Abstract
A stable and persistent Hepatitis C virus (HCV) replication cell culture model was developed to examine clearance of viral replication during long-term treatment using interferon-α (IFN-α), IFN-λ, and ribavirin (RBV). Persistently HCV-infected cell culture exhibited an impaired antiviral response to IFN-α+RBV combination treatment, whereas IFN-λ treatment produced a strong and sustained antiviral response that cleared HCV replication. HCV replication in persistently infected cells induced chronic endoplasmic reticulum (ER) stress and an autophagy response that selectively down-regulated the functional IFN-α receptor-1 chain of type I, but not type II (IFN-γ) or type III (IFN-λ) IFN receptors. Down-regulation of IFN-α receptor-1 resulted in defective JAK–STAT signaling, impaired STAT phosphorylation, and impaired nuclear translocation of STAT. Furthermore, HCV replication impaired RBV uptake, because of reduced expression of the nucleoside transporters ENT1 and CNT1. Silencing ER stress and the autophagy response using chemical inhibitors or siRNA additively inhibited HCV replication and induced viral clearance by the IFN-α+RBV combination treatment. These results indicate that HCV induces ER stress and that the autophagy response selectively impairs type I (but not type III) IFN signaling, which explains why IFN-λ (but not IFN-α) produced a sustained antiviral response against HCV. The results also indicate that inhibition of ER stress and of the autophagy response overcomes IFN-α+RBV resistance mechanisms associated with HCV infection.
Hepatitis C virus (HCV) infects more than 170 million people worldwide and is one of the leading causes of chronic liver disease, liver cirrhosis, and hepatocellular carcinoma in the United States.1,2 Combination therapy using interferon-α (IFN-α), ribavirin (RBV), and a protease inhibitor is the current standard of care for HCV genotype 1 infection.3,4 Although this triple combination therapy has significantly improved the sustained virological response of chronic HCV 1a infection, the treatment response has not improved significantly among prior nonresponders to pegylated interferon and RBV.5,6 Several studies have suggested that the risk of HCV-induced liver cirrhosis and hepatoma is substantially reduced in patients who clear HCV infection and achieve a sustained virological response.7,8 The poor sustained virological response with triple therapy in patients who are nonresponders to the combination of IFN-α and RBV (IFN-α+RBV) is a major unsolved problem in treating chronic hepatitis C. The mechanism of HCV resistance under these conditions is not well understood. A better understanding of the mechanism of HCV clearance by IFN-α and RBV could lead to improvements in treatment for such patients and reduce the burden of liver cirrhosis and hepatoma.
The availability of highly efficient cell culture systems suitable for study of HCV has enabled molecular studies of IFN-α antiviral mechanisms against HCV. A series of publications from our laboratory with others have verified that the JAK–STAT pathway induced by IFN-α is critical for the HCV antiviral mechanism in cell culture models.9–11 Studies performed over the last several years indicate that IFN-α signaling is controlled by a number of factors, including suppressor of cytokine signaling (SOCS) family members SOCS1 and SOCS3, ubiquitin-specific peptidase 18 (USP18), the protein inhibitor of activated STAT1 (PIAS1), and protein phosphatase 2A (PP2A).12 Although RBV is used in combination with IFN-α to treat patients with HCV infection, the mechanisms by which many patients develop resistance to RBV are not well understood. One report indicated that reduced RBV uptake by HCV-infected cells contributed to an impaired antiviral response.13 However, no previous systematic studies have investigated how the IFN-α and RBV synergistic antiviral mechanisms are impaired during chronic HCV infection.
Recent clinical studies indicate that the overall success of triple combination therapy depends on the initial patient response to combined IFN-α+RBV treatment and on host genetic polymorphisms of the IFN-λ gene (IFNL3; alias IL-28B).14,15 The importance of the IL-28B genotype and the IFN-λ system in the clearance of HCV replication is not well understood. Elucidating the factors determining HCV clearance by type I and type III IFNs and RBV, using a stable and persistently HCV-infected cell culture system, should open new approaches to improving the sustained virological response among nonresponders.
We developed a stable and persistently HCV-infected Huh-7.5 cell culture system to examine the contribution of viral and host-cell factors in the mechanisms of HCV clearance induced by long-term antiviral treatment using IFN-α+RBV or IFN-λ. Here, we show that HCV replication in the persistently HCV-infected cell culture induces endoplasmic reticulum (ER) stress and an autophagy response that selectively impairs IFN-α+RBV–mediated viral clearance. On the other hand, our results indicate that exogenous IFN-λ treatment leads to HCV clearance in persistently HCV-infected cells in culture. Furthermore, our results suggest that targeting chronic ER stress and HCV-induced autophagy may overcome IFN-α and RBV resistance mechanisms in chronic HCV infection.
Materials and Methods
Cell Lines, Infectious Clones, Plasmids, Antibodies, and Chemicals
Huh-7 and Huh-7.5 human hepatocellular carcinoma cells, a kind gift from Charles M. Rice (Rockefeller University, New York), were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Life Technologies, Carlsbad, CA) and supplemented with nonessential amino acids, sodium pyruvate, and 10% (v/v) fetal bovine serum (FBS). Huh-7 cells with defective JAK–STAT signaling (cell line R-24-1), described previously,9 and a human fibroblast cell line (2H-11) were maintained as above. Cells were grown at 37°C in a 5% CO2–enriched atmosphere within a humidified incubator. The Renilla luciferase reporter–based pJFH-ΔV3-Rluc clone used in our experiment has been described previously.16 The following were obtained commercially: IFN-α (EMD Merck, Billerica, MA); IFN-λ (IL-29; PeproTech, Rocky Hill, NJ); Torin 1 (Selleck Chemicals, Houston, TX); RBV, Acridine Orange, 4-phenylbutyric acid (PBA), thapsigargin (TG), and hydroxychloroquine (HCQ) (Sigma-Aldrich, St. Louis, MO); [3H]cytidine, [3H]RBV (Moravek Biomedicals, Brea, CA); and plasmids p5xATF6-GL3, pSTAT1-GFP (Addgene, Cambridge, MA), pSTAT2-GFP (a gift from Hansjörg Hauser, GBF-National Research Institute for Biotechnology, Braunschweig, Germany), and pISRE-luciferase (provided by Stephen Goodbourn, St. George's Hospital and Medical School, University of London, London, UK). siRNAs against PERK, IRE1α, ATF6, and ATG7 (Life Technologies) were as described previously.17 Synthetic siRNAs targeted to 5′UTR of HCV genome (si321 and si359) were from Life Technologies.18 Antibodies specific for IFNAR1 (Biogen Idec, Cambridge, MA), IFNAR2, IFNGR1, CNT1, ATF6 (Santa Cruz Biotechnology, Santa Cruz, CA), IFN-λ receptor IL10Rβ (R&D Systems, Minneapolis, MN), β-actin, GAPDH, p-STAT1, STAT1, p-STAT2, STAT2, LC3, p62, beclin 1, BiP, IRE1α, p-eIF2α, PERK, ATG7 (Cell Signaling Technology, Danvers, MA), ENT1 (Abgent, San Diego, CA), HCV Core protein (Thermo Fisher Scientific, Waltham, MA), and anti-Renilla luciferase (EMD Merck, Billerica, MA) were obtained from the respective manufacturers.17,18
Development of a Stable Infected Huh-7.5 Cell Culture System
Huh-7.5 cells were transfected with 20 μg of in vitro transcribed HCV RNA prepared from clone pJFH-ΔV3-Rluc (HCV genotype 2a), and culture-derived infectious HCV stocks were prepared from the supernatants of Huh-7.5 cells using a protocol described previously.19 In brief, Huh-7.5 cells were infected with multiplicity of infection (MOI) 0.1 JFH-ΔV3-Rluc virus overnight, and on the next day the infected culture was washed with PBS and then incubated with 10 mL of DMEM containing 10% FBS (v/v). Infected Huh-7.5 cells were cultured long-term by splitting at a 1:10 ratio at 6-day intervals. Replication of HCV in the infected cell culture at each interval was confirmed by measuring the Renilla luciferase activity or the HCV RNA level by RT-qPCR.
Interferon and RBV Treatment and HCV Quantification
To calibrate and compare the biological activity and antiviral efficacy of IFN-α and IFN-λ (for which concentration is expressed in different units), we first determined the 90% inhibitory concentration (IC90) of these two cytokines. Huh-7.5 cells were infected with MOI = 0.1 JFH-ΔV3-Rluc virus overnight. The next day, the infective virus was removed, and cells were treated with different concentrations of IFN-α or IFN-λ. After 72 hours, the IC90 was determined to be 100 IU/mL for IFN-α and 10 ng/mL for IFN-λ (as discussed further under Results). In the next step, we compared the treatment response of IFN-α and IFN-λ in the HCV-infected cell culture model. Huh-7.5 cells were infected with MOI = 0.1 JFH-ΔV3-Rluc reporter virus overnight. In one set of experiments, IFN-α and RBV were added after 48 hours of infection, to determine the antiviral effect in a cell culture system. After 48 hours of infection, cells were treated with 2.5 × IC90, 5 × IC90, or 10 × IC90 IFN-α or IFN-λ, or with 40 μg/mL RBV or 2.5 × IC90 + 40 μg/mL IFN-α+RBV. After 72 hours, half of the cells were used for measuring the antiviral effect and the remaining cells were seeded for a second round of treatment. The antiviral effects of IFN-α, IFN-λ, RBV alone, and the IFN-α+RBV combination were measured at 3-day intervals. In another set of experiments, infected cells were cultured for 1 week to increase the level of viral replication. On day 8, cells were treated with RBV or IFN-α+RBV as described. The sustained antiviral effect was measured for an extended time period. Five consecutive treatments were given at 6-day intervals for up to 37 days. Antiviral activity was assessed after each treatment. Luciferase activity of cell lysates was measured using a Renilla luciferase assay system kit (Promega, Madison, WI). Total protein concentration of the lysate was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific). Renilla luciferase activity was normalized to protein concentration.
RT-qPCR
An RT-qPCR assay for HCV quantification was performed according to a protocol described previously.18 In brief, 2 μg of cellular RNA or total RNA from 1 mL of conditioned culture medium was used to amplify the 5′UTR of the HCV genome using sense primer 5′-TCTTCACGCAGAAAGCGTCTA-3′ (60–80; HCV/S) and antisense primer 5′-CGGTTCCGCAGACCACTATG-3′ (157–138; HCV/AS). The probe 5′-/56-FAM/TGAGTGTCG/ZEN/TGCAGCCTCCAGGA/3IBκFQ/-3′ (Integrated DNA Technologies, Coralville, IA), labeled at the 5′ ends with a 6-carboxyfluorescein (FAM) fluorophore reporter molecule and ZEN-Iowa Black FQ (IBFQ) double quenchers, was used to reduce the background and increase signal. The RT-qPCR assay was performed in 20 μL containing 10 μL of iQ supermix (Bio-Rad Laboratories, Hercules, CA), 0.25 μmol/L of each primer and probe, and 4 μL of cDNA product obtained from the reverse-transcription reaction. The amplification was performed using a standard program: 48°C for 30 minutes and 95°C for 10 minutes, followed by 45 cycles of 95°C for 15 seconds (denaturation) and 60°C for 1 minute (annealing and extension). HCV cDNA standards were used, starting at 1 × 109 copies of virus and decreasing in 10-fold serial dilutions. Amplification, data acquisition, and analysis were performed using a CFX96 real-time PCR system with CFX Manager software version 1.0 (Bio-Rad Laboratories).
Western Blot Analysis
Cells were lysed in ice-cold radioimmunoprecipitation assay buffer. Total protein content in the lysate was quantified using a protein assay kit (Bio-Rad Laboratories). An equal amount of protein from each sample was mixed in 4× SDS loading buffer. Proteins were separated using NuPAGE 12% gels (Life Technologies) and then were transferred onto a nitrocellulose membrane (GE Healthcare, Little Chalfont, UK). The membrane was blocked with filtered blocking solution [10 mmol/L Tris-buffered saline (pH 7.4), 0.1% (v/v) Tween 20, 5% (v/v) nonfat dried milk] for 2 hours with gentle shaking at room temperature. Next, the membrane was washed with 10 mmol/L Tris-buffered saline (pH 7.4) containing 0.1% Tween 20 (v/v), three times for 5 minutes each. The primary antibody was diluted (according to the manufacturer’s instructions) in blocking reagent [10 mmol/L Tris-buffered saline (pH 7.4), 0.1% Tween 20 (v/v), 5% nonfat dried milk (w/v) or 5% bovine serum albumin (w/v)], added to the membrane, and incubated at 4°C overnight with gentle shaking. The next day, the membrane was washed with wash buffer three times for 5 minutes each. The horseradish peroxidase–conjugated secondary antibody was diluted in blocking reagent [10 mmol/L Tris-buffered saline (pH 7.4), 0.1% Tween 20 (v/v), 5% nonfat dried milk (w/v)], added to the membrane, and incubated at room temperature for 2 hours with gentle shaking. The membrane was again washed with wash buffer three times for 5 minutes each. Enhanced chemiluminescence ECL detection reagent (GE Healthcare) was then added to the membrane according to the manufacturer’s instructions. The membrane was then exposed to chemiluminescence film (GE Healthcare).
