Abstract
RNA interference (RNAi) is a ubiquitous mechanism of eukaryotic gene regulation that can be exploited for specific gene silencing. Retroviruses have been successfully used for stable expression of short hairpin RNAs in mammalian cells, leading to persistent inhibition of gene expression by RNAi. Here, we apply retrovirus-mediated RNAi to epithelial Madin-Darby canine kidney cells, whose properties limit the applicability of other RNAi methods. We demonstrate efficient suppression of a set of 13 target genes by retroviral coexpression of short hairpin RNAs and a selectable marker. We characterize the resulting knockdown cell populations with regard to composition and stability, and examine the usefulness of proposed guidelines for choosing RNAi target sequences. Finally, we show that this system can be used to simultaneously target two genes, giving rise to double knockdowns. Thus, retrovirus-mediated RNAi is a convenient method for gene silencing in Madin-Darby canine kidney cells, and is likely to be applicable to virtually any mammalian cell.
RNA interference (RNAi) is an evolutionarily ancient mechanism of gene regulation in eukaryotes (1). It is triggered by double-stranded RNA (dsRNA) and leads to degradation or translational repression of mRNAs containing complementary sequences. The machinery that mediates RNAi not only protects cells from viruses and transposons, but is also implicated in the regulation of a growing number of cellular processes (2, 3). Beyond its natural roles, RNAi offers a unique avenue for the analysis of gene function because it can be used as a powerful tool for gene silencing (4).
The specificity of RNAi is determined by 21- to 23-nt RNA duplexes, referred to as micro-RNAs (miRNAs) or small interfering RNAs (siRNAs), depending on their origin. miRNAs are generated from endogenous precursors, which form hairpin structures with stretches of dsRNA. These are cleaved by the ribonuclease Dicer to produce mature miRNAs. After unwinding, one of the strands becomes incorporated into the RNA-induced silencing complex (RISC) and guides the destruction or repression of complementary mRNAs. siRNAs arise from viral or other exogenous dsRNA, but they use the same mechanism to effect mRNA degradation (3).
siRNAs can be introduced into mammalian cells by various means (5, 6). First, chemically synthesized siRNAs can be transfected into cells (7). Although synthetic siRNAs can achieve effective and very rapid “knockdowns,” their use is limited to cells that can be transfected at high rates. In addition, their effects are transient. To circumvent the latter problem, so-called short hairpin RNAs (shRNAs) can be expressed from stably transfected plasmids (8, 9). Like miRNA precursors, shRNAs form hairpin structures, which are cleaved by Dicer to produce siRNAs. However, the generation of shRNA-expressing cell lines is time-consuming and involves clonal selection. An alternative way to achieve stable integration of shRNA expression cassettes into the genome of cells is delivery via retroviruses (10-13). Their production is fast and simple and, through incorporation of the vesicular stomatitis virus G protein into their envelope, they are able to infect almost any dividing mammalian cell (14). If expression of shRNAs is combined with expression of a selectable marker, populations in which every cell contains at least one active copy of the retroviral genome can readily be obtained (10-12). Furthermore, the high efficiency of retroviral transduction avoids the caveats associated with clonal selection.
The epithelial Madin-Darby canine kidney (MDCK) cell line is able to establish a polarized monolayer in vitro that resembles the in vivo properties of simple epithelia. MDCK cells are therefore widely used as a model system to study cell polarity and polarized membrane trafficking (15, 16). We sought to inhibit the expression of 13 genes, all of which have been proposed to play a role in mediating protein transport to the plasma membrane. However, MDCK cells are difficult to transfect by standard methods. Moreover, the establishment of a polarized monolayer takes at least 3 days after plating on a suitable support, by which time the effects of transient transfection have usually deteriorated. Because these circumstances largely preclude the use of synthetic siRNAs, we decided to use retroviruses to introduce shRNAs into MDCK cells.
In this paper, we modify, characterize, and extend an existing system for the retroviral delivery of shRNAs into mammalian cells (13). We demonstrate that with this system many genes can be efficiently inhibited in MDCK cells. We report on the composition and stability of the resulting retrovirus-transduced populations of knockdown cells, as well as on the characteristic features of functional compared to nonfunctional siRNAs. Finally, we show that retrovirus-mediated RNAi can be used to simultaneously target two genes, thus generating “double knockdowns.”
