Abstract
The colonization of land by tetrapod ancestors is one of the major questions in the evolution of vertebrates. Despite intense molecular phylogenetic research on this problem during the last 15 years, there is, until now, no statistically supported answer to the question of whether coelacanths or lungfish are the closest living relatives of tetrapods. We determined DNA sequences of the nuclear-encoded recombination activating genes (Rag1 and Rag2) from all three major lungfish groups, the Australian Neoceratodis forsteri, the South American Lepidosiren paradoxa and the African lungfish Protopterus dolloi, and the Indonesian coelacanth Latimeria menadoensis. Phylogenetic analyses of both the single gene and the concatenated data sets of RAG1 and RAG2 found that the lungfishes are the closest living relatives of the land vertebrates. These results are supported by high bootstrap values, Bayesian posterior probabilities, and likelihood ratio tests.
Since the discovery in 1938 of the “living fossil,” the coelacanth Latimeria chalumnae, a representative of a group of lobe-finned fish thought to have gone extinct ≈80 million years ago (1, 2), there has been remarkable interest in this legendary fish by the public and scientists alike. However, the evolutionary relationships of the coelacanths to the other two living groups of lobe-finned fish (Sarcopterygii), the lungfish (Dipnoi), and the land vertebrates (Tetrapoda) remain debated until today. Since its discovery, many comparative morphologists and paleontologists considered the coelacanth to be the closest living relative of the land vertebrates (1-4), although the lungfish were historically thought, and continued to be thought by some researchers, to hold that claim. More recently, however, several analyses began to challenge the sistergroup relationship between the coelacanth and the tetrapods, first on morphological and paleontological grounds (5-7) and later based on molecular phylogenetic analyses (8, 9). Palaeontological studies are limited to studying morphological features, and strong phylogenetic inferences are often hindered because of missing data in incomplete fossils. The most recent paleontological evidence demonstrated that the lungfish represent an ancient lineage and that several of the features defining this group remained highly conserved throughout the entire evolutionary history of land vertebrates (10). The majority of palaeontological studies published during the last decade suggest that lungfish (Dipnoi) are the closest living relatives of the tetrapods or, alternatively, that coelacanths and lungfish form a monophyletic group that is equally closely related to the land vertebrates (11, 12).
A wealth of molecular phylogenetic studies addressed the tetrapod origin question, first based on mitochondrial DNA data by using partial gene sequences, single genes, or a few genes, and more recently, based on complete mitochondrial genomes (9, 13-18). Most of these mitochondria-based molecular phylogenetic studies favored the lungfish as the closest living relatives of tetrapods. Also studies based on nuclear data sets, e.g., on rRNA (16) and myelin DM20 (19), tackled this problem. The ML tree of the DM20 genes (19) also favored a sistergroup relationship between lungfish and tetrapods, although the support for this topology was only suggestive and, therefore, could not definitively lay the problem to rest. Molecular phylogenetic studies allow the estimation of the strength of the statistical support for particular hypotheses based on different tree-building methods. Clearly, not all phylogenetic methods are equally powerful or reliable; however, it is generally agreed that when several phylogenetic methods converge on the same topology, that can be taken as added evidence in support of a particular hypothesis. Moreover, ML-based likelihood ratio tests (LRTs) can test the significance between alternative competing topologies (20-23).
An alternative set of molecular characters to DNA sequences or amino acid residues are those that are expected to show low levels of homoplasy, such as the presence/absence of intronic sequences and indels in coding sequences. This type of phylogenetic marker has been applied to the tetrapod orgin question as well (24, 25). A single amino acid deletion in the aminoterminal region of the Rag2 gene was found to be absent in tetrapods and in the African lungfish Protopterus, but was present in the coelacanth, in ray-finned fish, and in sharks (Chondrichthyes). This synapomorphic deletion seems to be favoring a lungfish tetrapod sistergroup relationship (25). However, because indels have been shown to be prone to homoplasy over long evolutionary time spans (26), additional markers are required to support the relationship suggested by the deletion in the Rag2 sequence (24, 25).