Nuclear Translocation Assay
Naïve or persistently infected Huh-7.5 cells were split in a 12-well cell culture plate (Thermo Fisher Scientific) at a density of 2 × 104 cells per well. After 20 hours, cells were transfected with 0.2 μg of STAT1-GFP or STAT2-GFP plasmid DNA using FuGENE 6 (Roche Diagnostics, Indianapolis, IN) transfection reagent. At 48 hours, both pSTAT1-GFP and pSTAT2-GFP transfected cells were treated with 500 IU/mL IFN-α (5 × IC90). The translocation of GFP was monitored using fluorescence microscopy (Olympus IX70 microscope); images were captured using an Olympus DP-71 digital camera.
RBV Uptake
RBV uptake into persistently HCV-infected cells was measured using a previously described protocol,13 with minor modifications. Infected cells were harvested after trypsin–EDTA treatment and were washed twice using DMEM growth medium supplemented with 10% FBS. Equal numbers of infected cells (1 × 106) were resuspended in 1 mL of growth medium in sterile polystyrene, round-bottom tubes with caps. RBV uptake assays were initiated by the addition of 100 μL of cell culture medium supplemented with either 7.2 μL of 5 μmol/L [3H]RBV or 12.5 μL of [3H]cytidine in a CO2 incubator at 37°C with continuous shaking. After 30 minutes of incubation, cells were treated with 1 mL of ice-cold PBS and were placed on ice for 5 minutes to stop the reaction. After this step, cells were washed twice with 1 mL of ice-cold PBS. Cell pellets were treated with 100 μL of cold lysis buffer [10 mmol/L Tris-HCl (pH 8.0), 10 mmol/L NaCl, 1.5 mmol/L MgCl2, and 0.1% NP-40 (v/v)]. A 40-μL aliquot of soluble protein lysate was mixed with 1 mL of scintillation fluid, and radioactivity was measured using a liquid scintillation analyzer (PerkinElmer, Walton, MA). Uptake values were expressed as counts per minute per cell.
Evaluation of ER Stress and Autophagy Response in HCV Cell Culture
Activity of the ATF6 protein, which functions as an ER stress sensor, was measured in HCV-infected culture. Initially, Huh-7.5 cells were infected with MOI = 0.1 JFH-ΔV3-Rluc virus overnight. The next day, infectious cell culture supernatant was removed and p5xATF6-GL3 plasmid containing the firefly reporter gene was transfected with FuGENE 6 (Roche Diagnostics) transfection reagent in both uninfected and infected Huh-7.5 cells. ATF6–firefly luciferase activity was measured in a kinetic manner for ≤10 days. In the same lysates, HCV–Renilla luciferase activity was also measured, to confirm HCV infection (data not shown). The autophagy response due to HCV replication in the culture was assessed by measuring the autophagy-related hallmark proteins LC3, p62, and beclin 1 by Western blotting and immunohistochemistry. A published protocol20 was followed to visualize acidic autophagolysosome formation by fluorescence microscopy, using Acridine Orange staining. In brief, cells were stained in DMEM containing 5 μg/mL Acridine Orange for 15 minutes. The cells were washed three times in PBS and examined by fluorescence microscopy. The color change of Acridine Orange from green to red was monitored under an Olympus IX70 microscope (40× objective lens) and a DP-71 digital camera. The exposure time was 200 ms. The manufacturer’s cell imaging software was used for the acquisition of microscopic images.
Electron Microscopy
Uninfected and persistently infected Huh-7.5 cells were pelleted, rinsed, and resuspended in 3% glutaraldehyde fixative (Sigma-Aldrich). Cell pellets were postfixed in 1% osmium tetroxide and dehydrated with an ethyl alcohol series. Samples were infiltrated and embedded in eponate 12 resin and polymerized at 60°C for 24 hours. Thin sections (70 nm thickness) of the samples were placed on copper grids. Cells were examined using a G2 F30 Tecnai transmission electron microscope (TEM) at 200 kV (FEI, Hillsboro, OR). The numbers of autophagic vacuoles per field per cell were counted.
Immunostaining for HCV Core
Infected Huh-7.5 cells with or without IFN treatment were mounted onto a glass slide via the cytospin centrifugation method. The cells were washed twice with 10 mmol/L PBS (pH 7.4) for 5 minutes. The cells were fixed in chilled acetone for 15 minutes and then permeabilized by treatment with Reveal Decloaker RTU pretreatment reagent (RV 100; Biocare Medical, Concord, CA) for 25 minutes at its boiling point. Slides were then cooled to room temperature for 25 minutes. Blocking was performed using Background Sniper blocking reagent (BS966; Biocare Medical) for 10 minutes at room temperature. The cells were incubated with monoclonal anti-Core antibody (Pierce hepatitis C virus Core antigen-specific mouse monoclonal antibody, Ma1-080; Thermo Fisher Scientific, Rockford, IL) diluted 1:200 with Da Vinci Green diluent (PD900; Biocare Medical) for 1 hour at room temperature. After the primary antibody incubation, the cells were washed three times in Tris-buffered saline (pH 8.0), and incubated with MACH 4 mouse probe (UP534; Biocare Medical) for 10 minutes. The cells were then incubated with MACH 4 horseradish peroxidase polymer (MRH534; Biocare Medical) for 30 minutes, and washed with Tris-buffered saline three times. Next, the cells were treated with diaminobenzidine chromogen (Dako, Carpinteria, CA) for 5 minutes. The slides were counterstained with hematoxylin for 30 seconds and Tacha’s bluing solution (HTBLU; Biocare Medical) for 30 seconds and then were dehydrated, mounted, and observed by light microscopy.
Cell Surface Expression and Quantification of IFN Receptors
Cell surface expression of IFN-α, IFN-γ, and IFN-λ receptors (IFNAR1, IFNγR1, and IL10Rβ, respectively) in uninfected and persistently infected cells was examined at 4°C using a published protocol.21 1 × 106 Huh-7 cells were washed in ice-cold DMEM containing 1% (v/v) FBS. Cells were incubated for 1 hour at 4°C with primary antibody (1:25 dilution) specific for IFN-α, IFN-γ, or IFN-λ receptors with gentle shaking. Cells were then washed once with DMEM containing 1% (v/v) FBS and then incubated with Alexa Fluor 488 (green)–conjugated secondary antibody (dilution of 1:50) (Life Technologies) for 30 minutes at 4°C with shaking. Finally, cells were washed twice with DMEM containing 1% (v/v) FBS and an aliquot of cell suspension was examined for IFN-receptor expression by confocal microscopy at the Tulane National Primate Center (Covington, LA). The surface GFP expressions of the IFN-α, IFN-γ, or IFN-λ receptors were quantified by flow cytometric analysis. Three independent experiments were performed.
Sorting of IFN-α Sensitive and Resistant HCV-Infected Cells by Flow Cytometry
Persistently infected Huh-7.5 cells (45 days of infection) that did not clear HCV replication after two consecutive treatments with 2.5 × IC90 IFN-α were separated by flow cytometry after intracellular HCV Core protein staining. Infected cells were fixed for 10 minutes with 2% (w/v) paraformaldehyde in PBS. After this step, cells were washed once using DMEM containing 5% (v/v) FBS, and then were treated with 1 mL of 1× fluorescence-activated cell sorting (FACS) permeabilizing solution [0.5% (w/v) saponin in PBS] at room temperature. Cells were then incubated with a monoclonal antibody to HCV Core protein for 60 minutes at room temperature. After this step, cells were washed twice with DMEM containing 5% (v/v) FBS and incubated with Alexa Fluor 488–conjugated secondary antibody (1:50) (Life Technologies) diluted in DMEM containing 5% (v/v) FBS. Finally, cells were washed twice, pelleted, resuspended in PBS, and analyzed by FACS. IFN-α–sensitive (HCV Core-) and resistant (HCV Core+) cells were separated by flow sorting on a FACSAria system (BD Bioscience, San Jose, CA). Sorted cells were lysed, and expression levels of IFNAR1 in IFN-α–sensitive and resistant cells were determined by Western blotting.
Statistical Analysis
All measurements were made at least in triplicate (n = 3). To compare means within groups, we performed one-factor analysis of variance using GraphPad Prism software version 5.01 (GraphPad Software, La Jolla, CA). We assumed that all measurements have normal probability distributions, which is expected for these types of data. When the overall P value for the analysis of variance analysis was significant (P < 0.05), we applied Dunnett’s post hoc test to compare control samples with experimental samples.22 For comparisons between multiple groups, each analyzed with analysis of variance, we used the Bonferroni correction23 to determine a revised cutoff for statistical significance that gives a combined 5% type I error probability.
Results
Development of a Stable and Persistently Infected Huh-7.5 Cell Culture System
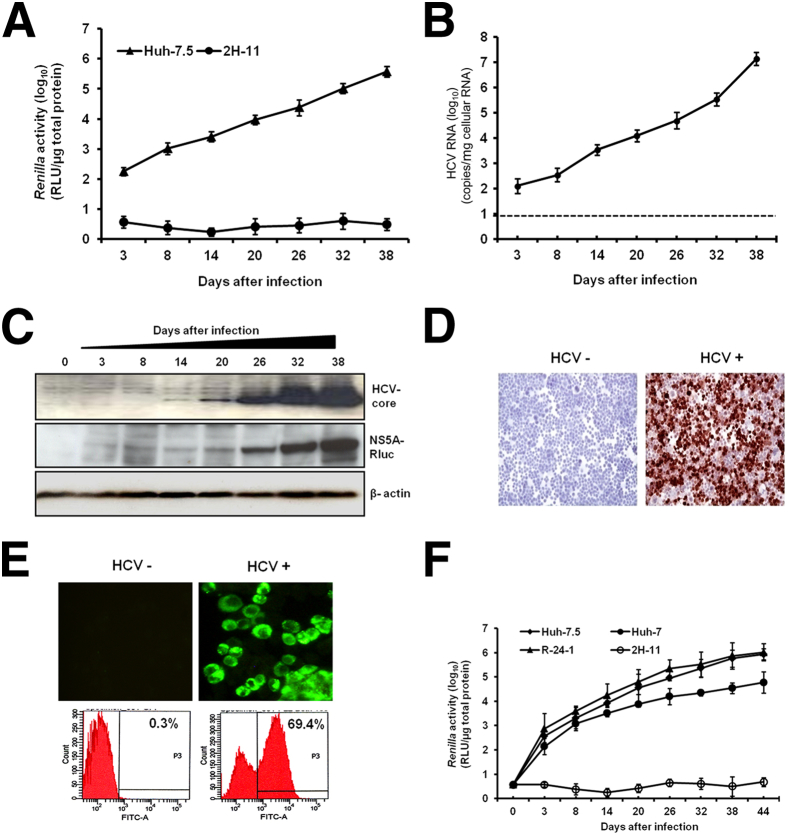
Huh-7.5 cells were infected with JFH-ΔV3-Rluc and then cultured continuously by splitting cells at 6-day intervals. HCV replication was confirmed by the measurement of Renilla luciferase activity of the cell lysate, as well as by determining HCV RNA titer using RT-qPCR (Figure 1, A and B). On day 3, the titer of HCV in the infected culture was 102 copies/μg of cellular RNA, which gradually increased to 107 copies/μg of cellular RNA after 38 days. These results were also confirmed by measuring the expression of HCV Core protein and HCV NS5A–Renilla luciferase fusion protein by Western blot analysis. The expression of HCV Core protein and NS5A–Renilla luciferase fusion protein in the infected cells increased over 38 days (Figure 1C), and these results were verified by quantification of band intensity (Supplemental Figure S1).
Figure 1.
