Abstract
Next-generation sequencing (NGS) is becoming a common approach for clinical testing of oncology specimens for mutations in cancer genes. Unlike inherited variants, cancer mutations may occur at low frequencies because of contamination from normal cells or tumor heterogeneity and can therefore be challenging to detect using common NGS analysis tools, which are often designed for constitutional genomic studies. We generated high-coverage (>1000×) NGS data from synthetic DNA mixtures with variant allele fractions (VAFs) of 25% to 2.5% to assess the performance of four variant callers, SAMtools, Genome Analysis Toolkit, VarScan2, and SPLINTER, in detecting low-frequency variants. SAMtools had the lowest sensitivity and detected only 49% of variants with VAFs of approximately 25%; whereas the Genome Analysis Toolkit, VarScan2, and SPLINTER detected at least 94% of variants with VAFs of approximately 10%. VarScan2 and SPLINTER achieved sensitivities of 97% and 89%, respectively, for variants with observed VAFs of 1% to 8%, with >98% sensitivity and >99% positive predictive value in coding regions. Coverage analysis demonstrated that >500× coverage was required for optimal performance. The specificity of SPLINTER improved with higher coverage, whereas VarScan2 yielded more false positive results at high coverage levels, although this effect was abrogated by removing low-quality reads before variant identification. Finally, we demonstrate the utility of high-sensitivity variant callers with data from 15 clinical lung cancers.
Molecular testing is gaining an increasing role in the diagnosis and management of cancer. In recent years, numerous studies have shown that specific somatic mutations and the mutational status of certain genes can either inform prognosis (eg, BRCA1/2 in breast/ovarian cancer; IDH1 in glioblastoma; and KIT, DNMT3A, IDH1/2, FLT3 ITD, and NPM1 in acute myeloid leukemia) or predict response to targeted therapies (KRAS/EGFR antibody therapy and KIT/tyrosine kinase inhibitors).1–8 The expanding catalog of clinically relevant mutations has led to the development of high-throughput assays for detecting somatic mutations directly from cancer specimens by combining targeted capture with next-generation sequencing (NGS) platforms. These approaches can provide comprehensive mutational profiling across a large number of genes simultaneously, with greatly increased efficiency and decreased cost as compared with conventional sequencing methods.9 To date, this approach has been successfully implemented in several clinical laboratories for detection of somatic mutations in cancer.10,11
Despite their promise, sequencing-based assays for detecting somatic mutations in cancer pose unique technical challenges compared with those used to detect constitutional variants. Cancer specimens from small biopsy specimens may contain few tumor cells, and both small and large specimens may contain substantial amounts of normal tissue, stromal components, and inflammatory cells. This can result in dilution of tumor DNA with that from nontumor cells, and thus even somatic mutations present in every tumor cell are present at low frequencies in the sample DNA. In addition, recent studies of both solid and hematologic cancers have demonstrated remarkable genetic heterogeneity; although tumors are typically derived from a single founding clone defined by somatic variants present in every tumor cell, there are often multiple tumor cell subpopulations with additional somatic variants.12–16 Also, somatically acquired aneuploidies and copy number variation are common in many solid tumor types, and gains or loss of genetic material can alter the observed allele fraction of sequence variants in these regions.17 NGS-based detection methods can capture low-frequency mutations because they provide a digital readout of sequence variants and high sequencing redundancy from hundreds to thousands of individual DNA molecules. However, a priori detection of low-frequency mutations (ie, detection of variants at non-hotspot positions in which the previous probability of a variant is low) relies on methods that are able to differentiate true variants from noise such as sequencing errors and alignment artifacts. In the absence of such algorithms, large numbers of false positive variants may be called because the inherent error rate of NGS platforms alone can approach 1%. Currently, many popular NGS analysis programs are designed for constitutional genome analysis in which variants are expected to occur in either 50% (heterozygous) or 100% (homozygous) of reads.18,19 These previous probabilities are often built into the detection algorithms, and variants with variant allele frequencies (VAFs) falling too far outside the expected range for homozygous and heterozygous variants may be considered of poor quality and not be called because of the high likelihood that they are false positive rather than inherited variants. Several groups have developed experimental20,21 and/or bioinformatics22–27 approaches for low-frequency variant detection that have been shown to detect variants at frequencies of 0.1% and lower. These methods typically require specialized library preparation, spiked-in control samples, or other modifications to standard NGS laboratory protocols to detect variants with <2% VAF. Moreover, some have been used only for low-frequency variant detection using whole genome or PCR enrichment strategies, and experience with application of these methods to data from other enrichment methods, such as hybridization capture, are limited.
