Abstract
Fragile X syndrome and associated disorders are characterized by the number of CGG repeats and methylation status of the FMR1 gene for which Southern blot (SB) historically has been required for analysis. This study describes a simple PCR-only workflow (mPCR) to replace SB analysis, that incorporates novel procedural controls, treatment of the DNA in separate control and methylation-sensitive restriction endonuclease reactions, amplification with labeled primers, and two-color amplicon sizing by capillary electrophoresis. mPCR was evaluated in two independent laboratories with 76 residual clinical samples that represented typical and challenging fragile X alleles in both males and females. mPCR enabled superior size resolution and analytical sensitivity for size and methylation mosaicism compared to SB. Full mutation mosaicism was detected down to 1% in a background of 99% normal allele with 50- to 100-fold less DNA than required for SB. A low level of full mutation mosaicism in one sample was detected using mPCR but not observed using SB. Overall, the sensitivity for detection of full mutation alleles was 100% (95% CI: 89%–100%) with an accuracy of 99% (95% CI: 93%–100%). mPCR analysis of DNA from individuals with Klinefelter and Turner syndromes, and DNA from sperm and blood, were consistent with SB. As such, mPCR enables accurate, sensitive, and standardized methods of FMR1 analysis that can harmonize results across different laboratories.
Diverse developmental, mental, and reproductive disorders are associated with both the number of cytosine-guanine-guanine (CGG) repeats and the methylation status of the fragile X mental retardation-1 (FMR1, NM_002024.4) gene.1–3 Excessive CGG repeat expansion is directly linked with hypermethylation of the gene through an epigenetic mechanism that is distinct from X chromosome inactivation and developmentally timed after lyonization.4 Because the FMR1 protein (FMRP) is a master regulator of genes involved in synaptic plasticity,5 the intellectual and behavioral consequences of quantitative FMR1 silencing are profound. Methylation of full mutation expansions (>200 CGG), however, can be incomplete, and less severe phenotypes may be associated with methylation mosaicism.6–8 In premutation alleles (55 to 200 CGG) the number of CGG can influence the risks and phenotype of fragile X–associated tremor/ataxia syndrome (FXTAS, OMIM 300623),9 fragile X–associated primary ovarian insufficiency (FXPOI, OMIM 300624),10,11 and autism spectrum disorders.12,13 Methylation status or X-inactivation in females may further influence the risk and phenotype of these conditions even if the results reported are still inconclusive.10,11,14 These premutation alleles are relatively common in the general population, occurring in 1 in 130 to 250 women and in 1 in 250 to 810 men, as reported in the United States,15,16 suggesting a broader need for FMR1 characterization in the general population. Differences in methylation status have also been reported between DNA from whole blood compared to skin fibroblasts, which may be closer in cellular origin to brain and more reflective of phenotype.17 Thus, it is critical to accurately and reliably assess the CGG repeat length and spectrum of methylation characteristics in individuals with FMR1 premutation and full mutation expansions, and to enable analysis of alternative sample types rather than peripheral blood.
Southern blot (SB) analysis is currently the gold standard method for determining size and methylation status in expanded FMR1 alleles. However, this procedure is severely limited by the amount of genomic DNA material that is required, a tedious workflow, and variable sensitivity. Disadvantages of SB include low resolution and the inability to accurately size premutation and normal alleles. Therefore, most clinical laboratories currently rely on a combination of PCR and SB analysis because of the technical limitations and the specific pitfalls of each method that, if used alone, could induce potential misinterpretation of the genotype.18,19
Various alternatives to SB analysis have been reported that use bisulfite or enzymatic pretreatment of DNA before PCR to obtain methylation status.20–25 These methods have been typically restricted to the analysis of male samples because of the inefficiency of PCR or confounding presence of two X chromosomes in females.24,25 Alternative methylation markers have been proposed but lack direct association with the number of CGG repeats.26 Concurrent assessment of CGG repeat length and of allele-specific methylation status in both males and females has been demonstrated.23 This methylation PCR (mPCR) method was based on the analysis of DNA treated with methylation-sensitive endonucleases before FMR1 gene-specific PCR.27 The results were concordant with SB analysis across a range of genotypes. However, a reference control was incorporated that overlapped with samples having alleles of 38 to 42 CGG, and the approach lacked additional controls that would benefit routine testing in a clinical laboratory environment.
Herein, we report the inclusion of novel procedural controls and a simplified workflow for mPCR that advance FMR1 analysis without the need for SB analysis. We compare results between methods using a range of challenging clinical samples obtained from two European laboratories. We demonstrate concordance and improved detection of methylation and size mosaicism relative to SB analysis. The ability to analyze novel sample types and samples with aneuploidy that might be encountered during standard fragile X testing is presented. Consequently, this report provides the first interlaboratory validation of a PCR-based FMR1 assay that can accurately assess repeat length and methylation status in both males and females (including all premutation and full mutation alleles), and thus support routine fragile X testing without the requirement for SB analysis.
Materials and Methods
Sample Cohort and Characterization
Residual DNA samples, previously tested for fragile X status at either the Laboratory of Human Genetics, Galliera Hospital in Genoa, Italy (GH), or the Laboratory for Diagnostic Genome Analysis of the Department of Clinical Genetics at the Leiden University Medical Center (LUMC) in the Netherlands, were selected for analysis. Genomic DNA was isolated at respective sites from peripheral blood using their standard methods. The 42 samples from the GH comprised 30 females and 12 males representing 7 normal, 1 intermediate, 9 premutation, 17 full mutation, and 5 full mutation mosaics (with pre- and full mutation alleles). This cohort also included a sample with a chromosome Xq27.3/q28 deletion, Klinefelter syndrome (47,XXY), and Turner syndrome (45,X). Matched DNA samples isolated from blood and sperm were analyzed. Residual DNA samples from LUMC comprised 8 males and 26 females representing 6 normal (including 3 homozygous females), 4 intermediate, 13 premutations, and 11 full mutations. Each laboratory performed testing on their laboratory’s DNA samples. PCR analysis for repeat length (repeat primed PCR) was done using AmplideX FMR1 PCR reagents (Asuragen, Austin, TX)28,29 or an alternate method30 as indicated. SB analysis was based on EcoRI/EagI digestion at GH and HindIII/EagI digestion at LUMC according to previously published methods.1,31
Cell-Line DNA Samples, Controls, and Methylation Standards
Cell-line DNA samples and controls were obtained from Asuragen or the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research (Coriell Institute, Camden, NJ). A summary of the samples used in this study, listed according to the source and the range of CGG repeats are provided in Table 1. Two cell-line DNA samples were mixed to form a 20% 940 CGG sensitivity control using 16 ng/μL NA06895 (23 CGG) and 4 ng/μL NA09237 (approximately 940 CGG). Four cell-line DNA samples were mixed to formulate a six-allele process control representing alleles for 20, 29, 31, 54, 120, and approximately 199 CGG.27 An additional pooled cell-line control, AmplideX process control (Asuragen), was used to generate peaks corresponding to 18, 30, 32, 56, 85, 116, and >200 CGG from a single PCR. The CGG repeat lengths for alleles up to 120 CGG were verified using DNA sequencing and used to calibrate CGG repeat length sizing on different capillary electrophoresis (CE) platforms. Full mutation methylation standards were formulated as a mixture of gene-specific PCR products of NA04025 (645 CGG)27 that were methylated using HpaII methyltransferase (New England Biolabs, Ipswich, MA) and mixed with unmethylated amplicons at approximately 6000 copies/μL in a background of 5 ng/μL NA06905. These standards comprised alleles at 23 and 78 CGG and amplicons >200 CGG at different mass ratios of methylated and unmethylated DNA. Expected results for methylation standards at 0%, 50%, and 100% methylation were successfully obtained at both laboratories before clinical sample testing.
