) signaling required for embryonic patterning. Further, deficiencies of specific sulfations within the HS disaccharide unit cause signaling defects. For example, mice mutant for Hs2st 2-O-sulfotransferase lack 2-O-sulfated uronic acid and exhibit multiple signaling defects leading to lethal kidney agenesis (8, 9). Inhibition of Drosophila HS 6-O-sulfotransferase gene expression by RNA interference reduces FGF signaling activity and disrupts the primary branching of the tracheal system (10), and treatment of cultured cells with chlorate blocks HS sulfation, resulting in defects in bone morphogenetic protein, Wnt, and FGF signaling (11–13). HS sulfation, therefore, has regulatory functions in multiple signaling pathways in the embryo. Biological mechanisms for regulation of HS sulfation in embryos and the biochemical functions of specific sulfate groups in signaling are not well understood.
Recently, a class of evolutionarily conserved HS 6-O endosulfatases has been identified that modify HS 6-O sulfation and developmental signaling (14–18). A second, closely related family member, Sulf2, has been identified in mammals and birds (ref. 15 and unpublished data). Sulf1 exhibits structural and enzymatic features distinct from known glucosamine-6-O-sulfatases (GlcNR6Sase), which are lysosomal exosulfatases that hydrolyze the terminal 6-O-sulfate groups for HS degradation (19). By contrast, Sulf1 is secreted through the Golgi complex and is docked on the cell surface, where it functions as a 6-O-endosulfatase, with substrate specificity for trisulfated IdoA2S-GlcNS6S disaccharide units of HS/heparin (14–16). The avian ortholog, QSulf1, is required for Wnt-dependent gene expression in muscle progenitor cells of the quail embryo (14) through specific 6-O-sulfations of cell surface HS proteoglycans that decrease the binding affinity between HS and Wnt ligands to promote Frizzled receptor activity (16).
FGF ligand–receptor interactions and FGF signaling require sulfated HS (20–23). HS chains containing trisulfated disaccharide units greatly promote FGF2–FGF receptor 1 (FGFR1) binding and signaling (24, 25), although FGF2 can bind to FGFR1 in the absence of HS in cell binding assays (26) and in crystallographic studies (27). Among the sulfate groups on HS, sulfation at the 6-O position of glucosamine residues is required for FGF2–FGFR1 and FGF4–FGFR1 interactions and signaling (22, 24, 25). Although distinct sequences and sulfation patterns in HS chains are required for FGF ligand and receptor binding, 6-O-sulfation of HS is crucial for FGF signaling activity (25, 28).
In this study, we have investigated the function of QSulf1 in the control of FGF signaling. FGFs and FGFRs (FGF receptors) play critical roles in many developmental and disease processes, including angiogenesis and cancer (29). Sulf1 is expressed in embryonic cell lineages controlled by multiple signals, including FGF (14, 17), and recent studies reveal that the human Sulf1 ortholog, HSulf1, can down-regulate FGF-dependent extracellular signaling-regulated kinase (ERK) kinase activity in human cancer cells (18). We now report that QSulf1 can inhibit FGF2- and FGF4-dependent mesoderm induction in Xenopus embryos and FGF2-induced angiogenesis in chick embryos. QSulf1 suppresses FGF2 signaling in these embryonic tissues by enzymatically modifying the 6-O-sulfation of cell surface HS or soluble heparin. Biochemical experiments also reveal that QSulf1-mediated 6-O-desulfation of HS reduces the formation of FGF2–HS–FGFR ternary complexes by inhibiting the interaction between FGFR1 with FGF2. These studies provide evidence that QSulf1 is a negative regulator of HS-mediated FGF signaling.
Materials and Methods
Plasmids, mRNAs, and Recombinant Proteins. Full-length QSulf1 cDNA was subcloned into pAG-myc and pCS2 vectors for mammalian cell expression and in vitro synthesis of QSulf1 mRNA, respectively. pCS2-XFGFR1K562E (30) and pFGFR1c-AP (31) were gifts from Robert Friesel and Alan Rapraeger. The drug-inducible iFGFR1 was activated with AP20187 (provided by Ariad Pharmaceuticals, Cambridge, MA) (32). Human recombinant FGF2 protein was purchased from Sigma, and Xenopus FGF4 (eFGF) was produced with pET-XeFGFi (33). mRNAs were synthesized by using the mMessage kit (Ambion, Austin, TX) and quantified by using a spectrometer. Active QSulf1 and catalytically inactive QSulf1(C-A) proteins were purified from stably transfected 293T cells (16).