Establishment of a stable and persistently infected HCV replication system in Huh-7.5 cells. Cells were infected with MOI = 0.1 JFH-ΔV3-Rluc virus overnight. The infected cells were cultured in DMEM with 10% (v/v) FBS, with passage every 6 days. A: The Renilla luciferase activity of infected cell lysates measured up to 38 days in culture indicates HCV replication after infection. The human fibroblast cell line (2H-11) was used as a negative control. B: HCV RNA level in the infected culture over 38 days. The dotted line indicates the limit of detection of the assay. C: HCV Core and NS5A–Rluc protein levels by time after infection, measured by Western blotting, with β-actin as a loading control. D: Immunocytochemical staining of uninfected (HCV−) and infected (HCV+) Huh-7.5 cells, using a monoclonal antibody specific for HCV Core protein; positive staining is reddish-brown. E: HCV Core protein was detected by immunofluorescence; the presence of HCV+ cells was confirmed by flow cytometric analysis and is reported as the percentage of HCV+ cells in the infected culture. F: Infectivity assay of culture supernatants was performed using three different hepatic cell lines (R-24-1 is an IFN-α–resistant Huh-7 cell line) and one nonhepatic cell line (2H-11). Cells were infected with 1 mL of MOI = 0.1 culture supernatant overnight, washed, and then cultured with growth medium. Cells were passaged at 6-day intervals, and Renilla luciferase activity of cell lysates was measured. Data are expressed as means ± SD. Original magnification: ×20 (D); ×40 (E). FITC, fluorescein isothiocyanate; RLU, relative light units.
The number of cells infected with HCV in the culture was determined by measuring Core protein expression by immunocytochemical staining (Figure 1D). Using a fluorochrome-labeled secondary antibody, the percentage of HCV Core+ cells was quantified using flow cytometric analysis. Approximately 70% of the cells in the culture were infected with HCV (Figure 1E). The persistently infected cells secreted infectious virus particles as confirmed by a multicycle infectivity assay using three different liver-derived cell lines and one non–liver-derived cell line. Huh-7 cells with defective JAK–STAT signaling (R-24-1) supported high-level HCV replication, compared with other Huh-7 cells. The human fibroblast cell line 2H-11 used as a negative control did not support HCV infection (Figure 1F). The viral titer of Huh-7.5 infected cells passaged for 13 rounds (P13) was much higher, compared with the original JFH1 that has not been passaged (P0) (Supplemental Figure S2).
Persistently Infected HCV Cell Culture Is Partially Responsive to IFN-α and RBV but Completely Responsive to IFN-λ
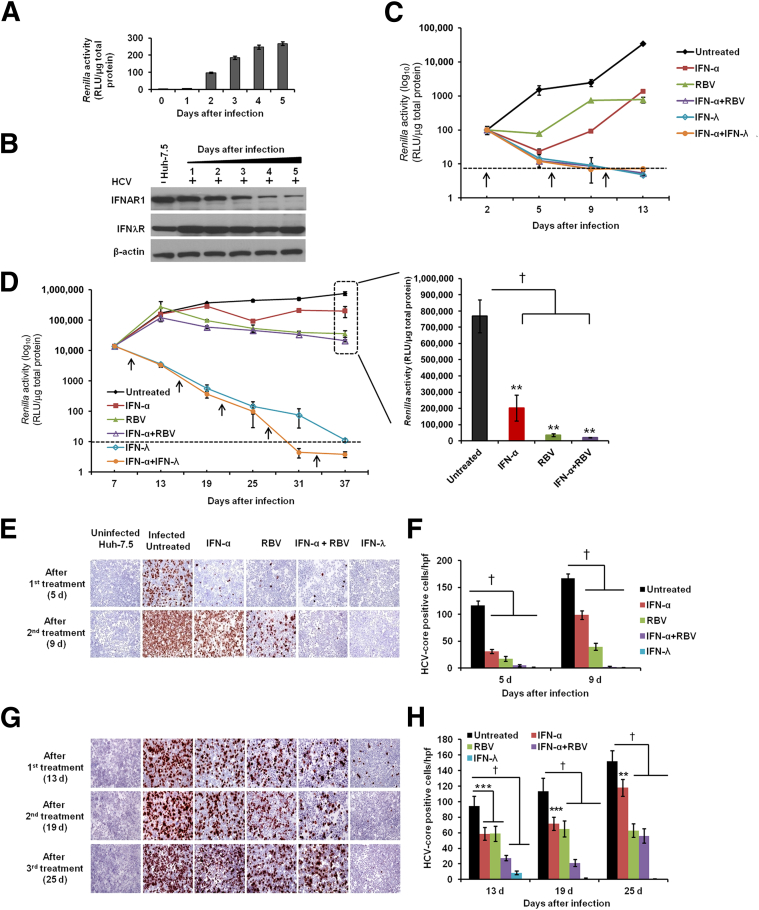
The IC90 of IFN-α and IFN-λ was determined, to allow calibration and comparison of their biological activity. We found that the IC90 for IFN-α, 100 IU/mL, was equivalent to 10 ng/mL of IFN-λ (Supplemental Figure S3A). This result is in agreement with published data.24 The antiviral treatment response of IFN-α, RBV, and IFN-λ in the HCV-infected culture was examined over 37 days. Huh-7.5 cells infected with JFH-ΔV3-Rluc recombinant virus showed increased Renilla luciferase expression (Figure 2A). HCV infection selectively down-regulated the expression of the IFN-α receptor, IFNAR1, but not of the IFN-λ receptor (Figure 2B). The HCV-infected culture was more sensitive to the IFN-α+RBV combination treatment, but relatively insensitive to either agent singly (Figure 2, C and D). Infected cells treated with IFN-α+RBV or with IFN-α+IFN-λ exhibited a decrease in Renilla luciferase activity, which fell to below the detection limit after 9 days (Figure 2C), as well as a decrease in HCV RNA level (data not shown). The antiviral effect of IFN-α+RBV was examined at multiple time points over 37 days. Results of three different measurements revealed that IFN-α, RBV, and IFN-α+RBV all inhibited HCV replication (P < 0.0001), but did not clear the virus (Figure 2D), indicative of a persistently infected state. Using a HCV subgenomic replicon model, we have previously observed that IFN-λ has marked antiviral activity in IFN-α–resistant cell strains (unpublished data). We therefore compared the antiviral activity of IFN-λ with that of IFN-α+RBV using our persistently infected Huh-7.5 cell culture model.
Figure 2.
HCV replication in persistently infected Huh-7.5 cell culture is partially resistant to IFN-α and RBV but not to IFN-λ treatment. A: Infection kinetics of culture supernatant (without passage) in Huh-7.5 cells was determined (MOI = 0.1) by measuring the Renilla luciferase activity, normalized to total protein. B: Levels of IFN-α and IFN-λ receptors (IFNAR1 and IFNλR) in the infected cells were measured by Western blotting, with β-actin as internal control. C: Antiviral response of IFN-α, RBV, and IFN-λ in short-term infected culture. Huh-7.5 cells were infected with cell culture–derived MOI = 0.1 HCV and treated with IFN-α, RBV, or IFN-λ, alone or in combination. The treatment was given every 96 hours (arrows). Antiviral activity was determined by measuring Renilla luciferase activity of cell lysates. D: Antiviral response of 2.5 × IC90 IFN-α, 40 μg/mL RBV, and 10 × IC90 IFN-λ in persistently infected culture. Arrows indicate treatments. To show the partial inhibition of HCV replication by IFN-α and RBV after five consecutive treatments (37 days), viral titer is graphed in greater detail at the left. E and F: HCV Core protein (brown) was detected by immunocytochemistry in 2-day-infected culture (E) and HCV Core+ cells (F) were counted in 10 different high-power fields and compared with untreated controls. G and H: HCV Core protein was detected in persistently infected culture (G) and HCV Core+ cells (H) were counted in 10 different high-power fields and compared with untreated controls. Data are expressed as means ± SD. ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001. Original magnification, ×40 (E and G). hpf, high-power field (×40).
Consistent with our previous findings, Renilla luciferase activity showed sustained and significant inhibition in the cultures treated with IFN-λ1 (IL-29) (Figure 2D). Moreover, IFN-α+IFN-λ inhibited HCV replication to a greater extent than either single agent alone, and levels at days 31 and 37 were below the assay detection limit. The luciferase-based HCV replication assay results were confirmed by measuring HCV RNA levels by RT-qPCR, using primers targeted to the 5′ UTR (data not shown). The antiviral activity of IFN-α and RBV was also confirmed by immunostaining of infected cells before and after treatment, using a monoclonal antibody to HCV Core protein in both short-term infected (Figure 2, E and F) and persistently infected culture (Figure 2, G and H). The immunostaining results were in agreement with the Renilla luciferase–based results, confirming the sustained antiviral activity of IFN-λ against persistent HCV infection.
IL-28A/B also inhibited HCV replication in the persistently infected cell culture in a similar capacity as IL-29 (data not shown). However, neither IFN-α nor IFN-α+RBV cleared HCV replication in our persistently infected cell culture model. Having determined that 100 IU/mL IFN-α is equivalent to 10 ng/mL IFN-λ, we compared the antiviral effects of the two interferons separately, with persistently infected cells treated with 2.5 × IC90, 5 × IC90, or 10 × IC90 of IFN-α or IFN-λ. The antiviral effect was compared over three consecutive treatments by the measurement of Renilla luciferase and Core protein immunostaining. Results of both assays (Supplemental Figure S3, B and D) confirmed that the antiviral effect of IFN-λ was significantly stronger than IFN-α when both were used at equivalent concentrations (P < 0.01, P < 0.001, and P < 0.0001). HCV Core protein expression was absent in 10 × IC90 IFN-λ–treated cells, but not in 10 × IC90 IFN-α–treated cells (Supplemental Figure S3, B–D). At equivalent 10 × IC90 concentration, IFN-λ showed significantly stronger inhibition and HCV clearance, compared with IFN-α.
The impaired clearance of HCV by IFN-α was not related to a lack of biological activity, because the same IFN-α preparation tested against an S3-GFP stable replicon cell line exhibited significant concentration-dependent antiviral activity (Supplemental Figure S3E). There is a possibility that long-term passaged virus-infected cells could have selected a population of cells that is less responsive to IFN-α. Τo address this concern, we prepared a cured cell line by removing HCV, using repeated treatment with a combination of two siRNAs (si321 and si359). The expression of IFN receptors (mainly IFNAR1 and IFNλR) and JAK–STAT signaling (mainly STAT1 and STAT2) in the cured cell line was restored to a level similar to that in the uninfected Huh-7.5 cells. Reinfection of the same cured cell line with HCV exhibited down-regulated expression of IFNAR1 and impaired JAK–STAT signaling (Supplemental Figure S4).
Persistently HCV-Infected Huh-7.5 Cell Culture Develops ER Stress and Autophagy Response
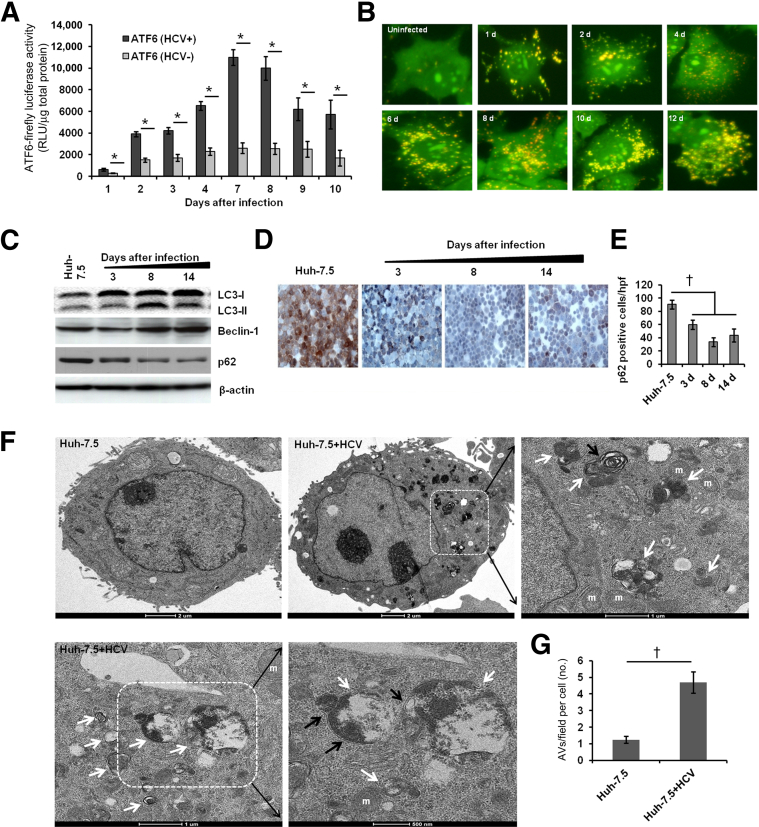
We have previously shown that initiation of the ER stress response in cells after HCV infection activates IRE1α, PERK, and p-eIF2α.9 In the present study, ATF6-luciferase activity showed that persistently HCV-infected culture also induced a chronic ER stress response. Persistently infected cells exhibited significantly higher ATF6-luciferase activity, compared with uninfected culture, over 10 days (Figure 3A). Recent reports indicate that a cellular autophagy response is activated secondary to ER stress as a cell-survival defense mechanism.25,26
Figure 3.
