Abstract
Recent years have seen development and implementation of anticancer therapies targeted to particular gene mutations, but methods to assay clinical cancer specimens in a comprehensive way for the critical mutations remain underdeveloped. We have developed UW-OncoPlex, a clinical molecular diagnostic assay to provide simultaneous deep-sequencing information, based on >500× average coverage, for all classes of mutations in 194 clinically relevant genes. To validate UW-OncoPlex, we tested 98 previously characterized clinical tumor specimens from 10 different cancer types, including 41 formalin-fixed paraffin-embedded tissue samples. Mixing studies indicated reliable mutation detection in samples with ≥10% tumor cells. In clinical samples with ≥10% tumor cells, UW-OncoPlex correctly identified 129 of 130 known mutations [sensitivity 99.2%, (95% CI, 95.8%–99.9%)], including single nucleotide variants, small insertions and deletions, internal tandem duplications, gene copy number gains and amplifications, gene copy losses, chromosomal gains and losses, and actionable genomic rearrangements, including ALK-EML4, ROS1, PML-RARA, and BCR-ABL. In the same samples, the assay also identified actionable point mutations in genes not previously analyzed and novel gene rearrangements of MLL and GRIK4 in melanoma, and of ASXL1, PIK3R1, and SGCZ in acute myeloid leukemia. To best guide existing and emerging treatment regimens and facilitate integration of genomic testing with patient care, we developed a framework for data analysis, decision support, and reporting clinically actionable results.
The era of precision oncology began in 1998 with the approval of the anti- human epidermal growth factor receptor 2 (HER2) monoclonal antibody, trastuzumab, for the treatment of HER2-positive breast cancer.1 At the same time, an immunohistochemistry-based diagnostic test (HercepTest; Dako, Glostrup, Denmark) was approved for the identification of tumors that express HER2, necessary to ascertain which patients are eligible for trastuzumab treatment. This advance was followed by the introduction of erlotinib, a small molecule tyrosine kinase inhibitor against epidermal growth factor receptor (EGFR), which has proven useful in patients with non-small cell lung cancer with activating EGFR mutations.2–4 More recently, two U.S. Food and Drug Administration–approved drugs that also require a genomic sequence-based companion diagnostic have advanced into late-stage clinical trials: vemurafenib, which targets metastatic malignant melanoma harboring the BRAF V600E mutation5 , and crizotinib, which has shown efficacy against non-small cell lung cancers that have ALK rearrangements.6 Clinical trials for additional agents directed against specific genes or mutations are currently underway, and are expected to progressively increase the repertoire of targeted cancer therapies available.
These successes and accumulated discoveries of potential cancer driver mutations through the use of exome and whole-genome sequencing7–12 raise important questions about the long-term practicality of existing clinical diagnostics for the molecular characterization of cancers. As new targeted therapies are approved for molecular subtypes, and more genes with prognostic value are identified, the number of single-gene tests needed to adequately classify a tumor subtype increases, with the consequences of potentially exhausting available tissue specimens and of driving up health care costs. Yet, despite the concerns for increased risk and health care expense associated with additional tissue acquisition for molecular testing, validated clinical diagnostics suitable for assaying multiple genes and different classes of mutations in a multiplexed fashion remain lacking. Most currently available multiplexed clinical assays examine only a limited number of specific sites in a relatively small number of genes.13,14 More recently, next-generation sequencing assays have been developed for detecting cancer-associated mutations in clinical specimens in a more comprehensive manner, but these assays have only been validated on a small number of tumor types (breast, colon, and prostate).15–17 As assays of this type become more widespread, a framework for identifying, interpreting, and reporting actionable variants will be required for this technology to reach its full potential as a clinical diagnostic test.
Here, we describe our development and clinical validation of a targeted massively parallel sequencing assay for 194 cancer-relevant genes, UW-OncoPlex, designed as a comprehensive diagnostic test for mutational events of all types in an efficient and cost-effective manner. The assay is intended to allow the most complete and informative molecular characterization of a wide variety of clinical specimens, and is scalable to large numbers of additional genes in the future. Our assay improves on earlier approaches, most importantly by expanding the spectrum of mutations detectable to include complex genomic rearrangements and copy number variants (CNVs), in addition to greater sensitivity for all variants. We also develop an accompanying data interpretation and decision support network to inform patient prognoses and therapeutic options.
Materials and Methods
DNA Samples
DNA samples used for validation experiments were derived from 98 different tissue samples, including colon cancer, melanoma, acute myeloid leukemia (AML), myeloproliferative disorders, chronic myeloid leukemia, lung cancer, gastrointestinal stromal tumor, and other neoplasms (Supplemental Table S1). All samples had prior molecular characterization of one or more mutations by a clinically validated targeted assay(s) in a Clinical Laboratory Improvement Amendments–certified laboratory. Clinical testing was performed using a combination of Sanger sequencing (KIT, MPL, PDGFRA, RUNX1), melting-curve analysis (BRAF, EGFR p.L858R, IDH1, IDH2, KRAS), allele-specific PCR (JAK2, BCR-ABL), fluorescence in situ hybridization (ALK, MLL, PML-RARA, ROS1), capillary electrophoresis fragment length polymorphism [CEBPA, EGFR exon 19 insertions and deletions (indels), FLT3-ITD], and DNA microarray (single-gene and chromosomal-scale deletions). Solid tissue samples (n = 41) were formalin-fixed, paraffin-embedded (FFPE); samples from hematological malignancies (n = 57) were fresh frozen cells from peripheral blood or bone marrow. DNA was extracted from all sample types using the Gentra Puregene DNA Isolation Kit (Catalog #158489; Qiagen, Valencia, CA). H&E-stained slides were reviewed before DNA extraction for all FFPE samples, and when feasible, macrodissection of tumor areas was performed to enrich tumor cellularity. Tumor cellularity in macrodissected areas was estimated by review of H&E-stained slides for FFPE specimens or quantified by flow cytometry for hematological specimens. No tumor enrichment was performed for hematological malignancy samples. Estimation of tumor cellularity was not available for eight samples (Supplemental Table S1). Collectively, these 98 samples comprised a validation panel that included multiple known single nucleotide variant (SNV), indel, translocation, inversion, and copy number variation (CNV) mutations. In addition, HapMap DNAs for NA12878 (selected as a reference sample) and NA18545 (selected because of a known heterozygous BCL2L11 intron 2 deletion polymorphism) were obtained from Coriell Cell Repositories (Camden, NJ). Clinical specimens were obtained in accordance with the declaration of Helsinki and the ethics guidelines of the human subjects division of the University of Washington and the University of Chicago.
For mixing studies, control DNA was derived from peripheral blood lymphocytes of a normal control subject. Before mixing, real-time PCR was performed on each sample to ensure the DNA quality/PCR amplification efficiency was equal for both the patient sample and diluent DNA.
Library Construction, Gene Capture, and Massively Parallel Sequencing
Sequencing libraries were prepared from DNA samples as described elsewhere.18 When available, 3 μg of DNA was used. The minimum input requirement was 750 ng of DNA, which did not result in loss of assay performance. For samples at or near the minimum input requirement, additional pre-capture PCR cycles (up to 10 cycles in total) were performed to generate the 500 ng required for hybridization. Libraries were hybridized to a custom design of complementary RNA (cRNA) biotinylated oligonucleotides targeting the exons of 194 genes (Table 1 and Supplemental Table S2) and select introns of genes involved in genomic rearrangements (Supplemental Table S3), for a total of >850 kb of targeted DNA. Post-capture PCR incorporated primers containing a unique 6-bp index as performed previously,18 allowing multiplexing of multiple samples onto the same sequencing lane.
