Abstract
Large-magnitude numerical distinctions (>10-fold) among drug responses of genetically contrasting cancers were crucial for guiding the development of some targeted therapies. Similar strategies brought epidemiological clues and prevention goals for genetic diseases. Such numerical guides, however, were incomplete or low magnitude for Fanconi anemia pathway (FANC) gene mutations relevant to cancer in FANC-mutation carriers (heterozygotes). We generated a four-gene FANC-null cancer panel, including the engineering of new PALB2/FANCN-null cancer cells by homologous recombination. A characteristic matching of FANCC-null, FANCG-null, BRCA2/FANCD1-null, and PALB2/FANCN-null phenotypes was confirmed by uniform tumor regression on single-dose cross-linker therapy in mice and by shared chemical hypersensitivities to various inter-strand cross-linking agents and γ-radiation in vitro. Some compounds, however, had contrasting magnitudes of sensitivity; a strikingly high (19- to 22-fold) hypersensitivity was seen among PALB2-null and BRCA2-null cells for the ethanol metabolite, acetaldehyde, associated with widespread chromosomal breakage at a concentration not producing breaks in parental cells. Because FANC-defective cancer cells can share or differ in their chemical sensitivities, patterns of selective hypersensitivity hold implications for the evolutionary understanding of this pathway. Clinical decisions for cancer-relevant prevention and management of FANC-mutation carriers could be modified by expanded studies of high-magnitude sensitivities.
High-impact clinical trials and targeted therapies have historically resulted from observing high-magnitude numerical comparisons of drug sensitivities between genetically distinct, matched cell lines (the pharmacogenetic windows)1 in in vitro studies. Examples are provided by early studies of the drugs, imatinib (Gleevec; Novartis, Basel, Switzerland), for chronic myeloid leukemia, eliciting a 10× to 20× hypersensitivity in BCR-ABL cancer lines,2 and gefitinib (Iressa; AstraZeneca, London, UK), eliciting a 10× sensitivity in epidermal growth factor receptor–mutant cancers.3 These represented qualitative, not merely quantitative, differences.
Fanconi anemia (FA), a rare recessive disease characterized by life-threatening diverse clinical features including cancer susceptibility, is caused by biallelic or hemizygous mutations in any one of 16 FANC genes.4–6 The FANC pathway genes function together in a conserved manner to repair damaged DNA by homologous recombination.4 Heritable and somatic mutations inducing clinical cancer risk among FANC mutation carriers (heterozygotes) are most commonly seen in the FANC gene, BRCA2/FANCD1, leading to the development of BRCA2-deficient cancers. Commonly used chemotherapy drugs against these malignancies include the inter-strand cross-linking agents (ICLs), having pharmacogenetic windows of 10× to 15× for all FANC-null cancers,7,8 and poly (ADP-ribose) polymerase (PARP) inhibitors, having >>25× sensitivity in some FANC-null genotypes.9,10 These also represent qualitative differences. Although these drugs are used clinically, the epidemiological, evolutionary, and clinical implications of their high-magnitude pharmacogenetic windows are still unclear.
PARP enzymes are involved in base-excision repair, a key pathway in single-strand break repair.11 Inhibition of PARP induces synthetic lethality in homologous recombination–defective backgrounds or in cells lacking homologous recombination–related proteins.9,10,12 As a result, patients with tumor-specific BRCA1 or BRCA2 deficiencies have responded to PARP inhibition-based therapy.13 Mechanisms of intrinsic resistance to these therapies explain why some cancers with BRCA2 mutations do not respond to these therapies14–16 and why pharmacogenetic discrepancies are seen when using the same drug in different cell lines or in different model systems.
Because these drugs or agents are not naturally occurring, this raises questions regarding evolution and disease epidemiological characteristics, specifically, what pattern of injury may have driven the evolution of BRCA2 and other cancer-relevant FANC genes. It remains unclear whether all FANC-null cancer states are essentially similar in their pharmacogenetic windows (ie, the quantitative differences when compared with FANC-competent matched cells). Caution may be warranted, for individual differences could pose a significant clinical problem. Such differences might arise from secondary cellular compensatory mechanisms that surface when different genes are inactivated.
A possible clue to the evolutionary aspects of FANC-relevant cancer genes was suggested by the studies of the naturally occurring aldehydes, formaldehyde and acetaldehyde. Formaldehyde occurs naturally in human plasma at a concentration ranging from 13 to 97 μmol/L.17 When tested in two isogeneic FANC-null cancer cell lines developed by us (null for FANCC and FANCG), formaldehyde elicited a low-magnitude hypersensitivity.18 Formaldehyde also induced chromosomal aberrations in a human B-cell line and low-magnitude hypersensitivities in FANC-null avian DT40 cell lines.19
Acetaldehyde is mandatorily formed during ethanol oxidative degradation and is degraded by acetaldehyde dehydrogenase (ALDH2). The response to acetaldehyde has not been studied in human, or specifically in FANC-null, cancer cells or in cells null for the particular FANC genes most relevant to human inherited cancer risks (BRCA2/FANCD1 and PALB2/FANCN). As with formaldehyde, a modest quantitative hypersensitivity (fivefold or lower) of acetaldehyde was shown in chicken DT40 cells genetically deficient for some FANC genes.20 Although not measured as a numerical pharmacogenetic window, confirmation was provided when marrow failure was observed after acetaldehyde production induced by ethanol administration in an FANC-null, ALDH2-deficient mouse.21 Alcohol and its catabolic intermediate, acetaldehyde, have epidemiological links with increased incidence of certain cancer types, such as upper aerodigestive tract, breast, and pancreas, and also stomach, specifically in Asian men having an ALDH2 mutation.22–24 Ethanol consumption, mutations impairing alcohol oxidation, ALDH2 gene function associated with the Asian flushing syndrome on alcohol ingestion, and elevated acetaldehyde levels vary significantly across populations.
An opportunity, therefore, existed to explore human cancer cell lines having defined null states for FANC genes. Genes relevant to the FANC-deficient cancers, when occurring in FANC mutation carriers, include PALB2, BRCA2, FANCC, and FANCG. Herein, we generated a four-gene, cancer-null panel, including engineering PALB2-null matched cancer cells. We identified numerically robust, high-magnitude pharmacogenetic windows using multiple engineered clones null for BRCA2 and PALB2 genes. A single dose of mitomycin C (MMC) initiated rapid tumor regression in xenografts, a dramatic response reflecting in vivo the large in vitro pharmacogenetic differences. These models may also be useful for the rarer FANC genotypes, because it is unlikely that clinical trials could be performed for each genotype. The lessons from preclinical models of multiple FANC pathway genotypes could provide the wanted insights. Our studies reinforce a pathway-based strategy by comparing chemical hypersensitivities of matched syngeneic pairs of cell lines deficient in clinically relevant genes. The findings herein of qualitative, high-magnitude numerical distinctions suggest implications for the pathway evolution, disease epidemiological characteristics, and therapeutic strategies for patients.
Materials and Methods
Cell Lines and Cell Culture
DLD1 cells were obtained from ATCC (Manassas, VA) and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics at 37°C and 5% CO2. p53R cells, carrying a p53-binding site driving a luciferase reporter,25 were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% FBS, antibiotics, and 20 mmol/L HEPES. CAPAN1 cells were cultured in Iscove’s modified Dulbecco’s medium, supplemented with 10% FBS and antibiotics.