HCV replication induces ER stress and an autophagy response. A: Uninfected (HCV−) and infected (HCV+) Huh-7.5 cells were transfected with ATF6–firefly luciferase reporter plasmid using FuGENE 6 transfection reagent. Luciferase activity in the cell lysates was measured over 10 days. B: Acridine Orange staining shows the induced autophagy response in Huh-7.5 cells infected with HCV. Cells without autophagy show green fluorescence. Cells with autophagy show accumulation of orange-red cytoplasmic autophagic vacuoles. C: Induction of autophagy in the infected cells at different time points was assessed by measuring autophagy-related proteins by Western blotting. D: The level of p62 (brown cytoplasmic staining) in persistently infected cells was measured by immunohistochemistry. E: The p62+ cells in 10 different high-power fields (×40) were counted at 3, 8 and 14 days and compared with uninfected control (Huh-7.5). F: TEM analysis of ultrastructure of uninfected (Huh-7.5) and persistently infected (Huh-7.5+HCV) hepatocytes. Representative high-power micrographs of autophagic vacuoles (AVs) present in the persistently infected cells (white arrow) show atypical double-membrane AVs (black arrow). Boxed regions are shown at higher magnification in the panel to the right. G: During TEM studies, 10 cytoplasmic fields in a grid were captured randomly in each cell. AV numbers per field per cell were compared between uninfected and persistently infected cells. Data are expressed as means ± SD. ∗P < 0.05; †P < 0.0001. Original magnification: ×40 (B and D). m, mitochondria.
We also investigated whether persistent HCV replication can induce the formation of autophagosomes, which then progress to autophagolysosomes through fusion with acidic lysosomes. We used Acridine Orange staining to examine this process (Figure 3B). There was a significant increase in the number of orange-colored autophagolysosomes in HCV-infected cells (Supplemental Figure S5), but the uninfected Huh-7.5 cells exhibited cytoplasmic and nuclear green fluorescence without induction of autophagy (Figure 3B). Induction of an autophagy response in the persistently HCV-infected culture was examined using assays described previously.20 Among these, LC3 is used as a specific marker for induction of autophagy; the processing of LC3-I into LC3-II is assessed by Western blotting. Infected cells were harvested at 3, 8, and 14 days and processing of LC3-I into LC3-II was examined. Induction of autophagy and conversion of LC3-1 into LC3-II was increased in the infected cells at 8 and 14 days (Figure 3C). The protein levels for one of the critical autophagy genes, beclin 1 (BECN1), was also induced in the persistently infected cells at 8 and 14 days (Figure 3C and Supplemental Figure S6). The expression of p62 protein was reduced significantly in the infected cells in a time-dependent manner, which indicated that HCV replication induced an autophagy response that selectively degraded p62 protein (Figure 3, D and E). The presence of HCV-induced autophagic vacuoles and the ultrastructure of Huh-7.5 cells with or without 14 days of virus infection were examined by TEM-based ultrastructural analysis. We quantified double-membrane autophagosomes by counting 10 HCV-infected and 10 uninfected Huh-7.5 cells using electron microscopy (Figure 3F). Significantly higher numbers of autophagic vacuoles were present in the persistently infected cells, compared with uninfected cells (Figure 3G). Taken together, these results provide evidence that HCV infection results in ER stress and autophagy responses.
Chronic ER Stress and the Autophagy Response Impair the Antiviral Action of IFN-α
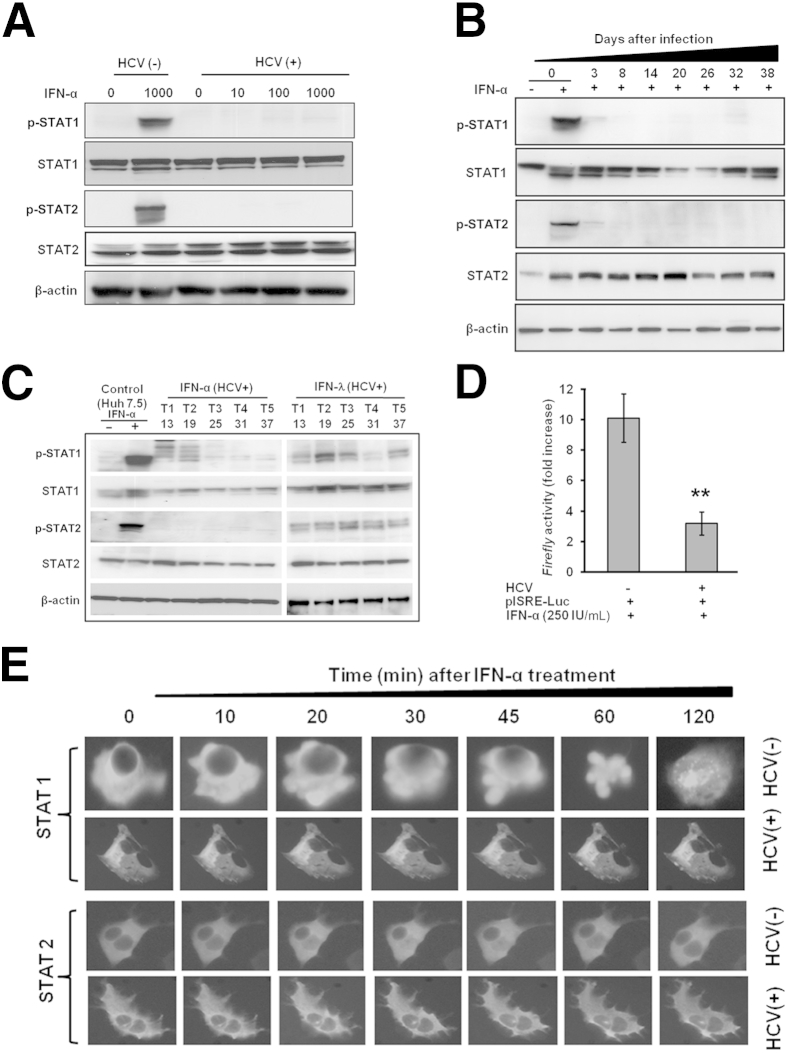
A series of reports using stable replicon cell lines have provided evidence that activation of JAK–STAT signaling is critical for the successful antiviral effects of IFN-α.9,27,28 Activation of JAK–STAT signaling can be detected by the measurement of IFN-α–induced STAT1 and STAT2 phosphorylation. Phosphorylation of STAT1 and STAT2 after 30 minutes of treatment with 10 × IC90 IFN-α was blocked in persistently infected cells for up to 38 days (Figure 4, A and B). Total STAT1 levels varied over the time course, but total STAT2 was essentially unaffected (Figure 4B). The levels of STAT1 and STAT2 proteins in infected cells were also measured by Western blotting, after five consecutive treatments (T1 to T5) with IFN-α or IFN-λ. The levels of p-STAT1 and p-STAT2 were rescued when the infected cells were treated with IFN-λ, but were undetectable after treatment with IFN-α (Figure 4C). IFN-α–mediated activation of STAT1 and STAT2 proteins was verified using an ISRE–firefly luciferase assay. ISRE promoter activity was significantly reduced (P = 0.01) in persistently infected cells (Figure 4D). To determine whether the defective STAT1 and STAT2 phosphorylation impairs nuclear translocation of STAT1 and STAT2, pSTAT1-GFP and pSTAT2-GFP plasmids were transiently transfected into uninfected and infected Huh-7.5 cells. The impaired nuclear translocation of STAT1 and STAT2 was observed only in cells persistently infected with HCV (Figure 4E).
Figure 4.
Persistent HCV replication impairs IFN-α–induced phosphorylation of STAT1 and STAT2 proteins, their nuclear translocation, and ISRE-luciferase promoter activation. A: Uninfected (HCV−) and infected cells (HCV+) were treated with 0, 0.1 × IC90, 1 × IC90, and 10 × IC90 IFN-α for 30 minutes. Equal protein amounts were separated on SDS-PAGE gels, and Western blotting analysis was performed using antibodies to p-STAT1, STAT1, p-STAT2, STAT2, and β-actin. B: Phosphorylation of STAT proteins in the persistently infected cells without or with IFN-α over 38 days was measured 30 minutes after treatment by Western blotting. C: HCV-infected cells were treated with 2.5 × IC90 IFN-α or 10 × IC90 IFN-λ. Five consecutive treatments (T1 to T5) were given, and STAT protein levels were measured 30 minutes after each treatment by Western blotting. D: IFN-α induced STAT1 and STAT2 activity in uninfected and infected Huh-7.5 cells. An equal number of uninfected and infected cells were transfected with ISRE-luciferase plasmid. After 3 hours, cells were treated with 2.5 × IC90 IFN-α; on the next day, cells were lysed and luciferase activity was measured. The presence of HCV replication was associated with significantly reduced ISRE-luciferase promoter activity. E: Nuclear translocation of pSTAT1-GFP and pSTAT2-GFP in the uninfected (HCV−) and persistently infected (HCV+) cell culture over 120 minutes was monitored by fluorescence microscopy. Data are expressed as means ± SD. ∗∗P < 0.01. Original magnification: ×40 (E).
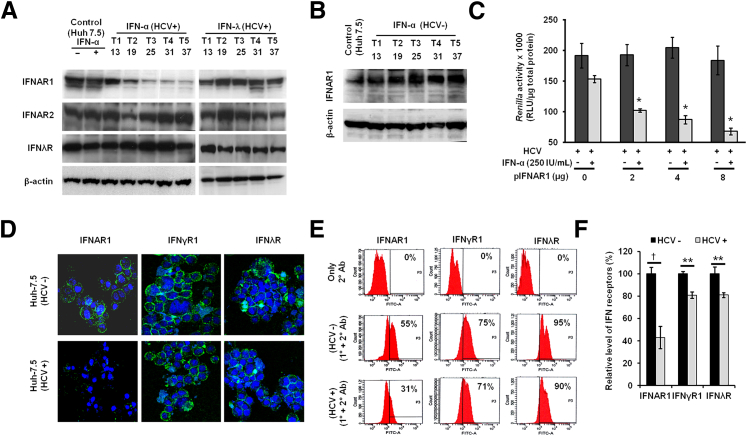
We examined whether the loss of cell surface expression of IFNAR1 affects JAK–STAT signaling in persistently infected cells. The expression level of IFNAR1 in the persistently HCV-infected cell culture system was examined by Western blotting. The levels of IFNAR1, IFNAR2, and the IFN-λ receptor in the persistently infected Huh 7.5 cells were also measured, by Western blotting, after five consecutive treatments (T1 to T5) with 2.5 × IC90 IFN-α or 10 × IC90 IFN-λ (Figure 5A). The reduction of IFNAR1 expression was greater in the untreated HCV-infected culture (Figure 2B) than in the IFN-α–treated HCV-infected culture (Figure 5A), because IFN-α treatment significantly inhibits the HCV replication that could have restored the expression level of IFNAR1. This conclusion was confirmed by measuring the levels of IFNAR1 between untreated and treated HCV-infected culture (Supplemental Figure S3F). The levels of IFNAR1 decreased over time, and this could be reversed by treatment with IFN-λ but not with IFN-α. The expression of IFNAR1 did not change in uninfected Huh-7.5 cells that were repeatedly treated with IFN-α for 37 days (Figure 5B), eliminating the possibility that the loss of IFNAR1 expression was due to antiproliferative effects of IFN-α. Furthermore, re-expression of ectopic IFNAR1 in the persistently infected cells significantly increased the antiviral activity of IFN-α (Figure 5C). The effect of HCV replication in the persistently infected cell culture on the cell surface expression of IFNAR1, IFNγR1, and IL10Rβ (IFNλR) was examined by confocal microscopy at day 37 (Figure 5D). The above-mentioned type I, type II, and type III interferon receptors were expressed constitutively in uninfected cells; in persistently infected cells, however, IFNAR1 expression was decreased to a significantly greater extent than the other two receptors (Figure 5, E and F).