Table 1.
Summary of the Control Samples and Cell Lines Used in this Study, Listed According to the Source and the Range of CGG Repeats
| Component | Coriell cat# | CGG repeats | Used in the study |
|---|---|---|---|
| Process control | NA20239 | 20, 199 | Pooled mixture of cell lines used to calibrate repeat sizing between instruments and laboratories |
| NA07541 | 29, 31 | ||
| NA20230 | 54 | ||
| NA06891 | 120 | ||
| 20% 940 | NA06895 | 23 | Mock mosaic sample used for assessment of instrument sensitivity |
| NA09237 | >200 | ||
| Methylation standards | NA06905 | 23, 78 | Carrier DNA background for methylation standards |
| NA04025 | >200 (645 CGG) | PCR products from this cell line were treated with HpaII methyltransferase and blended to create methylation standards | |
| Digestion and reference controls | DigCtrl | ∼212 bp (“−6 CGG”) | Plasmid DNA spiked into each sample to be detected in FAM but decreased in HEX |
| RefCtrl | ∼230 bp (“0 CGG”) | Plasmid DNA spiked into each sample | |
| AmplideX process control | 45913 | 18, 30, 32, 56, 85, 1, 16, and >200 | Prepooled cell-line mixture generating alleles used for instrument repeat sizing correction and as a routine batch control |
FMR1 mPCR Assay
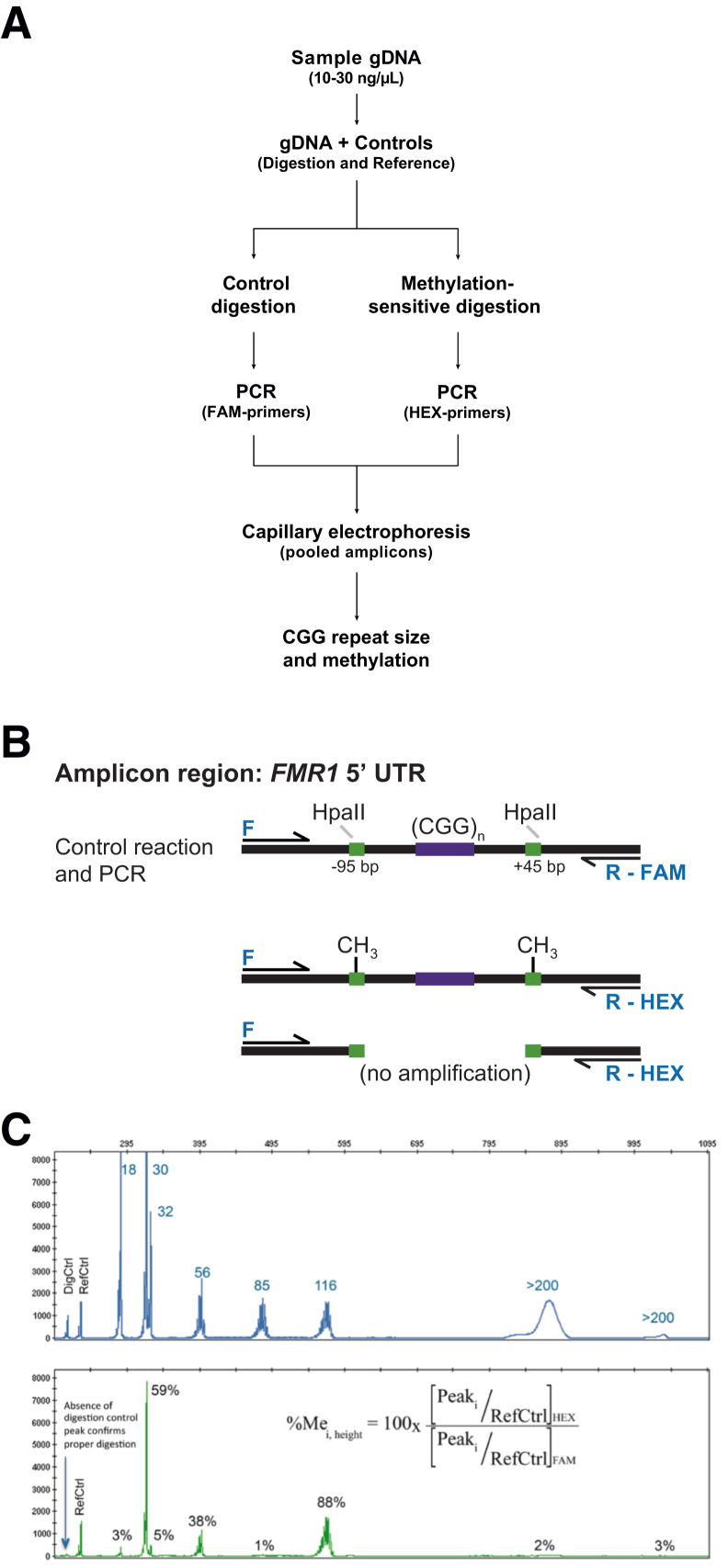
DNA samples were analyzed for methylation status and CGG repeat length using the AmplideX FMR1 mPCR reagents (Asuragen) according to the manufacturer’s recommended protocol. An overview of the workflow, a representative schematic of the effect of digestion with HpaII, and an example data profile for the AmplideX process control are shown in Figure 1. Briefly, 8 μL of 10 to 30 ng/μL DNA samples were premixed with two plasmids: a digestion control (DigCtrl) and PCR reference control (RefCtrl). This premixture was separately aliquoted to a control or methylation-sensitive digestion reaction. Restriction digestion, PCR, and capillary electrophoresis were performed as previously described.23 All alleles were detected using FAM-labeled primers, but only the proportion of the protected methylated allele was available for PCR using HEX-labeled primers (Figure 1B). Lack of methylation at either HpaII site resulted in digestion and thus no amplification.