Table 1.
Gene Panel
| Tier 1: Currently actionable | Tier 2: Actionable in the near future | Tier 3: Frequently mutated | Germline pharmacogenomics | ||||
|---|---|---|---|---|---|---|---|
| ABL1 | KIF5B | ABL2 | MAPK1 | APC | PTCH1 | ABCB1 | SLC22A2 |
| AKT1 | KIT | AKT2 | MC1R | BAK1 | PTPN11 | ABCC2 | SLCO1B3 |
| ALK | KRAS | AKT3 | MCL1 | BCL2 | PTPRD | ABCC4 | SOD2 |
| ASXL1 | MAP2K1 | ATM | MDM2 | CDH1 | RB1 | ABCG2 | SULT1A1 |
| AURKA | MET | AURKB | MDM4 | CDKN2A | RICTOR | C1orf144 | TPMT |
| BAP1 | MLL | BCOR | MEN1 | CREBBP | RPS14 | COMT | TYMS |
| BCR | KRAS | CBL | MITF | CRLF2 | SF1 | CYP1B1 | UGT1A1 |
| BCL2L11 | MPL | CBLB | MLH1 | CSF1R | SF3B1 | CYP2C19 | UMPS |
| BRAF | NKX2-1 | CDK6 | MRE11A | CTNNB1 | SMAD2 | CYP2C8 | |
| BRCA1 | NPM1 | CHEK1 | MSH2 | EPHA3 | SMAD3 | CYP2D6 | |
| BRCA2 | NRAS | CHEK2 | MSH6 | EPHA5 | SMAD4 | CYP3A4 | |
| CCND1 | PDGFRA | ERBB3 | MYC | EPHB6 | SMARCA4 | CYP3A5 | |
| CCNE1 | PIK3CA | ERBB4 | MYCN | ETV6 | SMARCB1 | DPYD | |
| CDK4 | PML | FBXW7 | NF2 | EZH2 | SPRY4 | EIF3A | |
| CDK8 | PTEN | FGFR1 | NOTCH1 | FGFR3 | SRC | ERCC2 | |
| CEBPA | RARA | FLT1 | PAX5 | GAB2 | TFG | ESR1 | |
| DDR2 | ROS1 | FLT4 | PDGFRB | GATA1 | TGFBR2 | ESR2 | |
| DNMT3A | RET | GATA2 | PIK3R1 | GNAS | TRRAP | FCGR1A | |
| EGFR | STK11 | GNA11 | PMS2 | GRIN2A | U2AF1 | FCGR2A | |
| EML4 | TP53 | GNAQ | RAF1 | HNF1A | U2AF65 | FCGR3A | |
| EPHB2 | VHL | GRM3 | RUNX1 | IL7R | ZRSR2 | GSTP1 | |
| ERBB2 | HDAC4 | SMO | JAK1 | GUCY1A2 | |||
| FGFR2 | HRAS | SRSF2 | MAP2K4 | ITPA | |||
| FGFR4 | IGF1R | SUZ12 | MUTYH | LRP2 | |||
| FLT3 | IKZF1 | TSC1 | MYCL1 | MAN1B1 | |||
| HIF1A | JAK3 | TSC2 | NF1 | MTHFR | |||
| IDH1 | KDM6A | TET2 | NOTCH2 | NQO1 | |||
| IDH2 | KDR | TYR | PBRM1 | NRP2 | |||
| JAK2 | MAP2K2 | WT1 | PRPF40B | SLC22A1 | |||
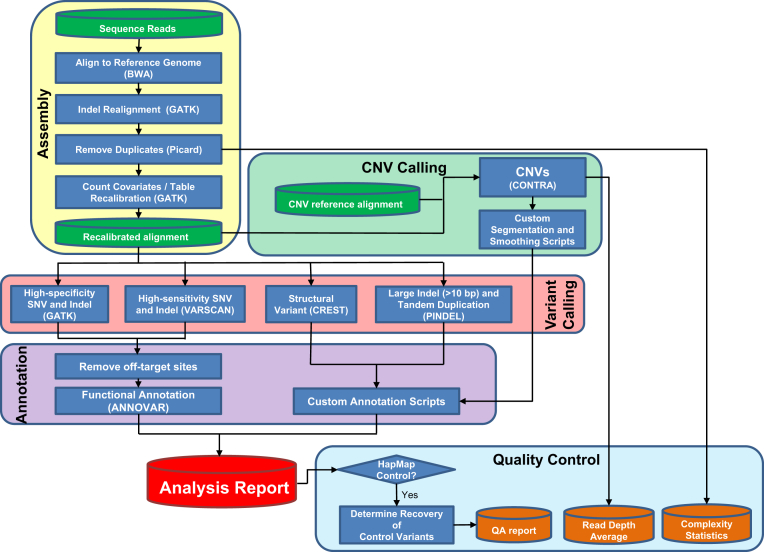
DNA sequencing was performed on a HiSeq2000 sequencing system (Illumina, San Diego, CA) with 2 × 101-bp, paired-end reads as described,18 and on a MiSeq (Illumina) with 2 × 150-bp, paired-end reads according to the manufacturer’s instructions. The average fraction of on-target reads was 0.423. The minimum gene-level average coverage was set to 50×, with genes below this threshold reported as failed. The minimum acceptable average coverage for the entire panel was set at 150×, and the minimum library complexity (the fraction of unique DNA fragments sequenced) was set at 20%. Up to 24 samples were multiplexed onto a single lane of the HiSeq sequencer (average of 540× coverage after removal of duplicate reads) (Figure 1), or up to two samples were multiplexed for a MiSeq run (average of 512× coverage after removal of duplicate reads) (Figure 1).
Figure 1.
Scheme showing the data analysis pipeline. QA, quality assurance.
Data Analysis
The data analysis pipeline is diagrammed in Figure 1. Initial read mapping against the human reference genome (hg19/GRCh37) and alignment processing was performed using BWA version 0.6.1-r104 (http://sourceforge.net/projects/bio-bwa/files, last accessed November 28, 2011)19 and SAMtools version 0.1.18 (http://sourceforge.net/projects/samtools/files, last accessed September 2, 2011),20 respectively. Sample-level, fully local indel realignment was then performed using GATK version 1.621 (Broad Institute, Cambridge, MA). Duplicate reads were removed using PICARD version 1.72 (http://picard.sourceforge.net, last accessed December 5, 2011). Quality score recalibration was then performed using GATK. This realigned and recalibrated alignment was used for all subsequent analyses. SNV and indel calling was performed through the GATK Universal Genotyper using default parameters and VarScan version 2.3.2 (http://sourceforge.net/projects/varscan, last accessed October 31, 2011).22 For indel calling through VarScan, the minimum variant frequency was set to 0.01 reads, and the minimum number of variant reads was set to 4, whereas for SNV calling, the minimum variant frequency was set to 0.03, and the minimum number of variant reads was set to 5, with default parameters for all other settings. We found that GATK variant calls supported by high-quality scores (>500) were also identified by VarScan. Variants of interest called by VarScan alone were sometimes accepted, but because of the algorithm’s relatively low specificity, such calls were limited to specific known actionable mutations or to indel mutations >3 bp in length that were supported by four or more variant reads passing the population frequency filters (as described below). Variants identified by VarScan alone were manually reviewed at the laboratory director level using the Integrated Genomics Viewer23 version 5.64 (Broad Institute) to assess the quality of base calls, the mapping quality for the reads, and the overall read depth at the site.