Figure 5.
Persistent HCV replication results in down-regulation of IFNAR1. A: HCV infection selectively down-regulates the IFNAR1 receptor. HCV-infected cells repeatedly treated with 2.5 × IC90 IFN-α or 10 × IC90 IFN-λ. Five consecutive treatments (T1 to T5, day 13 to day 37) were given, and after each treatment the expression of IFNAR1, IFNAR2, and IL10Rβ (IFNλR) was measured by Western blotting, with β-actin as loading control. B: Uninfected Huh-7.5 cells (HCV−) were repeatedly treated with IFN-α, and IFNAR1 was measured at the five time points by Western blotting, with β-actin as loading control. C: Overexpression of IFNAR1 by transient transfection increased the IFN-α antiviral response against HCV in the infected cell culture (P < 0.02). Antiviral action of IFN-α was assessed by measuring Renilla luciferase activity 48 hours after treatment and compared with untreated controls. D: The surface expression of IFN-α (IFNAR1), IFN-γ (IFNγR1), and IFN-λ (IL10Rβ) receptors in uninfected and HCV-infected cells was evaluated by confocal microscopy. E: The expression of IFNAR1, IFNγR1, and IL10Rβ in normal (HCV−) and persistently infected (HCV+) Huh-7.5 cells was quantified by FACS analysis. Cells incubated with secondary antibody were used as an experimental control. Percentages indicate the proportion of cells expressing IFN receptors. Data are representative of three independent experiments. F: Quantitative expression of IFNAR1, IFNγR1, and IFNλR was determined by FACS analysis. Data are expressed as means ± SD. ∗P < 0.05, ∗∗P < 0.01, and †P < 0.0001. Original magnification: ×60 (D). Ab, antibody; 1°, primary; 2°, secondary.
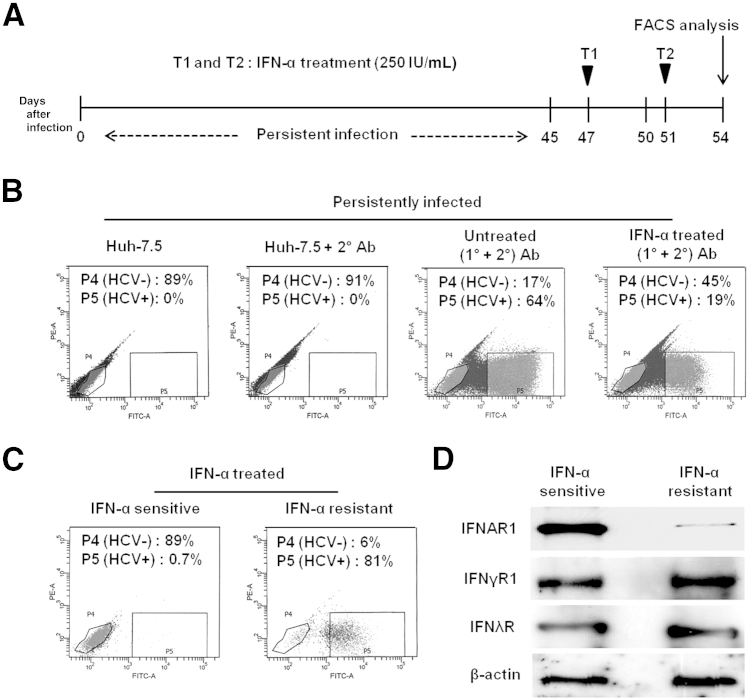
We further verified our hypothesis, that the lack of cell surface expression of IFNAR1 might cause the IFN-α resistance, by analyzing HCV+ and HCV− cell populations after flow sorting (Figure 6). Persistently infected cells were treated with 2.5 × IC90 IFN-α on days 47 and 51, and cell sorting after HCV Core protein staining was performed on day 54 (Figure 6A). Treatment with IFN-α induced HCV clearance to 45% at P4 (compared with 17% for untreated cells); nearly one third of IFN-α–treated infected cells (19% out of 64%) did not clear HCV at P5 (Figure 6B). The specificity of sorted cells was verified; sorted cells were exclusively either HCV− (IFN-α sensitive) or HCV+ (IFN-α resistant) (Figure 6C). Protein extracts of flow-sorted HCV+ and HCV− cell populations were examined for the expression of IFNAR1, IFNγR1, IFNλR, and β-actin by Western blotting (Figure 6D). The IFN-α–resistant cells had a notably lower level of IFNAR1 than the IFN-α–sensitive cells. The faint IFNAR1 band seen in the resistant cells (Figure 6D) could be due to the presence of 6% of HCV− cells separated by flow sorting. The expression of IFNγR1, IFNλR, and β-actin between the two cell populations was similar.
Figure 6.
Persistent HCV infection impairs antiviral action of IFN-α via down-regulation of IFNAR1. A: Experimental design. Persistently infected Huh-7.5 cells (45 days of infection) received two consecutive treatments (T1 and T2) with 2.5 × IC90 IFN-α, and then HCV+ and HCV− cells were sorted by using FACS. B: Detection of HCV in untreated and IFN-α–treated cells. HCV Core protein was labeled by immunofluorescence technique using green fluorescence secondary antibody and then detected by flow cytometry. Persistently infected cells (Huh-7.5) and cells with only secondary antibody (Huh-7.5 + 2° Ab) were used as controls. C: After IFN-α treatment, HCV− (IFN-α–sensitive) and HCV+ (IFN-α–resistant) cell populations were separated by FACS. D: The level of IFN receptors between IFN-α–sensitive and resistant populations was measured by Western blotting, with β-actin as internal control. PE, phycoerythrin.
Reduced Expression of Nucleoside Transporter and Impaired RBV Uptake in Persistently Infected Cells
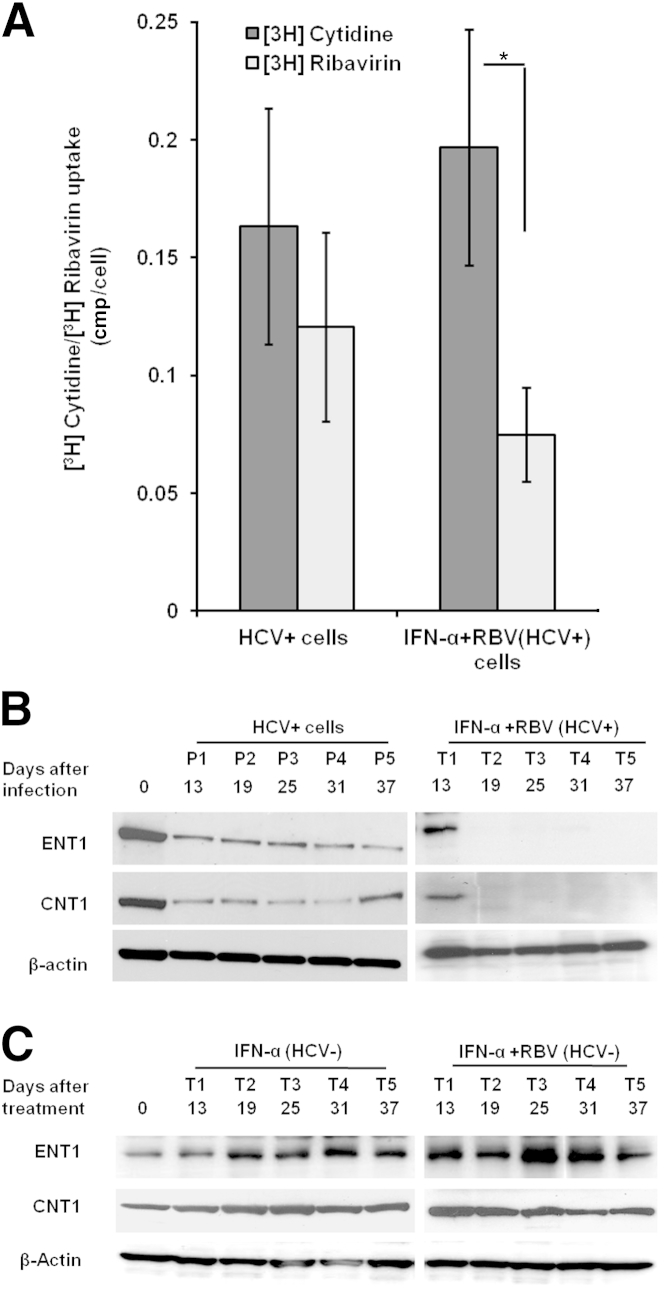
RBV plays a crucial role in the treatment of chronic HCV infection, although the mechanism or mechanisms by which many patients develop resistance to RBV are not well understood. Ibarra and Pfeiffer13 reported that reduced uptake by HCV-infected cells contributes to an impaired antiviral response. They demonstrated that RBV uptake was reduced in the infected cells, although transporter levels were unaltered. In the present study, we investigated whether reduced cellular import of RBV could have caused its treatment failure in persistently HCV-infected cells. Persistently infected cells showed 26% less RBV uptake with respect to cytidine (used as a control), but the difference was not statistically significant. Uptake was reduced by up to 62% in the IFN-α+RBV–resistant cell strain (Figure 7A). The significant reduction of RBV uptake in the IFN-α+RBV–resistant cell strain (P = 0.02) could have been due to the fact these cultures had previously been exposed to 40 μg/mL RBV. RBV is transported into cells via the nucleoside transporters ENT1 and CNT1.29 The expression of the ENT1 and CNT1 nucleoside transporters in the infected and uninfected cells was examined by Western blotting. The expression of ENT1 and CNT1 was notably reduced in the IFN-α+RBV–resistant cells, compared with the HCV-infected cells (Figure 7B). The expression level of ENT1 and CNT1 in uninfected Huh-7.5 cells did not change significantly when treated with IFN-α or with IFN-α+RBV (Figure 7C).
Figure 7.
Impaired RBV uptake and reduced expression of nucleoside transporters in an IFN-α- and RBV-resistant cell strain. A: [3H]Cytidine and [3H]RBV uptake assay. Persistently infected cells were repeatedly treated with IFN-α and RBV for 37 days to produce an IFN-α+RBV–resistant strain of the Huh-7.5 cell line. Persistently infected Huh-7.5 cells were used as a control. Cells were incubated in medium containing [3H]RBV and [3H]cytidine (as a control). [3H]RBV uptake was increased, relative to control (P = 0.02), as determined by scintillation counting. The data were normalized to account for cell numbers. B: Western blot analysis of nucleoside transporters (ENT1 and CNT1) using equal amount of protein lysates from uninfected Huh-7.5 cells, HCV-infected Huh-7.5 cells, and IFN-α+RBV–resistant persistently infected cells at five treatment times (T1 to T5, day 13 to day 37). C: The level of transporters (ENT1 and CNT1) after prolonged IFN-α and IFN-α+RBV treatment in uninfected cells. Huh-7.5 cells received five consecutive treatments (day 13 to day 37) of IFN-α and IFN-α+RBV, and the transporter levels were measured by Western blotting. Data are expressed as means ± SD. ∗P < 0.05.