In an attempt to resolve the tetrapod origin question, we amplified large fragments of the recombination activating genes (Rag1 and Rag2), two highly conserved nuclear markers in jawed vertebrates. The deduced amino acid sequences corresponding to most of the Rag1 and Rag2 genes were analyzed separated as well as concatenated.
Materials and Methods
Species Used in This Study and DNA Extraction. All genomic DNA samples were extracted from deep-frozen tissue (-80°C) by using standard protocols (27). The DNA was air dried and then dissolved in an appropriate volume of TE (pH 8) at a concentration of <0.5 mg/ml.
Genes Sequenced and PCR Amplification. For this study, novel Rag1 and Rag2 sequences were determined for the following lobe-finned fish: the Australian lungfish Neoceratodis forsteri, the South American lungfish Lepidosiren paradoxa, and the African lungfish Protopterus dolloi, as well as the Indonesian coelacanth Latimeria menadoensis. Previous work demonstrated that both Rag1 and Rag2 genes possess no intron sequences in the land vertebrates, the lungfish, and the coelacanth (24, 25). All PCR amplifications were performed by using total genomic DNA. PCR reactions performed on DNA of the Indonesian coelacanth Latimeria menadoensis worked more reliably than those for the three different lungfishes, probably because of their enormous genomes, which are in the range of 100 billion bp, i.e., ≈35 times bigger than the human genome. The PCR amplifications were done in a final volume of 50 μl, routinely with 35 cycles with 1.5 units of a mixture of the Taq polymerase (Red-Taq, Sigma) and a Pwo polymerase (Roche Applied Science) with proofreading activity (50:1). The fragments were sequenced in both directions.
For the amplification of the most conserved region of the Rag2 gene (≈400 amino acid residues), a newly designed set of primers were used that were located slightly internal to those previously used (25). The first 30 and approximately the last 100 aa of the Rag2 gene are rather variable and hard to align and were not amplified. For RAG1 most of the more highly conserved second one-half of the protein was initially amplified by using either the universal primers described in ref. 28 or newly designed PCR primers against the highly conserved QYHKMYR and HCDIGNA regions of the gene. For later amplifications, sequence-specific primers were used for each of the species that were located close to the borders of the known fragment and directed toward the amino and carboxyl terminus in combination with universal primers directed against highly conserved areas of the molecule.
Phylogenetic Analyses. In addition to the eight newly determined sequences, more relevant orthologous RAG1 and RAG2 sequences were selected from GenBank. All sequences were aligned by clustalx (29), and the alignment was refined by eye by using the ED option of the must package (30). The alignment was unambiguous because the sequences are highly conserved among jawed vertebrates. All data sets were analyzed with all four major phylogenetic methods, distance-based methods (neighbor-joining, NJ), maximum parsimony (MP), maximum likelihood (ML) by using quartet puzzling (QP), and a Bayesian approach (Ba). The bootstrap support for individual branches of the trees were obtained by using 2,000 bootstrap replicates and the minimum evolution approach based on γ parameter-corrected distances as implemented in the program package mega 2 (31). Bootstrap support based on 2,000 replicates by the MP method was inferred with the program paup* 4b10, using the options 10 times random addition and tree bisection and reconnection (32). QP support values based on ML analyses were estimated with tree-puzzle (33). Ba based on mrbayes 2.01 were calculated; the posterior probabilities obtained are given in the form of percentage values (34). Site-specific likelihood values as well as the corresponding absolute likelihood values were estimated with the codeml program of the paml package (35). Likelihood ratio tests were implemented in the consel package and were used to test the statistical significance of alternative topologies with approximately unbiased (AU) tests as implemented in the consel package (36).
Not all phylogenetic methods perform equally well, and they differ in their robustness with regard to several potential phylogenetic pitfalls such as differences in molecular clock rates (37, 38). Recent simulation models also indicated that Bayesian posterior probabilities tend to be too “liberal” when compared to bootstrap values from ML analyses (37). However, we performed all typically used phylogenetic methods on these data sets to further test the level of support for the alternative phylogenetic hypotheses.