Materials and Methods
Antibodies. The polyclonal caveolin-1 N20 antibody was from Santa Cruz Biotechnology, and the transferrin receptor antibody was from Zymed. The rab8 and rab11 antibodies were from BD Transduction Laboratories. The antibody against TI-VAMP (17) was kindly provided by Thierry Galli (INSERM U536, Paris), the annexin 2 HH7 antibody (18) was a gift from Volker Gerke (Institute for Medical Biochemistry, University of Münster, Münster, Germany), the syntaxin 3 antibody (19) was a gift from Vesa Olkkonen (National Public Health Institute, Helsinki), and hybridoma cells producing the gp135 antibody (20) were supplied by George Ojakian (Department of Anatomy and Cell Biology, State University of New York Downstate Medical Center, Brooklyn). The annexin 13b, annexin 13-2, and VIP36 antibodies, as well as the monoclonal gp114 antibody, have been characterized (21-23). The antibody against VIP17/MAL (referred to as VIP17) was generated in our laboratory.
Plasmids. To create pRVH1-puro, the puromycin resistance gene from pSVpaX1 (24) (provided by Frank Buchholz, Max Planck Institute of Molecular Cell Biology and Genetics) was amplified by PCR, introducing XbaI and NotI restriction sites in the process. The PCR product was cloned into the XbaI/NotI site of pRVH1 (13) (provided by Ruslan Medzhitov, Howard Hughes Medical Institute, Yale University, New Haven, CT), replacing the CD4 gene present in the original vector. By using the same procedure, pRVH1-hygro was created by inserting the hygromycin resistance gene from pRAD54B-hyg (25) (provided by Kiyoshi Miyagawa, Department of Molecular Pathology, University of Hiroshima, Hiroshima, Japan) into pRVH1. pMD.G, which expresses the vesicular stomatitis virus G (VSV-G) protein under control of the human cytomegalovirus immediate-early (CMV) promoter, was provided by Richard Mulligan (Harvard Medical School, Boston). pSUPER (9), which contains the human H1 promoter, was provided by Reuven Agami (The Netherlands Cancer Institute, Amsterdam).
Target Sequence Selection and Cloning. Because a complete dog genome sequence is not yet available, the coding sequence of several target genes had to be reconstructed from partial sequences, which were obtained from the National Center for Biotechnology Information Canis familiaris trace archive by using the discontiguous MEGA BLAST search tool (www.ncbi.nlm.nih.gov/blast/tracemb.shtml). Unique sequences conforming to either AAN19 as suggested by Elbashir et al. (26) or to GN20 were selected. Target sequences containing EcoRI or XhoI sites (which would interfere with the subsequent cloning), TTT at their 3′ end (which would create a cluster of transcription-terminating thymidines), or stretches of more than three identical bases (which have been proposed to reduce RNAi efficiency) were avoided. Oligonucleotides encoding shRNAs directed against the different target genes were designed and cloned into pSUPER according to Brummelkamp et al. (9). The shRNA expression cassette was then transferred into the XhoI/EcoRI site of pRVH1-puro or pRVH1-hygro.
Virus Production. The human Phoenix gag-pol packaging cell line (www.stanford.edu/group/nolan/retroviral_systems/phx.html; obtained from the American Type Culture Collection with authorization by Garry Nolan, School of Medicine, Stanford University, Stanford, CA) was kept in high-glucose DMEM (4.5 g/liter) containing 10% FCS, 2 mM glutamine, and 100 units/ml penicillin and streptomycin. Nearly confluent cells in six-well plates were transfected with 4 μg of pRVH1-puro or pRVH1-hygro and 0.4 μg of pMD.G per well by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Twenty-four hours posttransfection, the medium was changed to low-glucose DMEM (1 g/liter) with the same supplements as above (1 ml per well), and placed at 32°C. Forty-eight hours posttransfection, the medium was collected, centrifuged for 5 min at 200 × g to remove cell debris, and used for infection. For producing virus stocks, Phoenix cells in 10-cm dishes were transfected with 24 μg of retroviral vector and 2.4 μg of pMD.G, the medium was changed to low-glucose DMEM (5 ml per dish) 24 h posttransfection, and cells were placed at 32°C. Batches of virus containing supernatant were then collected every 24 h for up to 7 days. The supernatant was passed through a 0.45-μm syringe filter and frozen in liquid nitrogen. Where indicated, the virus was concentrated immediately before infection as described (13).
Target Cell Transduction. MDCK strain II cells were cultured in MEM with 5% FCS, 2 mM glutamine, and 100 units/ml penicillin and streptomycin. For infection, 1.5 × 105 cells were mixed with 450 μl of virus containing supernatant in the presence of 4 μg/ml polybrene (Sigma), seeded into a well of a 12-well plate, and incubated at 32°C. Twelve hours postinfection, the medium was changed to normal culture medium, and cells were returned to 37°C. Thirty-six to 48 h postinfection, cells were trypsinized and seeded into a well of a six-well plate in the presence of 4 μg/ml puromycin or 800 μg/ml hygromycin (both from BD Biosciences). Selection was done for 48 or 60 h, respectively. Six days postinfection, the knockdown efficiency was assayed by quantitative RT-PCR or immunoblotting. For double knockdowns, cells were sequentially infected with RVH1-puro virus, selected with puromycin, infected with RVH1-hygro virus, and selected with hygromycin as above.