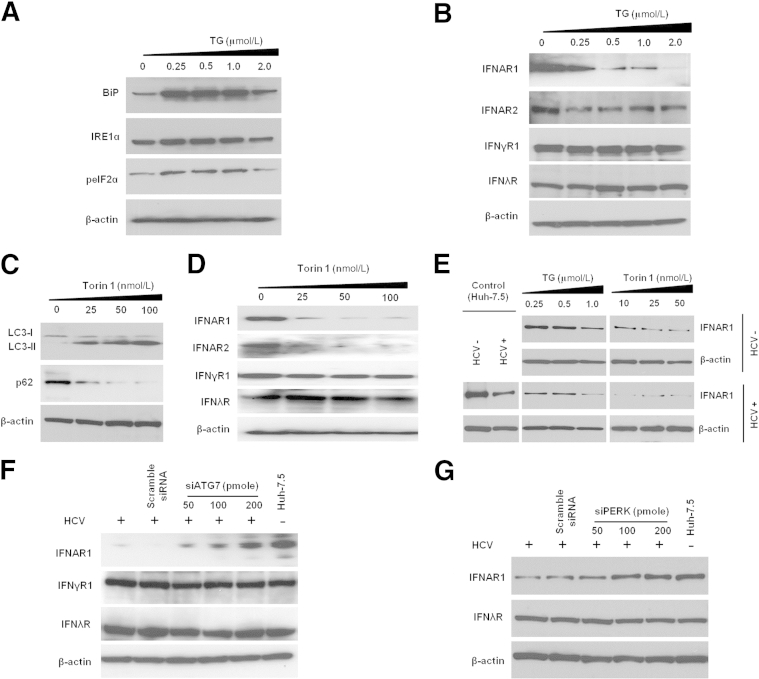
Induction of ER Stress and Autophagy Response Selectively Down-Regulates IFNAR1
To investigate the relationship between ER stress and autophagy and IFN receptors, we first induced ER stress and autophagy response with the known inducers TG and Torin 1, respectively, in uninfected Huh-7.5 cells (Figure 8, A and C). ER stress and autophagy induction impaired expression of type 1 IFN receptors (IFNAR1 and IFNAR2) (Figure 8, B and D). We performed TG and Torin 1 titration experiments in HCV-infected and uninfected Huh-7.5 cells to compare IFNAR1 expression by Western blotting. Degradation of IFNAR1 was significantly higher if infected cells were treated with either TG or Torin 1, indicating that the mechanisms of IFNAR1 regulation are linked to ER stress and autophagy response associated with HCV infection (Figure 8E). In another approach, we verified that knockdown of the autophagy-related gene ATG7 and the ER stress kinase gene PERK (EIF2AK3) by siRNA rescued the expression of IFNAR1 in persistently infected cells (Figure 8, F and G). Taken together, these results provide evidence for the relationship of ER stress and autophagy with IFNAR1 down-regulation.
Figure 8.
Induction of ER stress and autophagy response selectively down-regulated the expression of IFN-α receptor 1 measured by Western blotting. A: Huh-7.5 cells were treated with TG, a known ER stress inducer, and the unfolded protein response–related proteins BiP, IRE1α, and peIF2α were measured. B: The level of IFN receptors with increasing concentration of TG was measured. C: Huh-7.5 cells were treated with increasing concentration of Torin 1, a known autophagy inducer, and the autophagy response was assessed by measuring LC3II and p62 levels. D: The level of IFN receptors with increasing concentration of Torin 1 was measured. E: TG and Torin 1 titrations in HCV-infected cells and uninfected cells. IFNAR1 down-regulation was greater in HCV-infected cells treated with autophagy inducer (Torin 1) or ER stress inducer (TG). F: Knockdown by siRNA of ATG7, one of the autophagy genes, rescued IFNAR1 level. Increasing concentration of siATG7 was transfected in the persistently infected Huh-7.5 cells. After 72 hours, cells were lysed and the level of IFN receptors was measured. G: Silencing PERK restores the expression of IFNAR1. siPERK in increasing concentrations was transfected to the persistently infected Huh-7.5 cell. After 72 hours, cells were lysed and the expression of IFNAR1 and IFNλR was measured.
Silencing of ER Stress and the Autophagy Response Overcome IFN-α Resistance in Persistently Infected Cells
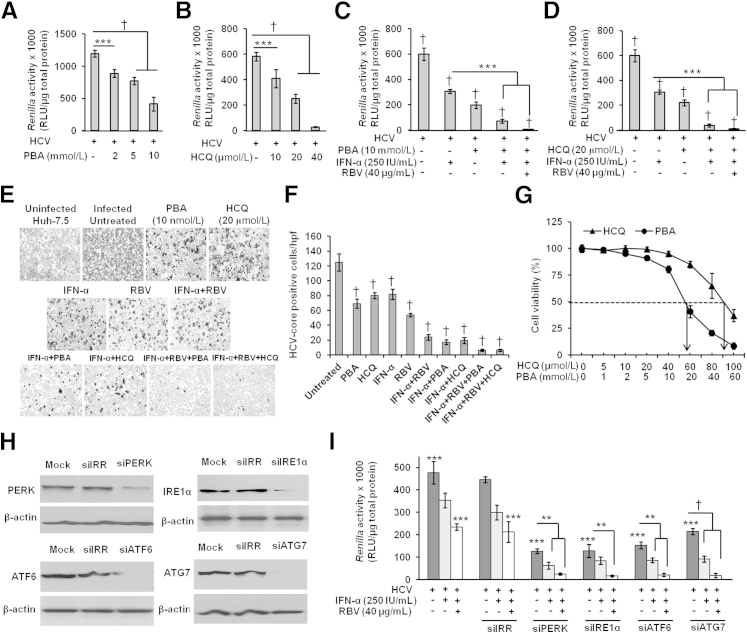
To further investigate the role of ER stress and the induced autophagy response in effecting defective JAK–STAT signaling and IFN-α resistance, persistently HCV-infected cells were pretreated with inhibitors of either ER stress (PBA) or autophagy (HCQ) for 24 hours; the antiviral activity of IFN-α was determined at 72 hours after treatment, by measuring Renilla luciferase activity. Both PBA and HCQ significantly inhibited HCV replication in a dose-dependent manner (Figure 9, A and B). We examined whether treatment of the HCV-infected culture with PBA or HCQ in combination with IFN-α, or with IFN-α+RBV, can clear HCV replication effectively. IFN-α significantly inhibited HCV replication in the presence of either PBA or HCQ (Figure 9, C and D). The results were confirmed by measuring HCV Core protein expression after immunostaining (Figure 9, E and F). The concentrations of PBA and HCQ used in the present study were not toxic to cells, as determined using an MTT assay (Figure 9G). In a different approach, genes for three cellular ER stress sensors (PERK, IRE1α, and ATF6) and one of the autophagy genes, ATG7, were silenced using siRNA; silencing was confirmed by Western blotting (Figure 9H). The siRNAs were transfected into the HCV-infected cell culture and, after 3 hours, cells were treated with 2.5 × IC90 IFN-α and 40 μg/mL RBV. The antiviral activity of IFN-α and of IFN-α+RBV improved significantly after these genes were silenced (Figure 9I).
Figure 9.
Inhibition of ER stress and autophagy response by chemical inhibitors significantly improves the IFN-α and RBV antiviral effect against HCV in persistently infected cells. A and B: Pretreatment with ER stress inhibitor (PBA) or autophagy inhibitor (HCQ) significantly inhibits HCV replication. Equal numbers of persistently infected cells were treated with different concentrations of PBA (0 to 10 mmol/L) and HCQ (0 to 40 μmol/L) overnight. The next day, cells were incubated with fresh medium; after 72 hours, HCV replication was assessed by measuring Renilla luciferase activity. C–E: Pretreatment with PBA and HCQ significantly improved antiviral activity of IFN-α, alone or in combination with RBV, inhibiting HCV replication. Equal numbers of persistently infected cells were treated overnight with 10 mmol/L PBA or 20 μmol/L HCQ. The next day, cells were incubated with fresh medium and treated with 250 IU/mL IFN-α alone or in combination with 40 μg/mL RBV. After 72 hours, HCV replication was assessed by measurement of Renilla luciferase activity (C and D) and HCV Core protein expression by immunostaining (E). F: HCV Core+ cells in 10 different high-power fields (×40) were counted and compared with untreated control. G: Cell viability with PBA or HCQ treatment was assessed by MTT assay. Huh-7.5 cells were treated overnight with different concentrations of PBA (0 to 60 mmol/L) or HCQ (0 to 100 μmol/L). After 72 hours, MTT assay was performed. The IC50 (dashed line) of PBA and of HCQ is indicated by an arrow. H and I: Silencing of ER stress sensors (PERK, IRE1α, ATF6) and the autophagy gene (ATG7) significantly inhibited HCV replication and also improved the antiviral action of IFN-α and RBV. Equal numbers of persistently infected cells were split in a six-well plate. The next day, cells were transfected with the respective siRNAs, mock transfection, or siIRR. After 3 hours of transfection, cells were treated with IFN-α alone or in combination with RBV. After a further 72 hours, a portion of the cells was lysed, and silencing efficacies of the siRNAs were determined by Western blotting (H) and HCV replication was assessed by measuring Renilla luciferase activity (I). All experiments were performed in triplicate. Data are expressed as means ± SD. ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001.
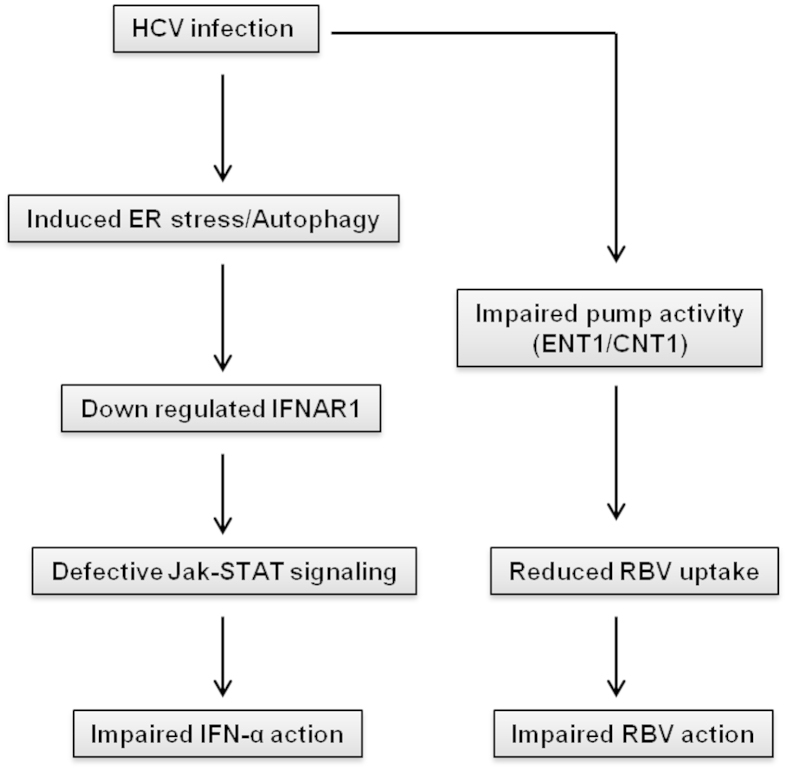
The silencing of ATG7 inhibits autophagy, which is why it restores the expression of IFNAR1. We observed that inhibiting autophagy by silencing ATG7 to some extent also inhibits HCV replication and Renilla luciferase activity. This is consistent with previous report that inhibition of autophagy inhibits HCV replication.17 Cells transfected with irrelevant siRNA (siIRR) also showed some reduction in Renilla luciferase activity, but these differences were not statistically significant in comparison with ATG7-transfected cells (Figure 9I). These experiments are suggestive of a causal relationship between ER stress and the autophagy response and the mechanisms underlying the HCV resistance to IFN-α+RBV. Based on these results, we propose a model of how HCV replication in the persistently infected cell culture model impairs the IFN-α and RBV antiviral mechanism (Figure 10).
Figure 10.
Schematic of impairment of IFN-α and RBV signaling in persistently HCV-infected culture.
Discussion
We developed a stable and persistently HCV-infected Huh-7.5 cell culture system to study long-term antiviral treatment response using IFN-α, RBV, and IFN-λ. Intracellular HCV replication and virus secretion in the persistently infected cell culture is stable for an extended period (more than a year). We showed that a very high percentage of cells in the culture are infected with HCV, which can be passaged over a number of generations. We propose that this cell culture model can be used to address a number of questions related to virus–host interaction in the persistence of HCV and pathogenesis.
Various host-related and virus-related factors have been proposed to be responsible for the mechanisms of HCV resistant to IFN-α and RBV.12 It has been observed in clinical studies that chronic HCV patients with a high viral load have a significantly lower sustained virologic response to IFN-α+RBV antiviral therapy,30–32 but the mechanism or mechanisms underlying the resistance are not well understood. Our cell culture study results indicate that HCV replication in the persistently infected cell culture cannot be completely eliminated by IFN-α or RBV alone, nor even with the IFN-α+RBV combination. In contrast, IFN-λ (IL-29) treatment at an equivalent concentration achieved a sustained antiviral response, leading to viral clearance in the persistently infected cell culture.