Results and Discussion
Both single RAG1 and RAG2 and the concatenated RAG1/RAG2 data sets (of 1,078 and 1,209 amino acid positions) were used in the phylogenetic analyses. In all three phylogenetic trees (Figs. 1, 2, 3), the three lungfishes, Australian, South American, and African, form a monophyletic group that is more closely related to the tetrapods than the coelacanth Latimeria menadoensis that forms the next basal lineage. In the longest concatenated data set, the sequence of the Australian lungfish was eliminated because its Rag1 sequence is shorter and thus limited in the number of amino acid positions that were available for phylogenetic analyses.
Fig. 1.
Phylogenetic tree inferred by using the ML method from 22 species and 689 amino acid positions from the Rag1 gene. To test the robustness of the internal nodes, 2,000 bootstrap replicates each for minimum evolution and MP (TBR, 10 times random addition) were performed. Furthermore, ML estimates were obtained either from tree-puzzle in the form of QP support values or from mrbayes in the form of posterior probabilities (Ba) given in percentage.
Fig. 2.
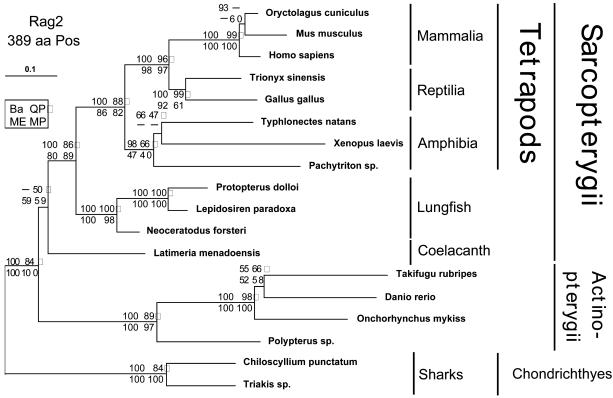
Phylogenetic tree inferred by using the ML method from 18 species and 389 amino acid positions from the Rag2 gene. The same phylogenetic analyses were done as described in Fig. 1.
Fig. 3.
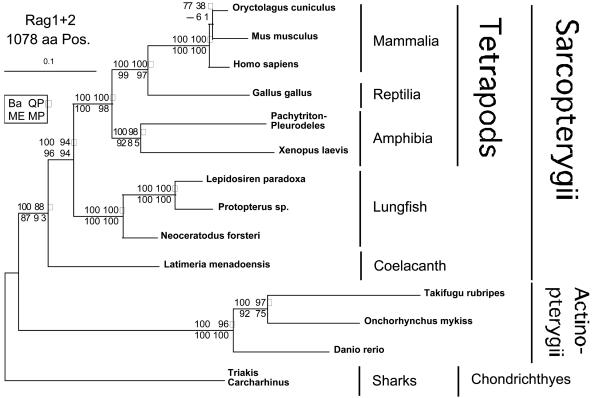
Phylogenetic tree inferred by using the ML method from 14 species and 1,078 amino acid positions from the concatenated RAG1 and RAG2 data sets. The same phylogenetic analyses were done as described in Fig. 1. Note that (i) the sequence named salamander is a fusion between the RAG1 sequence of Pleurodeles and the RAG2 sequence of Pachytriton and (ii) the outgroup sequence named sharks is a fusion between the RAG1 sequence of Carcharhinus and the RAG2 sequence of Triakis.