PINDEL version 0.2.424 was used to identify tandem duplications and indels >10 bp in length. Structural variants were identified using CREST version 1.0.25
For CNV analysis, copy number states for individual probes were initially called using CONTRA version 2.0.3 (http://sourceforge.net/projects/contra-cnv/files, last accessed July 24, 2012)26 with reference to a CNV control comprising reads from two independent rounds of library preparation and sequencing of the HapMap individual NA12878. CNV calls were made at the resolution of individual exons using custom Perl scripts. On a gene-by-gene basis, read-depth statistics were calculated from baits originating in the same exon. Adjacent exons were merged into larger segments if the read depths of their component baits were not significantly different (P > 0.0001) by Student’s t-test, and read-depth statistics were recalculated for the larger segments. Segments were required to have an average adjusted log2 ratio compared to the reference sample of ±0.3 to be considered as a possible CNV event. To facilitate identification of meaningful segments, each segment was also evaluated using a score, calculated by multiplying the number of bases in the segment by the absolute value of the average adjusted log2 ratio, and by which higher-scoring segments tended to be more credible than lower-scoring segments. CNV plots were visualized using the R package ggplot2 (http://ggplot2.org, last accessed August 15, 2012),27 with graphical overlay of segments having average adjusted log2 ratios ±0.3 and scores exceeding 1500. We defined copy number gain as a predicted copy number of 3 or 4, and amplification as a predicted copy number of 5 or higher.
SNV and indel data were annotated with gene-based annotation, conservation scores, predicted effects at the protein level, and population frequency using ANNOVAR (BIOBASE, Wolfenbüttel, Germany),28 and output from other programs was annotated and formatted using custom scripts to facilitate review. ANNOVAR was also used to annotate the frequency of variants in a database obtained from sequencing of patients internally. We initially filtered variant calls based on population frequency data from both the 1000 Genomes project29 and our internal variant frequency database, excluding variants (other than clinically flagged variants) occurring in greater than 1% of any population as likely germline variation or recurrent sequencing artifact. Clinically important mutations that are suspected to be germline based on allelic ratio, or based on prior genetics knowledge (such as a known founder mutation) are reported with a statement indicating that if there is clinical suspicion that mutation may be germline, additional testing is indicated.
The quality of sequencing libraries was routinely evaluated on the basis of information about library complexity, sequence read depth, and the fraction of reference variants in NA12878 (as described in Results) recovered in positive controls.
Genomic Microarray Analysis
For validation of CNV calling, seven FFPE samples and one HEME sample were tested in parallel by genomic microarray (Supplemental Table S1). Samples were characterized using SurePrint G3 Cancer CGH+SNP 4×180K microarray (Agilent Technologies, Santa Clara, CA) with probes targeting 657 cancer-associated genes and genomic regions and probes tiled across the genome to allow profiling of DNA copy number and copy neutral aberrations, such as loss of heterozygosity, according to the manufacturer’s protocols. Samples were normalized against manufacturer-provided sex-matched reference standards (HapMap samples NA12891 and NA12878).
Results
Selection of the Gene Panel
Genes included in UW-OncoPlex were chosen to meet criteria for one of three tiers corresponding to current clinical actionability (Table 1). Tier 1 genes harbor mutations that are currently clinically actionable, either by providing information about sensitivity or resistance to specific targeted therapies or by providing prognostic information related to patient outcome. Tier 2 genes are expected to be actionable in the near future, with targeted therapies currently under active development in clinical trials. Tier 3 genes are recognized as recurrently mutated in cancers, but robust prognostic information or targeted therapies are not yet available. We assigned each gene to the lowest tier in which it fulfilled requirements for one or more cancers. Additionally, we included a panel of genes influencing drug metabolism and clearance. Because of the less well-defined therapeutic implications of variants in these genes, we are not yet reporting results from this part of the panel.
Construction of a Tumor Sequencing Data Analysis Pipeline
We formulated a data analysis pipeline (Figure 1) based on the Genome Analysis Toolkit (GATK) best practices guidelines (http://www.broadinstitute.org/gatk/guide/best-practices, last accessed December 19, 2011), following recommended methods for genome alignment and recalibration before variant calling. Although GATK includes software tools able to robustly identify SNVs and indels, it employs a Bayesian likelihood model to predict genotype, which assumes a diploid genome. This model is problematic for tumor DNA, which may demonstrate a skewed allele balance secondary to a variety of factors. Indeed, in experimental samples, we found GATK variant calling alone was insufficient to reliably identify some variants in experimental tumor samples, and therefore, we added a more sensitive genotype-calling algorithm, VarScan, which identifies any variant in a heuristic fashion if there are sufficient supporting reads.30 Identifying variants by two separate approaches allows for redundancy, and thus robustness. To facilitate interpretation and review of called variants, SNVs and indels were annotated with respect to functional effects and population frequencies. We integrated additional software tools to identify other forms of genomic variation aside from SNVs and relatively small indels. PINDEL24 has previously proven useful in detecting otherwise difficult-to-call internal tandem duplications (ITD) in FLT331 and was therefore used in our study as a means to identify both ITD and large indels (exceeding 10 bases in length). Genomic structural variants, including translocations, inversions, and large deletions, were identified using CREST,25 which reanalyzes partially mapped (soft-clipped) reads to identify sequence fragments resulting from genomic rearrangements. Copy number variation was performed with CONTRA26 and a series of custom segmentation and visualization scripts. Integration of these genomic analysis techniques into the same analysis pipeline allows for identification of all types of genetic mutations found in cancers.
Limit of Detection
An important consideration of the assay is the successful detection of low-level mutations in patient samples. Even after tumor enrichment steps such as macrodissection, patient samples are still usually a heterogeneous mixture of tumor and non-neoplastic cells derived from surrounding tissue or from reactive infiltrate, which may skew the representation of mutant alleles. Further, cancers are heterogeneous in nature,32 and a mutation of interest may exist in only a subset of tumor cells.
To evaluate the limit of mutation detection for UW-OncoPlex, we performed serial mixing studies of tumor DNA with non-neoplastic control DNA and determined how effectively various forms of known heterozygous mutations were recovered over a range of serial dilutions (Table 2). A 4-bp insertion mutation in NPM1 was detectable at the highest tumor dilution examined (1:16), which represented approximately 3% tumor cells because the starting sample had 45% tumor cellularity. FLT3-ITD was also successfully identified at the limits of detection for the highest tumor dilution tested (1:16). An SNV mutation in DNMT3A was detectable at a 1:8 dilution of tumor to normal DNA, but was not found at the 1:16 dilution. The limit of detection was therefore at approximately 6% tumor cellularity, although the precise limit of detection at each locus is likely to be influenced by aneuploidy and by other factors such as GC content and variation in read depth. We therefore conservatively estimate that the assay is capable of reliably recovering a heterozygous mutation when present in >10% of cells in the original sample. A separate mixing study using an FFPE sample harboring an EGFR exon 19 deletion mutation produced similar results (Supplemental Table S4).
Table 2.