Quantitative RT-PCR. RNA isolation, cDNA synthesis, and quantitative RT-PCR were done as described (27). The levels of ubiquitin mRNA were used as an internal standard to determine relative mRNA levels of the target genes in control and knockdown cells. Detailed information on PCR conditions and primer sequences is available on request.
Immunofluorescence and Confocal Analysis. Immunostaining of MDCK cells was performed as described (22). Images were acquired with a Leica TCS SP2 laser scanning confocal microscope (Leica, Bensheim, Germany) with ×20 magnification. Identical settings were used for control and knockdown cells.
Internal Energy Calculation. Internal energy profiles of siRNA duplexes, as well as the duplex stabilities at the 5′ ends of sense and antisense strands, were calculated with rnastructure 3.71 software by using the oligowalk program (ref. 28; www.bioinfo.rpi.edu/applications/mfold). Starting from the 5′ end of the antisense strand, pentamer hybridization energies along the length of the siRNA duplexes were determined. Depending on the target sequence format (AAN19 or GN20), the duplexed regions were 19 or 21 nt long, giving 15 or 17 pentamer hybridization energies. These were then used to construct internal energy profiles. This procedure is similar to the one used by Khvorova et al. (29), except that the sequences were not extended at the 3′ end. The duplex stabilities at the 5′ ends of sense and antisense strands were also calculated on the basis of pentamer hybridization energies.
Results
Inhibition of Gene Expression by Retrovirus-Mediated RNAi. Our set of target mRNAs consisted of 13 transcripts. Between one and six target sequences were selected per transcript, giving rise to a total of 37 target sequences (Table 2, which is published as supporting information on the PNAS web site). Corresponding shRNAs were designed according to Brummelkamp et al. (9) (Fig. 7, which is published as supporting information on the PNAS web site) and introduced into the retroviral vector RVH1-puro. Besides the H1 promoter, which drives the expression of shRNAs, RVH1-puro harbors a puromycin resistance gene controlled by the CMV promoter. Retroviruses, pseudotyped with the VSV-G envelope protein for enhanced stability and efficient infection of MDCK cells, were produced and typically yielded transduction rates of ≈70%. After puromycin selection to eliminate nontransduced cells, the RNAi-mediated depletion of target mRNAs was measured by quantitative RT-PCR. With the best target sequence for each gene, mRNA reductions in the range of 70-95% were achieved (Table 1). These results were highly reproducible as indicated by the standard deviations calculated from at least three independent experiments in each case.
Table 1. RNAi-mediated reduction of target gene mRNA levels.
| Target gene | mRNA reduction, % |
|---|---|
| rab11b | 96 ± 1 |
| rab8a | 93 ± 4 |
| VIP17 | 92 ± 2 |
| Annexin 13 | 91 ± 2 |
| Caveolin-1 | 90 ± 3 |
| TI-VAMP | 90 ± 4 |
| MAL2 | 88 ± 3 |
| Annexin 2 | 81 ± 4 |
| VIP36 | 81 ± 5 |
| rab11a | 77 ± 6 |
| Annexin 13b | 74 ± 4 |
| KIFC3 | 73 ± 4 |
| Syntaxin 3 | 71 ± 2 |
Target gene mRNA levels were determined by quantitative RT-PCR. Levels in knockdown cells are given as percent mRNA reduction relative to the levels in control cells transduced with empty RVH1-puro. Standard deviations are from at least three independent experiments in each case. Note that annexin 13a cannot be targeted individually because annexin 13b comprises the entire annexin 13a sequence. The annexin 13 knockdown therefore affects both isoforms.
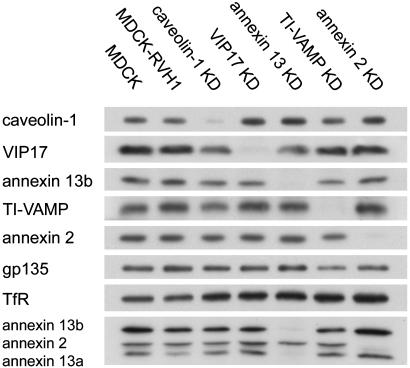
Immunoblotting showed that the reduced mRNA levels of caveolin-1, VIP17, annexin 13, TI-VAMP, and annexin 2 corresponded to strong and specific reductions of the target protein levels, whereas the empty RVH1-puro had no effect (Fig. 1). Specific reductions were also observed in VIP36, annexin 13b, and syntaxin 3 knockdown cells (data not shown). To assess how well mRNA and protein depletion correlated, we analyzed the reduced protein levels of caveolin-1, TI-VAMP, annexin 2, annexin 13b, and syntaxin 3 in more detail by immunoblotting with serial dilutions. Except for caveolin-1, where the protein was reduced by ≈80% compared to an mRNA reduction of 90%, the protein levels of the other four genes were depleted at least as strongly as their mRNA levels (data not shown). In the case of caveolin-1, which forms the characteristic striated coat on the cytoplasmic surface of caveolae, this could result from a very stable pool of the protein. For lack of antibodies we were unable to analyze the protein levels of MAL2 and KIFC3, whereas in the case of rab8a/b and rab11a/b the available antibodies recognized both isoforms. However, we assume that also for these genes, the mRNA depletion translated into a corresponding reduction of the protein levels.