Animal Cap Assays. Xenopus laevis embryos obtained by standard protocols (34, 35) were injected into the animal pole at the one-cell stage with mRNAs and then cultured until stage 8–9 in 0.1× MMR (35) plus gelatin (100 ng/ml), recombinant proteins, AP20187, and heparin as specified in the figure legends. Ten animal caps were collected for each experimental group. Data shown are representative of at least three independent experiments. The use of Xenopus and quail embryos was approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Western Blot Analysis for ERK1/2. Animal caps were lysed in 15 μl of buffer [80 mM β-glycerophosphate/20 mM EGTA/1 mM DTT/15 mM MgCl2/20 mM Hepes, pH 7.5/protease inhibitors (Roche)]. Fourteen and 2 μl of the samples were resolved on SDS/PAGE (Bio-Rad), transferred, and subsequently probed for diphosphorylated ERK1/2 (Dp-ERK1/2) and total ERK1/2, respectively. Primary antibodies include mouse anti-ERK1/2 (1:4,000, Sigma) and mouse anti-Dp-ERK1/2 (1:2,000, Sigma). Signals were detected by peroxidase-conjugated secondary antibodies and quantified with imagequant (Molecular Dynamics).
RT-PCR Assays. Total RNA was purified from stage 11 cultured animal caps and embryos by using an RNAqueous kit (Ambion) and quantified by spectrometer. RT-PCR for EF1α, Brachyury, and MyoD expression was performed as described (36). Data shown are representative of five independent assays.
FGF2-Heparin Beads Binding Assay. Heparin conjugated to acrylic beads (Sigma) was digested with QSulf1 or QSulf1(C-A) mutant protein at 37°C overnight on a shaker (16) and then collected by centrifuging. After washing with Hanks' balanced saline solution (HBSS, Invitrogen), heparin beads were divided into aliquotes and incubated with various amounts of FGF2 in a 50-μl binding reaction containing 20 μl of beads. After 30 min at room temperature, heparin beads were washed and FGF2 bound to heparin beads was analyzed by Western blotting. Data shown are representative of three independent experiments.
In Vitro FGF2 Binding to FGFR1c-AP. FGFR1c-alkaline phosphatase (AP) protein was obtained from the conditioned medium of 293T cells transfected with pFGFR1c-AP, 48 h after switching to serum-free DMEM/F12 (Invitrogen). Protein in conditioned medium was quantified by colorimetric dye concentrate assay (Bio-Rad). The purification of QSulf1 and enzymatic digestion of heparin were as described (16). The binding assay mixtures [200 μl total volume containing 10 ng of FGF2, 10 ng of FGFR1c-AP, and varying amounts of heparin pretreated with either QSulf1 or QSulf1(C-A) in HBSS] were incubated for 30 min at room temperature. Complexes were immunoprecipitated after 2-h incubation with 10 μl of a slurry of anti-AP antibody coupled to agarose beads (Sigma). FGF2 bound to FGFR1c-AP was resolved by SDS/10%PAGE, detected by Western blotting, and quantified by using imagequant. Dilution of anti-FGF2 antibody was 1:2,000 (Sigma).
Chorioallantonic Membrane Angiogenesis Assay. Fertilized chicken eggs were incubated in a humidified 38°C oven for 10 days. Filter papers (0.25 cm2) soaked in 10 μl of PBS containing 20 ng of FGF2 with control heparin or QSulf-1-digested heparin (200 ng) were applied to an avascular area on the chorioallantonic membrane exposed through a window in the shell. The eggs were sealed with tape and incubated for 3 additional days. The chorioallantonic membrane was then excised adjacent to the filters, fixed, and examined under the microscope to count numbers of blood vessel branches on each filter (24). Angiogenesis was scored from 1 (low) to 4 (high) according to Friedlander et al. (37). A score of 1 equals 0–2 branches per membrane and a score of 4 equals 20–25 branches per filter.
Results and Discussion
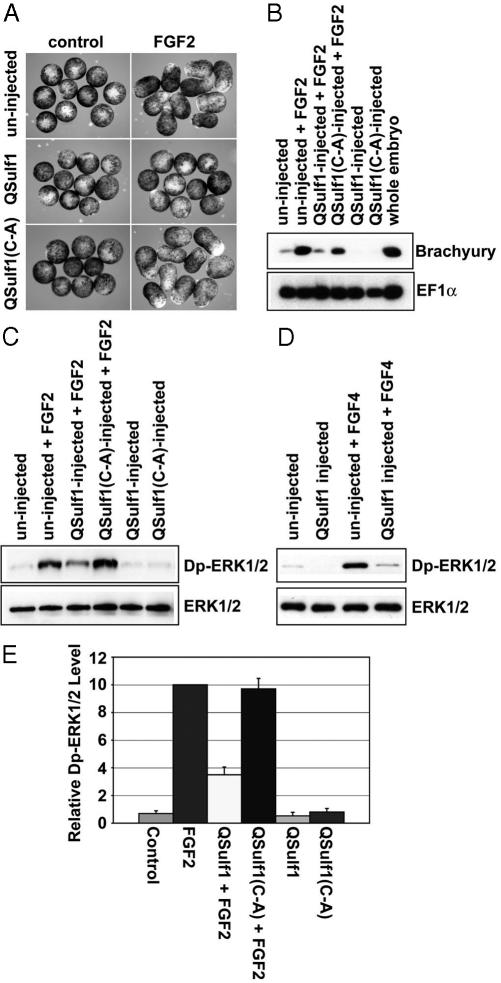