Limits of Mutation Detection for Three Variants
| Sample | Variant reads at each dilution |
||
|---|---|---|---|
| FLT3-ITD | NPM1 | DNMT3A | |
| Undiluted∗ | 120 | 78 | 85 |
| 1:2 | 80 | 59 | 67 |
| 1:4 | 27 | 21 | 19 |
| 1:8 | 25 | 16 | 18 |
| 1:16 | 3 | 6 | ND |
Mutations: FLT3-ITD c.1804_1805ins24bp, NPM1 c.860_863dup, DNMT3A c.1627G>T (p.G543C).
ND, not detected.
45% tumor cellularity undiluted; acute myeloid leukemia (AML) sample HEME49 used for mixing study.
Sensitivity
We first evaluated the performance of UW-OncoPlex against a panel of clinical samples containing known mutations of various types, including point mutations (SNVs), indels, CNVs, and gene rearrangements. All mutations had been previously identified through routine clinical testing (Table 3 and Supplemental Table S1). When restricting the analysis to the 82 samples with ≥10% tumor cellularity, deep sequencing identified 90 of 91 [98.9% (95% CI, 94.0%–99.8%)] of expected SNV and indel mutations previously identified by single-gene testing methods (Table 3). The single missed mutation was a FLT3-ITD approximately 72-bp insertion variant in an AML sample (Supplemental Table S1). Although the sample had 22% blasts by flow cytometry, the FLT3-ITD was detected at a very low allelic ratio of 0.03 by capillary electrophoresis, suggesting that the mutation was present in only a small subset of tumor cells.
Table 3.
Validation of Previously Known Mutations in 98 Clinical Samples
| Mutation class | Gene/region | Mutation | Mutations recovered |
|
|---|---|---|---|---|
| Tumor cellularity ≥10% | Tumor cellularity <10%∗ | |||
| SNV | BRAF | p.V600E | 8/8 | |
| SNV | BRAF | p.V600K | 2/2 | |
| SNV | CEBPA | p.E59X | 1/1 | |
| SNV | EGFR | p.L858R | 5/5 | 0/1 |
| SNV | IDH1 | Codon 132 | 5/5 | 1/1 |
| SNV | IDH2 | p.R140Q | 4/4 | |
| SNV | JAK2 | p.V617F | 1/1 | 5/6 |
| SNV | KIT | p.K638E | 1/1 | |
| SNV | KIT | p.D812Y | 1/1 | |
| SNV | KIT | p.D816V | 0/1 | |
| SNV | KRAS | Codon 12 or 13 | 5/5 | 2/2 |
| SNV | MPL | p.W515L | 1/1 | |
| SNV | PDGFRA | p.D842V | 2/2 | |
| SNV | RUNX1 | p.Y260X | 1/1† | |
| Indel | CEPBA | C-terminal ins | 2/2 | 0/1 |
| Indel | EGFR | Exon 19 del | 4/4 | |
| Indel | EGFR | Exon 19 ins | 1/1 | |
| Indel | EGFR | Exon 20 ins | 1/1 | |
| Indel | FLT3 | Tandem dup (ITD) | 23/24‡ | 3/5 |
| Indel | KIT | Exon 11 del/ins | 3/3 | |
| Indel | NPM1 | p.W288Cfs∗12 | 20/20 | 4/4 |
| CNV | AKT2 | Amplification | 1/1 | |
| CNV | BCL2L11 | 2.9-kb del intron 2 | 1/1† | |
| CNV | BRAF | Copy gain | 1/1 | |
| CNV | CDKN2A | Copy loss | 3/3 | |
| CNV | Chr1, 2, 3, 6, 15, 18, 22 | Copy losses | 1/1 | |
| CNV | Chr1 | Copy gain | 1/1 | |
| CNV | Chr7 | Copy loss | 2/2 | |
| CNV | Chr8 | Trisomy | 1/1 | |
| CNV | Chr9 | Copy loss | 1/1 | |
| CNV | Chr14 | Trisomy | 1/1 | |
| CNV | ChrX | Copy loss | 1/1 | |
| CNV | EGFR | Copy gain | 1/1 | |
| CNV | FGFR1 | Copy gain | 3/3 | |
| CNV | GNAQ | Copy loss | 1/1 | |
| CNV | HIF1A | Copy loss | 1/1 | |
| CNV | MDM2 | Amplification | 1/1 | |
| CNV | MET | Copy gain | 2/2 | |
| CNV | MITF | Copy gain | 1/1 | |
| CNV | MYC | Copy gain | 1/1 | |
| CNV | PIK3CA | Copy gain | 1/1 | |
| CNV | SMAD4 | Copy loss | 1/1 | |
| CNV | SMO | Copy gain | 1/1 | |
| SV | ALK-EML4 | Gene fusion | 5/5 | |
| SV | BCR-ABL | Gene fusion | 1/1 | |
| SV | MLL-MLLT3 | Gene fusion | 2/2 | 0/2 |
| SV | PML-RARA | Gene fusion | 2/2 | |
| SV | ROS1 | Gene inversion | 1/1 | |
| Total | 129/130 | 16/24 | ||
| Sensitivity | 99.2% | 67% | ||
| (95% CI) | (95.8%–99.9%) | (47%–82%) | ||
SV, structural variation.
<10% cellularity includes samples with unknown tumor cellularity.
Mutations in these two samples were germline.
The FLT3-ITD mutation not recovered was detected by capillary electrophoresis at an allelic ratio of 0.03, suggesting the mutation was present in only a small subset of tumor cells.
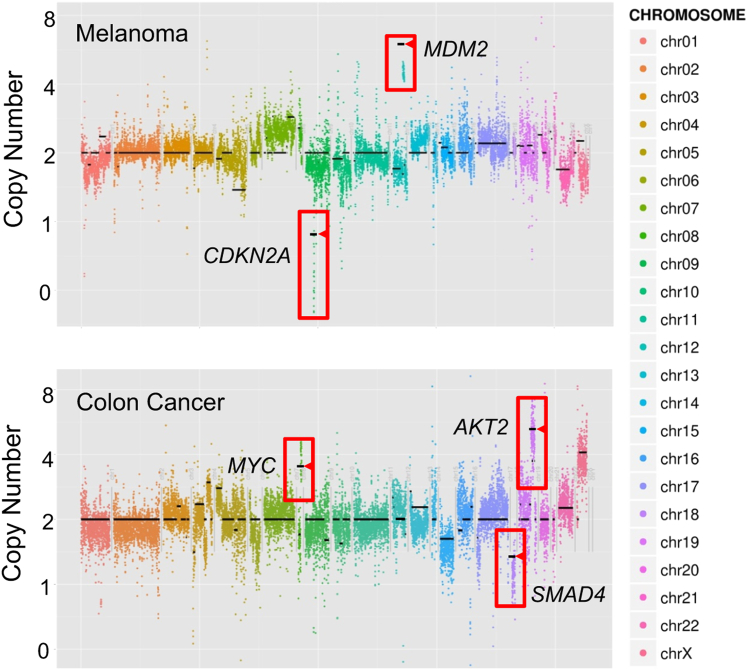
Importantly, in samples with ≥10% tumor cellularity the assay detected known structural variants, including gene rearrangements validated by fluorescence in situ hybridization and CNVs measured by array genomic hybridization (Table 3 and Figure 2). All known CNV events [28 of 28, 100% (95% CI, 88%–100%)] were recovered, including the BCL2L11 (BIM) 2.9-kb intron 2 deletion polymorphism that is associated with tyrosine kinase inhibitor resistance, and gene amplifications of AKT2 and MDM2 (Figure 2).33 All known gene rearrangements [11 of 11, 100% (95% CI, 74%–100%)] were accurately recovered, including five ALK-EML4 rearrangements, two PML-RARA gene fusions, two MLL-MLLT3 gene fusions, one ROS1 inversion, and one BCR-ABL gene fusion (Table 3).