Figure 1.
A: Methylation PCR (mPCR) workflow. DNA samples were prepared for methylation assessment by mixing with procedural controls and treating with separate methylation-sensitive restriction and control reactions followed by PCR using different dye labels. The HEX- and FAM-labeled amplicons were pooled and sized using capillary electrophoresis. B: Schematic representation of the amplicon region and outcomes of digestion and PCR. The PCR primers spanned two HpaII restriction sites 95 bp from the beginning and another 45 bp from the end of the repeat region using a forward (F) primer and either a FAM- or HEX-labeled reverse primer: R-FAM or R-HEX, respectively. Products from the control digestion were intact and amplified using FAM-labeled primers, but after digestion with HpaII, only alleles fully methylated at both sites were amplified using HEX-labeled primers. Lack of methylation at either site resulted in no amplification. C: Example of two-color data profiles for a multi-allele control. Electropherograms in the FAM channel yielded the total FMR1 profile for that sample and determination of CGG repeats. The signal intensity in the HEX channel corresponded to the methylated component of that allele amplicon. The percent methylation was determined using the ratio of peak heights for each in both channels (equation) normalized to the peak height of the reference control (RefCtrl), one of the plasmid DNA controls spiked into each PCR. Reduction of signal in the HEX channel for the digestion control (DigCtrl), another plasmid DNA spiked into each reaction, correlated with activity of HpaII with expected reduction of signal in the HEX channel. gDNA, genomic DNA; UTR, untranslated region.
Data Analysis
Electropherograms were analyzed using GeneMapper version 4.1 (Life Technologies, Carlsbad, CA). The DigCtrl was formulated with unmethylated HpaII sites and yielded PCR products detected at 212 bp. PCR products from the RefCtrl were detected at 230 bp. Both amplicons were outside the range of a minimally sized FMR1 allele (233 bp or 1 CGG) and did not interfere with sample detection (Figure 1C). Omission of HpaII in the digestion reaction was observed as a high signal in the HEX channel for the DigCtrl, and consequent lack of digestion and higher percent methylation for unmethylated alleles. Samples were analyzed when the ratio of HEX to FAM in the DigCtrl was <0.15 (>85% digested) and the ratio of HEX to FAM peak heights for the RefCtrl amplicons were between 0.5 and 3. The CGG repeat length was derived for alleles up to 200 CGG using a linear fit to the process control amplicons versus size in bp. Full mutation alleles were identified as >200 CGG. The percent methylation for each peak, %Mei,height, was calculated as the normalized ratio of peak heights in the HEX channel to FAM channel for each allele amplicon according to the equation:
| (1) |
where Peaki corresponds to the height of each allele amplicon peak and RefCtrl corresponds to the height of the reference control peak in each color. Methylation percentages >100% were reported as 100%. The percent activation on an allele was calculated as 1 − %Me.
Results
This study was structured to assess mPCR in two independent laboratories with respect to: i) analytical sensitivity and detection of low-abundance full mutation mosaicism; ii) concordance with SB analysis; and iii) methylation assessment of novel sample types.
Analytical Sensitivity and Detection of Size and Methylation Mosaicism
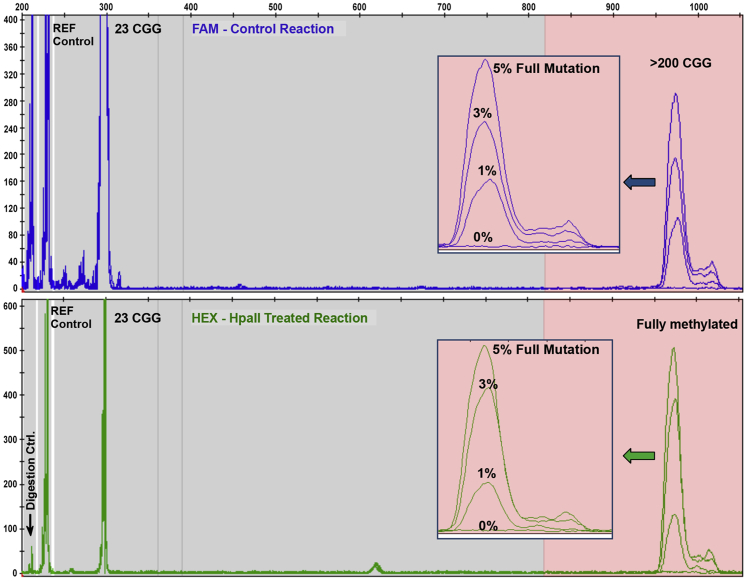
The sensitivity of the method was evaluated over a range of DNA inputs and for detection of low-abundance full mutation mosaicism. In a titration of a mosaic full mutation allele, as little as 1% of a mass fraction of a 645 CGG allele in the background of a 23 CGG allele was observed (Figure 2). Thus, 800 pg (242 haploid copies) of a full mutation allele could be detected in a total of 80 ng of DNA. mPCR analysis of 20 to 320 ng (2.5 to 40 ng/μL) DNA from males and females was also assessed. The signal intensity of full mutation alleles was similar above 20 ng (10 ng each for the HEX and FAM PCRs) without a significant change in percent methylation. The optimal range for mPCR was 40 to 160 ng of input DNA, which was still approximately 50- to 100-fold less than required for SB analysis. For normal alleles at or above 40 ng, the signal was generally saturating or >6000 relative fluorescence units (Supplemental Figure S1). In these cases, methylation status of the normal allele was either obtained from a second CE injection of 2 kV for 10 seconds or listed as not determined.
Figure 2.
Methylation PCR (mPCR) sensitivity for the detection of a full mutation mosaic allele. Peak profiles for a titration of NA04025 (male full mutation, 645 CGG) mixed into a background of NA06895 (normal male, 23 CGG) show detection to as low as 1% mass fraction of the 645-CGG allele in the background of a 99% mass fraction of a normal sample (800 pg of a full mutation allele in a total of 80 ng of DNA mixture).
mPCR and SB Analysis of Representative Clinical Samples
A set of 76 clinical samples, including different sources of DNA and individuals with X chromosome aneuploidies, was analyzed using mPCR and SB. These samples highlighted important distinctions between mPCR and SB analysis with regard to sensitivity, resolution, and data formats. Representative comparisons for eight samples, arranged according to full mutation type and sex, are shown in Figure 3.