Fig. 1.
RNAi-mediated reduction of target gene protein levels. Equal amounts of protein from MDCK cells, MDCK cells transduced with empty RVH1-puro (MDCK-RVH1), and knockdown (KD) cells were analyzed by immunoblotting. gp135 and transferrin receptor (TfR) were used as controls. Note that the annexin 13-2 antibody recognizes annexin 13a, annexin 13b, and annexin 2 (lowermost blot), and that both annexin 13a and b are depleted in annexin 13 knockdown cells.
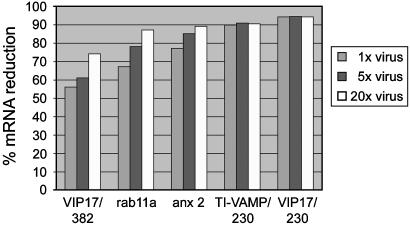
Raising the retrovirus titer might improve depletion of a target mRNA because increasing the number of integration events leads to stronger shRNA expression and, subsequently, to higher intracellular siRNA concentrations. To test to what extent target mRNA depletion can be augmented in this way, different viruses with knockdown efficiencies ranging from 55% to 95% were concentrated by pelleting. No significant loss of infectivity was caused by the pelleting procedure alone, because virus resuspended in the original volume performed as well as untreated virus. Substantially improved knockdowns could be obtained with concentrated virus (Fig. 2). This enhancement was most pronounced for viruses achieving weak or intermediate knockdowns, whereas higher titers did not improve the knockdown in the case of viruses that yielded mRNA reductions of ≥90% already without concentrating. This finding indicates that removal of the remaining mRNA is prevented by factors other than insufficient levels of shRNA expression. Nevertheless, these results show that increasing the virus titer can often be used to improve knockdowns.
Fig. 2.
Improvement of knockdowns by increasing virus titers. Five viruses expressing different shRNAs (VIP17/382, rab11a, anx2, TI-VAMP/230, and VIP17/230; anx 2, annexin 2) were pelleted by centrifugation and resuspended in 1, 0.2, or 0.05 times the original volume, resulting in 1, 5, or 20 times the original virus concentration. Percent mRNA reductions relative to the levels in control cells transduced with empty RVH1-puro were measured as in Table 1.
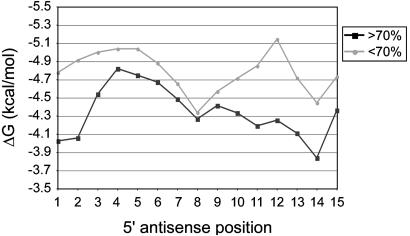
Properties of Functional Versus Nonfunctional Target siRNAs. The above data indicate that the retroviral RNAi system is able to strongly and specifically inhibit gene expression. However, to date no well defined rules have been established for the selection of efficient target sequences, making this step an obstacle to routine application of RNAi in mammalian cells. Our set of 37 target sequences is relatively small, making it difficult to derive selection guidelines from it. Yet, the data set can be used to test previous suggestions for the selection of target sequences, such as the proposal that the free energy profile of siRNAs is predictive of their efficiencies (29). Therefore, we subjected the siRNA duplexes that presumably arise from Dicer-mediated processing of the expressed shRNAs (Fig. 7) to the same kind of analysis carried out by Khvorova et al. (29). siRNAs were grouped in two categories, functional siRNAs (>70% mRNA reduction, n = 21) and nonfunctional siRNAs (<70% reduction, n = 16). The average energy profiles of functional and nonfunctional siRNAs were calculated starting from the 5′ antisense end of the duplexes. We found that nonfunctional siRNAs have a more stable 5′ antisense end than functional siRNAs. In addition, nonfunctional siRNAs are more stable in the central region between positions 10 and 13 (Fig. 3). These results are consistent with earlier findings (29, 30). We then compared the relative stabilities of the two ends of siRNA duplexes. In the case of nonfunctional siRNAs, the two ends were of nearly equal duplex stability (ΔG = 0.17 kcal/mol). Functional siRNAs on the other hand had less stable 5′ antisense ends (ΔG = 0.7 kcal/mol). This finding is in agreement with the idea that a less stable 5′ antisense end leads to preferential incorporation of the antisense strand into the RNA-induced silencing complex (RISC) and hence to efficient mRNA degradation (29, 31). Finally, we analyzed the frequency with which each of the four nucleotides occurred at each position of functional and nonfunctional siRNAs. We found that adenosine is overrepresented at position 11 of the antisense strand of functional siRNAs, because it occurred in 52% of the functional siRNAs compared to 31% of the nonfunctional siRNAs. Strikingly, all seven siRNAs that achieved mRNA reductions of 90% or more had an adenosine at position 11 of the antisense strand. Because mRNA cleavage invariably takes place between the nucleotides complementary to position 10 and 11 of the antisense strand (32), this finding is consistent with the proposal that the RISC preferentially cleaves mRNA 3′ of uridine (33). The fact that these trends manifest themselves so clearly even in our small data set indicates that the efficiency of siRNAs strongly depends on the characteristic features of functional siRNAs described above.