RAG1 Data Set. The first data set consists of 22 RAG1 sequences (689 amino acid positions) (Fig. 1) for four mammals, eight reptiles, and two amphibians (in total 14 tetrapods), in addition to the four newly determined sequences. Three ray-finned fish (Actinopterygii) and a shark sequence were used as outgroups. The RAG1 data set is able to resolve all major relationships within the jawed vertebrates (Gnathostomata), most of them with high bootstrap values. The support from the Bayesian analysis is in most cases 100% (P = 1); the lowest value is 91% for the monophyly of lobe-finned fish (including tetrapods), the Sarcopterygii. The ML-based method (QP) shows generally high support values for all inferred branches, with three exceptions: (i) the nodes supporting the monophyly of lungfishes, (ii) the node supporting the sistergroup relationship of tetrapods and lungfishes, and (iii) the node supporting the monophyly of the Sarcopterygii (44%). Part of the problem can be explained by the surprising result that tree-puzzle supports an obviously artificial monophyletic group of Neoceratodus and the coelacanth with the highest value of 48% in this region of the tree. It is known that the tree-puzzle program is rather sensitive to pronounced differences in evolutionary rates because of the quartet approach (38). The African and the South American lungfishes evolve quite fast, and the Australian lungfish and the coelacanth sequences evolve comparatively slower. This constellation of pronounced differences in evolutionary rates may lead to an artificial grouping of sequences with similar evolutionary speed (usually the slowly evolving ones). Often a more basal position of the fast evolving lineages due to long branch attraction effects will result, because these faster sequences are “pulled” toward the faster evolving sequences at the root, i.e., outgroup of the tree. This explains why the highest support of QP among basal Sarcopterygii (48%) seems to favor a clearly incorrect grouping of Neoceratodus and the coelacanth, again supporting the notion (38) that QP might not be the most appropriate method for this phylogenetic problem. The bootstrap support obtained by the two remaining methods, γ-corrected distance-based minimum evolution and MP, is in general quite high.
RAG2 Data Set. The RAG2 data set consists of 18 sequences (389 amino acid positions) (Fig. 2). In addition to the four new sequences of lobe-finned fishes, there are three mammalian, two reptilian, and three amphibian sequences (in total eight tetrapods) included in the analyses of the RAG2 data set. The most basal ray-finned fish, Polypterus, three ray-finned fish, as well as two sharks (order Galeoidea) form the outgroup. The RAG2 tree has exactly the same topology as the tree obtained with the RAG1 data set, with two minor exceptions: (i) the relationships among mammals differ from the topology of the RAG1 tree (the relationship indicated here is probably correct) (39-42) and (ii) and the relationships among the ray-finned fish. The latter is likely caused by an aberrant amino acid composition of the trout sequence (data not shown). The relationships among the three amphibian orders (caecilians, frogs, and salamanders) are notoriously difficult to resolve because of their early origin, relative fast evolutionary rates, and divergence in rapid succession from a common ancestor (39). Therefore, it is not surprising that the RAG2 data set is not able to resolve their interrelationships with certainty (except Ba 98%, supporting the monophyly of the amphibians). We find low but consistent support for the monophyly of the Sarcopterygii (Fig. 2). The support in favor of the nonmonophyly of the Sarcopterygii is in terms of Bayesian inference rather low as also for the amphibian interrelationships (66%) and the likely incorrect grouping of Danio and Takifugu (55%).
The Concatenated RAG1 and RAG2 Data Sets. The smaller concatenated RAG1 and RAG2 data set contains a total of 1,078 amino acid positions but only 14 species, because in most taxa only one of the two Rag sequences was determined. To maximize the number of species, we combined closely related species in two concatenated data sets. The RAG1 sequence of Pleurodeles (family Salamandridae) and the RAG2 sequence of Pachytriton of the same family were concatenated and termed “salamander,” the second concatenate included the RAG1 sequence of Carcharhinus (order Carcharhiniformes) and the RAG2 sequence of Triakis, belonging to the same order, and was named “sharks.”
The phylogenetic tree resulting from these analyses (Fig. 3) was identical in topology to the one from the RAG1 data set. In almost all cases, support for the nodes is higher, especially in the basal nodes, which are important for the tetrapod origin question. The topology within the teleost fishes now agrees with the traditional taxonomy, with Danio (Ostariophysi) representing the basal lineage to the two euteleostian fishes. This topology is highly supported by all four phylogenetic methods used; although the MP value is somewhat lower with 75% bootstrap support. The concatenated data set shows solid support (100% bootstrap support) by all four methods for the monophyly of the lungfish and the monophyly of the tetrapods (Fig. 3). Most importantly, the combined RAG1 + RAG2 data set provides high support of, on average, >95% bootstrap support for a monophyletic group consisting of the lungfishes and the tetrapods, the Choanata (Dipnoi + Tetrapoda). Finally, the monophyly of the Sarcopterygii, which was not recovered with particularly high support based on the RAG2 data set, is now highly supported, consistently by all four phylogenetic methods.