Targeted Disruption of PALB2 by Homologous Recombination
We disrupted the PALB2 gene according to the technique described.26 The targeting construct excised exon 8 of the PALB2 gene such that a frameshift and a stop codon were generated. We used a promoter-trap method in which the targeting construct contained the selection marker, neomycin.27 The construct consisted of two homology arms (HAs) flanking a central element, pSEPT, which contained a splice acceptor, an internal ribosomal entry sequence, coding sequences of the neomycin transferase gene, and a polyadenylation signal sequence. The pSEPT element was flanked by LoxP sites. The HAs were ligated to pAAV (Stratagene, Santa Clara, CA). The targeting construct was cotransfected with pRC and pHelper into HEK293 cells using Lipofectamine (Invitrogen, Grand Island, NY). Virus was harvested from the HEK293 cells after 48 hours, and a DLD1 clone (number 6, isolated by limiting dilution) was infected. Infected cells were distributed to 96-well plates by limiting dilution. Neomycin-resistant clones were screened after 3 weeks by PCR using a primer (Table 1) inside the selection cassette and a primer outside of the HAs to detect homologous integrations. The selection cassette was removed from clones by Cre-mediated excision. A heterozygous PALB2 clone (PALB2+/−), identified and confirmed by using primers (Table 1) on either side of the deleted region, was used when removing the second allele with the same exon 8–targeting construct. Clones having biallelic disruption of PALB2 were identified by PCR screening, as previously described. The neomycin cassette was again removed.26
Table 1.
Primer Sequences
| Primer name | Sequence | Use |
|---|---|---|
| LF | 5′-AACCTCCCCAGGCTCAGTAA-3′ | LHA forward-screening primer |
| LR | 5′-AAATCCTCCTCGTTTTTGGA-3′ | LHA reverse-screening primer |
| RF | 5′-CAGGTTCAGGGGGAGGTGTG-3′ | RHA forward-screening primer |
| RR | 5′-ATGTCTGGCTTCCACCTCACTAAC-3′ | RHA reverse-screening primer |
| CF | 5′-CTTTACACAGAGGTGCCCAAT-3′ | Forward primer to genotype PALB2-null cells |
| CR | 5′-CTCCCAGGTTCAAGCGACT-3′ | Reverse primer to genotype PALB2-null cells |
| RT-PCR | ||
| Forward | 5′-AGTGCCATGTTTTGGGAAAG-3′ | RT-PCR to confirm PALB2 exon 8 deletion |
| Reverse | 5′-TCCATCTTCTGCAAACGTCA-3′ | RT-PCR to confirm PALB2 exon 8 deletion |
LHA, left homology arm; RHA, right homology arm.
Plasmid Purification, RNA and Genomic DNA Isolation, PCR, RT-PCR, and Sequencing
Plasmid DNA was purified (Qiagen, Valencia, CA). Genomic DNA (crude) was released from cancer cells either by using the Lyse-N-Go PCR Reagent (Pierce Biotechnology, Rockford, IL) or by purification (Qiagen). For most PCRs, the touchdown method was used. RNA was isolated (RNeasy Mini Kit, Qiagen). cDNA was made from RNA (Superscript III; Invitrogen). PCR products were resolved on agarose gels using lithium boric acid electrophoresis (Faster Better Media LLC, Hunt Valley, MD).28 Automated sequencing was performed by Macrogen USA (Rockville, MD).
Protein Isolation and Western Blot Analysis
Cells were washed once in PBS and lysed by radioimmunoprecipitation assay buffer supplemented with protease inhibitor mixture (Catalog No. 1183617001; Roche Applied Science, Indianapolis, IN), followed by sonication. Protein concentrations were determined (DC protein assay; Bio-Rad, Philadelphia, PA). Samples were boiled with loading buffer and resolved by SDS-PAGE. Proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon; Millipore, Billerica, MA) and detected with anti-PALB2 antibody and horseradish peroxidase–linked anti-rabbit secondary antibody (both from Santa Cruz Biotechnology, Santa Cruz, CA). Membranes were developed using chemiluminiscence (Catalog No. WBKLS0500; Millipore).
Cell Cycle Analysis
A total of 1 × 106 cells were treated with MMC for 48 hours, washed once in PBS, fixed in 70% ethanol at −20°C for at least 30 minutes, washed again with PBS, and incubated in 0.2 mL of propidium iodide solution [0.1% Triton X-100, 1× PBS, 200 μg/mL RNase A (Sigma, St. Louis, MO), and 20 μg/mL propidium iodide (Sigma)] at 37°C for 1 hour. The cells were diluted with 1 mL PBS (Gibco, Grand Island, NY) for flow cytometry (FACScalibur; BD Biosciences, San Jose, CA). CellQuest Pro software version 5.2.1 (BD Biosciences) was used to interpret cell cycle profiles.
Treatment of Cells and Cell Quantitation
A total of 1000 and 2000 cells were plated per well for wild-type (WT) and null cultures, respectively. The cells were allowed to adhere and, after 24 hours, were exposed to various drugs: melphalan, MMC, cisplatin, etoposide, camptothecin (Sigma-Aldrich, St. Louis, MO), KU0058948 (synthesized as needed), and biological metabolites (formaldehyde, acetaldehyde, glyoxal solution, acrolein, butyraldehyde, crotonaldehyde, benzaldehyde, glutaraldehyde, phenylacetaldehyde, cinnamaldehyde, acetaldehyde dimethyl acetal, and aminoacetaldehyde dimethyl acetal; Sigma-Aldrich). After 6 days, the cells were washed and lysed in 40 μL 0.03% SDS, and 0.5% Picogreen (Molecular Probes, Grand Island, NY) was added. Fluorescence was measured, and the relative cell numbers were calculated, defining the untreated samples as 1. One or two independent replications of the experiments were performed per drug, with each graphed data point reflecting the average readings from six wells in a given representative experiment.
Synthesis of KU0058948
The PARP inhibitor, KU0058948, was synthesized after a reported protocol (WO 2004080976) in five steps. Briefly, 2-carboxy benzaldehyde was first converted into 3-oxo-1,3-dihydro-isobenzofuranyl phosphonate dimethyl ester, which was then treated with 2-fluoro-5-formyl benzonitrile to afford the 2-fluoro-5-(3-oxo-1,3-dihydro isobenzofuranylidene methyl) benzonitrile intermediate as a mixture of E/Z isomers. This mixture was then refluxed with NaOH, followed by the addition of hydrazine hydrate to replace the isobenzofuranyl ring with a dihydrophthazine moiety. The generated intermediate was then used for a tetramethyluronium hexafluorophosphate–mediated coupling with t-butyl homopiperazine carboxylate, which yielded the desired compound on acidic workup in an acceptable overall yield.
Chromosome Breakage Assay
BRCA2−/−, PALB2−/−1, and parental cells were treated with and without 0.1 μg/mL diepoxybutane (DEB), or with and without 1 mmol/L acetaldehyde [the inhibitory concentration of 50% (IC50) concentration range in BRCA2−/−, PALB2−/− lines] for 3 days, after which medium was changed. Cells were allowed to recover for 1 day, then treated with 1 μg/mL colcemid and harvested according to standard protocols. For each sample, approximately 30 metaphases (only 10 for acetaldehyde treated because of extensive damage) were analyzed by the cytogenetics facility at The Kennedy Krieger Institute (Baltimore, MD). In untreated parental DLD1 cells and derived clones, a significant minority (<40%) of cells were hyperdiploid.
Irradiation of Cells and Colony Formation Assay
Cells were plated, allowed to adhere, and exposed to 1.3 Gy/min 137Cs γ rays (Gammacell 40 Exactor; MDS Nordion, Inc., Ottawa, ON, Canada). Cells were subsequently incubated for 7 days or until colonies appeared, fixed, and stained (10% neutral-buffered formalin, 1:500 crystal violet). All macroscopically visible colonies were counted. Experiments were independently replicated. Each sample was tested at two different cell concentrations, each performed in duplicate. The number of cells plated was 10,000 and 100,000 for 0-, 2-, 4-, 6-, 8-, and 10-Gy exposures.
Estimation of Doubling Time of Cells
Cells were plated at a concentration of 2.5 × 104 cells in 30 25-cm2 flasks. Cells were washed with 1× PBS, followed by trypsinization. Cells were disaggregated in 1 mL PBS and counted by hemocytometer. At each time point, two independent samples were counted. Counting was done every 6 hours, from 0 to 90 hours (total of 15 time points). The proliferation curves were plotted, and the doubling times of the parental and PALB2-null cells were estimated.
p53-Luciferase Reporter Assays
p53 Responses were assessed by the p53R luciferase reporter assay25 using Steady-Glo (Promega, Madison, WI), according to the manufacturer’s protocol. Briefly, FANC-competent cells were plated in triplicate, incubated with acetaldehyde (Catalog No. W200344; Sigma-Aldrich) for 18 hours, and then lysed for a luciferase assay. We used the PerkinElmer (Waltham, MA) Microbeta Trilux plate reader to measure luminescence.