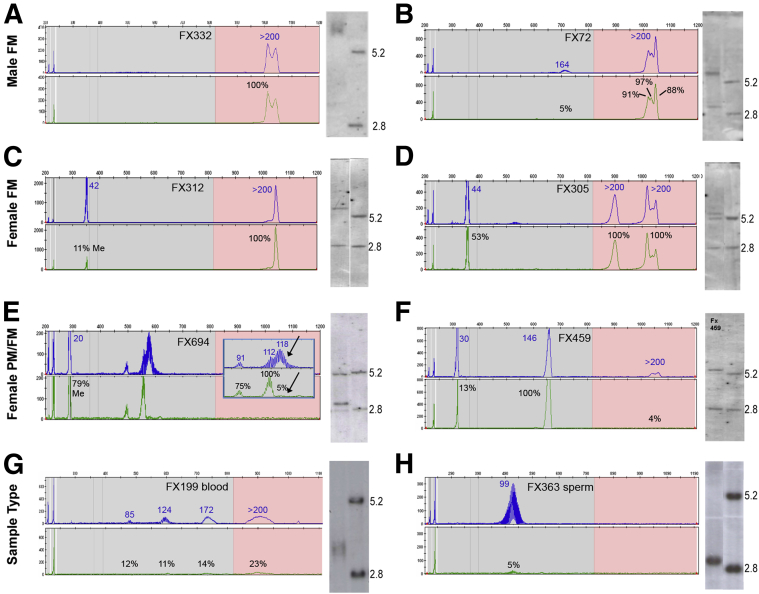
Figure 3.
Methylation PCR (mPCR) electropherograms and Southern blot images for male full mutation (FM), female FM, female premutation (PM), and mosaic samples and matched sources of DNA. Male, fully methylated full mutation allele (A); male with FM/PM mosaicism (B); female, FM with skewed X-inactivation (C); female, FM with random X-inactivation (D); female, PM with specific pattern of size and methylation mosaicism obscured using SB, with arrows highlighting difference in size and methylation states for the larger allele (inset) (E); female sample showing higher resolution and sensitivity of detection for a FM allele not detected using SB (F); matched blood (G); and sperm sample for a male with full mutation allele in the blood but premutation in the sperm (H). SB images include a normal female sample illustrating 2.8 and 5.2 kb bands, respectively.
A fully methylated full mutation allele in a male sample with an estimated 400 to 660 CGG repeats by SB analysis was detected as >200 CGG and 100% methylation using mPCR (Figure 3A). Similarly, a male full mutation sample with premutation mosaicism was detected with approximately 5% methylation of a 164 CGG mosaic allele consistent with the weaker premutation-sized band detected using SB (Figure 3B). In DNA from females, both the normal and full mutation alleles were easily detected using mPCR along with assessment of the normal allele methylation status. An example of highly skewed activation on the normal allele (11% methylation) and methylation on the full mutation allele (100%) was observed (Figure 3C). This result was consistent with an absence of the 5.2 kb band on the SB and the methylated band in the full mutation region. In comparison, a female sample was detected with 53% methylation of the normal allele, indicating random X-inactivation, whereas the full mutation allele from this sample showed 100% methylation, consistent with SB analysis (Figure 3D). Consequently, mPCR provided very similar data for full mutation alleles as SB analysis.
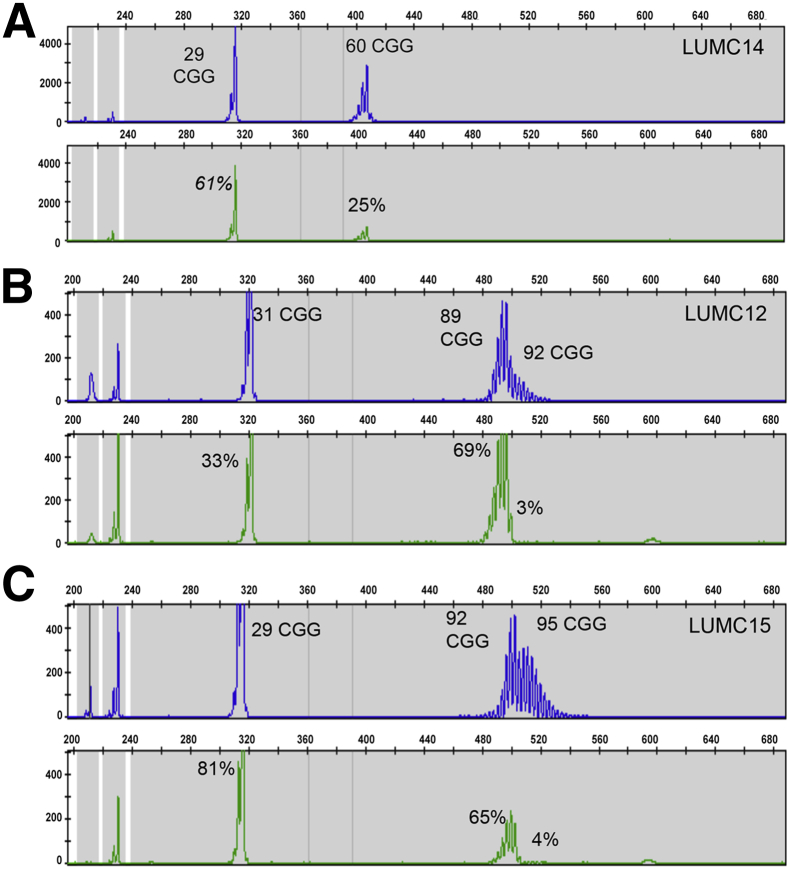
Female premutation alleles were often characterized by two distinct distributions of size and methylation states (Figure 4). In contrast to a single distribution of peaks with a uniform change in methylation (Figure 4A), many premutation alleles had a set of fully methylated peaks of one size distribution followed by unmethylated peaks of longer size distribution (Figure 4, B and C). This level of mosaicism, observed in 14 of 22 females, was unresolved using SB analysis. For example, mPCR analysis yielded repeat lengths of 91 and 112 CGG that were mostly methylated and peaks centered at 118 CGG that were unmethylated (Figure 3E). SB analysis of DNA from another female revealed only a fully methylated premutation band, because of skewed X-inactivation, whereas the mPCR revealed also the presence of an unmethylated expansion >200 CGG (4%) undetectable by SB (Figure 3F). Therefore, when comparing to SB, partial methylation was estimated as the methylation from several representative peaks in the electropherogram. This effect could be subtle. The unmethylated region of the premutation allele could have very low signal intensity (Supplemental Figure S2) or be indistinct by SB analysis because low sizing resolution for premutation alleles. Thus, mPCR provided superior analysis of CGG size and methylation mosaicism in these apparently more complex female premutation alleles.
Figure 4.