QSulf1 Suppresses FGF2 Signaling and Mesoderm Induction. The Xenopus animal cap assay was used to study QSulf1 activity in FGF signaling. Untreated explants of the animal pole of the Xenopus blastula form ectodermal derivatives (35), whereas FGF-treated explants form elongated and differentiated mesoderm (34), providing a convenient FGF signaling assay system. QSulf1 and catalytically inactive QSulf1(C-A) (14) were expressed in animal caps by injecting in vitro synthesized mRNAs into Xenopus embryos at the one-cell stage, followed by isolation of animal pole explants from blastula stage embryos. Expression of QSulf1 or QSulf1(C-A) alone did not induce morphological changes or expression of mesodermal markers. However, QSulf1 suppressed both FGF2-induced tissue elongation (Fig. 1A) and mesodermal differentiation, as assayed by expression of Brachyury (Fig. 1B). QSulf1 expression also suppressed the phosphorylation of ERK1/2, which are direct targets of FGF2 signaling (Fig. 1 C and E). By contrast, enzymatically inactive QSulf1(C-A) did not block FGF2-induced tissue elongation or Brachyury expression and had no effect on the ERK1/2 phosphorylation (Fig. 1 A–C and E). QSulf1 similarly inhibits mesoderm formation induced by FGF4 (eFGF), a FGF isoform normally active in the Xenopus embryo (Fig. 1D) (34). These data, therefore, establish that QSulf1 acts upstream of ERK1/2 phosphorylation to inhibit FGF signaling. Significantly, the inhibitory activity of QSulf1 on FGF signaling contrasts with its positive regulatory activity on Wnt signaling (16), indicating that QSulf1 has dual regulatory functions and implying that FGF and Wnt signaling have different HS 6-O-sulfation requirements.
Fig. 1.
QSulf1 suppresses FGF-induced mesoderm induction in Xenopus animal cap assays. Xenopus embryos were injected with QSulf1 or inactive QSulf1(C-A) mRNAs (750 pg) at the one-cell stage. Animal caps were dissected at stage 8–9 and cultured to assess mesoderm induction and ERK1/2 activation. (A) Untreated animal cap explants form epidermis and assume a spherical shape, but caps elongate and form mesodermal tissues when induced with FGF. Expression of QSulf1, but not catalytically inactive QSulf1(C-A), blocks FGF2-induced animal cap elongation. (B) Animal caps were cultured until stage 11 before isolating RNA to assess gene expression by RT-PCR. QSulf1 blocked mesodermal gene expression (Brachyury) induced by FGF2 (30 ng/ml), whereas catalytically inactive QSulf1(C-A) had no effect. The slight reduction of Brachyury expression in this experiment was not significant when averaged in five independent experiments. EF1α was used as a gel-loading control and whole embryo RNA was used as a positive control for expression of mesoderm genes. (C and D) For assays of ERK1/2 phosphorylation, animal caps were cultured for 1 h in the presence or absence of FGF2 or FGF4 (30 ng/ml), and protein extracts were prepared for Western blot analysis using antibodies against diphosphorylated ERK1/2 (Dp-ERK1/2) and total ERK1/2 proteins. Total ERK1/2 levels were used to monitor gel loading. FGF2-induced Dp-ERK1/2 phosphorylation is suppressed by QSulf1, but not by enzymatically inactive QSulf1(C-A) (C). QSulf1 also suppressed FGF4-induced Dp-ERK1/2 activation (D). (E) Phosphorylation of ERK1/2 shown in panel C was quantified and presented as the mean with standard error from four independent experiments. The relative Dp-ERK1/2 levels were calculated by measuring the ratios of Dp-ERK1/2 to total ERK1/2, and then normalizing to an FGF2-induced level that was arbitrarily assigned a value of 10. QSulf1 repression of FGF2-induced Dp-ERK1/2 was statistically significantly different from FGF2 control (P = 0.001), whereas QSulf1(C-A) had no effect on FGF2 signaling activity (P = 0.72).
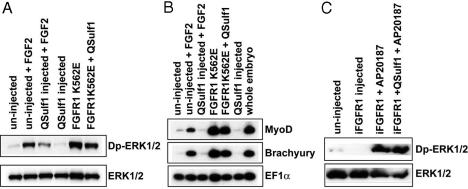
QSulf1 Functions Upstream of FGFR1 Receptor to Modify Extracellular HS. We then investigated whether QSulf1 functions upstream of FGFR1 to inhibit FGF signaling, as predicted from its activity as an extracellular 6-O-endosulfatase (16). For these studies, QSulf1 was coexpressed in animal caps with two constitutively active mutant forms of FGFR1. FGFR1K562E has a mutation in its intracellular tyrosine kinase domain and is constitutively active independent of HS-mediated receptor dimerization (30), and iFGFR1 lacks the extracellular ligand-binding domain, but has a membrane-targeting, amino-terminal myristylation sequence and two mutated FKBP12 domains that bind the synthetic drug AP20187 to promote rapid receptor dimerization and activation in response to AP20187 (32). These constitutively activated FGFR1s were predicted to be resistant to QSulf1 inhibition of FGF signaling if QSulf1 functions by modifying cell surface HS to control receptor dimerization. In support of this mode of action, we found that both FGFR1K562E and iFGFR1 are insensitive to the inhibitory activity of QSulf1 in FGF signaling, as assayed by Dp-ERK1/2 activation (Fig. 2 A and C) and induction of mesodermal genes (Fig. 2B).