Xenograft Models
Six cell lines (1 × 106 cells per injection for DLD1 parental, PALB2+/−, PALB2−/−1, PALB2−/−2, and BRCA2−/− and 1.5 × 106 cells per injection for CAPAN1) were injected s.c. into the two flanks of female athymic nude mice, aged 4 to 5 weeks (Charles River Laboratories, Frederick, MD) with RPMI 1640 medium and Matrigel (BD Biosciences) [1 to 1.5 × 106 cells in 100 μL of RPMI 1640 medium (Gibco) and Matrigel; 1:1]. Twelve mice were inoculated for each cell line. Tumors were treated when each reached a size of 150 to 200 mm3, defined as day 0; four received no treatment. One untreated tumor of each group was harvested on day 1; eight were treated with MMC, of which one each was harvested on days 1, 4, and 7. The drug was administered once i.p. at 5 mg/kg on day 0. The length and width of tumors were measured with a caliper. Tumor volume was calculated as follows: Tumor Volume = 1/2 × (Length × Width2).
Care of animals was in accord with institutional guidelines.
Histopathological Examination
Harvested tumor tissue was fixed in buffered formalin until it was processed for histopathological characteristics.
Statistical Analysis
The mean (x), SD, and SEM were computed for replicated experiments. The 90% CIs of the fold difference (pharmacogenetic window) between parental and null cells for three experimental replicates of the drug sensitivity assays were calculated using the following formula:
| (1) |
Results
Targeted Disruption of PALB2
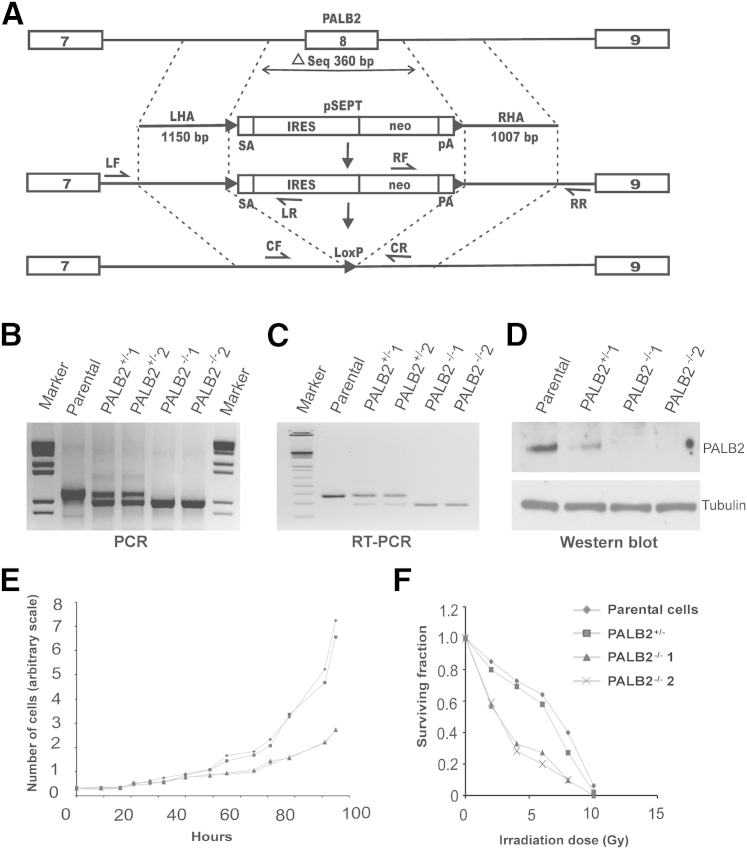
We disrupted the PALB2 gene in DLD1 cells by serially deleting exon 8 in both alleles (Figure 1A). We limited our attempts of generating PALB2-null cells to DLD1 parental cells (clone 6) only, based on prior experience29,30 documenting a failure to isolate BRCA2-null clones in four other tested parental lines. We screened 80 neomycin-resistant clones after targeting the first allele and obtained six PALB2+/− clones. After targeting the second allele and screening 270 neomycin-resistant recombinants, we obtained two independent PALB2−/− clones, termed PALB2−/−1 and PALB2−/−2. PCR (Figure 1B), RT-PCR (Figure 1C), and Western blot analyses (Figure 1D) confirmed the hemizygous and homozygous gene disruptions. Of the 270 screened clones, we identified 22 that had integration in the PALB2 locus, of which 20 had re-integrated the construct into the already inactivated allele. The ratio of clones (10:1) integrated into the disrupted allele to the ones integrated into the WT allele suggested that PALB2, such as BRCA2, was also a lethal gene, or had loss of fitness, when null (Supplemental Table S1).30
Figure 1.
Structural and functional evidence of PALB2 gene disruption. A: Targeting scheme. Primer pairs CF + CR were used to confirm deletion of exon 8 in both alleles. IRES, internal ribosomal entry sequence; LF + LR and RF + RR, left and right homology arm screening primers; LHA and RHA, left and right homology arms; neo, coding sequence of neomycin transferase; pA, polyadenylation signal sequence; solitary numbers label exons; SA, splice acceptor. All primer sequences are provided in Table 1. B: PCR detected the WT and deleted alleles using genomic DNA as a template. C: RT-PCR detected the WT and truncated mRNA transcripts. D: Immunoblots detected the presence or absence of PALB2 protein. E: Proliferation curves. The population growth of parental, hemizygous, and PALB2-null cells was compared. F: Colony-formation assays after ionizing radiation. Each point represented the average of duplicate measurements for each cell line in a representative experiment (of two experiments).
Functional Validation of PALB2−/− Cells
The two PALB2-null clones proliferated slowly compared with the PALB2-hemizygous and parental cells. The doubling time was estimated at 27 to 28 hours for null clones and 19 to 20 hours for parental and hemizygous clones. The slow proliferation (Figure 1E) of the null cells was a stable feature over successive passages. We assessed the effect of PALB2 deficiency on cell survival after 137Cs γ-irradiation using colony-formation assays. We observed a twofold to threefold decrease in the survival of irradiated PALB2-null cells compared with the parental line (Figure 1F). On treatment with the cross-linker MMC for 48 hours, the PALB2-deficient cells became arrested at the G2/M phase of the cell division cycle. By using flow cytometry, we determined the fractions of cells at the different stages of the cell cycle 48 hours after treatment with MMC (Supplemental Figure S1). A pronounced G2/M arrest (>40% cells) was observed in PALB2−/− cells at the lower concentration of 20 nmol/L, similar to the effect on parental cells at 200 nmol/L (ie, a pharmacogenetic window of 10× in this assay).