These results are quite different from those of studies performed using subgenomic replicon cell lines or acutely HCV-infected cell culture models. In our persistently HCV-infected cell culture, expression of nucleoside transporters (ENT1, CNT1) is reduced, which explains why RBV treatment alone was not effective for clearing HCV replication in this cell culture. To find an explanation why HCV replication is not eliminated even after repeated treatment with IFN-α, we looked at the modulation of the cell surface expression of IFNAR1 (type I IFN receptor). We found that the HCV-induced ER stress and autophagy response selectively repair type I interferon signaling, but not type III. This finding is supported by evidence that ER stress down-regulates IFNAR1 expression.33
A number of studies have shown that the unfolded protein response due to ER stress and the autophagy response both play an important role for HCV replication and for suppression of antiviral signaling in infected cells.9,33–36 Hepatic ER stress37 and an increased autophagic response38 have been reported in liver of chronically HCV-infected patients, relative to healthy liver. The present study demonstrates a causal link between chronic ER stress and autophagy and the negative expression of IFNAR1 and impaired antiviral response. This hypothesis is supported by results indicating that inhibiting the ER stress and autophagy responses improved the antiviral response of IFN-α+RBV. The mechanisms of HCV persistence in the infected culture involve new rounds of infection, as well as carriage from previously infected cells. We suspect, therefore, that HCV replication is not uniform in the culture, which could explain why a significant number of persistently infected cells have cleared HCV after IFN-α treatment but a portion of the infected cell population remains nonresponsive to IFN-α. In support of this notion, IFNAR1 expression was drastically low in the sorted IFN-α–resistant cell population, compared with IFN-α–sensitive cells. This has been verified for other virus models in which only high doses of active or inactive HSV induce IFNAR1 phosphorylation and degradation.39 Based on these data, we propose that infected cells supporting only a very high level of viral replication experience a loss of cell surface expression of IFNAR1. We also propose that this persistently HCV-infected cell culture model can be used to investigate the significance of other mechanisms described in the literature relating HCV resistance to IFN-α.
Interestingly, we found that IFN-λ has a strong and sustained antiviral effect against HCV, compared with IFN-α and RBV. IFN-λ, alone or in combination with IFN-α, cleared HCV replication in the persistently HCV-infected cell culture model. These results suggest that type III IFN (IFN-λ) may be important in controlling HCV infections in humans. The relevance of this result is supported by a number of clinical studies indicating that IL-28B genotype assessment provides a predictor of sustained virologic response to IFN-α+RBV therapy. The underlying mechanisms of how the IL-28B genotype controls HCV clearance are not clear. The IFN-λ family includes at least three members: IFN-λ1, IFN-λ2, and IFN-λ3, which are encoded by the IL-29, IL-28, and IL-28B genes (IFNL1, IFNL2, and IFNL3, respectively). We speculate that the production of IFN-λ in chronically infected HCV patients may be linked to IL-28B gene polymorphisms. This hypothesis is supported by reports that IFN-λ production is regulated by IL-28B gene polymorphisms.40,41
Our present results are also supported by reports indicating that the IFN-λ antiviral response against HCV is stronger than that of IFN-α.24,42 Rice and colleagues42 showed that IFN-λ has a distinct gene induction signature and signal transduction profile against HCV. A recent study by Bartenschlager and colleagues24 showed that persistently infected cell culture is resistant to IFN-α and that IFN-λ showed a stronger antiviral response than IFN-α. Lambda interferons are produced during HCV infection and are therefore considered to play an important role in the innate immune response against HCV. How IFN-λ1 contributes to the antiviral control of chronic HCV infection in humans is unclear. Our results show that a persistently infected cell culture model develops resistance to IFN-α and RBV because of defective JAK–STAT signaling and impaired RBV uptake. The fact that IFN-λ clears HCV replication in the persistently infected cell culture warrants further studies to elucidate the novel antiviral mechanisms of IFN-λ against HCV.
Acknowledgments
We thank Mallory Heath and Michelle E. McCarthy for critically reviewing this manuscript; Charles M. Rice for providing Huh-7 and Huh-7.5 cells; Jibao He and Susan Magliato (Tulane University) for TEM analysis; Krzysztof Moroz (Tulane University) for photographing immunostaining slides; Xavier Alvarez (Tulane Primate Center) and Krzysztof Reiss (Louisiana State University) for confocal images; and Mary Price (Louisiana Cancer Research Consortium) for FACS analysis.
Footnotes
Supported by grants from NIH (CA127481, CA089121, and AI103106), by funds from the Louisiana Cancer Research Consortium (LCRC), and by bridge funding from Tulane University Health Sciences Center (S.D.).
Supplemental Data
Cells were infected with MOI = 0.1 JFH-ΔV3-Rluc virus overnight and the infected cells were passaged every 6 days, to 38 days after infection. HCV Core and NS5A–Rluc protein levels were measured by Western blotting, with β-actin as a loading control. Band intensities were quantified using ImageJ software version 1.48f (NIH, Bethesda, MD).
Comparison of viral load between highly passaged (P13) and unpassaged (P0) culture supernatant. Supernatant was serially diluted in serum-free medium, and 1 mL of each diluted infection was added to the Huh-7.5 cells. Cells were infected with equal volume (1 mL) of undiluted culture supernatants (both P0 and P13) with MOI = 0.1. Cells were infected for 24 hours and after 5 days of infection, HCV-RNA level was quantified by RT-qPCR. ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001.
Comparison of antiviral potencies of IFN-α and IFN-λ against HCV in persistently infected cell culture. A: Huh-7.5 cells were infected with MOI = 0.1 JFH-ΔV3-Rluc virus for 24 hours and then were treated with different concentrations of IFN-α or IFN-λ. After 72 hours, cells were lysed, Renilla luciferase activity was measured, and IC90 was determined for IFN-α (α 0.01–2000 IU/mL) and IFN-λ (λ 0.01–200 ng/mL). B: Persistently infected Huh-7.5 cells were treated with 2.5 × IC90, 5 × IC90, and 10 × IC90 IFN-α or IFN-λ. Cells received three consecutive treatments (T1 to T3) at 6-day intervals. After each treatment, antiviral efficacy of IFN-α and IFN-λ was determined by the measurement of Renilla luciferase activity.The results indicate that antiviral effects of IFN-λ are stronger than those of IFN-α. C: The antiviral effects of IFN-α and IFN-λ after a second treatment was confirmed by immunohistochemistry for HCV Core protein expression. D: Quantification of HCV Core+ cells in 10 different high-power fields (×40), compared with untreated control. Both assays (B and D) confirmed that the antiviral effect of IFN-λ is significantly stronger than IFN-α when used at equivalent concentrations. HCV Core protein expression essentially absent in 10 × IC90 IFN-λ–treated cells. E: Antiviral activity of IFN-α in a subgenomic replicon cell line. HCV subgenomic replicon cells (S3-GFP) received two consecutive treatments with 0 to 250 IU/mL of IFN-α at 72-hour intervals. Antiviral activity was determined by measuring GFP+ cells by flow cytometric analysis. F: Comparison of IFNAR1 expression between untreated HCV-infected Huh-7.5 culture and culture that have been repeatedly treated with IFN-α. Persistently infected cells were cultured without or with 2.5 × IC90 IFN-α treatment repeated at 6-day intervals. The expression of IFNAR1 in the cell lysates at the four time points (day 13 to day 31) was examined by Western blot analysis. ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001. RLU, relative light units.
Multiple-passage, long-term persistent infection did not select a population of cells less sensitive to IFN-α. A: Persistently infected Huh-7.5 cells (65 days of infection) were treated with a combination of siRNAs against HCV (100 pmole each of si321 and si359). After four consecutive treatments (arrows), HCV Renilla luciferase activity fell below the detection limit (dotted line). B: HCV-free cells (cured Huh-7.5) were cultured for three weeks, and Renilla luciferase activity was measured to confirm the complete clearance of HCV replication. The levels of IFN receptors and the proteins involved in the JAK–STAT pathway were compared between uninfected Huh-7.5 cells and cured Huh-7.5 cells. Reinfection of the same cured Huh-7.5 cells led to down-regulation of IFNAR1 and induced defective JAK–STAT signaling. Huh-7.5 and cured Huh-7.5 cells were infected with MOI = 0.1 HCV for 5 days. Expression levels of IFNAR1, p-STAT1, and p-STAT2 were measured by Western blotting.
Acridine Orange staining shows the induced autophagy response in Huh-7.5 cells due to HCV replication with time. Cells with autophagy show accumulation of orange-red cytoplasmic autophagic vacuoles. The orange-red autophagic vacuoles in 10 different cells were counted under ×40 magnification and compared with uninfected control (Huh-7.5). ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001.
Induction of autophagy in HCV-infected Huh-7.5 cells at 3, 8, and 14 days after infection was assessed by measuring autophagy-related proteins by Western blotting. Band intensity was quantified using ImageJ software.
Supplemental Data
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2013.10.005.
References
- 1.Alter M.J. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shepard C.W., Finelli L., Alter M.J. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558–567. doi: 10.1016/S1473-3099(05)70216-4. [DOI] [PubMed] [Google Scholar]
- 3.Butt A.A., Kanwal F. Boceprevir and telaprevir in the management of hepatitis C virus-infected patients. Clin Infect Dis. 2012;54:96–104. doi: 10.1093/cid/cir774. [DOI] [PubMed] [Google Scholar]
- 4.Dore G.J., Matthews G.V., Rockstroh J. Future of hepatitis C therapy: development of direct-acting antivirals. Curr Opin HIV AIDS. 2011;6:508–513. doi: 10.1097/COH.0b013e32834b87f8. [DOI] [PubMed] [Google Scholar]
- 5.Halfon P., Locarnini S.J. Hepatitis C virus resistance to protease inhibitors. Hepatology. 2011;55:192–206. doi: 10.1016/j.jhep.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 6.Pawlotsky J.M. Treatment failure and resistant with direct-acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53:1742–1751. doi: 10.1002/hep.24262. [DOI] [PubMed] [Google Scholar]
- 7.Aronsohn A., Reau N. Long-term outcomes after treatment with interferon and ribavirin in HCV patients. J Clin Gastroenterol. 2009;43:661–671. doi: 10.1097/MCG.0b013e31819f66e2. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida H., Arakawa Y., Sata M., Nishiguchi S., Yano M., Fujiyama S., Yamada G., Yokosuka O., Shiratori Y., Omata M. Interferon therapy prolonged life expectancy among chronic hepatitis C patients. Gastroenterology. 2002;123:483–491. doi: 10.1053/gast.2002.34785. [DOI] [PubMed] [Google Scholar]
- 9.Datta S., Hazari S., Chandra P.K., Samara M., Poat B., Gunduz F., Wimley W.C., Hauser H., Koster M., Lamaze C., Balart L.A., Garry R.F., Dash S. Mechanisms of HCV’s resistance to IFN-alpha in cell culture involve expression of functional IFN-alpha receptor 1. Virol J. 2011;8:351. doi: 10.1186/1743-422X-8-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazari S., Chandra P.K., Poat B., Datta S., Garry R.F., Foster T.P., Kousoulas G., Wakita T., Dash S. Impaired antiviral activity of interferon alpha against hepatitis C virus 2a in Huh-7 cells with a defective Jak-Stat pathway. Virol J. 2010;7:36. doi: 10.1186/1743-422X-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hazari S., Taylor L., Haque S., Garry R.F., Florman S., Luftig R., Regenstein F., Dash S. Reduced expression of Jak-1 and Tyk-2 proteins leads to interferon resistance in hepatitis C virus replicon. Virol J. 2007;4:89. doi: 10.1186/1743-422X-4-89. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Lemon S.M. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem. 2010;285:22741–22747. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ibarra K.D., Pfeiffer J.K. Reduced ribavirin antiviral efficacy via nucleoside transporter-mediated drug resistance. J Virol. 2009;83:4538–4547. doi: 10.1128/JVI.02280-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rauch A., Kutalik Z., Descombes P., Cai T., Di Iulio J., Mueller T., Bochud M., Battegay M., Bernasconi E., Borovicka J., Colombo S., Cerny A., Dufour J.F., Furrer H., Günthard H.F., Heim M., Hirschel B., Malinverni R., Moradpour D., Müllhaupt B., Witteck A., Beckmann J.S., Berg T., Bergmann S., Negro F., Telenti A., Bochud P.Y., Swiss Hepatitis C Cohort Study. Swiss HIV Cohort Study Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroeneterology. 2010;138:1338–1345. doi: 10.1053/j.gastro.2009.12.056. [DOI] [PubMed] [Google Scholar]