Fig. 3.
Internal energy profiles of functional versus nonfunctional siRNAs. Pentamer hybridization energies were calculated along the length of the siRNA duplexes starting from the 5′ end of the antisense strands. The average pentamer hybridization energies (ΔG) for the first 15 positions of functional siRNAs (>70% mRNA reduction) and nonfunctional siRNAs (<70% mRNA reduction) are shown.
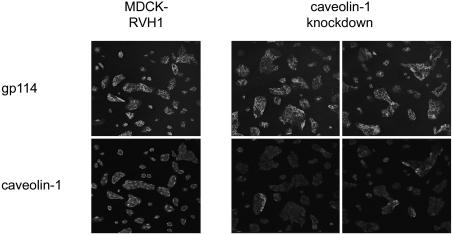
Composition and Stability of Retrovirus-Transduced Cell Populations. Both mRNA and protein levels provide a measure for the average knockdown, but they do not indicate how the residual protein is distributed in a population of knockdown cells. We therefore examined the distribution of the residual caveolin-1 protein in knockdown cells by using immunofluorescence. There was a strong overall reduction in the caveolin-1 signal in knockdown cells compared to control cells, whereas the expression of the plasma membrane protein gp114 was unaffected. However, the expression of caveolin-1 in knockdown cells was quite heterogeneous and occasionally as strong as in control cells (Fig. 4). Using a virus directed against caveolin-1 in which the puromycin resistance gene was replaced by GFP, we observed that the correlation between the extent of caveolin-1 depletion and GFP expression is not strict (data not shown). This finding indicates that the activities of the H1 and the CMV promoter of RVH1-puro, which drive expression of the shRNAs and the puromycin resistance gene, are not tightly linked, despite their immediate neighborhood. This would explain why a small number of puromycin-resistant cells retain substantial expression of the targeted protein. In addition, mutations in the shRNA expression cassette, which might occur during reverse transcription of the retroviral genome by the error-prone viral reverse transcriptase, could contribute to this phenomenon.
Fig. 4.
Distribution of residual protein in a knockdown cell population. Caveolin-1 and the plasma membrane protein gp114 were visualized by immunofluorescence in caveolin-1 knockdown cells and control cells transduced with empty RVH1-puro (MDCK-RVH1).
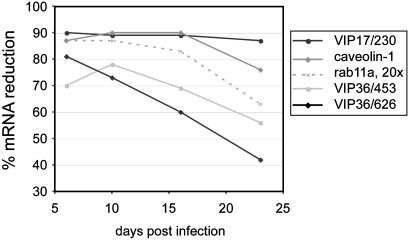
If RNAi-mediated depletion of a protein caused a growth disadvantage, cells with low levels of that protein would gradually be eliminated from a heterogeneous cell population, thus limiting the time during which experiments can be performed. To test the stability of knockdowns within populations of retrovirus-transduced cells, we performed time-course experiments. We followed the mRNA levels of VIP17, caveolin-1, rab11a, and VIP36 in continuously dividing knockdown cell populations for over 3 weeks after infection. In all cases, the mRNA reduction decayed over time (Fig. 5). Reselecting with puromycin did not restore the knockdowns, ruling out that this decline is due to cells that have lost or inactivated the integrated retroviral construct. These results show that the knockdowns are not stable within a population of transduced cells. Instead, they slowly decay over time in a manner that may correlate with the severity of the growth disadvantage caused by the knockdown. However, no major changes occurred within the first 10 days after the end of the initial selection, especially in the case of very strong knockdowns.
Fig. 5.