Cell Proliferation on Drug Treatment
The role of PALB2 in DNA damage-response and in replication fork maintenance is indicated by the hypersensitivity of the PALB2-deficient cells to DNA cross-linking agents. We estimated cell survival after a 6-day exposure of the parental, heterozygous, and null cells to various relevant drugs. PALB2−/− cells had increased sensitivity to the ICL agents melphalan (20×), MMC (17×), and cisplatin (15×) (Supplemental Figure S2), and were hypersensitive to the tested topoisomerase inhibitors etoposide (8×) and camptothecin (6×) (Supplemental Figure S2). Three replicate experiments were done for MMC treatment, and the 90% CIs of the pharmacogenetic window between the parental and null cells were found to be 13× to 22× for PALB2−/−1 cells and 19× to 27× for BRCA2−/− cells. Naturally derived BRCA2-null CAPAN1 cells31 had intermediate hypersensitivities (Supplemental Figure S3), consistent with the lower drug sensitivities previously noted for these cells.32
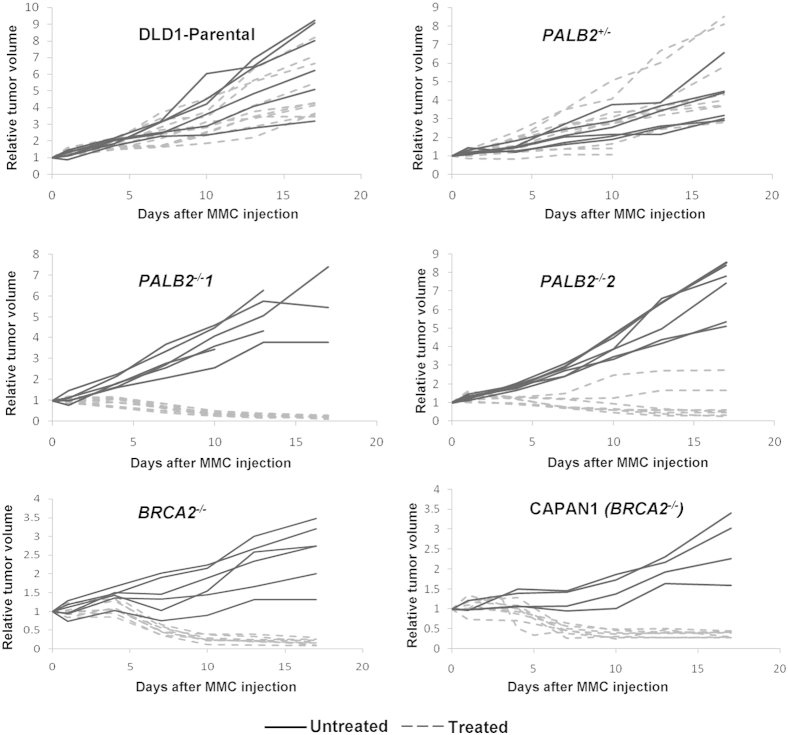
In Vivo Confirmation of Therapeutic Validity and Comparability of FANC Defects
Xenograft models using PALB2−/− and BRCA2−/− cancer cell lines were explored because these distal pathway genes command the greatest therapeutic and clinical interest. Mice were treated with a single dose of 5 mg/kg MMC. The DLD1 parental cell line and PALB2+/− did not respond to MMC treatment (Figure 2). In contrast, PALB2−/− cell lines showed sustained tumor regression through days 18 to 20 (Figure 2). The xenograft responses to MMC mirrored the in vitro hypersensitivity patterns. We also tested two BRCA2-null cell lines, CAPAN1 (a naturally occurring BRCA2-mutated cell line) and an engineered cell line, BRCA2−/−. These cell lines also had sustained tumor regression on MMC treatment (Figure 2). This confirmed that these cell lines had the characteristic phenotype previously reported for other FANC-null cancer cells.33
Figure 2.
Tumor regression in FA-deficient xenografts after MMC treatment. In vivo treatment of xenografts with a single dose of MMC. PALB2- and BRCA2-deficient xenografts (as indicated), parental xenografts, and PALB2-hemizygous xenografts were established and grown to an initial volume of 150 to 200 mm3 before initiation of treatment. Final xenograft volume was expressed as relative to initial volume. Mice were treated with a single 5 mg/kg i.p. dose of MMC.
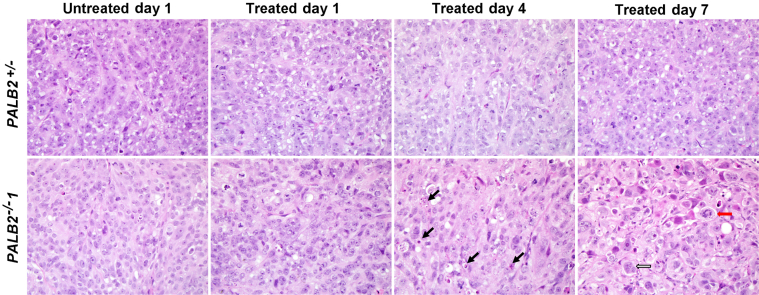
Histological sections of xenografted tissues from untreated mice and mice 1, 4, and 7 days after treatment with mitomycin C were reviewed for the extent of mitotic figures, apoptosis, confluent necrosis, hemorrhage, and fibrosis. In all xenografts, tissues at day 1 did not have appreciable differences from that seen in untreated tumors. However, by day 4, all xenografts null for PALB2 (PALB2−/−1 and PALB2−/−2) and BRCA2 (CAPAN1 and BRCA2−/−) had morphological changes characterized by multiple apoptotic bodies, cellular enlargement lacking an increase in nuclear/cytoplasmic ratio, prominent nucleoli, multinucleation, and enlarged eosinophilic cells containing chromosomes in disarray (Figure 3). These changes were multifocal within a given xenograft at day 4 and extensive at day 7 after treatment. Fibrosis and confluent necrosis were not typical in these xenografts, although CAPAN1 cells had a hyalinized and inflamed stroma by day 7. By contrast, no morphological changes were observed in tumors of the FANC-WT cell line, DLD1, or the heterozygous PALB2-mutant line, PALB2+/− (Figure 3).
Figure 3.
Tumor histological characteristics. Morphological features of pancreatic cancer xenografts treated with MMC. Representative photomicrographs of H&E-stained histological sections of untreated xenografts and xenografts treated at 1, 4, and 7 days were obtained. A PALB2+/− cell line had a heterozygous PALB2 mutation, and PALB2−/−1 had a homozygous PALB2 deletion. Black arrows indicate apoptotic bodies/debris; red arrow, a ballooned cell with disorganized chromosomes; white arrow, a multinucleated cell in PALB2−/−1. By contrast, no morphological changes were seen in treated PALB2+/− tumors.
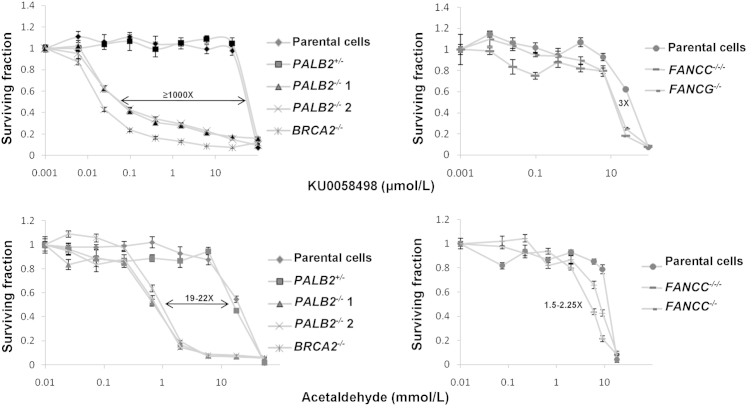
High-Magnitude Hypersensitivities to PARP Inhibition and Acetaldehyde
Both BRCA2−/− and PALB2−/− cell lines had extreme hypersensitivities (≥1000×) to the positive control condition, PARP inhibition (KU0058948) (Figure 4). They also had high-magnitude, 19× to 22× hypersensitivities to acetaldehyde (Figure 4), but no differential responses with formaldehyde or glyoxal solution (Table 2). Three replicate experiments were done with KU0058948 and acetaldehyde, and the 90% CIs of the pharmacogenetic window between the parental and null cells using KU0058948 were found to be 831× to 1057× for PALB2−/−1 cells and 1066× to 1199× for BRCA2−/− cells. The 90% CIs with acetaldehyde treatment were 15× to 29× for PALB2−/−1 cells and 15× to 23× for BRCA2−/− cells. We used p53R cells to explore whether acetaldehyde produced DNA strand breaks in cultured FANC-WT cells. Our results indicate that acetaldehyde induced a weak p53 response in p53R cells, inferring no major toxicity in FANC-competent cells through initial DNA strand breaks. Both BRCA2−/− and PALB2−/− genotypes were tested using several other aldehydes (a two- to nine-carbon series) (Table 2), including acrolein, butyraldehyde, crotonaldehyde, benzaldehyde, glutaraldehyde, phenylacetaldehyde, cinnamaldehyde, acetaldehyde dimethyl acetal, and aminoacetaldehyde dimethyl acetal. We explored structure-activity relationships centered around acetaldehyde. A series of aliphatic and aromatic aldehydes (one to nine carbons) were examined in the hope that a higher molecular weight might lead to a less volatile lead candidate with a similar biological response. Because aldehydes are also well known to be unstable and yet exceptional cross-linking agents, we also tried to replace the active aldehyde moiety with a protected aldehyde synthon (acetaldehyde dimethyl acetal and aminoacetaldehyde dimethyl acetal) capable of generating the active functionality on degradation. None of these treatments elicited differential responses between WT and null cells (Table 2). They were either uniformly toxic or benign to all cell types. We tested the sensitivities of the proximal genes, FANCC and FANCG, with the PARP inhibitor, KU0058948, and with the biological metabolites, acetaldehyde and formaldehyde; there was a low-magnitude 3× difference with PARP inhibition, and 2× and 3× differences with acetaldehyde (Figure 4) and formaldehyde (Table 2), respectively.