LRTs. The consel package was used to determine AU LRTs (36). Other LRTs (Kishino-Hasegawa and Shimodaira-Hasegawa) and the rell bootstrap proportions were also calculated with consel and are reported in Table 1. Three alternative topologies were tested, representing all possible rooted solutions for the relationships of the three sarcopterygian groups (e.g., see refs. 6, 9, and 17). Topology one, which always corresponds to the best (ML) tree, identifies the lungfishes as the closest living relatives of tetrapods; topology two is the sistergroup relationship between the coelacanth and the tetrapods; and topology three is the sistergroup relationship between the lungfishes and the coelacanth. The results obtained by the LRTs for these three alternative topologies from the two concatenated RAG1 and RAG2 data sets both in the presence and the absence of the sequence from the Australian lungfish Neoceratodus are summarized in Table 1. Initially, we performed only two tests, one with the shorter data set (1,078 aa) and all 14 sequences and one with the longer data set (1,209 aa) (13 species) without the Neoceratodus sequence. As shown in Table 1, we obtained a nonsignificant LRT result by using the shorter data set and all 14 sequences. However, the longer data set without the Neoceratodus sequence significantly rejected the two alternative topologies at a similar significance level for all three LRTs applied, i.e., the Kishino-Hasegawa, Shimodaira-Hasegawa, and AU tests. By adding more sequence positions (131 aa) from a somewhat less conserved region of the RAG1 protein, we also obtained significant statistical support for the hypothesis that lungfishes are the closest relatives of land vertebrates.
Table 1. LRTs as implemented in the consel package.
| Tree | In L | ΔIn L | ±SE | rell bootstrap proportion | Kishino-Hasegawa probability | Shimodaira-Hasegawa probability | AU probability |
|---|---|---|---|---|---|---|---|
| 14 sequences with Neoceratodus | |||||||
| 1 | −13284.8 | 0.913 | |||||
| 2 | −13296.7 | −11.9 | 7.97 | 0.050 | 0.067 | 0.071 | 0.092 |
| 3 | −13297.3 | −12.5 | 7.82 | 0.037 | 0.061 | 0.064 | 0.066 |
| 13 sequences without Neoceratodus | |||||||
| 1 | −12946.4 | 0.956 | |||||
| 2 | −12961.6 | −15.2 | 8.01 | 0.015 | 0.036 | 0.036 | 0.034 |
| 3 | −12961.0 | −14.6 | 8.21 | 0.029 | 0.042 | 0.042 | 0.044 |
| 14 sequences with Neoceratodus | |||||||
| 1 | −16190.6 | 0.919 | |||||
| 2 | −16202.2 | −11.6 | 7.41 | 0.045 | 0.065 | 0.066 | 0.069 |
| 3 | −16202.4 | −11.8 | 7.36 | 0.035 | 0.061 | 0.062 | 0.067 |
| 13 sequences without Neoceratodus | |||||||
| 1 | −15852.9 | 0.979 | |||||
| 2 | −15867.4 | −14.5 | 7.45 | 0.013 | 0.033 | 0.033 | 0.027 |
| 3 | −15866.6 | −13.7 | 7.67 | 0.026 | 0.043 | 0.043 | 0.036 |
Two different concatenated data sets were analyzed in the presence or absence of the slowly evolving sequences from Neoceratodus. The first data set (upper half) contains a total 1,078 amino acid positions with 674 (14) or 657 (13) different site patterns and either 14 (with Neoceratodus) or 13 (without) sequences, respectively. All three possible alternative topologies were compared; the favored ML tree topology corresponds always to the first hypothesis, i.e., that lungfish are the closest living relatives of tetrapods. For the different groups the following single-letter abbreviations were used. T, tetrapods; L, lungfish; C, coelacanth; A, Actinopterygii; S, sharks. Statistically significant values below the 5% (P = 0.05) threshold are shown in bold for the different LRTs. 1. ((((T,L), C), A), S), 2. ((((T,C), L), A), S), 3. (((T, (L, C)), A), S). The second data set (lower half) has a total of 1,209 amino acid positions (131 aa more), but the number of different site patterns is 792 (14) or 770 (13) for either 14 or 13 sequences (without Neoceratodus), respectively. The proportionally higher number of site patterns indicates that the additional portion of the amino acid sequences is more variable. The topologies tested are the same as in the first data set.