- 15.Suppiah V., Moldovan M., Ahlenstiel G., Berg T., Weltman M., Abate M.L., Bassendine M., Spengler U., Dore G.J., Powell E., Riordan S., Sheridan D., Smedile A., Fragomeli V., Müller T., Bahlo M., Stewart G.J., Booth D.R., George J. IL-28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100–1104. doi: 10.1038/ng.447. [DOI] [PubMed] [Google Scholar]
- 16.Liu S., Nelson C.A., Xiao L., Lu L., Seth P.P., Davis D.R., Hagedorn C.H. Measuring antiviral activity of benzimidazole molecules that alters IRES RNA structure with an infectious hepatitis C virus chimera expressing Renilla luciferase. Antiviral Res. 2011;89:54–63. doi: 10.1016/j.antiviral.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sir D., Chen W.L., Choi J., Wakita T., Yen T.S., Ou J.H. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandra P.K., Kundu A.K., Hazari S., Chandra S., Bao L., Ooms T., Morris G.F., Wu T., Mandal T., Dash S. Inhibition of hepatitis C virus replication by intracellular delivery of multiple siRNAs by nanosomes. Mol Ther. 2012;20:1724–1736. doi: 10.1038/mt.2012.107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Chandra P.K., Hazari S., Poat B., Gunduz F., Prabhu R., Liu G., Burioni R., Clementi M., Garry R.F., Dash S. Intracytoplasmic stable expression of IgG1 antibody targeting NS3 helicase inhibits replication of highly efficient hepatitis C virus 2a clone. Virol J. 2010;7:118. doi: 10.1186/1743-422X-7-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M., Tan W., Zhou J., Leow J., Go M., Lee H.S., Casey P.J. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem. 2008;283:18678–18684. doi: 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- 21.Henriques C.M., Rino J., Nibbs R.J., Graham G.J., Barata J.T. IL-7 induces rapid clathrin-mediated internalization and JAK3-dependent degradation of IL-7Ralpha in T cells. Blood. 2010;115:3269–3277. doi: 10.1182/blood-2009-10-246876. [DOI] [PubMed] [Google Scholar]
- 22.Dunnett C.W. A multiple comparison procedure for comparing several treatments with a control. J Am Stat Assoc. 1955;50:1096–1121. [Google Scholar]
- 23.Abdi H. Bonferroni test. Encyclopedia of Measurement and Statistics. In: Salkind N.J., editor. Sage; Thousand Oaks, CA: 2007. pp. 103–107. [Google Scholar]
- 24.Bauhofer O., Ruggieri A., Schmid B., Schirmacher P., Bartenschlager R. Persistence of HCV in quiescent hepatic cells under conditions of an interferon-induced antiviral response. Gastroenterology. 2012;143:429–438.e8. doi: 10.1053/j.gastro.2012.04.018. [DOI] [PubMed] [Google Scholar]
- 25.Merquiol E., Uzi D., Mueller T., Goldenberg D., Nahmias Y., Xavier R.J., Tirosh B., Shibolet O. HCV causes chronic endoplasmic reticulum stress leading to adaptation and interference with the unfolded protein response. PLoS One. 2011;6:e24660. doi: 10.1371/journal.pone.0024660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J.A., Urano F., Imaizumi K. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9230. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poat B., Hazari S., Chandra P.K., Gunduz F., Alvarez X., Balart L.A., Garry R.F., Dash S. Intracellular expression of IRF-9 Stat fusion protein overcome defective Jak-Stat signaling and inhibits HCV RNA replication. Virol J. 2010;7:265. doi: 10.1186/1743-422X-7-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poat B., Hazari S., Chandra P.K., Gunduz F., Balart L.A., Alvarez X., Dash S. SH2 modified STAT1 induces HLA-1 expression and improves IFN-γ signaling in IFN-α resistant HCV replicon cells. PLoS One. 2010;5:e13117. doi: 10.1371/journal.pone.0013117. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Kong W., Engel K., Wang J. Mammalian nucleoside transporters. Curr Drug Metab. 2004;5:63–84. doi: 10.2174/1389200043489162. [DOI] [PubMed] [Google Scholar]
- 30.Fried M.W., Shiffman M.L., Reddy K.R., Smith C., Marinos G., Gonçales F.L., Jr., Häussinger D., Diago M., Carosi G., Dhumeaux D., Craxi A., Lin A., Hoffman J., Yu J. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975–982. doi: 10.1056/NEJMoa020047. [DOI] [PubMed] [Google Scholar]
- 31.Manns M.P., McHutchison J.G., Gordon S.C., Rustgi V.K., Shiffman M., Reindollar R., Goodman Z.D., Koury K., Ling M., Albrecht J.K. Peginterferon alfa-2a plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958–965. doi: 10.1016/s0140-6736(01)06102-5. [DOI] [PubMed] [Google Scholar]
- 32.Reddy K.R., Shiffman M.L., Rodriguez-Torres M., Cheinquer H., Abdurakhmanov D., Bakulin I., Morozov V., Silva G.F., Geyvandova N., Stanciu C., Rabbia M., McKenna M., Thommes J.A., Harrison S.A., PROGRESS Study Investigators Induction pegylated interferon alfa-2a and high dose ribavirin do not increase SVR in healthy patients with genotype 1a and high viral loads. Gastroenterology. 2010;139:1972–1983. doi: 10.1053/j.gastro.2010.08.051. [DOI] [PubMed] [Google Scholar]
- 33.Liu J., HuangFu W.C., Kumar K.G., Qian J., Casey J.P., Hamanaka R.B., Grigoriadou C., Aldabe R., Diehl J.A., Fuchs S.Y. Virus-induced unfolded protein response attenuates anti-viral defenses via phosphorylation-dependent degradation of type I interferon receptor. Cell Host Microbe. 2009;5:72–83. doi: 10.1016/j.chom.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ke P.Y., Chen S.S. Activation of the unfolded protein response and autophagy after hepatitis C virus infection suppresses innate antiviral immunity in vitro. J Clin Invest. 2011;121:37–56. doi: 10.1172/JCI41474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shrivastava S., Raychoudhury A., Steele R., Ray R., Ray R.B. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011;53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shrivastava S., Bhanja Chowdhury J., Steele R., Ray R., Ray R.B. Hepatitis C virus up regulates beclin1 for induction of autophagy and activates mTOR signaling. J Virol. 2012;86:8705–8712. doi: 10.1128/JVI.00616-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asselah T., Estrabaud E., Bieche I., Lapalus M., De Muynck S., Vidaud M., Saadoun D., Soumelis V., Marcellin P. Hepatitis C: viral and host factors associated with non-response to pegylated interferon plus ribavirin. Liver Int. 2010;30:1259–1269. doi: 10.1111/j.1478-3231.2010.02283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rautou P.E., Cazals-Hatem D., Feldmann G., Mansouri A., Grodet A., Barge S., Martinot-Peignoux M., Duces A., Bièche I., Lebrec D., Bedossa P., Paradis V., Marcellin P., Valla D., Asselah T., Moreau R. Changes in autophagic response in patients with chronic hepatitis C virus infection. Am J Pathol. 2011;178:2708–2715. doi: 10.1016/j.ajpath.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian J., Zheng H., Huangfu W.C., Liu J., Carbone C.J., Leu N.A., Baker D.P., Fuchs S.Y. Pathogen recognition receptor signaling accelerates phosphorylation-dependent degradation of IFNAR1. PLoS Pathog. 2011;7:e1002065. doi: 10.1371/journal.ppat.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langhans B., Kupfer B., Braunschweiger I., Arndt S., Schulte W., Nischalke H.D., Nattermann J., Oldenburg J., Sauerbruch T., Spengler U. Interferon-lambda serum levels in hepatitis C. J Hepatol. 2011;54:859–965. doi: 10.1016/j.jhep.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 41.Shi X., Pan Y., Wang M., Wang D., Li W., Jiang T., Zhang P., Chi X., Jiang Y., Gao Y., Zhong J., Sun B., Xu D., Jiang J., Niu J. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinase population. PLoS One. 2012;7:e37054. doi: 10.1371/journal.pone.0037054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marcello T., Grakoui A., Barba-Spaeth G., Machlin E.S., Kotenko S.V., MacDonald M.R., Rice C.M. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroeneterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells were infected with MOI = 0.1 JFH-ΔV3-Rluc virus overnight and the infected cells were passaged every 6 days, to 38 days after infection. HCV Core and NS5A–Rluc protein levels were measured by Western blotting, with β-actin as a loading control. Band intensities were quantified using ImageJ software version 1.48f (NIH, Bethesda, MD).
Comparison of viral load between highly passaged (P13) and unpassaged (P0) culture supernatant. Supernatant was serially diluted in serum-free medium, and 1 mL of each diluted infection was added to the Huh-7.5 cells. Cells were infected with equal volume (1 mL) of undiluted culture supernatants (both P0 and P13) with MOI = 0.1. Cells were infected for 24 hours and after 5 days of infection, HCV-RNA level was quantified by RT-qPCR. ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001.
Comparison of antiviral potencies of IFN-α and IFN-λ against HCV in persistently infected cell culture. A: Huh-7.5 cells were infected with MOI = 0.1 JFH-ΔV3-Rluc virus for 24 hours and then were treated with different concentrations of IFN-α or IFN-λ. After 72 hours, cells were lysed, Renilla luciferase activity was measured, and IC90 was determined for IFN-α (α 0.01–2000 IU/mL) and IFN-λ (λ 0.01–200 ng/mL). B: Persistently infected Huh-7.5 cells were treated with 2.5 × IC90, 5 × IC90, and 10 × IC90 IFN-α or IFN-λ. Cells received three consecutive treatments (T1 to T3) at 6-day intervals. After each treatment, antiviral efficacy of IFN-α and IFN-λ was determined by the measurement of Renilla luciferase activity.The results indicate that antiviral effects of IFN-λ are stronger than those of IFN-α. C: The antiviral effects of IFN-α and IFN-λ after a second treatment was confirmed by immunohistochemistry for HCV Core protein expression. D: Quantification of HCV Core+ cells in 10 different high-power fields (×40), compared with untreated control. Both assays (B and D) confirmed that the antiviral effect of IFN-λ is significantly stronger than IFN-α when used at equivalent concentrations. HCV Core protein expression essentially absent in 10 × IC90 IFN-λ–treated cells. E: Antiviral activity of IFN-α in a subgenomic replicon cell line. HCV subgenomic replicon cells (S3-GFP) received two consecutive treatments with 0 to 250 IU/mL of IFN-α at 72-hour intervals. Antiviral activity was determined by measuring GFP+ cells by flow cytometric analysis. F: Comparison of IFNAR1 expression between untreated HCV-infected Huh-7.5 culture and culture that have been repeatedly treated with IFN-α. Persistently infected cells were cultured without or with 2.5 × IC90 IFN-α treatment repeated at 6-day intervals. The expression of IFNAR1 in the cell lysates at the four time points (day 13 to day 31) was examined by Western blot analysis. ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001. RLU, relative light units.
Multiple-passage, long-term persistent infection did not select a population of cells less sensitive to IFN-α. A: Persistently infected Huh-7.5 cells (65 days of infection) were treated with a combination of siRNAs against HCV (100 pmole each of si321 and si359). After four consecutive treatments (arrows), HCV Renilla luciferase activity fell below the detection limit (dotted line). B: HCV-free cells (cured Huh-7.5) were cultured for three weeks, and Renilla luciferase activity was measured to confirm the complete clearance of HCV replication. The levels of IFN receptors and the proteins involved in the JAK–STAT pathway were compared between uninfected Huh-7.5 cells and cured Huh-7.5 cells. Reinfection of the same cured Huh-7.5 cells led to down-regulation of IFNAR1 and induced defective JAK–STAT signaling. Huh-7.5 and cured Huh-7.5 cells were infected with MOI = 0.1 HCV for 5 days. Expression levels of IFNAR1, p-STAT1, and p-STAT2 were measured by Western blotting.
Acridine Orange staining shows the induced autophagy response in Huh-7.5 cells due to HCV replication with time. Cells with autophagy show accumulation of orange-red cytoplasmic autophagic vacuoles. The orange-red autophagic vacuoles in 10 different cells were counted under ×40 magnification and compared with uninfected control (Huh-7.5). ∗∗P < 0.01, ∗∗∗P < 0.001, and †P < 0.0001.
Induction of autophagy in HCV-infected Huh-7.5 cells at 3, 8, and 14 days after infection was assessed by measuring autophagy-related proteins by Western blotting. Band intensity was quantified using ImageJ software.