Figure 4.
Chemical sensitivities among an FANC-deficient cancer cell panel using compounds eliciting divergences among FANC-null models. Cell populations after treatment with PARP I inhibitor, KU0058948, and the biological metabolite, acetaldehyde, at indicated concentrations compared with untreated cells. The IC50 ratios (pharmacogenetic windows) comparing parental with FANC-deficient cells were determined from an average of three independent experiments; they are displayed within graphs of representative experiments. Error bars indicate SEM of six replicate wells in a representative experiment.
Table 2.
Relative Chemical Sensitivities of FANC-Null Genotypes
| Treatment | Class | Pharmacogenetic windows (IC50 of null cells)∗ |
|||
|---|---|---|---|---|---|
| PALB2−/− | BRCA2−/− | FANCC−/−/− | FANCG−/− | ||
| Melphalan | Cross-linker | 20× (0.25 μmol/L) | 25׆ | 14׆ | 14׆ |
| Mitomycin C | Cross-linker | 17× (10 nmol/L) | 23׆ | 13׆ | 12׆ |
| Cisplatin | Cross-linker | 15× (0.6–0.7 μmol/L) | 16׆ | 9׆ | 7׆ |
| KU0058948 | PARP inhibitor | 1000× (0.075 μmol/L) | 1133× (0.022 μmol/L) | 3× | 3× (17 μmol/L) |
| Etoposide | Topoisomerase II inhibitor | 8× (50–60 nmol/L) | 10׆ | — † | — † |
| Camptothecin | Topoisomerase I inhibitor | 6× (0.625 nmol/L) | 6׆ | ND | ND |
| Formaldehyde | 1-C aldehyde | — | — | 2.8× (2 μmol/L) | 2.8× (2 μmol/L) |
| Acetaldehyde | 2-C aldehyde | 22× (0.7 mmol/L) | 19× (∼1 mmol/L) | — | 2.4× (5.3 mmol/L) |
| Other tested aldehydes‡ | 2-9C aldehydes | — | — | ND | ND |
ND, not done; —, less than twofold window.
Windows were defined as the ratio of the IC50 values of parental cells and gene-knockout cells; data are from the graphical results of Figure 4 and Supplemental Figure S2.
Data from our prior reports.8,29
See Results.
Chromosomal Breakage and Instability
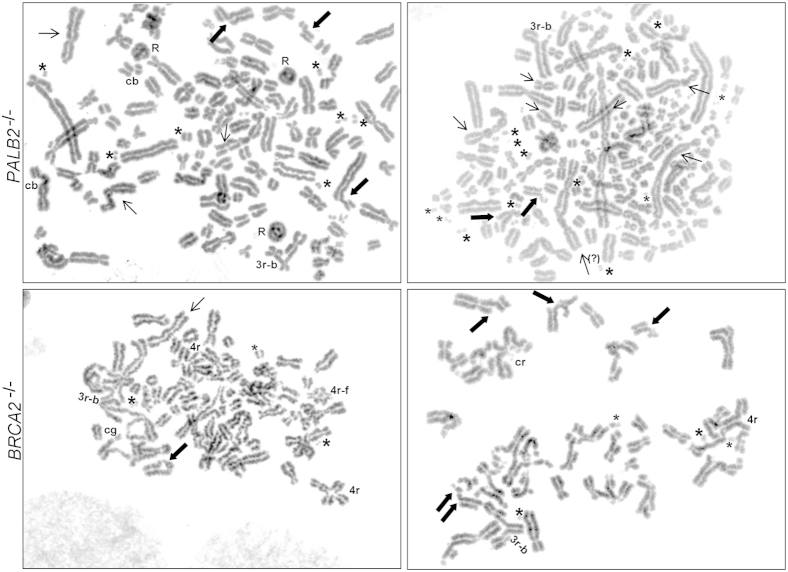
BRCA2−/− and PALB2−/− cells displayed increased chromosomal instability, which was further enhanced after treatment with DEB and acetaldehyde. Treatment-induced breakage serves as a conventional diagnostic test for the FA syndrome. The number of breaks/gaps scored per cell was 0.1, 0.5, and 1.3, respectively, for the untreated parental, BRCA2−/−, and PALB2−/−1 cells. The number of breaks and gaps per cell scored on DEB treatment were 0.03, 5.2, and 4.2, respectively, for the parental, BRCA2−/−, and PALB2−/−1 cells (Supplemental Figures S4 and S5, A and B). For acetaldehyde-treated cells, there were no breaks in parental cells but many breaks in the BRCA2−/− and PALB2−/−1 cells (Figure 5). Numerous fragments were seen in these cell lines, along with rings, broken or intact triradials, quadriradials, and other complex radials, indicating breakage and rejoining of chromatids and/or chromosomes (Figure 5). BRCA2−/− and PALB2−/− cells, therefore, displayed much higher rates of induced chromosomal aberrations than parental cells.
Figure 5.
Metaphase chromosomes after acetaldehyde exposure. Acetaldehyde exposure to PALB2−/− and BRCA2−/− cells promoted widespread chromosomal aberrations. Thick arrows indicate chromatid breaks; thin arrows, dicentric chromosomes; asterisks, fragments. cb, chromosome breaks; cg, chromosome gaps; R, rings; 3r, triradials; 3r-b, broken triradials; 4r, quadriradials; 4r-f, quadriradials with fragments.
Discussion
Multigene Cancer Panels for Preclinical Studies Provide Novelty and Cancer Modeling
Our multigene panel consisting of engineered FANCC-, FANCG-, BRCA2/FANCD1-, and PALB2/FANCN-null human cancer cell lines had the shared phenotypes typical of FANC pathway defects, including hypersensitivities to the ICL agents and γ-radiation in vitro, and comparable tumor regression in xenograft models when treated with a single dose of MMC. The same panel had divergences of hypersensitivities for some chemicals, however, including the epidemiologically important ethanol metabolite, acetaldehyde, the PARP inhibitor, KU0058948, and the topoisomerase II inhibitor, etoposide. High-magnitude pharmacogenetic windows were observed. These were distinguished from prior work in avian and noncancer mammalian cells, which had low-magnitude (<5×) changes.19,20,34 A panel of cancer cells, also novel in being tumorigenic, enables chemotherapeutic animal trials of pharmacogenetically targeted agents and could aid preclinical therapeutic explorations.
FANC-Null Status Is Insufficient to Convey Every FANC-Related Hypersensitivity to Cancer Cells
The large numerical discrepancies found in the current work clearly challenge a loosely held, but conventional, dogma that FANC-null cells might largely share a set of common chemical hypersensitivities used to make some clinical decisions. Our results advance the idea that knowledge of divergent phenotypes may be useful to dissect differing functions in proximal and distal FANC genes and in anticipating rational treatment of patients with differing FANC genotypes. This idea was promoted by Gallmeier and Kern32 using a natural cell line, supported by Hucl et al29 using experimental cancer lines, and with divergences of lower magnitude, echoed in prior studies of noncancer cells.34 Our high-magnitude divergences of hypersensitivities seen for acetaldehyde, a PARP inhibitor, and etoposide may be gene-dependent, affected by clonal variation or by the choice of the tumor cells under study. Notably, the same considerations apply to real human tumors in a setting of personalized therapy. Thus, panel-based strategies comparing the sensitivities of matched, engineered genotypes with differing chemical probes suggest insights for genetic epidemiology, prevention strategies, and exploration of novel therapeutic options because of the discrepant hypersensitivity patterns being unambiguous, emerging in multiple knockout clones, and remaining stable in these models. Our work supported concerns that the mere knowledge of a patient’s FANC genotype may be insufficient to predict the success of a broad FANC-targeted, personalized cancer therapy.