Examples of size and methylation mosaic patterns in female premutation alleles detected using methylation PCR (mPCR). A and B: An example profile, LUMC14 (A), wherein the entire premutation allele is partially methylated compared to B, a profile in which the premutation allele is split into a group of fully methylated peaks and a group of unmethylated peaks. C: A similar profile highlighting size and methylation mosaicism in these alleles.
mPCR analysis of matched blood and sperm DNA, and samples with other karyotypes also yielded results consistent with SB analysis. DNA from an unmethylated full mutation male, observed as a smear <5.2 kb by SB, was detected as a broad range of lower signal unmethylated peaks using mPCR (Figure 3G). These peaks were separated by 3 bp each and extended into the full mutation range of >200 CGG. Repeat analysis of this sample yielded the same result, and neither the control DNA nor other clinical DNA samples with a full mutation allele manifested this peak pattern. A sample of DNA isolated from sperm from the same patient was detected with an unmethylated premutation centered at 99 CGG in concordance with the unmethylated band detected using SB (Figure 3H). Even though the methylation profiles were similar between methods, acquisition of the mPCR profiles required substantially less DNA than SB analysis required.
Samples with X chromosome aneuploidies could be flagged for further testing using mPCR. A female sample with a chromosome del(X)(q27.3q28) deletion was detected as a single, completely unmethylated peak of 37 CGG using mPCR. Similarly, a female sample with Turner syndrome, 45,X, was detected as an unmethylated, single-peak allele of 29 CGG (Supplemental Figure S3A). Apparent homozygosity of the 29 CGG allele was excluded because an unmethylated peak would be inconsistent with the profile of a normal homozygous female. These results were consistent with SB analysis and distinct from other female homozygous samples in our study. Analysis of a male sample with Klinefelter syndrome, 47,XXY, was observed as a fully methylated full mutation allele and a fully methylated 29 CGG allele of high signal intensity (Supplemental Figure S3B). Critically, the presence of the normal allele did not confound detection of a full mutation allele in the same male sample and yielded an mPCR profile distinct from methylation mosaicism.
Concordance Assessment between mPCR and SB Analysis
Results of SB analysis, repeat length, and percent methylation using mPCR for 76 clinical samples are listed in Supplemental Table S1. mPCR results were categorically concordant to SB analysis for 75 of 76 samples. In one case (GH-FX549), an unmethylated full mutation allele was detected using mPCR in a female sample previously identified with only a large fully methylated premutation allele (Figure 3F). Thus, if considering SB as the reference method, mPCR had a sensitivity of 100% [95% confidence interval (CI): 89%–100%] and specificity of 99% (95% CI: 93%–100%) for full mutation allele detection. For control DNA tested at both sites, the sensitivity and specificity were 100%. The activation ratio, calculated as the unmethylated ratio on the normal allele for four normal females was 0.47 compared to 0.70 for 11 female full mutation samples. Of the 33 full mutation samples in our study that were fully methylated by SB, 29 had methylation percentages by mPCR detected as >90%. Two samples were detected with 88% and 79% methylation, respectively, and another with full mutation peaks ranging from 87% to 100% methylation. In another sample (GH-FX630/10—data not shown), mPCR yielded 47% methylation on a full mutation allele indicated as fully methylated using SB analysis; however, the SB had low-intensity bands not allowing partial methylation of this allele to be ruled out. The additional findings in the clinical samples (eg, unmethylated or partially methylated full mutations) were considered components of a complex genotype. Differences in detection reflected the higher sensitivity of mPCR than SB analysis.
Discussion
The growth in referrals for fragile X testing and recent advances in therapies for fragile X syndrome place a higher demand on the throughput, efficiency, and interlaboratory accuracy of fragile X testing. The molecular analysis of FMR1 has always been quite time consuming and low throughput because of the reliance on a combination of PCR and SB. Moreover, multiple procedural variants of SB analysis can influence its sensitivity and may result in miscalls of full mutation status.32 Recent advances in targeted therapies for fragile X have shown a predictive response based on the degree of FMR1 methylation. Improvements in hallmark symptoms of fragile X syndrome were observed for individuals with a fully methylated full mutation allele but not for those with mosaicism.33 This link between response to therapy and accurate assessment of methylation makes reliable and sensitive detection of these features paramount.
In this study, a previously published methylation PCR method,23 improved with the incorporation of novel procedural controls and a modified workflow for routine FMR1 was assessed. This mPCR method was validated by two independent European laboratories using a range of challenging clinical samples. mPCR enabled superior size resolution and analytical sensitivity for size and methylation mosaicism compared to SB, while providing nearly perfect categorical concordance across 76 residual clinical samples. The observed shift in activation ratio for female full mutation alleles, even though limited in number, was consistent with data from SB analysis and previously published results.23,34 Furthermore, highly skewed activation of the X chromosome harboring a premutation allele was resolved using mPCR for all samples in our study (Supplemental Table S1).
Our approach represents a fundamental improvement over the workflow and reagents required for SB analysis. In our laboratories, the total time for SB analysis was a minimum of 4.5 days, with 1.5 days for digestion and agarose gel, 2 days for overnight SB, labeling, and overnight hybridization followed by washing, and 1 day for film exposure. This process required maintenance and disposal of different stock solutions, buffers, and radiolabeled probes. By contrast, the digestion and PCR steps of mPCR could be completed within 6 hours using an overnight run on the CE for complete analysis of 48 to 96 samples within 24 hours. Only eight reagent vials for digestion and PCR, and a ROX-labeled ladder were required, without a need for radioactive or chemiluminescent probes. The incorporation of novel controls helped improve reliability relative to SB analysis. A mixture of cell-line DNA samples representing different CGG repeats and methylation states, similar to the control shown in Figure 1C, were tested as a single-well batch control (data not shown). The reference control provided an indication and internal normalizer for efficiency between the HEX- and FAM-labeled PCRs. The digestion control provided an internal quality metric of digestion efficiency for each sample. Although SB can be used to estimate the size of full mutation alleles, we elected to report these as >200 CGG, which was the sizing resolution limit of CE analysis. This approach is consistent with sizing inaccuracy of SB35 and the understanding that the methylation status, and not the magnitude of CGG repeats, is associated with phenotype.2 However, a simple agarose gel of the FAM-labeled PCR products can be used to obtain sizing of full mutation alleles,27 and we have begun to explore semiautomated or precast gel systems to enable this approach.
The 50- to 100-fold reduced DNA input required by mPCR compared to SB can enable the analysis of alternative clinical specimens than peripheral blood with potential benefits for sample procurement, sample management, and/or phenotype association studies. For example, the ratio of full mutation to premutation alleles in skin fibroblasts, which are ectodermal and closer to brain cells in origin, might be a better indicator of psychological impairment than the ratio in blood cells.17,36 Collection of DNA from alternative sources such as saliva or buccal swabs can also reduce the costs, storage, and shipping burdens associated with whole-blood DNA. Because fear of phlebotomy can be the most significant reason for refusing to participate in a clinical research study,37 the ability to obtain alternative samples for fragile X testing may encourage broader participation in important research studies.