Streamlined protocols for standard NGS library preparation and target enrichment using hybridization capture are now relatively common and have made it attractive for clinical molecular laboratories to adapt these methods for detection of somatic mutations directly from cancer samples. However, there are few data on the performance of this workflow for detecting low-frequency variants in deep coverage sequence data from a single sample (ie, not a tumor–normal sample pair) using common NGS variant detection tools, which are widely used and cited but have typically been developed for detection of inherited variants in low-coverage NGS data. In the present study, we used a laboratory-derived dilution series of well-characterized HapMap DNA samples to determine the performance of common analysis tools in detecting low-frequency variants in hybridization capture NGS data. We tested three widely used programs, SAMtools (version 0.1.18)), Genome Analysis Toolkit (GATK; UnifiedGenotyper version 2.3), and VarScan2 (version 2.3.5; compared in Supplemental Table S1), and one high-sensitivity algorithm that creates instrument-run and context-dependent error models (SPLINTER; version 6t) to analyze high-coverage (>1000×), hybridization capture NGS data for 26 cancer genes from mixed HapMap samples with a range of expected minor variant frequencies of 25% to 2.5%.18,19,25,27 The performance of these tools was assessed using a set of gold standard single nucleotide variants (SNVs) obtained by sequencing the pure HapMap samples individually. We also examined the effect of various coverage depths on low-frequency allele detection and tested each program using a set of 15 routine lung adenocarcinoma specimens to gauge the practical effect of applying these tools to clinical samples. Our results showed that there is considerable variability in the sensitivity for low-frequency variants across the programs tested and that some of the most popular tools perform poorly at even moderate VAFs. However, we found that GATK performed well for variants with VAFs ≥10% and that SPLINTER and VarScan2 showed good sensitivity even at the lowest VAF tested, <5%, with acceptable positive predictive value (PPV) in clinically interpretable regions.
Materials and Methods
DNA Extraction
HapMap DNA samples used in the present study were obtained from the Coriell Cell Repository (Coriell Institute for Medical Research, Camden, NJ) as cell lines and included samples NA17989, NA18484, NA18507, NA18872, and NA19127. Cells were grown under standard conditions, and DNA was prepared using the QIAamp DNA Micro Kit (Qiagen, Inc., Valencia, CA) per the manufacturer’s instructions. The DNA concentration for each sample was determined using a Qubit fluorometer (Life Technologies Corp., Grand Island, NY). Mixed samples were created by mixing DNA from one sample (NA17989; Chinese ethnicity) with each of the four others (NA18484, NA18507, NA18872, and NA19127; all of Yoruban ethnicity) in the amounts given in Table 1. DNA from the 15 formalin-fixed, paraffin-embedded (FFPE) lung adenocarcinomas was extracted as previously described.28
Table 1.
Description and Sequencing Results for Pure and Mixed HapMap Samples
| Sample | Input DNA, μg (% of mix)‡ | Expected VAF, %§ | Observed VAF, %¶ | Sequencing results∗ |
Gold standard SNVs† |
|
|---|---|---|---|---|---|---|
| Reads (millions) | Coverage (mean) | Total (unique) | ||||
| Pure Samples | ||||||
| NA17989 | 1 | 14.8 | 1417 | 231 (NA) | ||
| NA18484 | 1 | 16.8 | 1158 | 294 (113) | ||
| NA18507 | 1 | 20.4 | 1796 | 277 (101) | ||
| NA18872 | 1 | 17.7 | 1619 | 278 (115) | ||
| NA19127 | 1 | 21.9 | 1816 | 254 (98) | ||
| Mixed Samples | ||||||
| NA18484_50 | 0.5 (50) | 25 | 26.0 | 24.6 | 1469 | |
| NA18507_50 | 0.5 (50) | 25 | 25.5 | 21.8 | 1493 | |
| NA18872_50 | 0.5 (50) | 25 | 26.1 | 28 | 1583 | |
| NA19127_50 | 0.5 (50) | 25 | 25.5 | 26.1 | 1200 | |
| NA18484_20 | 0.2 (20) | 10 | 11.5 | 22.2 | 1594 | |
| NA18507_20 | 0.2 (20) | 10 | 10.5 | 19.9 | 1476 | |
| NA18872_20 | 0.2 (20) | 10 | 12.6 | 21.5 | 1529 | |
| NA19127_20 | 0.2 (20) | 10 | 9.8 | 16 | 1229 | |
| NA18484_10 | 0.1 (10) | 5 | 6.2 | 23 | 1822 | |
| NA18507_10 | 0.1 (10) | 5 | 9.2 | 19.7 | 1598 | |
| NA18872_10 | 0.1 (10) | 5 | 6.5 | 21.6 | 1817 | |
| NA19127_10 | 0.1 (10) | 5 | 5.6 | 19.5 | 1667 | |
| NA18484_5 | 0.05 (5) | 2.5 | 4.4 | 24.6 | 1969 | |
| NA18507_5 | 0.05 (5) | 2.5 | 4.9 | 22.7 | 1929 | |
| NA18872_5 | 0.05 (5) | 2.5 | 4.1 | 24 | 1909 | |
| NA19127_5 | 0.05 (5) | 2.5 | 3.3 | 19.8 | 1649 | |
SNVs, single nucleotide variants; VAFs, variant allele fractions.