Comparison of Formaldehyde with Acetaldehyde in Human Cancers
Both formaldehyde and acetaldehyde are natural human metabolites known to damage DNA and to elicit hypersensitive responses in FANC-null cells. In our model, the formaldehyde pharmacogenetic window was 2× to 3×, whereas that of acetaldehyde could be a log larger, at 19× to 22×. These pharmacogenetic windows implied that the BRCA2 and PALB2 genes in our syngeneic matched cell line pairs protected against one third to one half of the toxicity caused by formaldehyde, but near 96% of the toxicity caused by acetaldehyde. This suggests that acetaldehyde may have had the larger role in driving the evolutionary selection operating on the BRCA2 and PALB2 genes. Intriguingly, studies of inherited syndromes and cancer epidemiological features imply that the FANC gene system may be adequately optimized to protect certain organs, such as the ductal epithelia of pancreas and breast, oral cavity, and upper aerodigestive tract (among others), whose risk of cancer is elevated by both ethanol intake and FANC gene mutations. The consequences of acetaldehyde damage might unite those two types of observation.
Acetaldehyde-Induced DNA Damage and Potential Therapeutic Options
Acetaldehyde, an intermediate metabolite of ethanol breakdown, is carcinogenic.35 Genotoxicity was also reported when cells from an FA patient were exposed to acetaldehyde, and chromosomal aberrations appeared under conditions in which cells of a healthy person experienced little effect.36 The best-studied DNA adduct from acetaldehyde is N2-ethyl-2′-deoxyguanosine,37 which is increased in liver DNA obtained from ethanol-treated rodents and in white blood cells from human alcohol abusers. The carcinogenic potential of this product is unclear. A different adduct, 1,N2-propano-2′-deoxyguanosine, formed from acetaldehyde in the presence of histones and other basic molecules,38 may be responsible for some mutagenic and genotoxic effects of acetaldehyde.
Prior literature suggested that acetaldehyde produced ICLs39 and implicated a role of the FANC DNA-damage response network in protecting cells against these cross-links. Our finding of widely differing pharmacogenetic windows (19× to 22× in cells null for some FA genes and 2× for others) may dampen support for the ICL theory somewhat as a major mechanism of action of acetaldehyde, given that all FANC defects in our panel had similar levels of hypersensitivity when tested using other ICL agents. Nonetheless, our cytogenetic observations support a mechanism producing widespread chromosomal breakage in BRCA2−/− and PALB2−/− cancer cells exposed to acetaldehyde.
The IC50 for acetaldehyde in our PALB2- and BRCA2-null cells was ≥700 μmol/L. This concentration is not physiological in untreated humans. Data from human volunteers, however, indicate that local salivary acetaldehyde concentrations could reach 450 μmol/L at high blood alcohol concentrations, far higher than the genotoxic threshold.40 It is plausible that ethanol, acetaldehyde, or ethanol-disulfirum administration could generate acetaldehyde levels sufficient for producing differential effects on FANC-null cells arising in carriers of FANC-gene mutations, long-term effects that could occur far lower than the IC50 concentration.
Implications for Cancer Epidemiology and Prevention
Cancer risks from ethanol could be stratified better in epidemiological studies by determining the BRCA2 and PALB2 mutational gene status of individuals in studied populations. Cancer risks of BRCA2 and PALB2 carriers could be further stratified by metrics gauging the exposure to acetaldehyde and ethanol and by examining gene polymorphisms of the ADH and ALDH2 genes. The polymorphisms govern the tissue concentrations of acetaldehyde and, indirectly, the tendencies to ingest alcohol.
Potential interventions to reduce cancer mortality in the carrier population can be discussed. FA patients and carriers might experience increased risks of cancer on consumption of alcohol or foods containing high levels of acetaldehyde. Reports in the literature suggest the use of aldehyde antagonists or aldehyde dehydrogenase agonists.41–44
Chemical prophylaxis or even aldehyde-based chemotherapy for FANC-null neoplasms might, in theory, pose an alternative at least as attractive as the current use of prophylactic surgery in BRCA2 mutation carriers. Given the levels of acetaldehyde achieved in humans in voluntary and anecdotal settings,45,46 one could speculate possibly treating PALB2- and BRCA2-deficient precursor neoplasms or cancers with either local infusion of acetaldehyde or systemic ethanol/disulfirum. Conversely, long-term exposure to acetaldehyde should also selectively harm FA patients or FANC-mutation carriers.
Clinical Importance of Large-Magnitude Pharmacogenetic Windows
Prior studies documenting hypersensitivities of BRCA2-null states to a positive control condition (PARP inhibition) were comparable to our results. By using revertant mutants of the BRCA2 gene, the hypersensitivity of the naturally occurring CAPAN1 cancer line to PARP inhibition was 1300×, and to the cross-linker cisplatin, 12×.15 In CHO V8 BRCA2-null cells, approximately 1000× sensitivity from PARP inhibition was observed.9 Mouse ES cells deficient for BRCA2 had a 100× sensitivity to PARP inhibition.9
Chemical hypersensitivities of PALB2-deficient states were also reported. However, gene-knockdown models had underestimated the large magnitudes of hypersensitivities seen in true genetically null cells.47–54
Experimental tumor regression is a dramatic demonstration of high-magnitude pharmacogenetic windows. Regression was documented in naturally occurring FANCC-, BRCA2-, and PALB2-deficient cancer xenografts after single-dose ICL treatments.33,55 In our study, the engineered, syngeneic human PALB2-null cancer model produced a 16× hypersensitivity to MMC, an approximately 1000× hypersensitivity to PARP inhibition in the null cells in vitro, and strong tumor regression in vivo after a single dose of MMC.
To provide perspective for such numbers, the mutation-targeting drugs, gefitinib (Iressa) and imatinib (Gleevec), were shown to have wide pharmacogenetic windows (10× to 20×) in in vitro studies of the relevant matched cells.2,3 Large divergences of IC50 values, produced by different chemicals among FANC-null states, in our cellular models may likewise aid advances in the epidemiology, prevention, and therapy in the settings of FANC mutations.
Acknowledgments
We thank Dr. Nita Ahuja for discussions and advice and Dr. Denise Batista and her staff at The Kennedy Krieger Cytogenetics facility for the chromosomal breakage studies.
Footnotes
Supported by NIH grants CA62924 and CA123483 and The Everett and Marjorie Kovler Professorship in Pancreas Cancer Research (all to S.E.K.). The breakage studies were supported by The Kennedy Krieger Intellectual & Developmental Disabilities Research Center grant 5P30 HD024061-22 (Michael Cataldo, Director, Department of Behavioral Psychology, Kennedy Krieger Institute, Baltimore, MD).
Supplemental Data
Cell cycle profiles 48 hours after MMC treatment. PALB2-deficient cells become arrested at the G2/M region of the cell cycle profile.
Functional validation of PALB2-deficient cells by drug sensitivity. Cell population after treatment with ICL agents (melphalan, MMC, and cisplatin), a topoisomerase II inhibitor (etoposide), and a topoisomerase I inhibitor (camptothecin) at the indicated concentrations compared with untreated cells. The pharmacogenetic windows comparing parental with PALB2-deficient cells were averaged from three independent experiments, and are displayed on representative experiments. BRCA2-null cells served as a positive control. Error bars indicate SEM of six replicate wells in a single experiment for each drug.
Cell survival in naturally derived BRCA2-null CAPAN 1 cells. Cell survival after treatment of CAPAN1 cells (bold dotted lines) with MMC, KU0058498, and acetaldehyde at the indicated concentrations compared with other cell lines tested (no matched control cells for CAPAN1). Error bars indicate SEM of six replicate wells in a single experiment for each drug.