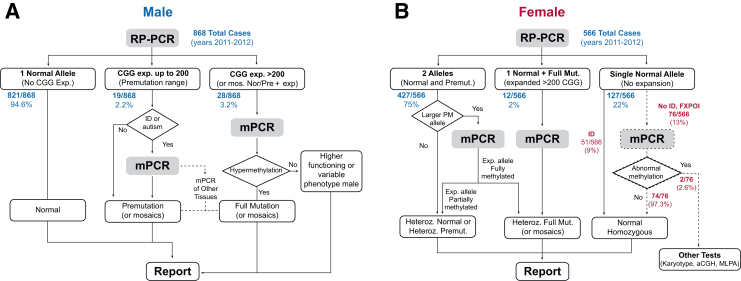
Incorporation of mPCR suggests standardization of fragile X testing schemes without the need for SB analysis (Figure 5). In this proposed scheme, all samples would be tested for repeat size using repeat primed PCR. mPCR would be performed on all males and females with an expanded allele >200 CGG. For males, mPCR analysis could be performed on all premutation samples, the approach taken at LUMC, or be dependent on the indication of intellectual disability (Figure 4A), the approach taken at the GH. For females with larger premutation alleles, mPCR could be used to better define the correct allele category based on size and degree of methylation. For example, in sample GH_FX 227, peaks corresponding to 32, 187, and >200 were detected by repeat primed PCR and mPCR (Supplemental Table S1), but only normal and approximately 200 were detected by SB. This distinction in size and methylation status, due to better size resolution of mPCR, may help refine classification or identify particular endophenotypes in females with these alleles.
Figure 5.
Proposed PCR-only workflow for routine FMR1 analysis in males (A) and females (B), including modeled outcomes for samples tested using the traditional workflow at Genoa Hospital. All samples >200 CGG can be reflexed to mPCR to assess methylation (or identify mosaicism or unmethylated expanded alleles). Male samples with premutation-range alleles can be assessed with mPCR based on the sample referral [intellectual disability (ID) or autism]. A similar first-step approach with females would yield expanded alleles >200 CGG for mPCR analysis along with samples with high premutation alleles for which mPCR would be performed. For those samples that show a single normal allele and for which the reason for referral is fragile X–associated primary ovarian insufficiency (FXPOI) (dashed pathway), mPCR can help find X abnormalities possibly linked to phenotype or infertility (such as Turner or deletions encompassing the FMR1 gene): the presence of only one unmethylated normal allele is inconsistent with an actual normal homozygosity, flagging the sample for further analyses. aCGH, array comparative genomic hybridization; MLPA, multiplex ligation-dependent probe amplification; RP-PCR, repeat primed PCR (AmplideX FMR1 PCR).
DNA from females indicated to be homozygous for a specific FMR1 repeat could also be analyzed for potential X-chromosome abnormalities (Figure 5B). Considering that an increasing number of females undergoing FMR1 testing are referred from infertility centers, mPCR may help identify particular features of premutation alleles or X chromosome abnormalities that might underlie the infertility. At the GH, 2 of 76 females (without intellectual disability and showing only one allele by repeat primed PCR) were found to have either an Xq deletion encompassing the FMR1 gene or Turner syndrome. Indeed, the sizing accuracy of mPCR compared to SB could provide tools to better understand the cutoff of full mutation allele sizes and links to phenotypes based on differences in methylation for alleles spanning the genotype range. Thus, mPCR represents an important technical advance in fragile X testing that can eliminate the need for SB analysis and meet growing demands for accurate characterization of the FMR1 gene.
Acknowledgments
We thank René Belfroid and Saskia Smith from the Laboratory for Diagnostic Genome Analysis (Leiden, the Netherlands) for technical assistance.
Footnotes
Supported by National Institute of Child Health & Human Development grant R44 HD060450-02 (S.F.-S., R.C., G.J.L., and A.G.H.) and by Galliera Genetic Bank–Italian Telethon grant (project no. GTB12001, to M.G., E.G., and D.A.C.).
M.G. and E.M.J.B. contributed equally to this work.
Disclosures: S.F.-S., R.C., G.J.L., and A.G.H. are employees of Asuragen, which manufactures the AmplideX reagents used in this study.
Supplemental Data
Peak height and methylation percentage as a function of DNA input for three female normal alleles and two male full-mutation (FM) alleles. The female samples were indicated by their paired allele for 23,112 CGG; 23,78 CGG; and 29 (>200 CGG). Only results for the normal allele were plotted. Signals on the normal allele were saturating above 40 ng, and lower injections were used as needed to determine methylation percentage. Full-mutation alleles, plotted for male samples, had higher signals and less variation in the percent methylation above 10 ng input. Data markers reflect results from independent replicate measurements.
Example of low signal intensity methylation mosaicism in a female premutation allele. This sample, LUMC_30, reveals a normal allele and fully methylated premutation (PM) allele. A stretch of approximately 30 CGG peaks of low intensity, present in the FAM, but not in the HEX channel, and more evident in the raw data, were unmethylated. The combination of peak ranges was concordant with a partially methylated premutation allele. This sample exemplifies the need to analyze different regions within female premutation samples.
Examples of mPCR data profiles and matched SB images for samples with X chromosome aneuploidy. A: Turner syndrome (45,X): the X monosomy is detected as a single, completely unmethylated peak consistent with the only 2.8-kb band seen by Southern Blot. Detection of a single unmethylated allele in a female rules out homozygosity for this repeat length and flags the sample for further analysis; B: Klinefelter syndrome (47,XXY) with skewed X-inactivation: the full-mutation allele was 100% methylated, whereas a normal allele in a male sample had high signal intensity and 100% methylation. These results were consistent with SB analysis and banding at 5.2 kb.