Sequencing results are shown for unique reads that mapped to the assay target regions with a mapping quality score >20.
Gold standard variants were identified and reviewed as described (see Materials and Methods). Total variants include only those in the targeted regions called by both the variant identification programs GATK and SAMtools and had a coverage of at least 50. Numbers in parentheses indicate unique heterozygous variants not present in NA17989 (ie, minor gold standard variants).
Mixed samples contained the indicated amount of DNA plus additional DNA from NA17989 to total 1 mg.
Expected VAF of unique heterozygous alleles relative to NA17989 for each minor sample.
Observed VAFs calculated from base counts at minor gold standard variant positions.
Library Preparation, Multiplex Sequencing, and Data Preprocessing
Illumina sequencing libraries were prepared using 1 μg genomic DNA following standard protocols as recommended by the manufacturer. DNA was first sheared to obtain approximately 300 bp fragments using a Covaris E210 instrument (Covaris, Inc., Woburn, MA), then end-repaired, ligated to indexed universal adapters for multiplex sequencing, and amplified via limited cycle PCR. Libraries were enriched using the Washington University comprehensive cancer set WUCaMP27 version 1.0 (Genomics and Pathology Services, Washington University, St. Louis, MO) (Supplemental Table S2) using Agilent SureSelect capture probes (Agilent Technologies, Santa Clara, CA). Quality control procedures for library preparation included verification of fragment size using an Agilent 2100 Bioanalyzer and post-capture real-time quantitative PCR quantification. All 21 enriched libraries (five pure HapMap samples and 16 mixed samples from four dilutions for each of four HapMap samples) were then pooled and sequenced on an Illumina HiSeq 2000 instrument (Illumina, Inc., San Diego, CA) using V3 chemistry to obtain paired-end 101-bp reads using standard sequencing parameters and on-instrument quality control procedures; pure libraries were sequenced together in one lane, and mixed libraries were pooled in equimolar amounts and sequenced in a separate lane. The resulting sequence data were demultiplexed and mapped to the hg19 human reference sequence using Novoalign version 2.08.02 (Novocraft Technologies, Sdn Bhd, Selangor, Malaysia). Mapped data were preprocessed to mark duplicate reads before all downstream variant identification analyses. Command line parameters for all processing steps are given in Supplemental Table S3.
Gold Standard Variant Identification
Sequence data from the five pure HapMap samples were analyzed using GATK (UnifiedGenotyper version 2.3) and SAMtools (version 0.1.18) using the Best Practices guidelines for read realignment and quality score recalibration of the data before variant identification with default parameters for each program (Supplemental Table S3) to obtain gold standard heterozygous and homozygous variant calls including SNVs and insertion/deletion variants (indels). Only variants called by both programs at positions with a coverage depth of at least 50 were included in this set, and variants called by only one program were discarded. All discordant, false positive, and low coverage (<50) positions in the gold standard datasets were masked from subsequent analyses. Because there was no agreement in the exact indel calls between SAMtools and GATK, all were excluded from this analysis; indel positions ±5 adjacent bp were also masked from all downstream analyses because most of these calls occurred in noncoding regions with low sequence complexity in which true polymorphisms may exist but which are challenging to resolve definitively, in particular across multiple variant callers. Heterozygous gold standard SNVs unique to the minor HapMap sample (termed minor gold standard variants) in each mixed sample were identified using custom scripts that executed the intersectBed utility of the BEDTools package.29 Inasmuch as no gold standard indels were identified using this approach, we identified all indels called by either GATK or SAMtools and selected 45 for validation via Sanger sequencing across all HapMap samples. Primers were designed flanking the putative indels for PCR amplification, followed by exonuclease and phosphatase treatment, Sanger sequencing, and capillary electrophoresis. Of these, five distinct indel variants were validated that were heterozygous and not present in the NA17989 HapMap sample and thus were used in the indel analysis of the mixed samples.