Metaphase chromosomes in PALB2-null cells after diepoxybutane exposure.
A and B: Metaphase chromosomes in BRCA2-null cells after diepoxybutane exposure.
Supplemental Data
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2013.09.023.
References
- 1.Hucl T., Gallmeier E., Kern S.E. Distinguishing rational from irrational applications of pharmacogenetic synergies from the bench to clinical trials. Cell Cycle. 2007;6:1336–1341. doi: 10.4161/cc.6.11.4359. [DOI] [PubMed] [Google Scholar]
- 2.Deininger M.W., Goldman J.M., Lydon N., Melo J.V. The tyrosine kinase inhibitor CGP57148B selectively inhibits the growth of BCR-ABL-positive cells. Blood. 1997;90:3691–3698. [PubMed] [Google Scholar]
- 3.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., Naoki K., Sasaki H., Fujii Y., Eck M.J., Sellers W.R., Johnson B.E., Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Kim H., D’Andrea A.D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012;26:1393–1408. doi: 10.1101/gad.195248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuster B., Knies K., Stoepker C., Velleuer E., Friedl R., Gottwald-Muhlhauser B., de Winter J.P., Schindler D. Whole exome sequencing reveals uncommon mutations in the recently identified Fanconi anemia gene SLX4/FANCP. Hum Mutat. 2013;34:93–96. doi: 10.1002/humu.22221. [DOI] [PubMed] [Google Scholar]
- 6.Bogliolo M., Schuster B., Stoepker C., Derkunt B., Su Y., Raams A., Trujillo J.P., Minguillón J., Ramírez M.J., Pujol R., Casado J.A., Banos R., Rio P., Knies K., Zúniga S., Benítez J., Bueren J.A., Jaspers N.G., Schärer O.D., de Winter J.P., Schindler D., Surrallés J. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92:800–806. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Heijden M.S., Brody J.R., Gallmeier E., Cunningham S.C., Dezentje D.A., Shen D., Hruban R.H., Kern S.E. Functional defects in the fanconi anemia pathway in pancreatic cancer cells. Am J Pathol. 2004;165:651–657. doi: 10.1016/S0002-9440(10)63329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallmeier E., Calhoun E.S., Rago C., Brody J.R., Cunningham S.C., Hucl T., Gorospe M., Kohli M., Lengauer C., Kern S.E. Targeted disruption of FANCC and FANCG in human cancer provides a preclinical model for specific therapeutic options. Gastroenterology. 2006;130:2145–2154. doi: 10.1053/j.gastro.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., Martin N.M., Jackson S.P., Smith G.C., Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 10.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 11.Wooster R., Weber B.L. Breast and ovarian cancer. N Engl J Med. 2003;348:2339–2347. doi: 10.1056/NEJMra012284. [DOI] [PubMed] [Google Scholar]
- 12.McCabe N., Turner N.C., Lord C.J., Kluzek K., Bialkowska A., Swift S., Giavara S., O’Connor M.J., Tutt A.N., Zdzienicka M.Z., Smith G.C., Ashworth A. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 2006;66:8109–8115. doi: 10.1158/0008-5472.CAN-06-0140. [DOI] [PubMed] [Google Scholar]
- 13.Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J., Ashworth A., Carmichael J., Kaye S.B., Schellens J.H., de Bono J.S. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 14.Rottenberg S., Jaspers J.E., Kersbergen A., van der Burg E., Nygren A.O., Zander S.A., Derksen P.W., de Bruin M., Zevenhoven J., Lau A., Boulter R., Cranston A., O’Connor M.J., Martin N.M., Borst P., Jonkers J. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105:17079–17084. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edwards S.L., Brough R., Lord C.J., Natrajan R., Vatcheva R., Levine D.A., Boyd J., Reis-Filho J.S., Ashworth A. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451:1111–1115. doi: 10.1038/nature06548. [DOI] [PubMed] [Google Scholar]
- 16.Barber L.J., Sandhu S., Chen L., Campbell J., Kozarewa I., Fenwick K., Assiotis I., Rodrigues D.N., Filho J.S., Moreno V., Mateo J., Molife L.R., De Bono J., Kaye S., Lord C.J., Ashworth A. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol. 2013;229:422–429. doi: 10.1002/path.4140. [DOI] [PubMed] [Google Scholar]
- 17.Heck H., Casanova M. The implausibility of leukemia induction by formaldehyde: a critical review of the biological evidence on distant-site toxicity. Regul Toxicol Pharmacol. 2004;40:92–106. doi: 10.1016/j.yrtph.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 18.Ridpath J.R., Nakamura A., Tano K., Luke A.M., Sonoda E., Arakawa H., Buerstedde J.M., Gillespie D.A., Sale J.E., Yamazoe M., Bishop D.K., Takata M., Takeda S., Watanabe M., Swenberg J.A., Nakamura J. Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 2007;67:11117–11122. doi: 10.1158/0008-5472.CAN-07-3028. [DOI] [PubMed] [Google Scholar]
- 19.Rosado I.V., Langevin F., Crossan G.P., Takata M., Patel K.J. Formaldehyde catabolism is essential in cells deficient for the Fanconi anemia DNA-repair pathway. Nat Struct Mol Biol. 2011;18:1432–1434. doi: 10.1038/nsmb.2173. [DOI] [PubMed] [Google Scholar]
- 20.Langevin F., Crossan G.P., Rosado I.V., Arends M.J., Patel K.J. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 21.Garaycoechea J.I., Crossan G.P., Langevin F., Daly M., Arends M.J., Patel K.J. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 22.Gapstur S.M., Jacobs E.J., Deka A., McCullough M.L., Patel A.V., Thun M.J. Association of alcohol intake with pancreatic cancer mortality in never smokers. Arch Intern Med. 2011;171:444–451. doi: 10.1001/archinternmed.2010.536. [DOI] [PubMed] [Google Scholar]
- 23.Longnecker M.P., Berlin J.A., Orza M.J., Chalmers T.C. A meta-analysis of alcohol consumption in relation to risk of breast cancer. JAMA. 1988;260:652–656. [PubMed] [Google Scholar]
- 24.Yokoyama A., Muramatsu T., Ohmori T., Yokoyama T., Okuyama K., Takahashi H., Hasegawa Y., Higuchi S., Maruyama K., Shirakura K., Ishii H. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19:1383–1387. doi: 10.1093/carcin/19.8.1383. [DOI] [PubMed] [Google Scholar]
- 25.Sohn T.A., Bansal R., Su G.H., Murphy K.M., Kern S.E. High-throughput measurement of the Tp53 response to anticancer drugs and random compounds using a stably integrated Tp53-responsive luciferase reporter. Carcinogenesis. 2002;23:949–957. doi: 10.1093/carcin/23.6.949. [DOI] [PubMed] [Google Scholar]
- 26.Rago C., Vogelstein B., Bunz F. Genetic knockouts and knockins in human somatic cells. Nat Protoc. 2007;2:2734–2746. doi: 10.1038/nprot.2007.408. [DOI] [PubMed] [Google Scholar]
- 27.Sedivy J.M., Dutriaux A. Gene targeting and somatic cell genetics–a rebirth or a coming of age? Trends Genet. 1999;15:88–90. doi: 10.1016/s0168-9525(98)01689-8. [DOI] [PubMed] [Google Scholar]
- 28.Brody J.R., Kern S.E. History and principles of conductive media for standard DNA electrophoresis. Anal Biochem. 2004;333:1–13. doi: 10.1016/j.ab.2004.05.054. [DOI] [PubMed] [Google Scholar]
- 29.Hucl T., Rago C., Gallmeier E., Brody J.R., Gorospe M., Kern S.E. A syngeneic variance library for functional annotation of human variation: application to BRCA2. Cancer Res. 2008;68:5023–5030. doi: 10.1158/0008-5472.CAN-07-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallmeier E., Hucl T., Calhoun E.S., Cunningham S.C., Bunz F., Brody J.R., Kern S.E. Gene-specific selection against experimental fanconi anemia gene inactivation in human cancer. Cancer Biol Ther. 2007;6:654–660. doi: 10.4161/cbt.6.5.3978. [DOI] [PubMed] [Google Scholar]