Stability of target gene mRNA reductions in knockdown cell populations. MDCK cells were transduced with different viruses (VIP17/230, caveolin-1, rab11a, VIP36/453, and VIP36/626). Target gene mRNA levels were monitored between days 6 and 23 postinfection. Percent mRNA reductions relative to the levels in control cells transduced with empty RVH1-puro were measured as in Table 1. Note that the rab11a virus was concentrated 20-fold before infection.
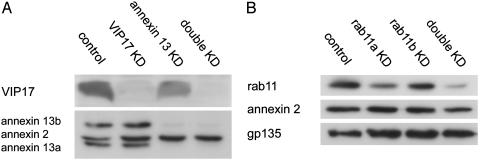
Generation of Double Knockdowns by Retrovirus-Mediated RNAi. To make simultaneous depletion of two proteins possible, we created a second retroviral vector, RVH1-hygro, which confers hygromycin instead of puromycin resistance. Viruses produced with this vector achieved the same knockdown efficiencies as RVH1-puro viruses, as verified for rab11a, VIP17, and VIP36 (data not shown). We then established a procedure for sequential transduction of MDCK cells with an RVH1-puro and an RVH1-hygro virus. In this way, the expression of two different proteins can be reduced to levels identical to those observed in the single knockdowns, as exemplified by the VIP17/annexin 13 and the rab11a/rab11b double knockdowns (Fig. 6).
Fig. 6.
RNAi-mediated reduction of target gene protein levels in single and double knockdown cells. Equal amounts of protein from MDCK cells transduced with empty RVH1-puro and empty RVH1-hygro (control) and single and double knockdown (KD) cells were analyzed by immunoblotting. Annexin 2 was used as control for the VIP17/annexin 13 knockdown (A), and annexin 2 and gp135 were used as controls for the rab11a/rab11b knockdown (B). Note that rab11a appears to be the major rab11 isoform in MDCK cells.
Discussion
In this paper, we have evaluated the usefulness of an RNAi system that combines retroviral transduction with selectable markers to generate single and double knockdowns in mammalian cells. We used this system because MDCK cells are difficult to transfect and because effective RNAi in polarized MDCK cells requires prolonged presence of siRNAs. However, the principal advantages of the retroviral system, namely the virtually unlimited host range, the high transduction rates, the speed and ease with which a population of knockdown cells can be generated, and the sustained expression of shRNAs, should make retroviruses a generally applicable and convenient tool for gene silencing.
Analysis of the residual protein in a population of knockdown cells revealed that, despite a strong overall reduction, there was a small number of cells with substantial levels of the targeted protein. Possibly as a result of this heterogeneity together with a growth disadvantage caused by the knockdown, mRNA reductions generally declined over time. However, the window of optimal gene inhibition was at least 10 days after selection in the case of strong knockdowns. This window should be sufficient to analyze resulting RNAi phenotypes, even if they relate to processes like cell polarization or differentiation. It should also be possible to perform rescue experiments, for example, with a mutant version of a targeted protein that is unaffected by the siRNA directed against the endogenous mRNA. Furthermore, it may not even be desirable to keep knockdown cells in culture for a long time because inhibiting the expression of a particular gene might induce compensatory mechanisms like up-regulation of genes that act as functional substitutes. A related concern is that the time needed to generate a drug-selected population of knockdown cells may already be long enough for such mechanisms to come into play. On the other hand, the retroviral system clearly has the capacity to produce phenotypes that agree with the proposed functions of our target genes in regulating membrane trafficking. For instance, knocking down VIP17, which has been shown to mediate transport to the apical surface of MDCK cells (34, 35), leads to mislocalization of the apical plasma membrane protein gp114 to the basolateral side of fully polarized cells. Also, inhibiting the expression of rab11 results in reduced transferrin uptake into MDCK cells (unpublished results). This finding could be due to a defect in the recycling of endocytosed transferrin receptor, in line with the role of rab11 in regulating trafficking through the recycling endosome (36).
Our results support the notion that any mRNA can be targeted by RNAi because we could substantially reduce expression of each of the 13 target genes. What remains unknown is whether knockdowns of >90% can be obtained for any given gene, and if so, how many target sequences will have to be tested to achieve this. Much will depend on refining the criteria for selecting target sequences. Several studies, including this one, have revealed certain characteristics of functional siRNAs. They are marked by a low stability of the 5′ antisense end compared to the 5′ sense end, an overall relatively low stability, and a preference for adenosine at position 11 of the antisense strand (29-31, 33). These features have already been used to construct an algorithm for the rational design of siRNAs that significantly improves the chances of choosing functional siRNAs (33). However, it is not clear how accurately the efficiency of an individual siRNA can be predicted. It is conceivable that factors unrelated to the properties of the siRNAs, like mRNA secondary structure, can interfere with target recognition by the RNA-induced silencing complex (RISC) (37). Such factors may explain why there are relatively recalcitrant transcripts like the syntaxin 3 mRNA, for which none of the six target sequences we tested gave a knockdown of more than ≈70%. An additional point to consider when introducing siRNAs into cells by means of shRNA expression is that not all shRNAs may be transcribed equally well. The U6 promoter, which has been used for expression of shRNAs (8), has a strict requirement for guanosine in the +1 position, whereas the H1 promoter used in this study is more flexible (5). Whether the H1 promoter has any preferences for certain bases downstream of the start site of transcription is not known.