Fig. 2.
Constitutively activated FGFR1s bypass QSulf1 inhibition of FGF2 and FGF4 signaling. FGFR1K562E is FGFR1 with an intracellular domain mutation conferring ligand-independent constitutive activity (A and B). iFGFR1 is a membrane-docked FGFR1 receptor with a deleted extracellular domain and an added intracellular domain to promote receptor dimerization and inducible activation in response to addition of AP20187 (C). QSulf1 mRNA (2 ng) was injected alone or coinjected with FGFR1K562E mRNA (100 pg) or iFGFR1 mRNA (20 pg) into one-cell stage Xenopus embryos. Animal caps were dissected at stage 8–9 for Western blot analysis of phoshorylated ERK1/2 (Dp-ERK1/2) and total ERK1/2 (ERK1/2) (A and C), and by RT-PCR to assay mesodermal gene expression (B). (A) Protein extracts were assayed 1 h after FGF2 treatment by Western blot analysis of Dp-ERK1/2. QSulf1 suppresses FGF2 (30 ng/ml) but not FGFR1K562E activation of Dp-ERK1/2. (B) QSulf1 suppresses FGF2, but not FGFR1K562E induction of mesodermal markers, Brachyury and MyoD. Whole-embryo samples provided a positive control for mesodermal gene expression. (C) AP20187 (1.25 μM) activation of iFGFR1 receptor induces Dp-ERK1/2 activation in the presence of QSulf1.
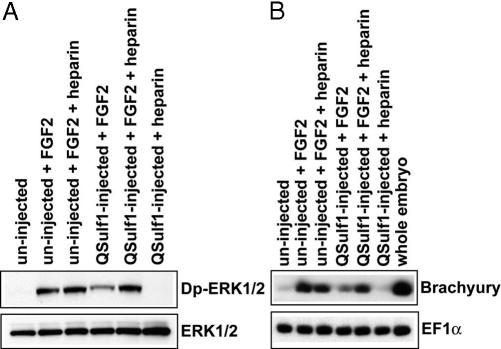
To test whether QSulf1 functions by modifying the sulfation of extracellular HS, we investigated whether exogenously added heparin could rescue FGF signaling inhibited by QSulf1 in the animal cap assay. QSulf1-injected animal caps were cultured in the presence or absence of heparin, a highly sulfated form of HS glycosaminoglycan. Heparin is a substrate for Sulf1-mediated 6-O-desulfation (15, 16) and can rescue FGF2 signaling in HS-deficient cells (21, 24). At concentrations (150–250 ng/ml) that rescue FGF2 signaling in cultured cells (38), exogenous heparin fully rescued QSulf1 inhibition of FGF2-mediated ERK1/2 activation and mesoderm induction (Fig. 3). Sulf1 also blocks ERK1/2 activation in response to heparin-dependent epidermal growth factor (EGF), but not to heparin-independent EGF in cultured ovarian cell lines (18). Together, these findings establish the specificity of Sulf1 function in HS-dependent signaling through enzymatically modification of the 6-O-sulfation of extracellular HS. Notably, uninjected animal caps treated with soluble heparin in the presence or absence of FGF2 are not stimulated further in FGF signaling, indicating that HS on the cell surface of embryonic animal caps is present in excess for FGFR1 activation and FGF signal transduction.
Fig. 3.
Exogenous heparin rescues the suppression of FGF2 signaling by QSulf1. QSulf1 mRNA was injected into one-cell stage Xenopus embryos (2 ng). Animal caps were dissected and cultured with soluble heparin at a concentration of 150 ng/ml. (A) Animal caps were collected after 1 h of FGF2 treatment. QSulf1 suppressed the ERK1/2 activation induced by FGF2 protein (30 ng/ml) and exogenous heparin rescued the suppression. Total ERK1/2 was assayed as a gel-loading control. (B) Animal caps were cultured until stage 11 to assess gene expression. QSulf1 suppression of mesodermal gene (Brachyury) expression was rescued by exogenous heparin. EF1α was a gel-loading control and the whole embryo RNA was a positive control for mesoderm gene expression.