Figure 2.
Copy number variants (CNVs) detection directly by UW-OncoPlex sequencing. Adjusted log2 ratio of read depth of sequencing data are plotted for individual baits (y axis) across captured genomic regions (x axis). Overlaid horizontal black lines indicate genomic microarray results in the target regions for the same sample, illustrating high concordance of CNV detection between UW-OncoPlex and genomic microarray. Amplifications or deletions of specific genes are indicated (boxes), with corresponding array CGH calls highlighted (arrowheads). Depicted are examples from a melanoma (FFPE06) and colon cancer sample (FFPE02).
Among the 17 samples with <10% or unknown tumor cellularity, the assay detected 16 of 24 known mutations [67% (95% CI, 47%–82%)] (Table 3). On the basis of these results and the limit of detection analysis, we established a threshold of 10% tumor cellularity for samples to be run on the assay.
Libraries prepared from FFPE material in this study did not differ substantially from those prepared from fresh DNA either with respect to the sensitivity of mutations recovered, the average sequencing read depth, or the fraction of on target reads. However, in some samples, FFPE-derived DNA required several more PCR cycles to generate adequate material for gene capture than an equivalent amount of DNA derived from fresh tissue, suggesting variable amounts of sample degradation in FFPE samples. It is expected that very highly degraded FFPE specimens would prove unsuitable for sequencing.
Comparison to a Reference Sample NA12878
To evaluate the performance characteristics of UW-OncoPlex against a well-characterized reference sample, we compared the variants detected for HapMap sample NA12878 against variants detected and analyzed independently by exome re-sequencing on the Illumina platform (1000 Genomes data set) and by whole-genome sequencing on the Complete Genomics platform (Complete Genomics data set), which uses entirely different sequencing chemistries. Within the regions targeted by UW-OncoPlex, 152 SNVs appeared in both published data sets. UW-OncoPlex analysis recovered 149 of 152 of these SNVs. The three variants not detected by UW-OncoPlex were all synonymous variants in NOTCH2. Given the number of paralogous sequences for this gene, we suspect these reported variants may represent false-positive results in both the 1000 Genomes and Complete Genomics datasets. In fact, at one of these three sites (chr1:120539837), only one read matched the SNV reported in the other data sets, with a mapping quality of zero.
In all, 149 credible SNVs were called in common by our pipeline and the two published data sets. In subsequent experiments, we used these variants as a quality control measure by evaluating their recovery in NA12878 as our run-specific control sample. We required that at least 148 of 149 variants were recovered in NA12878 for the run to be considered in control.
Reproducibility
To evaluate the reproducibility of the assay, we generated multiple sequencing libraries from HapMap sample NA12878 and analyzed them on eight separate sequencing runs. All 149 credible variants for NA12878 were detected in each run. In addition, repeat libraries were generated from a subset of 34 patient samples and sequenced on a separate HiSeq run by a different user, and all known mutations were successfully recovered (Supplemental Table S1).
Specificity Analysis Based on Detection of Mutations Not Previously Known
In the 98 samples in the validation series, UW-OncoPlex revealed more than 200 potentially actionable mutations that were not already known because the mutation region(s) were not previously tested. We estimated analytic specificity for UW-OncoPlex by confirmatory Sanger sequencing on a subset of these samples with mutations not previously known to be present. We examined 17 variants in three genes, and in all cases (17 of 17), Sanger sequencing validated the variant, yielding 100% specificity (95% CI, 82%–100%) (Supplemental Figures S1, S2, and S3 and Supplemental Table S5).
Detection of Novel Gene Rearrangements
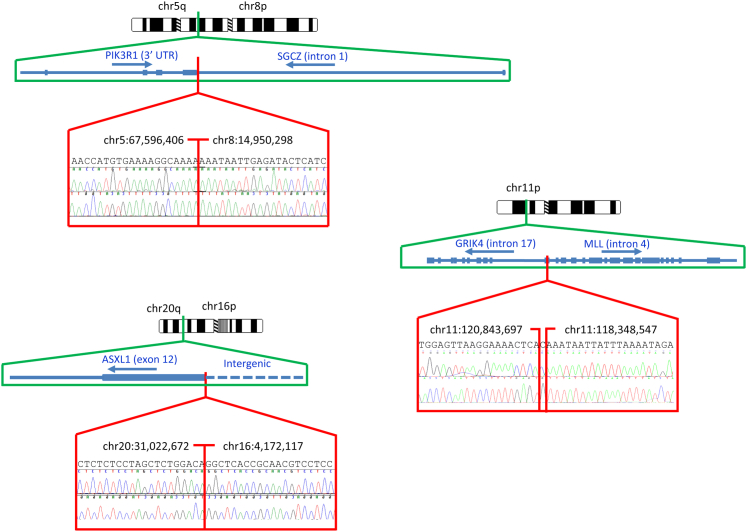
In the 98 samples in the validation series, UW-OncoPlex also revealed three novel gene rearrangements (Figure 3). In an AML sample, we detected a rearrangement predicted to disrupt the final exon (exon 12) of ASXL1 by transposing it into intergenic DNA. In a second AML specimen, we detected a translocation involving PIK3R1 and SGCZ predicted to result in disruption of both genes. In a melanoma sample, we detected a translocation involving MLL and GRIK4, also predicted to disrupt the coding sequence of both genes. All three gene rearrangement events were confirmed by bidirectional Sanger sequencing.
Figure 3.
Novel gene rearrangements detected. Chromosomal ideograms, gene-level schematics of events, and confirmatory bidirectional Sanger sequencing are displayed for three chromosomal rearrangements. Predicted sequences of rearrangements inferred from genomic sequence data are displayed above electropherograms, with exact chromosomal breakpoints indicated (Hg19 coordinates). UTR, untranslated region.
For each of these rearrangements, only one of the two genes was targeted for hybridization gene capture. These results indicate that it is possible to identify gene rearrangements by deep sequencing if only one of the two rearrangement partners is sequenced, greatly expanding the number of structural rearrangements that are potentially detectable by the assay.