References
- 1.Verkerk A.J.M.H., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P.A., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., Eussen B.E., van Ommen G.J.B., Blonden L.A.J., Riggins G.J., Chastain J.L., Kunst C.B., Galjaard H., Caskey C.T., Nelson D.L., Oostra B.A., Warren S.T. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 2.Rousseau F., Heitz D., Tarleton J., MacPherson J., Malmgren H., Dahl N., Barnicoat A., Mathew C., Mornet E., Tejada I., Maddalena A., Spiegel R., Schinzel A., Marcos J.A.G., Schorderet D.F., Schaap T., Maccioni L., Russo S., Jacobs P.A., Schwartz C., Mandel J.L. A multicenter study on genotype-phenotype correlations in the fragile X syndrome, using direct diagnosis with probe StB12.3: the first 2,253 cases. Am J Hum Genet. 1994;55:225–237. [PMC free article] [PubMed] [Google Scholar]
- 3.Hagerman R.J., Hagerman P.J. Testing for fragile X gene mutations throughout the life span. JAMA. 2008;300:2419–2421. doi: 10.1001/jama.2008.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willemsen R., Bontekoe C.J., Severijnen L.A., Oostra B.A. Timing of the absence of FMR1 expression in full mutation chorionic villi. Hum Genet. 2002;110:601–605. doi: 10.1007/s00439-002-0723-5. [DOI] [PubMed] [Google Scholar]
- 5.Irwin S.A., Galvez R., Greenough W.T. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 6.Grasso M., Faravelli F., Lo Nigro C., Chiurazzi P., Sperandeo M.P., Argusti A., Pomponi M.G., Lecora M., Sebastio G.F., Perroni L., Andria G., Neri G., Bricarelli F.D. Mosaicism for the full mutation and a microdeletion involving the CGG repeat and flanking sequences in the FMR1 gene in eight fragile X patients. Am J Med Genet. 1999;85:311–316. doi: 10.1002/(sici)1096-8628(19990730)85:3<311::aid-ajmg24>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 7.Stoger R., Genereux D.P., Hagerman R.J., Hagerman P.J., Tassone F., Laird C.D. Testing the FMR1 promoter for mosaicism in DNA methylation among CpG sites, strands, and cells in FMR1-expressing males with fragile X syndrome. PLoS One. 2011;6:e23648. doi: 10.1371/journal.pone.0023648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wohrle D., Salat U., Glaser D., Mucke J., Meisel-Stosiek M., Schindler D., Vogel W., Steinbach P. Unusual mutations in high functioning fragile X males: apparent instability of expanded unmethylated CGG repeats. J Med Genet. 1998;35:103–111. doi: 10.1136/jmg.35.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leehey M.A., Berry-Kravis E., Goetz C.G., Zhang L., Hall D.A., Li L., Rice C.D., Lara R., Cogswell J., Reynolds A., Gane L., Jacquemont S., Tassone F., Grigsby J., Hagerman R.J., Hagerman P.J. FMR1 CGG repeat length predicts motor dysfunction in premutation carriers. Neurology. 2008;70:1397–1402. doi: 10.1212/01.wnl.0000281692.98200.f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodega B., Bione S., Dalpra L., Toniolo D., Ornaghi F., Vegetti W., Ginelli E., Marozzi A. Influence of intermediate and uninterrupted FMR1 CGG expansions in premature ovarian failure manifestation. Hum Reprod. 2006;21:952–957. doi: 10.1093/humrep/dei432. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez-Revenga L., Madrigal I., Badenas C., Xuncla M., Jimenez L., Mila M. Premature ovarian failure and fragile X female premutation carriers: no evidence for a skewed X-chromosome inactivation pattern. Menopause. 2009;16:944–949. doi: 10.1097/gme.0b013e3181a06a37. [DOI] [PubMed] [Google Scholar]
- 12.Goodlin-Jones B.L., Tassone F., Gane L.W., Hagerman R.J. Autistic spectrum disorder and the fragile X premutation. J Dev Behav Pediatr. 2004;25:392–398. doi: 10.1097/00004703-200412000-00002. [DOI] [PubMed] [Google Scholar]
- 13.Clifford S., Dissanayake C., Bui Q.M., Huggins R., Taylor A.K., Loesch D.Z. Autism spectrum phenotype in males and females with fragile X full mutation and premutation. J Autism Dev Disord. 2007;37:738–747. doi: 10.1007/s10803-006-0205-z. [DOI] [PubMed] [Google Scholar]
- 14.Spath M.A., Nillesen W.N., Smits A.P., Feuth T.B., Braat D.D., van Kessel A.G., Yntema H.G. X chromosome inactivation does not define the development of premature ovarian failure in fragile X premutation carriers. Am J Med Genet. 2010;152A:387–393. doi: 10.1002/ajmg.a.33243. [DOI] [PubMed] [Google Scholar]
- 15.Jacquemont S., Hagerman R.J., Leehey M.A., Hall D.A., Levine R.A., Brunberg J.A., Zhang L., Jardini T., Gane L.W., Harris S.W., Herman K., Grigsby J., Greco C.M., Berry-Kravis E., Tassone F., Hagerman P.J. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- 16.Hantash F.M., Goos D.M., Crossley B., Anderson B., Zhang K., Sun W., Strom C.M. FMR1 premutation carrier frequency in patients undergoing routine population-based carrier screening: insights into the prevalence of fragile X syndrome, fragile X-associated tremor/ataxia syndrome, and fragile X-associated primary ovarian insufficiency in the United States. Genet Med. 2011;13:39–45. doi: 10.1097/GIM.0b013e3181fa9fad. [DOI] [PubMed] [Google Scholar]
- 17.Dobkin C.S., Nolin S.L., Cohen I., Sudhalter V., Bialer M.G., Ding X.H., Jenkins E.C., Zhong N., Brown W.T. Tissue differences in fragile X mosaics: mosaicism in blood cells may differ greatly from skin. Am J Med Genet. 1996;64:296–301. doi: 10.1002/(SICI)1096-8628(19960809)64:2<296::AID-AJMG13>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 18.Sherman S., Pletcher B.A., Driscoll D.A. Fragile X syndrome: diagnostic and carrier testing. Genet Med. 2005;7:584–587. doi: 10.1097/01.GIM.0000182468.22666.dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquemont S., Birnbaum S., Redler S., Steinbach P., Biancalana V. Clinical utility gene card for: fragile X mental retardation syndrome, fragile X-associated tremor/ataxia syndrome and fragile X-associated primary ovarian insufficiency. Eur J Hum Genet. 2011 doi: 10.1038/ejhg.2011.55. http://dx.doi.org/10.1038/ejhg.2011.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panagopoulos I., Lassen C., Kristoffersson U., Aman P. A methylation PCR approach for detection of fragile X syndrome. Hum Mutat. 1999;14:71–79. doi: 10.1002/(SICI)1098-1004(1999)14:1<71::AID-HUMU9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., Lum J.M., Yeo G.H., Kiing J., Tay S.K., Chong S.S. Simplified molecular diagnosis of fragile X syndrome by fluorescent methylation-specific PCR and GeneScan analysis. Clin Chem. 2006;52:1492–1500. doi: 10.1373/clinchem.2006.068593. [DOI] [PubMed] [Google Scholar]