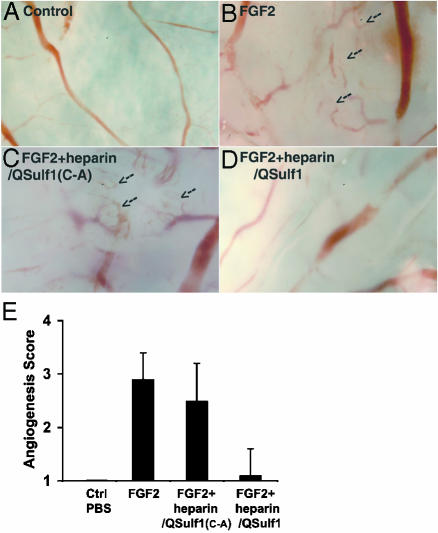
QSulf1-Modified Heparin Inhibits FGF2-Induced Angiogenesis. We next tested whether QSulf1 can modify heparin to produce extracellular inhibitors of FGF signaling. For these studies, a chorioallantonic membrane angiogenesis system was used, providing a sensitive and quantitative in vivo assay for FGF2 signaling. Angiogenesis was monitored by scoring blood vessel branching using a 1–4 scale, according to Friedlander et al. (37). Chorioallantoic membranes treated with control PBS were unbranched (score 1) (Fig. 4 A and E), whereas membranes treated with FGF2 formed extensive branches (score 2.9) (Fig. 4 B and E). Addition of QSulf1-desulfated heparin nearly completely blocked FGF-induced branching (score 1.1) (Fig. 4 C and E), whereas heparin treated with enzymatically inactive QSulf1(C-A) did not significantly inhibit FGF2-induced branching (score 2.5) (Fig. 4 D and E). Therefore, QSulf1 can modify soluble heparin to produce potent angiogenesis inhibitors.
Fig. 4.
QSulf1-treated heparin suppresses FGF2-induced angiogenesis. Filters soaked with FGF2, with or without heparin, were applied to the chorioallantonic membrane of day 10 chicken embryos. Blood vessel formation on the membrane was analyzed after 3 days. (A) Application of control PBS filter did not significantly induce formation of blood vessel branches. (B) Application of FGF2 (25 ng) filter induced extensive formation of blood vessel branches. (C) Application of filter containing FGF2 and QSulf1-treated heparin (200 ng) suppressed FGF2 induction of blood vessel branches. (D) Application of filter containing FGF2 and QSulf1(C-A)-treated heparin (200 ng) did not significantly affect FGF2 induction of blood vessel branches. Arrows indicate formation of blood vessel branches. (E) Quantification of angiogenesis. Angiogenesis was scored from 1 (low) to 4 (high) according to Friedlander et al. (37). Each experimental group included >14 embryos. The data are presented as the mean score and standard deviation of three independent experiments. The suppression of angiogenesis by QSulf1-treated heparin is statistically significant (P < 0.05), whereas QSulf1(C-A)-treated heparin did not significantly affect FGF2-induced angiogenesis (P = 0.82).
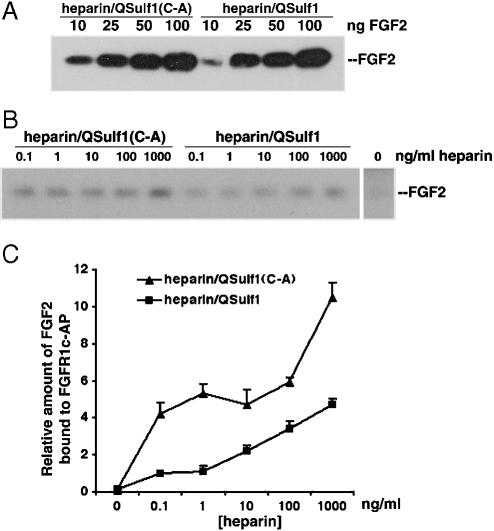
QSulf1-Modified Heparin Disrupts FGF2–FGFR1 Complex Formation. To investigate the mechanisms by which QSulf1-modified HS blocks FGF signaling, we investigated FGF2 binding to QSulf-1-modified heparin. Heparin conjugated to acrylic beads was digested with QSulf1 or catalytically inactive QSulf1(C-A) and then assayed for binding to an excess of FGF2, as quantified by Western blotting of bound FGF. We found that FGF2 binds equally well to QSulf1-treated heparin and QSulf1(C-A)-treated heparin (Fig. 5A), establishing that QSulf1 treatment does affect FGF2 binding to HS, consistent with previous findings that FGF2 binding to heparin requires 2-O- and not 6-O-sulfates (22, 24, 25), which are removed by QSulf1 (15, 16).
Fig. 5.
QSulf1-treated heparin reduces the binding of FGF2 to FGFR1, but not to heparin. (A) QSulf1 does not affect the binding of FGF2 to heparin. Heparin beads pretreated with QSulf1 or QSulf1(C-A) mutant protein were incubated with the indicated amount of FGF2. The heparin-bound FGF2 was analyzed by Western blotting. No significant difference was detected in FGF2 binding to QSulf1- and QSulf1(C-A)-treated heparin. (B) QSulf1 treatment of heparin reduces the formation of a FGF2–heparin–FGFR1 complex. QSulf1- or QSulf1(C-A)-treated soluble heparin at the indicated concentrations was incubated with FGF2 (10 ng/ml) and FGFR1c-AP (10 ng/ml). FGFR1c-AP was immunoprecipitated with an anti-AP antibody conjugated to agarose beads, and bound FGF2 was detected by Western blotting. (C) Quantification of FGF2 bound to FGFR1c-AP in the presence of QSulf1- or QSulf1(C-A)-treated heparin. The binding of FGF2 to FGFR1c-AP in the presence of QSulf1-treated heparin was reduced 2- to 5-fold compared with the binding in the presence of QSulf1(C-A)-treated heparin.