Two more complementary analyses were conducted to further evaluate the robustness of this result and the influence of the Neoceratodus sequences by (i) using the shorter concatenated data set without Neoceratodus and (ii) including the partial sequence of Neoceratodus into the larger concatenated data set. The influence of the slowly evolving Neoceratodus sequence on the LRTs is shown in Table 1. Although the inclusion of the Neoceratodus sequences weakens the strength of the phylogenetic analyses, the monophyly of the Sarcopterygii and, most importantly, the sistergroup relationship of tetrapods and lungfish are nonetheless supported by high bootstrap values for both sets of analyses even with the inclusion of the Neoceratodus sequence.
The phylogenetic analyses of two RAG proteins presented here were based on the biggest nuclear sequence data set collected so far on the tetrapod origin question. These data strongly support the hypothesis that the lungfishes and not the coelacanth are the closest relatives of the land vertebrates. This result emphasizes the importance of the study of all aspects of the biology and genomics of extinct and extant lungfish: our closest “fish” relatives.
Acknowledgments
We thank Dennis V. Lavrov and Hervé Philippe for helpful discussions and valuable suggestions on an earlier version of the manuscript, and we acknowledge helpful input from members of the Meyer laboratory. This research was supported by grants from the Deutsche Forschungsgemeinschaft (to H.B. and A.M.).
Abbreviations: Rag1 and Rag2, recombination-activating genes 1 and 2; ML, maximum likelihood; AU, approximate unbiased; MP, maximum parsimony; LRT, likelihood ratio tests; Ba, Bayesian approach.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY442925-AY442930).
References
- 1.Smith, J. L. B. (1939) Nature 143, 455-456. [Google Scholar]
- 2.Smith, S. L. B. (1939) Nature 143, 748-750. [Google Scholar]
- 3.Campbell, N. A. (1987) Biology (Cummings, Menlo Park, CA).
- 4.Romer, A. S. (1966) Vertebrate Paleontology (Univ. Chicago Press, Chicago).
- 5.Rosen, D. E., Forey, P. L., Gardiner, B. G. & Patterson, C. (1981) Bull. Am. Nat. Hist. Mus. 167, 159-276. [Google Scholar]
- 6.Meyer, A. & Dolven, S. I. (1992) J. Mol. Evol. 35, 102-113. [DOI] [PubMed] [Google Scholar]
- 7.Panchen, A. L. & Smith, T. R. (1987) Biol. Rev. Cambridge Philos. Soc. 62, 341-438. [Google Scholar]
- 8.Hedges, S. B., Hass, C. A. & Maxson, L. R. (1993) Nature 363, 501-502. [DOI] [PubMed] [Google Scholar]
- 9.Meyer, A. & Wilson, A. C. (1990) J. Mol. Evol. 31, 359-364. [DOI] [PubMed] [Google Scholar]
- 10.Reisz, R. R. & Smith, M. M. (2001) Nature 411, 548-550. [DOI] [PubMed] [Google Scholar]
- 11.Zhu, M. & Yu, X. (2002) Nature 418, 767-770. [DOI] [PubMed] [Google Scholar]
- 12.Zhu, M., Yu, X. & Ahlberg, P. E. (2001) Nature 410, 81-84. [DOI] [PubMed] [Google Scholar]
- 13.Yokobori, S., Hasegawa, M., Ueda, T., Okada, N., Nishikawa, K. & Watanabe, K. (1994) J. Mol. Evol. 38, 602-609. [DOI] [PubMed] [Google Scholar]
- 14.Cao, Y., Waddell, P. J., Okada, N. & Hasegawa, M. (1998) Mol. Biol. Evol. 15, 1637-1646. [DOI] [PubMed] [Google Scholar]