- 31.Goggins M., Schutte M., Lu J., Moskaluk C.A., Weinstein C.L., Petersen G.M., Yeo C.J., Jackson C.E., Lynch H.T., Hruban R.H., Kern S.E. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 32.Gallmeier E., Kern S.E. Absence of specific cell killing of the BRCA2-deficient human cancer cell line CAPAN1 by poly(ADP-ribose) polymerase inhibition. Cancer Biol Ther. 2005;4:703–706. doi: 10.4161/cbt.4.7.1909. [DOI] [PubMed] [Google Scholar]
- 33.van der Heijden M.S., Brody J.R., Dezentje D.A., Gallmeier E., Cunningham S.C., Swartz M.J., DeMarzo A.M., Offerhaus G.J., Isacoff W.H., Hruban R.H., Kern S.E. In vivo therapeutic responses contingent on Fanconi anemia/BRCA2 status of the tumor. Clin Cancer Res. 2005;11:7508–7515. doi: 10.1158/1078-0432.CCR-05-1048. [DOI] [PubMed] [Google Scholar]
- 34.Murai J., Huang S.Y., Das B.B., Renaud A., Zhang Y., Doroshow J.H., Ji J., Takeda S., Pommier Y. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72:5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lachenmeier D.W., Kanteres F., Rehm J. Carcinogenicity of acetaldehyde in alcoholic beverages: risk assessment outside ethanol metabolism. Addiction. 2009;104:533–550. doi: 10.1111/j.1360-0443.2009.02516.x. [DOI] [PubMed] [Google Scholar]
- 36.Obe G., Natarajan A.T., Meyers M., Hertog A.D. Induction of chromosomal aberrations in peripheral lymphocytes of human blood in vitro, and of SCEs in bone-marrow cells of mice in vivo by ethanol and its metabolite acetaldehyde. Mutat Res. 1979;68:291–294. doi: 10.1016/0165-1218(79)90160-5. [DOI] [PubMed] [Google Scholar]
- 37.Vaca C.E., Fang J.L., Schweda E.K. Studies of the reaction of acetaldehyde with deoxynucleosides. Chem Biol Interact. 1995;98:51–67. doi: 10.1016/0009-2797(95)03632-v. [DOI] [PubMed] [Google Scholar]
- 38.Sako M., Inagaki S., Esaka Y., Deyashiki Y. Histones accelerate the cyclic 1,N2-propanoguanine adduct-formation of DNA by the primary metabolite of alcohol and carcinogenic crotonaldehyde. Bioorg Med Chem Lett. 2003;13:3497–3498. doi: 10.1016/s0960-894x(03)00800-x. [DOI] [PubMed] [Google Scholar]
- 39.Theruvathu J.A., Jaruga P., Nath R.G., Dizdaroglu M., Brooks P.J. Polyamines stimulate the formation of mutagenic 1,N2-propanodeoxyguanosine adducts from acetaldehyde. Nucleic Acids Res. 2005;33:3513–3520. doi: 10.1093/nar/gki661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Homann N., Jousimies-Somer H., Jokelainen K., Heine R., Salaspuro M. High acetaldehyde levels in saliva after ethanol consumption: methodological aspects and pathogenetic implications. Carcinogenesis. 1997;18:1739–1743. doi: 10.1093/carcin/18.9.1739. [DOI] [PubMed] [Google Scholar]
- 41.Perez-Miller S., Younus H., Vanam R., Chen C.H., Mochly-Rosen D., Hurley T.D. Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat Struct Mol Biol. 2010;17:159–164. doi: 10.1038/nsmb.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Juliano C., Cossu M., Rota M.T., Satta D., Poggi P., Giunchedi P. Buccal tablets containing cysteine and chlorhexidine for the reduction of acetaldehyde levels in the oral cavity. Drug Dev Ind Pharm. 2011;37:1192–1199. doi: 10.3109/03639045.2011.563783. [DOI] [PubMed] [Google Scholar]
- 43.Brooks P.J., Enoch M.A., Goldman D., Li T.K., Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 2009;6:e50. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashimoto K., Pinkas G., Evans L., Liu H., Al-Hasan Y., Thompson L.P. Protective effect of N-acetylcysteine on liver damage during chronic intrauterine hypoxia in fetal guinea pig. Reprod Sci. 2012;19:1001–1009. doi: 10.1177/1933719112440052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Asmussen E., Boje O. The effect of alcohol and some drugs on the capacity for work. Acta Physiol Scand. 1948;15:109–113. doi: 10.1111/j.1748-1716.1948.tb00488.x. [DOI] [PubMed] [Google Scholar]
- 46.Beyeler C., Fisch H.U., Preisig R. The disulfiram-alcohol reaction: factors determining and potential tests predicting severity. Alcohol Clin Exp Res. 1985;9:118–124. doi: 10.1111/j.1530-0277.1985.tb05531.x. [DOI] [PubMed] [Google Scholar]
- 47.Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., Livingston D.M. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 48.Xia B., Dorsman J.C., Ameziane N., de Vries Y., Rooimans M.A., Sheng Q., Pals G., Errami A., Gluckman E., Llera J., Wang W., Livingston D.M., Joenje H., de Winter J.P. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 49.Zhang F., Fan Q., Ren K., Andreassen P.R. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buisson R., Dion-Cote A.M., Coulombe Y., Launay H., Cai H., Stasiak A.Z., Stasiak A., Xia B., Masson J.Y. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bleuyard J.Y., Buisson R., Masson J.Y., Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13:135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hayakawa T., Zhang F., Hayakawa N., Ohtani Y., Shinmyozu K., Nakayama J., Andreassen P.R. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123:1124–1130. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reid S., Schindler D., Hanenberg H., Barker K., Hanks S., Kalb R., Neveling K., Kelly P., Seal S., Freund M., Wurm M., Batish S.D., Lach F.P., Yetgin S., Neitzel H., Ariffin H., Tischkowitz M., Mathew C.G., Auerbach A.D., Rahman N. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 54.Xiao H., Zhang K.J., Xia B. Defects of FA/BRCA pathway in lymphoma cell lines. Int J Hematol. 2008;88:543–550. doi: 10.1007/s12185-008-0199-8. [DOI] [PubMed] [Google Scholar]
- 55.Villarroel M.C., Rajeshkumar N.V., Garrido-Laguna I., De Jesus-Acosta A., Jones S., Maitra A., Hruban R.H., Eshleman J.R., Klein A., Laheru D., Donehower R., Hidalgo M. Personalizing cancer treatment in the age of global genomic analyses: pALB2 gene mutations and the response to DNA damaging agents in pancreatic cancer. Mol Cancer Ther. 2011;10:3–8. doi: 10.1158/1535-7163.MCT-10-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell cycle profiles 48 hours after MMC treatment. PALB2-deficient cells become arrested at the G2/M region of the cell cycle profile.
Functional validation of PALB2-deficient cells by drug sensitivity. Cell population after treatment with ICL agents (melphalan, MMC, and cisplatin), a topoisomerase II inhibitor (etoposide), and a topoisomerase I inhibitor (camptothecin) at the indicated concentrations compared with untreated cells. The pharmacogenetic windows comparing parental with PALB2-deficient cells were averaged from three independent experiments, and are displayed on representative experiments. BRCA2-null cells served as a positive control. Error bars indicate SEM of six replicate wells in a single experiment for each drug.
Cell survival in naturally derived BRCA2-null CAPAN 1 cells. Cell survival after treatment of CAPAN1 cells (bold dotted lines) with MMC, KU0058498, and acetaldehyde at the indicated concentrations compared with other cell lines tested (no matched control cells for CAPAN1). Error bars indicate SEM of six replicate wells in a single experiment for each drug.
Metaphase chromosomes in PALB2-null cells after diepoxybutane exposure.
A and B: Metaphase chromosomes in BRCA2-null cells after diepoxybutane exposure.