We then examined whether QSulf1 affects FGF2–heparin–FGFR ternary complex formation. Heparin-mediated FGF2–FGFR1 binding was assayed by using a soluble FGFR1c containing the extracellular domain of FGFR1 fused to an AP tag (31). FGF2 and FGFR1c were incubated with increasing concentrations of QSulf1- or QSulf1(C-A)-treated heparin (0–1,000 ng/ml), and FGF2–heparin–FGFR ternary complexes were then immunoprecipitated with anti-AP antibody for Western blot analysis. Only a low level of FGF2 was bound to FGFR1c-AP in the absence of heparin (Fig. 5 B and C). Heparin treated with QSulf1 reduces binding by 2- to 5-fold, whereas heparin treated with mutant QSulf1(C-A) promotes binding of FGF2 to FGFR1c (Fig. 5 B and C), indicating that QSulf1 digestion reduces the capacity of heparin to promote ternary complex formation. QSulf1 has substrate specificity for desulfation of only a subset of 6-O-sulfated disaccharides in HS chains (16), which these findings show are required to promote FGF2–FGFR1 ternary complex formation (22, 24, 25).
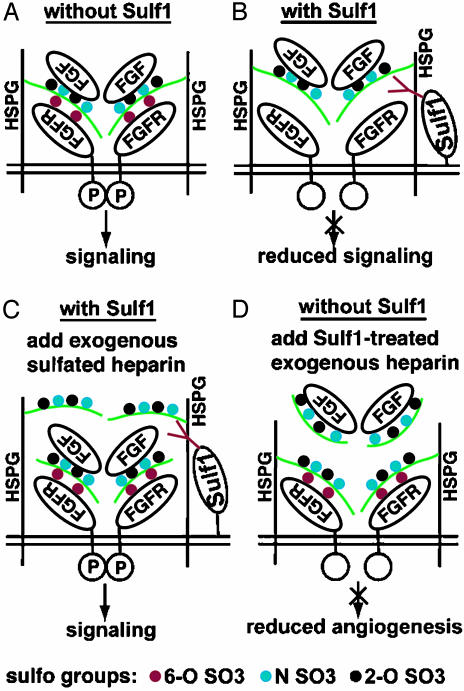
Our finding that QSulf1 blocks HS-mediated FGF2–heparin–FGFR1 ternary complex formation provides insight into the mechanism of QSulf1 inhibition of mesoderm induction and angiogenesis (Fig. 6). HS 2-O-sulfates on HS bind FGF2 ligand and 6-O-sulfates allow receptor binding/dimerization (24, 25) and formation of FGF2–HS–FGFR1 ternary complex (Fig. 6A). QSulf1 blocks FGF2 signaling by enzymatic removal of a subset of 6-O-sulfates on cell surface HS to inhibit ligand–receptor ternary complex formation and receptor dimerization (Fig. 6B). The inhibitory activity of QSulf1 in animal cap cells is rescued by exogenous soluble heparin, likely by providing heparin chains with 6-O-sulfated glucosamine residues to replace the 6-O-desulfated HS chains on QSulf1-expressing cells to allow FGF2–FGFR1 complex formation and receptor dimerization (Fig. 6C). QSulf1 enzymatically modifies the 6-O-sulfation of heparin to produce a potent soluble inhibitor of FGF2 signaling that can bind to FGF2, but not to FGFR1, thus competing with endogenous HS on the cell surface to block ternary complex formation and inhibit angiogenesis (Fig. 6D).
Fig. 6.
A model of FGF signaling regulation by Sulf1 through modulating FGF ligand–receptor interaction. A 2:2:2 model (two ligands, two HS chains, and two receptors) was proposed by others to illustrate ternary complex formation of FGF2–HS–FGFR1 during signaling (22, 40). (A) In the absence of Sulf1, sulfated cell surface HS promotes FGF ligand–receptor interaction, receptor dimerization, and activation of intracellular signaling. (B) Selective 6-O-desulfation of endogenous HS by QSulf1 reduces FGF ligand–receptor binding but has little effect on FGF ligand binding to HS. The reduced ligand–receptor interaction suppresses receptor dimerization and signaling. (C) Exogenous sulfated heparin rescues signaling by replacing QSulf1-desulfated endogenous cell surface HS and promoting FGF ligand–receptor interaction. (D) Sulf1 6-O-desulfated exogenous heparin has reduced affinity for FGFR1 but competitively binds to FGF2 and interferes with endogenous HS-mediated FGF2 angiogenic activity. The “P” in the intracellular domain of FGFR1 indicates phosphorylation and activation.