- 22.Dahl C., Guldberg P. A ligation assay for multiplex analysis of CpG methylation using bisulfite-treated DNA. Nucleic Acids Res. 2007;35:e144. doi: 10.1093/nar/gkm984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen L., Hadd A.G., Sah S., Houghton J.F., Filipovic-Sadic S., Zhang W., Hagerman P.J., Tassone F., Latham G.J. High-resolution methylation polymerase chain reaction for fragile X analysis: evidence for novel FMR1 methylation patterns undetected in Southern blot analyses. Genet Med. 2011;13:528–538. doi: 10.1097/GIM.0b013e31820a780f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nygren A.O., Lens S.I., Carvalho R. Methylation-specific multiplex ligation-dependent probe amplification enables a rapid and reliable distinction between male FMR1 premutation and full-mutation alleles. J Mol Diagn. 2008;10:496–501. doi: 10.2353/jmoldx.2008.080053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coffee B., Keith K., Albizua I., Malone T., Mowrey J., Sherman S.L., Warren S.T. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet. 2009;85:503–514. doi: 10.1016/j.ajhg.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Godler D.E., Slater H.R., Bui Q.M., Ono M., Gehling F., Francis D., Amor D.J., Hopper J.L., Hagerman R., Loesch D.Z. FMR1 intron 1 methylation predicts FMRP expression in blood of female carriers of expanded FMR1 alleles. J Mol Diagn. 2011;13:528–536. doi: 10.1016/j.jmoldx.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filipovic-Sadic S., Sah S., Chen L., Krosting J., Sekinger E., Zhang W., Hagerman P.J., Stenzel T.T., Hadd A.G., Latham G.J., Tassone F. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin Chem. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L., Hadd A., Sah S., Filipovic-Sadic S., Krosting J., Sekinger E., Pan R., Hagerman P.J., Stenzel T.T., Tassone F., Latham G.J. An information-rich CGG repeat primed PCR that detects the full range of fragile X expanded alleles and minimizes the need for southern blot analysis. J Mol Diagn. 2010;12:589–600. doi: 10.2353/jmoldx.2010.090227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juusola J.S., Anderson P., Sabato F., Wilkinson D.S., Pandya A., Ferreira-Gonzalez A. Performance evaluation of two methods using commercially available reagents for PCR-based detection of FMR1 mutation. J Mol Diagn. 2012;14:476–486. doi: 10.1016/j.jmoldx.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Tassone F., Pan R., Amiri K., Taylor A.K., Hagerman P.J. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J Mol Diagn. 2008;10:43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rousseau F., Heitz D., Biancalana V., Blumenfeld S., Kretz C., Boue J., Tommerup N., Van Der Hagen C., DeLozier-Blanchet C., Croquette M.F., Gilgenkrantz S., Jalbert P., Voelckel M.A., Oberle I., Mandel J.L. Direct diagnosis by DNA analysis of the fragile X syndrome of mental retardation. N Engl J Med. 1991;325:1673–1681. doi: 10.1056/NEJM199112123252401. [DOI] [PubMed] [Google Scholar]
- 32.Seneca S., Lissens W., Endels K., Caljon B., Bonduelle M., Keymolen K., De Rademaeker M., Ullmann U., Haentjens P., Van Berkel K., Van Dooren S. Reliable and sensitive detection of fragile X (expanded) alleles in clinical prenatal DNA samples with a fast turnaround time. J Mol Diagn. 2012;14:560–568. doi: 10.1016/j.jmoldx.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Jacquemont S., Curie A., des Portes V., Torrioli M.G., Berry-Kravis E., Hagerman R.J., Ramos F.J., Cornish K., He Y., Paulding C., Neri G., Chen F., Hadjikhani N., Martinet D., Meyer J., Beckmann J.S., Delange K., Brun A., Bussy G., Gasparini F., Hilse T., Floesser A., Branson J., Bilbe G., Johns D., Gomez-Mancilla B. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci Transl Med. 2011;3:64ra1. doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]
- 34.de Vries B.B., Wiegers A.M., Smits A.P., Mohkamsing S., Duivenvoorden H.J., Fryns J.P., Curfs L.M., Halley D.J., Oostra B.A., van den Ouweland A.M., Niermeijer M.F. Mental status of females with an FMR1 gene full mutation. Am J Hum Genet. 1996;58:1025–1032. [PMC free article] [PubMed] [Google Scholar]
- 35.Weck K.E., Zehnbauer B., Datto M., Schrijver I. Molecular genetic testing for fragile X syndrome: laboratory performance on the College of American Pathologists proficiency surveys (2001-2009) Genet Med. 2012;14:306–312. doi: 10.1038/gim.2011.11. [DOI] [PubMed] [Google Scholar]
- 36.MacKenzie J.J., Sumargo I., Taylor S.A. A cryptic full mutation in a male with a classical fragile X phenotype. Clin Genet. 2006;70:39–42. doi: 10.1111/j.1399-0004.2006.00634.x. [DOI] [PubMed] [Google Scholar]
- 37.Dlugos D.J., Scattergood T.M., Ferraro T.N., Berrettinni W.H., Buono R.J. Recruitment rates and fear of phlebotomy in pediatric patients in a genetic study of epilepsy. Epilepsy Behav. 2005;6:444–446. doi: 10.1016/j.yebeh.2005.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Peak height and methylation percentage as a function of DNA input for three female normal alleles and two male full-mutation (FM) alleles. The female samples were indicated by their paired allele for 23,112 CGG; 23,78 CGG; and 29 (>200 CGG). Only results for the normal allele were plotted. Signals on the normal allele were saturating above 40 ng, and lower injections were used as needed to determine methylation percentage. Full-mutation alleles, plotted for male samples, had higher signals and less variation in the percent methylation above 10 ng input. Data markers reflect results from independent replicate measurements.
Example of low signal intensity methylation mosaicism in a female premutation allele. This sample, LUMC_30, reveals a normal allele and fully methylated premutation (PM) allele. A stretch of approximately 30 CGG peaks of low intensity, present in the FAM, but not in the HEX channel, and more evident in the raw data, were unmethylated. The combination of peak ranges was concordant with a partially methylated premutation allele. This sample exemplifies the need to analyze different regions within female premutation samples.
Examples of mPCR data profiles and matched SB images for samples with X chromosome aneuploidy. A: Turner syndrome (45,X): the X monosomy is detected as a single, completely unmethylated peak consistent with the only 2.8-kb band seen by Southern Blot. Detection of a single unmethylated allele in a female rules out homozygosity for this repeat length and flags the sample for further analysis; B: Klinefelter syndrome (47,XXY) with skewed X-inactivation: the full-mutation allele was 100% methylated, whereas a normal allele in a male sample had high signal intensity and 100% methylation. These results were consistent with SB analysis and banding at 5.2 kb.