- 15.Zardoya, R. & Meyer, A. (1996) Proc. Natl. Acad. Sci. USA 93, 5449-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zardoya, R. & Meyer, A. (1996) Mol. Biol. Evol. 13, 933-942. [DOI] [PubMed] [Google Scholar]
- 17.Zardoya, R. & Meyer, A. (1997) Naturwissenschaften 84, 389-397. [DOI] [PubMed] [Google Scholar]
- 18.Zardoya, R., Cao, Y., Hasegawa, M. & Meyer, A. (1998) Mol. Biol. Evol. 15, 506-517. [DOI] [PubMed] [Google Scholar]
- 19.Tohyama, Y., Ichimiya, T., Kasama-Yoshida, H., Cao, Y., Hasegawa, M., Kojima, H., Tamai, Y. & Kurihara, T. (2000) Brain Res. Mol. Brain Res. 80, 256-259. [DOI] [PubMed] [Google Scholar]
- 20.Kishino, H. & Hasegawa, M. (1989) J. Mol. Evol. 29, 170-179. [DOI] [PubMed] [Google Scholar]
- 21.Goldman, N., Anderson, J. P. & Rodrigo, A. G. (2000) Syst. Biol. 49, 652-670. [DOI] [PubMed] [Google Scholar]
- 22.Ota, R., Waddell, P. J., Hasegawa, M., Shimodaira, H. & Kishino, H. (2000) Mol. Biol. Evol. 17, 798-803. [DOI] [PubMed] [Google Scholar]
- 23.Shimodaira, H. (2002) Syst. Biol. 51, 492-508. [DOI] [PubMed] [Google Scholar]
- 24.Venkatesh, B., Ning, Y. & Brenner, S. (1999) Proc. Natl. Acad. Sci. USA 96, 10267-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Venkatesh, B., Erdmann, M. V. & Brenner, S. (2001) Proc. Natl. Acad. Sci. USA 98, 11382-11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bapteste, E. & Philippe, H. (2002) Mol. Biol. Evol. 19, 972-977. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 28.Martin, A. P. (1999) Mol. Biol. Evol. 16, 996-1002. [DOI] [PubMed] [Google Scholar]
- 29.Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philippe, H. (1993) Nucleic Acids Res. 21, 5264-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar, S., Tamura, K., Jakobsen, I. B. & Nei, M. (2001) Bioinformatics 17, 1244-1245. [DOI] [PubMed] [Google Scholar]
- 32.Swofford, D. L. (1999) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4b10.
- 33.Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. (2002) Bioinformatics 18, 502-504. [DOI] [PubMed] [Google Scholar]
- 34.Huelsenbeck, J. P. & Ronquist, F. (2001) Bioinformatics 17, 754-755. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Z. (1997) Comput. Appl. Biosci. 13, 555-556. [DOI] [PubMed] [Google Scholar]
- 36.Shimodaira, H. & Hasegawa, M. (2001) Bioinformatics 17, 1246-1247. [DOI] [PubMed] [Google Scholar]
- 37.Suzuku, Y., Glazko, G. V. & Nei, M. (2002) Proc. Natl. Acad. Sci. USA 99, 16138-16143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ranwez, V. G., O. (2001) Mol. Biol. Evol. 18, 1103-1116. [DOI] [PubMed] [Google Scholar]
- 39.Meyer, A. & Zardoya, R. (2002) Annu. Rev. Ecol. Syst. 34, 311-338. [Google Scholar]
- 40.Madsen, O., Scally, M., Douady, C. J., Kao, D. J., DeBry, R. W., Adkins, R., Amrine, H. M., Stanhope, M. J., de Jong, W. W. & Springer, M. S. (2001) Nature 409, 610-614. [DOI] [PubMed] [Google Scholar]
- 41.Murphy, W. J., Eizirik, E., Johnson, W. E., Zhang, Y. P., Ryder, O. A. & O'Brien, S. J. (2001) Nature 409, 614-618. [DOI] [PubMed] [Google Scholar]
- 42.Murphy, W. J., Eizirik, E., O'Brien, S. J., Madsen, O., Scally, M., Douady, C. J., Teeling, E., Ryder, O. A., Stanhope, M. J., de Jong, W. W. & Springer, M. S. (2001) Science 294, 2348-2351. [DOI] [PubMed] [Google Scholar]