Although we show that QSulf1 is a negative regulator of FGF2 signaling, QSulf1 is a positive regulator of Wnt signaling through its activity to decrease the binding affinity of heparin for the Wnt ligand (16). QSulf1 is expressed in distinct patterns in multiple progenitor lineages in the early embryo, including somites, floor plate, neural tube, and kidney (14, 17). Developmental signals including Wnt and FGF are required for specification of these QSulf1-expressing progenitor lineages. QSulf1, therefore, can function as a “switch” to promote Wnt signaling and block FGF signaling during specification of QSulf1-expressing lineages. A diversity of animals have QSulf1 orthologs, including Caenorhabditis elegans, Drosophila, and human (14, 15), and an isoform, Sulf2, has been identified in vertebrates, indicating that Sulf enzymes are widely used regulators of FGF and Wnt signaling.
In addition to its regulatory functions in embryos, Sulf1 also is expressed in adult tissues and likely functions in pathophysiological processes such as cancer. HSulf1 expression is suppressed in ovarian cancer cells, and that HSulf1 overexpression in these cancer cells blocks ERK activation by FGF2 and epidermal growth factor and inhibits proliferation (18). The growth factor signaling functions of QSulf1 in living cells are based on its enzymatic activity and specificity for 6-O-desulfation of HS chains. QSulf1 also can enzymatically modify soluble heparin to produce potent chemical inhibitors of angiogenesis. These findings, and the specificity of QSulf1 for HS domains involved in ligand receptor interactions, indicate that QSulf1 enzyme will be a useful reagent to generate heparin-based compounds, both in vivo and in vitro, for use as therapeutic agents to promote stem cell production for tissue and organ regeneration and to control tumor cell growth and angiogenesis in the treatment of specific cancers (39).
Acknowledgments
We thank Dr. Robert Friesel for providing pCS2-XFGFR1 plasmids, Dr. Alan Rapraeger for pFGFR1c-AP plasmid, and Dr. Tom Pringle for providing information on the evolution of the sulfatase gene family (www.mad-cow.org/00/annotation_frames/tools/genbrow/sulfatases/sulfatases.html). This work was supported by National Institute of Child Health and Human Development grants (to C.P.E. and D.S.K.), National Institutes of Health Institutional Postdoctoral Fellowship DK07006 (to S.W.), and a National Institutes of Health postdoctoral fellowship (to X.A.).
Abbreviations: HS, heparan sulfate; FGF, fibroblast growth factor; FGFR1, FGF receptor 1; AP, alkaline phosphatese; QSulf1, quail Sulf1; ERK, extracellular signaling-regulated kinase.
References
- 1.Nybakken, K. & Perrimon, N. (2002) Biochim. Biophys. Acta 1573, 280–291. [DOI] [PubMed] [Google Scholar]
- 2.Sasisekharan, R., Shriver, Z., Venkataraman, G. & Narayanasami, U. (2002) Nat. Rev. Cancer 2, 521–528. [DOI] [PubMed] [Google Scholar]
- 3.Rapraeger, A. C. (2002) Methods Cell Biol. 69, 83–109. [DOI] [PubMed] [Google Scholar]
- 4.Esko, J. D. & Lindahl, U. (2001) J. Clin. Invest. 108, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin, X. & Perrimon, N. (1999) Nature 400, 281–284. [DOI] [PubMed] [Google Scholar]
- 6.Toyoda, H., Kinoshita-Toyoda, A., Fox, B. & Selleck, S. B. (2000) J. Biol. Chem. 275, 21856–21861. [DOI] [PubMed] [Google Scholar]
- 7.Luders, F., Segawa, H., Stein, D., Selva, E. M., Perrimon, N., Turco, S. J. & Hacker, U. (2003) EMBO J. 22, 3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullock, S. L., Fletcher, J. M., Beddington, R. S. & Wilson, V. A. (1998) Genes Dev. 12, 1894–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merry, C. L., Bullock, S. L., Swan, D. C., Backen, A. C., Lyon, M., Beddington, R. S., Wilson, V. A. & Gallagher, J. T. (2001) J. Biol. Chem. 276, 35429–35434. [DOI] [PubMed] [Google Scholar]
- 10.Kamimura, K., Fujise, M., Villa, F., Izumi, S., Habuchi, H., Kimata, K. & Nakato, H. (2001) J. Biol. Chem. 276, 17014–17021. [DOI] [PubMed] [Google Scholar]
- 11.Reichsman, F., Smith, L. & Cumberledge, S. (1996) J. Cell Biol. 135, 819–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izvolsky, K. I., Shoykhet, D., Yang, Y., Yu, Q., Nugent, M. A. & Cardoso, W. V. (2003) Dev. Biol. 258, 185–200. [DOI] [PubMed] [Google Scholar]