Many genes may be functionally redundant, as suggested by the large number of knockout mice without obvious phenotypes. Interfering with a particular function may therefore require removal of several gene activities. Furthermore, alternative pathways often exist in processes like membrane trafficking, so that even though a particular transport route is blocked, proteins still reach their correct destination by taking alternative routes. In this regard it is illuminating to note that despite extensive genetic screens for secretion mutants in yeast, only a few genes are known that regulate exocytic transport from the trans-Golgi compartment. Most likely, these were missed due to the existence of at least two pathways to the yeast plasma membrane that can compensate for each other (38). Studies of gene interactions, for example by epistasis experiments or synthetic lethality screens in yeast, also demonstrate the need to simultaneously suppress the activity of more than one gene. In mammalian cells, this can be achieved by RNAi with transient transfection of synthetic siRNAs or plasmids expressing shRNAs (26, 39). In principle, there is no reason why expression vectors could not be used to derive stably transfected double knockdown cell lines, but the disadvantages of this approach, namely the time required and the drawbacks of clonal selection, remain. The retroviral system avoids both problems. Double knockdowns can be produced within a week, and the high transduction rates ensure that effects arising from the integration into a particular locus of the host genome will be averaged out in a cell population.
The capacity of the RNAi machinery is not easily saturated, as shown by the facts that knockdowns can be improved by increasing the virus titer and that efficient double knockdowns can be generated. Triple or quadruple knockdowns might therefore be possible. By using a retroviral system, this could be achieved by introducing yet another selectable marker. Alternatively, retroviruses could be combined with adenoviruses, which have been successfully used for RNAi and can also infect nondividing cells like fully polarized MDCK cells (6). However, overloading the RNAi machinery may at some point compromise the regulatory functions of the cellular miRNAs, leading to unspecific side effects. Furthermore, exceedingly high levels of siRNAs might activate aspects of the interferon response (40), even though this defense mechanism is normally not activated by short double-stranded RNAs (7).
In summary, we have shown that the retroviral system we describe here can be used to efficiently generate single and double knockdowns in MDCK cells and presumably also in almost any other mammalian cell type. As already noted by others (11), the reagents to generate retroviruses are readily available in the scientific community, their handling is very straightforward, and thus the use of retroviruses as tools for gene silencing is open to any laboratory. In the near future, we are likely to witness a wave of reports on large-scale or even genomewide RNAi screens in mammalian cells with the most tractable cell types. Pinpointing the molecular functions of proteins and understanding their mechanisms of action will nevertheless often require their analysis in specialized model systems. For this, a broad repertoire of RNAi methods will be beneficial.
Supplementary Material
Acknowledgments
We thank Francis Stewart for critically reading the manuscript.
Abbreviations: MDCK, Madin-Darby canine kidney; miRNA, micro-RNA; RNAi, RNA interference; shRNA, short hairpin RNA; siRNA, small interfering RNA.