Data Interpretation and Clinical Reporting
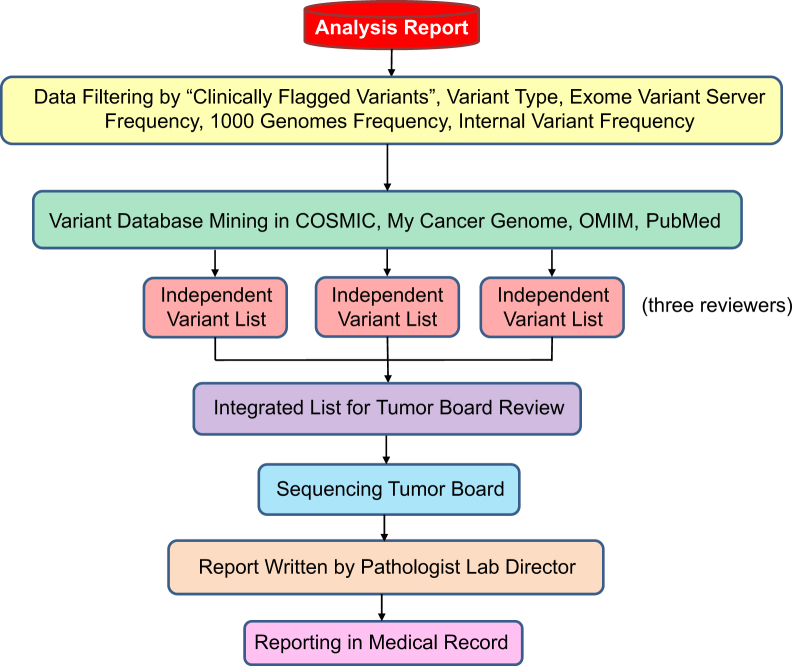
Next-generation sequencing uncovers large numbers of genetic variants, which pose challenges in analyzing clinical samples.34 To accurately interpret data from the UW-OncoPlex assay and subsequently convey that information to clinicians, we developed a framework for data interpretation and reporting. We first implemented a strategy to systematically identify the most important genomic variants and to interpret the potential for impacting patient prognosis and therapy (Figure 4). Actionable SNV and indel variants were evaluated first as clinically flagged variants, including mutations targeted by conventional clinical testing (for example, BRAF p.V600E), and which are specifically flagged by our data analysis pipeline. We next applied filters to exclude variants likely to be inconsequential (synonymous variants, variants in intergenic DNA, etc), and to remove variants present at >1% frequency in the general population (as surveyed by Exome Variant Server and 1000 Genomes data). We also filtered against frequency data from our own internal database of genomic variants observed during UW-OncoPlex testing, a strategy that has proven especially valuable, both by providing robust population frequency data for our specific patient population, and by eliminating recurrent sequencing or analysis artifacts. Variants of interest were then evaluated using a combination of database mining and literature review to further investigate the potential clinical impact in the context of current medical literature. CNV and structural rearrangement data were similarly analyzed, but due to the sporadic nature of these events, could only be examined on a case-by-case basis without prior filtering.
Figure 4.
Scheme showing the framework for variant interpretation and reporting.
At least three expert reviewers from the tumor sequencing board independently compiled lists of potentially reportable variants and reviewed the quality of the data that were being used to support the variant calls, by examining genotype quality score metrics and evaluating base call quality and mapping quality of primary sequence reads using the Integrated Genomics Viewer, as appropriate. The reviewers next surveyed the primary literature about the identified variants to inform recommendations made to the ordering provider. These independent lists were combined by the laboratory director and returned to the board for group review. Once the sequencing tumor board agreed on a final list of reportable variants, the lab director composed a formal report for the medical record describing the variants detected, the clinical action associated with each variant, and a short list of pertinent negative genes for which no mutations were detected.
The turnaround time for the entire assay from specimen acquisition to clinical reporting ranged from 2 to 8 weeks, with an average of 5 weeks.
Discussion
Continuing advances in cancer genomics and development of targeted therapies have begun to shift clinical paradigms from treating cancers of a specific type to treating cancers with specific genetic lesions. For example, targeted therapies exist for patients with lung cancers harboring mutations of EGFR or ALK and ROS1 rearrangements. The ability to profile every cancer specimen for a comprehensive set of clinically actionable mutations underpins the emerging vision of individualized oncology, allowing effective cancer treatments to be optimized on the basis of genotype alone. To this end, we have developed a framework for comprehensively interrogating actionable and near-actionable mutations in cancer samples using targeted gene capture and next-generation DNA sequencing. The method is cost effective and robust enough to integrate into routine clinical care. In contrast to the fixed content of commercial mutation hotspot panels such as Ion AmpliSeq (Life Technologies, Carlsbad, CA), our approach is more inclusive, interrogating for a large panel of genes, all exons, and where appropriate, intronic sequences frequently involved in gene rearrangement events. In addition, our data analysis pipeline permits detection of all classes of genomic mutation, from point mutations to structural rearrangements. Detection is very sensitive even in the context of tumor heterogeneity or impurity. Further, because this assay is comprehensive, we anticipate that routine testing using this approach will prove valuable as a discovery tool, permitting identification of additional mutations in specific cancer types that may inform the development of new therapeutic agents or provide prognostic information.
UW-OncoPlex proved effective with heterogeneous tumor samples, with very high sensitivity in samples with ≥10% tumor cellularity. In such samples, the assay recovered 99.2% (129 of 130) of known mutations (95% CI, 95.8%–99.9%). In samples of <10% or unknown tumor cellularity, the assay detected 67% (16 of 24) of mutations (95% CI, 47%–82%). This highlights the critical role of the pathologist in quality assessment of all samples before clinical testing to ensure adequate tumor cellularity for accurate sequencing results. In practice, we evaluate cellularity of all samples before UW-OncoPlex testing and append a caveat statement to reports for samples that are close to the 10% cellularity threshold (ie, between 10% and 20% cellularity), indicating that false-negative results for the specimen cannot be entirely excluded.
Conventional molecular testing for cancers requires prior knowledge or assumptions about the specific mutations that will be detected in a cancer of a particular type (eg, BRAF and KRAS testing for colorectal cancers, EGFR and ALK-EML4 testing for lung cancers). It is increasingly becoming appreciated that neoplasms may harbor mutations in genes classically found in tumors from different tissues of origin. Such mutations may serve as targets for effective, but noncanonical cancer therapies.35 Such unanticipated actionable mutations may prove valuable in selecting patient therapy regimens, yet current approaches to testing interrogate only sites that are specifically requested. UW-OncoPlex sequencing recovered many potentially actionable mutations in genes not targeted by routine clinical testing algorithms (Supplemental Table S1). Our findings illustrate the benefit of a comprehensive approach to cancer testing that provides genomic information about clinically actionable mutations in an unbiased fashion.
In addition to previously undetected substitution and indel mutations, sequencing of the validation samples revealed several novel structural variant mutations, including three novel gene rearrangements that may play an active role in cancer development (Figure 4). In one AML specimen, a rearrangement disrupting the final exon (exon 12) of ASXL1 was detected. Disruptive point mutations and frameshifts in ASXL1 exon 12 are found commonly in AML and are a marker for poor prognosis.36 To the best of our knowledge, disruption of ASXL1 by gene rearrangement has not previously been reported. In another case of AML, we detected a translocation involving SGCZ and the 3′ untranslated region of PIK3R1. SGCZ has a structural role in connecting cytoskeletal proteins with the extracellular matrix.37 P1K3R1 mutations have been implicated in diverse cancer types,38 typically gain-of-function changes that activate oncogenic PI3K signaling, and rarely dominant-negative mutations affecting PTEN stabilization.39 Disruption of the 3′ untranslated region in this case may affect mRNA stability or miRNA regulation of the transcript, with subsequent impact on PI3K signaling. Lastly, in a melanoma case, we detected a translocation involving MLL and GRIK4, which disrupted the coding sequence of both genes. MLL encodes a histone methyltransferase and is frequently activated in AML by partial tandem duplication or by forming gene fusions with over 100 translocation partners described to date, but has not been previously implicated in melanoma.40,41 GRIK4 is a glutamate receptor.42 The oncogenic roles of glutamate signaling pathways have become recently recognized in melanoma,43–45 and the functional significance of this fusion merits further investigation. The Sanger-validated recovery of unsuspected and/or unreported point mutations and structural rearrangements in many samples highlights the use of genomic diagnostic testing as a research and discovery tool.