- 13.Irie, A., Habuchi, H., Kimata, K. & Sanai, Y. (2003) Biochem. Biophys. Res. Commun. 308, 858–865. [DOI] [PubMed] [Google Scholar]
- 14.Dhoot, G. K., Gustafsson, M. K., Ai, X., Sun, W., Standiford, D. M. & Emerson, C. P., Jr. (2001) Science 293, 1663–1666. [DOI] [PubMed] [Google Scholar]
- 15.Morimoto-Tomita, M., Uchimura, K., Werb, Z., Hemmerich, S. & Rosen, S. D. (2002) J. Biol. Chem. 277, 49175–49185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ai, X., Do, A., Lozynska, O., Kusche-Gullberg, M., Lindahl, U. & Emerson, C. P., Jr. (2003) J. Cell Biol. 162, 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohto, T., Uchida, H., Yamazaki, H., Keino-Masu, K., Matsui, A. & Masu, M. (2002) Genes Cells 7, 173–185. [DOI] [PubMed] [Google Scholar]
- 18.Lai, J., Chien, J., Staub, J., Avula, R., Greene, E. L., Matthews, T. A., Smith, D. I., Kaufman, S. H., Roberts, L. R. & Shridhar, V. (2003) J. Biol. Chem. 278, 23107–23117. [DOI] [PubMed] [Google Scholar]
- 19.Kresse, H., Paschke, E., von Figura, K., Gilberg, W. & Fuchs, W. (1980) Proc. Natl. Acad. Sci. USA 77, 6822–6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yayon, A., Klagsbrun, M., Esko, J. D., Leder, P. & Ornitz, D. M. (1991) Cell 64, 841–848. [DOI] [PubMed] [Google Scholar]
- 21.Ornitz, D. M., Yayon, A., Flanagan, J. G., Svahn, C. M., Levi, E. & Leder, P. (1992) Mol. Cell. Biol. 12, 240–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlessinger, J., Plotnikov, A. N., Ibrahimi, O. A., Eliseenkova, A. V., Yeh, B. K., Yayon, A., Linhardt, R. J. & Mohammadi, M. (2000) Mol. Cell 6, 743–750. [DOI] [PubMed] [Google Scholar]
- 23.Pellegrini, L., Burke, D. F., von Delft, F., Mulloy, B. & Blundell, T. L. (2000) Nature 407, 1029–1034. [DOI] [PubMed] [Google Scholar]
- 24.Lundin, L., Larsson, H., Kreuger, J., Kanda, S., Lindahl, U., Salmivirta, M. & Claesson-Welsh, L. (2000) J. Biol. Chem. 275, 24653–24660. [DOI] [PubMed] [Google Scholar]
- 25.Pye, D. A., Vives, R. R., Turnbull, J. E., Hyde, P. & Gallagher, J. T. (1998) J. Biol. Chem. 273, 22936–22942. [DOI] [PubMed] [Google Scholar]
- 26.Roghani, M., Mansukhani, A., Dell'Era, P., Bellosta, P., Basilico, C., Rifkin, D. B. & Moscatelli, D. (1994) J. Biol. Chem. 269, 3976–3984. [PubMed] [Google Scholar]
- 27.Plotnikov, A. N., Hubbard, S. R., Schlessinger, J. & Mohammadi, M. (2000) Cell 101, 413–424. [DOI] [PubMed] [Google Scholar]
- 28.Jemth, P., Kreuger, J., Kusche-Gullberg, M., Sturiale, L., Gimenez-Gallego, G. & Lindahl, U. (2002) J. Biol. Chem. 277, 30567–30573. [DOI] [PubMed] [Google Scholar]
- 29.Ornitz, D. M. & Itoh, N. (2001) Genome Biol. 2, REVIEWS3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neilson, K. M. & Friesel, R. (1996) J. Biol. Chem. 271, 25049–25057. [DOI] [PubMed] [Google Scholar]
- 31.Allen, B. L., Filla, M. S. & Rapraeger, A. C. (2001) J. Cell Biol. 155, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pownall, M. E., Welm, B. E., Freeman, K. W., Spencer, D. M., Rosen, J. M. & Isaacs, H. V. (2003) Dev. Biol. 256, 89–99. [DOI] [PubMed] [Google Scholar]
- 33.Isaacs, H. V., Tannahill, D. & Slack, J. M. (1992) Development (Cambridge, U.K.) 114, 711–720. [DOI] [PubMed] [Google Scholar]
- 34.Slack, J. M., Darlington, B. G., Heath, J. K. & Godsave, S. F. (1987) Nature 326, 197–200. [DOI] [PubMed] [Google Scholar]
- 35.Yao, J. & Kessler, D. S. (2000) Methods Mol. Biol. 137, 169–178. [DOI] [PubMed] [Google Scholar]
- 36.Engleka, M. J., Craig, E. J. & Kessler, D. S. (2001) Dev. Biol. 237, 159–172. [DOI] [PubMed] [Google Scholar]
- 37.Friedlander, M., Brooks, P. C., Shaffer, R. W., Kincaid, C. M., Varner, J. A. & Cheresh, D. A. (1995) Science 270, 1500–1502. [DOI] [PubMed] [Google Scholar]
- 38.Fannon, M., Forsten, K. E. & Nugent, M. A. (2000) Biochemistry 39, 1434–1445. [DOI] [PubMed] [Google Scholar]
- 39.Folkman, J. (2002) Semin. Oncol. 29, 15–18. [DOI] [PubMed] [Google Scholar]
- 40.Wu, Z. L., Zhang, L., Yabe, T., Kuberan, B., Beeler, D. L., Love, A. & Rosenberg, R. D. (2003) J. Biol. Chem. 278, 17121–17129. [DOI] [PubMed] [Google Scholar]