References
- 1.Hannon, G. J. (2002) Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- 2.Denli, A. M. & Hannon, G. J. (2003) Trends Biochem. Sci. 4, 196-201. [DOI] [PubMed] [Google Scholar]
- 3.Bartel, D. P. (2004) Cell 116, 281-297. [DOI] [PubMed] [Google Scholar]
- 4.Paddison, P. J. & Hannon, G. J. (2003) Curr. Opin. Mol. Ther. 5, 217-224. [PubMed] [Google Scholar]
- 5.Dykxhoorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457-467. [DOI] [PubMed] [Google Scholar]
- 6.Wilson, J. A. & Richardson, C. D. (2003) Curr. Opin. Mol. Ther. 5, 389-396. [PubMed] [Google Scholar]
- 7.Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K. & Tuschl, T. (2001) Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- 8.Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J. & Conklin, D. S. (2002) Genes Dev. 16, 948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Science 296, 550-553. [DOI] [PubMed] [Google Scholar]
- 10.Paddison, P. J. & Hannon, G. J. (2002) Cancer Cell 2, 17-23. [DOI] [PubMed] [Google Scholar]
- 11.Devroe, E. & Silver, P. A. (2002) BMC Biotechnol. 2, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brummelkamp, T. R., Bernards, R. & Agami, R. (2002) Cancer Cell 2, 243-247. [DOI] [PubMed] [Google Scholar]
- 13.Barton, G. M. & Medzhitov, R. (2002) Proc. Natl. Acad. Sci. USA 99, 14943-14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yee, J. K., Friedmann, T. & Burns, J. C. (1994) Methods Cell Biol. 43, 99-112. [DOI] [PubMed] [Google Scholar]
- 15.Yeaman, C., Grindstaff, K. K. & Nelson, W. J. (1999) Physiol. Rev. 79, 73-98. [DOI] [PubMed] [Google Scholar]
- 16.Nelson, J. W. & Yeaman, C. (2001) Trends Cell Biol. 11, 483-486. [DOI] [PubMed] [Google Scholar]
- 17.Muzerelle, A., Alberts, P., Martinez-Arca, S., Jeannequin, O., Lafaye, P., Mazie, J. C., Galli, T. & Gaspar, P. (2003) Neuroscience 122, 59-75. [DOI] [PubMed] [Google Scholar]
- 18.Thiel, C., Osborn, M. & Gerke, V. (1992) J. Cell Sci. 103, 733-742. [DOI] [PubMed] [Google Scholar]
- 19.Riento, K., Galli, T., Jansson, S., Ehnholm, C., Lehtonen, E. & Olkkonen, V. M. (1998) J. Cell Sci. 111, 2681-2688. [DOI] [PubMed] [Google Scholar]
- 20.Ojakian, G. K. & Schwimmer, R. (1988) J. Cell Biol. 107, 2377-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecat, S., Verkade, P., Thiele, C., Fiedler, K., Simons, K. & Lafont, F. (2000) J. Cell Sci. 113, 2607-2618. [DOI] [PubMed] [Google Scholar]
- 22.Füllekrug, J., Scheiffele, P. & Simons, K. (1999) J. Cell Sci. 112, 2813-2821. [DOI] [PubMed] [Google Scholar]
- 23.Balcarova-Ständer, J., Pfeiffer, S. E., Fuller, S. D. & Simons, K. (1984) EMBO J. 3, 2687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchholz, F., Angrand, P. O. & Stewart, A. F. (1996) Nucleic Acids Res. 24, 3118-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyagawa, K., Tsuruga, T., Kinomura, A., Usui, K., Katsura, M., Tashiro, S., Mishima, H. & Tanaka, K. (2002) EMBO J. 21, 175-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elbashir, S. M., Harborth, J., Weber, K. & Tuschl, T. (2002) Methods 26, 199-202. [DOI] [PubMed] [Google Scholar]
- 27.Schuck, S., Soloaga, A., Schratt, G., Arthur, J. S. & Nordheim, A. (2003) BMC Mol. Biol. 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuker, M. (2003) Nucleic Acids Res. 31, 3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khvorova, A., Reynolds, A. & Jayasena, S. D. (2003) Cell 115, 209-216. [DOI] [PubMed] [Google Scholar]
- 30.Aza-Blanc, P., Cooper, C. L., Wagner, K., Batalov, S., Deveraux, Q. L. & Cooke, M. P. (2003) Mol. Cell 12, 627-637. [DOI] [PubMed] [Google Scholar]
- 31.Schwarz, D. S., Hutvagner, G., Du, T., Xu, Z., Aronin, N. & Zamore, P. D. (2003) Cell 115, 199-208. [DOI] [PubMed] [Google Scholar]
- 32.Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W. & Tuschl, T. (2001) EMBO J. 20, 6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W. S. & Khvorova, A. (2004) Nat. Biotechnol. 22, 326-330. [DOI] [PubMed] [Google Scholar]
- 34.Puertollano, R., Martin-Belmonte, F., Millan, J., de Marco, M. C., Albar, J. P., Kremer, L. & Alonso, M. A. (1999) J. Cell Biol. 145, 141-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheong, K. H., Zacchetti, D., Schneeberger, E. E. & Simons, K. (1999) Proc. Natl. Acad. Sci. USA 96, 6241-6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullrich, O., Reinsch, S., Urbe, S., Zerial, M. & Parton, R. G. (1996) J. Cell Biol. 135, 913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vickers, T. A., Koo, S., Bennett, C. F., Crooke, S. T., Dean, N. M. & Baker, B. F. (2003) J. Biol. Chem. 278, 7108-7118. [DOI] [PubMed] [Google Scholar]
- 38.Harsay, E. & Schekman, R. (2002) J. Cell Biol. 156, 271-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, J. Y., Taylor, J., DeRuiter, S. L., Vojtek, A. B. & Turner, D. L. (2003) Mol. Ther. 7, 228-236. [DOI] [PubMed] [Google Scholar]
- 40.Bridge, A. J., Pebernard, S., Ducraux, A., Nicoulaz, A. L. & Iggo, R. (2003) Nat. Genet. 34, 263-264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.