To effectively communicate results, it was necessary to develop robust strategies for interpreting and reporting variants identified by UW-OncoPlex. We adopted a strategy (Figure 4) involving a small (three- to six-member) sequencing tumor board that independently, then collaboratively, reviews testing results. Our reporting strategy is labor intensive and may not be practical for all laboratories, but we believe that the redundancy that overreading provides is both important and beneficial to the task of variant interpretation for such highly complex data. As such, we have instituted this strategy as our standard, and have made it a priority to complete these reviews within a timeline compatible with clinical reporting.
Next-generation sequencing technologies hold considerable promise for transforming clinical molecular testing of cancers, allowing comprehensive detection of actionable mutations irrespective of cancer type. As new actionable genes are identified, they can be added to subsequent iterations of the UW-OncoPlex panel with rapid assay revalidation, allowing the diagnostic to evolve as our knowledge of relevant cancer mutations improves. We also look forward to technical advancements that will further increase utility of the assay. Improvements in next-generation sequencing platforms and sequencing library preparation techniques46,47 will further decrease costs and improve throughput and accuracy. Novel data analysis tools may further improve the sensitivity of the assay without enhancement to the underlying sequence data itself. This study demonstrates that next-generation sequencing assays such as UW-OncoPlex are already capable of providing valuable predictive and prognostic information from clinical samples and can have an immediate impact on patient care.
Acknowledgments
We thank Daniel Sabath for reviewing the manuscript, Anne Thornton for help with library preparation and sequencing, and Deborah Barden, Rachel Slusher, Laura Akagi, Youly Welt, Tatyana Marushchak, Christine Lakey, Lena Murillo, Shelley Engelhard, Calista Ku, Deena Hanke, and Patty Callahan for help with preparing genomic DNA.
Footnotes
Supported by the Department of Defense Ovarian Cancer Research ProgramOC093285 (T.W.) and NIH grant R01CA157744 (T.W. and M.-C.K.).
C.C.P. and S.J.S. contributed equally to this work.
Supplemental Data
Confirmation of FLT3 point mutations. Single representative electropherograms from bidirectional sequencing are displayed for control and tumor samples.
Confirmation of DNMT3A mutations. Single representative electropherograms from bidirectional sequencing are displayed for control and tumor samples.
Confirmation of ASXL1 mutations. Single representative electropherograms from bidirectional sequencing are displayed for control and tumor samples.
References
- 1.Slamon D.J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J., Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 2.Lynch T.J., Bell D.W., Sordella R., Gurubhagavatula S., Okimoto R.A., Brannigan B.W., Harris P.L., Haserlat S.M., Supko J.G., Haluska F.G., Louis D.N., Christiani D.C., Settleman J., Haber D.A. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 3.Paez J.G., Janne P.A., Lee J.C., Tracy S., Greulich H., Gabriel S., Herman P., Kaye F.J., Lindeman N., Boggon T.J., Naoki K., Sasaki H., Fujii Y., Eck M.J., Sellers W.R., Johnson B.E., Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 4.Pao W., Miller V., Zakowski M., Doherty J., Politi K., Sarkaria I., Singh B., Heelan R., Rusch V., Fulton L., Mardis E., Kupfer D., Wilson R., Kris M., Varmus H. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci U S A. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman P.B., Hauschild A., Robert C., Haanen J.B., Ascierto P., Larkin J., Dummer R., Garbe C., Testori A., Maio M., Hogg D., Lorigan P., Lebbe C., Jouary T., Schadendorf D., Ribas A., O’Day S.J., Sosman J.A., Kirkwood J.M., Eggermont A.M., Dreno B., Nolop K., Li J., Nelson B., Hou J., Lee R.J., Flaherty K.T., McArthur G.A. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak E.L., Bang Y.J., Camidge D.R., Shaw A.T., Solomon B., Maki R.G., Ou S.H., Dezube B.J., Janne P.A., Costa D.B., Varella-Garcia M., Kim W.H., Lynch T.J., Fidias P., Stubbs H., Engelman J.A., Sequist L.V., Tan W., Gandhi L., Mino-Kenudson M., Wei G.C., Shreeve S.M., Ratain M.J., Settleman J., Christensen J.G., Haber D.A., Wilner K., Salgia R., Shapiro G.I., Clark J.W., Iafrate A.J. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wei X., Walia V., Lin J.C., Teer J.K., Prickett T.D., Gartner J., Davis S., Stemke-Hale K., Davies M.A., Gershenwald J.E., Robinson W., Robinson S., Rosenberg S.A., Samuels Y. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stark M.S., Woods S.L., Gartside M.G., Bonazzi V.F., Dutton-Regester K., Aoude L.G., Chow D., Sereduk C., Niemi N.M., Tang N., Ellis J.J., Reid J., Zismann V., Tyagi S., Muzny D., Newsham I., Wu Y., Palmer J.M., Pollak T., Youngkin D., Brooks B.R., Lanagan C., Schmidt C.W., Kobe B., MacKeigan J.P., Yin H., Brown K.M., Gibbs R., Trent J., Hayward N.K. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet. 2012;44:165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaev S.I., Rimoldi D., Iseli C., Valsesia A., Robyr D., Gehrig C., Harshman K., Guipponi M., Bukach O., Zoete V., Michielin O., Muehlethaler K., Speiser D., Beckmann J.S., Xenarios I., Halazonetis T.D., Jongeneel C.V., Stevenson B.J., Antonarakis S.E. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2012;44:133–139. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 10.Leary R.J., Kinde I., Diehl F., Schmidt K., Clouser C., Duncan C., Antipova A., Lee C., McKernan K., De La Vega F.M., Kinzler K.W., Vogelstein B., Diaz L.A., Jr., Velculescu V.E. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2:20ra14. doi: 10.1126/scitranslmed.3000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roychowdhury S., Iyer M.K., Robinson D.R., Lonigro R.J., Wu Y.M., Cao X., Kalyana-Sundaram S., Sam L., Balbin O.A., Quist M.J., Barrette T., Everett J., Siddiqui J., Kunju L.P., Navone N., Araujo J.C., Troncoso P., Logothetis C.J., Innis J.W., Smith D.C., Lao C.D., Kim S.Y., Roberts J.S., Gruber S.B., Pienta K.J., Talpaz M., Chinnaiyan A.M. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu G., Broniscer A., McEachron T.A., Lu C., Paugh B.S., Becksfort J., Qu C., Ding L., Huether R., Parker M., Zhang J., Gajjar A., Dyer M.A., Mullighan C.G., Gilbertson R.J., Mardis E.R., Wilson R.K., Downing J.R., Ellison D.W., Baker S.J. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacConaill L.E., Campbell C.D., Kehoe S.M., Bass A.J., Hatton C., Niu L., Davis M., Yao K., Hanna M., Mondal C., Luongo L., Emery C.M., Baker A.C., Philips J., Goff D.J., Fiorentino M., Rubin M.A., Polyak K., Chan J., Wang Y., Fletcher J.A., Santagata S., Corso G., Roviello F., Shivdasani R., Kieran M.W., Ligon K.L., Stiles C.D., Hahn W.C., Meyerson M.L., Garraway L.A. Profiling critical cancer gene mutations in clinical tumor samples. PLoS One. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su Z., Dias-Santagata D., Duke M., Hutchinson K., Lin Y.L., Borger D.R., Chung C.H., Massion P.P., Vnencak-Jones C.L., Iafrate A.J., Pao W. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13:74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harismendy O., Schwab R.B., Bao L., Olson J., Rozenzhak S., Kotsopoulos S.K., Pond S., Crain B., Chee M.S., Messer K., Link D.R., Frazer K.A. Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol. 2011;12:R124. doi: 10.1186/gb-2011-12-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagle N., Berger M.F., Davis M.J., Blumenstiel B., Defelice M., Pochanard P., Ducar M., Van Hummelen P., Macconaill L.E., Hahn W.C., Meyerson M., Gabriel S.B., Garraway L.A. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltran H., Yelensky R., Frampton G.M., Park K., Downing S.R., Macdonald T.Y., Jarosz M., Lipson D., Tagawa S.T., Nanus D.M., Stephens P.J., Mosquera J.M., Cronin M.T., Rubin M.A. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard C.C., Smith C., Salipante S.J., Lee M.K., Thornton A.M., Nord A.S., Gulden C., Kupfer S.S., Swisher E.M., Bennett R.L., Novetsky A.P., Jarvik G.P., Olopade O.I., Goodfellow P.J., King M.C., Tait J.F., Walsh T. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup: The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nature Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Mullighan C.G., Easton J., Roberts S., Heatley S.L., Ma J., Rusch M.C., Chen K., Harris C.C., Ding L., Holmfeldt L., Payne-Turner D., Fan X., Wei L., Zhao D., Obenauer J.C., Naeve C., Mardis E.R., Wilson R.K., Downing J.R., Zhang J. CREST maps somatic structural variation in cancer genomes with base-pair resolution. Nat Methods. 2011;8:652–654. doi: 10.1038/nmeth.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Lupat R., Amarasinghe K.C., Thompson E.R., Doyle M.A., Ryland G.L., Tothill R.W., Halgamuge S.K., Campbell I.G., Gorringe K.L. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–1313. doi: 10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickham H. Springer; New York: 2009. ggplot2: elegant graphics for data analysis. [Google Scholar]
- 28.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koboldt D.C., Chen K., Wylie T., Larson D.E., McLellan M.D., Mardis E.R., Weinstock G.M., Wilson R.K., Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spencer D.H., Abel H.J., Lockwood C.M., Payton J.E., Szankasi P., Kelley T.W., Kulkarni S., Pfeifer J.D., Duncavage E.J. Detection of FLT3 internal tandem duplication in targeted. short-read-length, next-generation sequencing data. J Mol Diagn. 2013;15:81–93. doi: 10.1016/j.jmoldx.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng K.P., Hillmer A.M., Chuah C.T., Juan W.C., Ko T.K., Teo A.S. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 34.Shendure J. Next-generation human genetics. Genome Biol. 2011;12:408. doi: 10.1186/gb-2011-12-9-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnittger S., Eder C., Jeromin S., Alpermann T., Fasan A., Grossmann V., Kohlmann A., Illig T., Klopp N., Wichmann H.E., Kreuzer K.A., Schmid C., Staib P., Peceny R., Schmitz N., Kern W., Haferlach C., Haferlach T. ASXL1 exon 12 mutations are frequent in AML with intermediate risk karyotype and are independently associated with an adverse outcome. Leukemia. 2013;27:82–91. doi: 10.1038/leu.2012.262. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler M.T., Zarnegar S., McNally E.M. Zeta-sarcoglycan, a novel component of the sarcoglycan complex, is reduced in muscular dystrophy. Hum Mol Genet. 2002;11:2147–2154. doi: 10.1093/hmg/11.18.2147. [DOI] [PubMed] [Google Scholar]
- 38.Quayle S.N., Lee J.Y., Cheung L.W., Ding L., Wiedemeyer R., Dewan R.W., Huang-Hobbs E., Zhuang L., Wilson R.K., Ligon K.L., Mills G.B., Cantley L.C., Chin L. Somatic mutations of PIK3R1 promote gliomagenesis. PLoS One. 2012;7:e49466. doi: 10.1371/journal.pone.0049466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheung L.W., Hennessy B.T., Li J., Yu S., Myers A.P., Djordjevic B., Lu Y., Stemke-Hale K., Dyer M.D., Zhang F., Ju Z., Cantley L.C., Scherer S.E., Liang H., Lu K.H., Broaddus R.R., Mills G.B. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov. 2011;1:170–185. doi: 10.1158/2159-8290.CD-11-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shen Y., Zhu Y.M., Fan X., Shi J.Y., Wang Q.R., Yan X.J., Gu Z.H., Wang Y.Y., Chen B., Jiang C.L., Yan H., Chen F.F., Chen H.M., Chen Z., Jin J., Chen S.J. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 41.Somervaille T.C., Cleary M.L. Grist for the MLL: how do MLL oncogenic fusion proteins generate leukemia stem cells? Int J Hematol. 2010;91:735–741. doi: 10.1007/s12185-010-0579-8. [DOI] [PubMed] [Google Scholar]
- 42.Szpirer C., Molne M., Antonacci R., Jenkins N.A., Finelli P., Szpirer J., Riviere M., Rocchi M., Gilbert D.J., Copeland N.G. The genes encoding the glutamate receptor subunits KA1 and KA2 (GRIK4 and GRIK5) are located on separate chromosomes in human, mouse, and rat. Proc Natl Acad Sci U S A. 1994;91:11849–11853. doi: 10.1073/pnas.91.25.11849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi K.Y., Chang K., Pickel J.M., Badger J.D., 2nd, Roche K.W. Expression of the metabotropic glutamate receptor 5 (mGluR5) induces melanoma in transgenic mice. Proc Natl Acad Sci U S A. 2011;108:15219–15224. doi: 10.1073/pnas.1107304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song Z., He C.D., Liu J., Sun C., Lu P., Li L., Gao L., Zhang Y., Xu Y., Shan L., Liu Y., Zou W., Gao H., Gao W. Blocking glutamate-mediated signalling inhibits human melanoma growth and migration. Exp Dermatol. 2012;21:926–931. doi: 10.1111/exd.12048. [DOI] [PubMed] [Google Scholar]
- 45.Martino J.J., Wall B.A., Mastrantoni E., Wilimczyk B.J., La Cava S.N., Degenhardt K., White E., Chen S. Metabotropic glutamate receptor 1 (Grm1) is an oncogene in epithelial cells. Oncogene. 2013;32:4366–4376. doi: 10.1038/onc.2012.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neiman M., Sundling S., Gronberg H., Hall P., Czene K., Lindberg J., Klevebring D. Library preparation and multiplex capture for massive parallel sequencing applications made efficient and easy. PLoS One. 2012;7:e48616. doi: 10.1371/journal.pone.0048616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rohland N., Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012;22:939–946. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of FLT3 point mutations. Single representative electropherograms from bidirectional sequencing are displayed for control and tumor samples.
Confirmation of DNMT3A mutations. Single representative electropherograms from bidirectional sequencing are displayed for control and tumor samples.
Confirmation of ASXL1 mutations. Single representative electropherograms from bidirectional sequencing are displayed for control and tumor samples.