Mixed-Sample Analysis
All data from mixed HapMap samples were analyzed using GATK (version 2.3), SAMtools (version 0.1.18), VarScan2 (version 2.3.5), and SPLINTER (version 6t), using custom scripts to execute each program. All programs were executed with default parameters except where indicated (Supplemental Table S3), although additional parameter configurations were also explored in an attempt to increase the sensitivity for low-frequency variants. Specifically, GATK and SAMtools parameters for internal down-sampling of reads to a prespecified depth (usually approximately 250), which could eliminate or reduce the frequency of minor variants simply through sampling, were set to a higher value (eg, we used values of at least 8000). We also discovered one parameter in the GATK program (version 2.0 or later) that intentionally down-weights variants that are at low frequency (the contamination parameter), which must be turned off to detect any low-frequency variants. For indel detection, we also changed the -minIndelCnt and -minIndelFrac parameters to 1 and 0.01, respectively. For all tools, only variants that passed default internal filters were considered (eg, variants with the str10 strand bias filter in VarScan2 were excluded); however, no additional filters were applied. Analysis using SPLINTER was performed as described previously.27 In brief, reads were mapped using Novoalign and processed using SPLINTER with a cycle cutoff of 60 and a run-specific error model generated from the M13 phage vector sequenced in the same lane as a separately indexed sample. SPLINTER cutoff values for variant identification are given in Supplemental Table S3. SPLINTER output was converted to variant cell format using a custom Perl script (available on request). In addition, in the event that SPLINTER reported multiple alleles at a single site, only the variant with the greatest VAF was considered. Parameters for all programs are given in Supplemental Table S3.
Performance analysis used the BEDTools intersectBed and custom awk commands (available on request) to identify the minor gold standard variants present among the variant calls for each mixed sample. Specificity analyses were performed subsequent to masking of discordant, indel, and low coverage positions identified during generation of the gold standard variant sets and excluded all indel calls made by each program.
Sensitivity was calculated as follows:
| (1) |
PPV was calculated as follows:
| (2) |
where the total gold standard variants included gold standard variants in both NA17989 and the minor sample in mixed samples, and the total variants called included all SNVs called after exclusion of any positions that were masked because of low coverage (<50), discordant calls, or indels ±5 bp in pure gold standard samples (see Gold Standard Variant Identification). Confidence intervals were calculated using the binomial distribution in R (version 2.15.1; R Project for Statistical Computing, http://www.r-project.org, accessed June 1, 2012).
Coverage analysis was performed using the Picard DownsampleSam function to randomly sample reads from the mixed dataset BAM files using sampling probabilities calculated as follows:
| (3) |
where desired coverage was 1500, 1250, 1000, 750, 500, 400, 200, or 100 and total coverage was the mean per-base coverage across the target region for each sample. In some mixed samples the highest coverage set was 1250 because the mean coverage in the original dataset was <1500. All down-sampled files (n = 121) were then analyzed as described to identify minor gold standard variants, and a custom Perl script was used to calculate the mean sensitivity for each mix (50%, 20%, 10%, or 5%) at positions with minimum observed coverages in bins of 100 (0 to 99, 100 to 199, and so on) across all mixed samples.
Read Quality Filtering
Quality filtering of reads was performed using a custom Perl script that accepted output from the SAMtools view command, which includes mapping quality, individual base qualities, and discrepancies with the reference sequence for each read. The script parsed this information and printed only lines corresponding to reads with a user-defined minimum mapping quality (set to 20), minimum base quality for all bases in the read (set to 20), and maximum number of discrepancies (set to 4).
Analysis of Lung Adenocarcinoma Samples
NGS data from formalin-fixed lung adenocarcinoma samples were previously generated using the same platform (WuCaMP27) as part of a separate study comparing formalin-fixed and fresh-frozen specimens for NGS analysis; tumor cellularity for these cases is shown in Supplemental Figure S1.28 These data were reanalyzed with the four variant identification programs using the same methods as described except only coding regions were analyzed and variants at VAF <2% were excluded. An additional analysis step included functional annotation of detected variants using the ANNOVAR software package (version 2012Oct23) using default parameters.30 A limited mixing study was also performed on two lung adenocarcinoma samples using 500 ng of two samples to generate a 50:50 mix sample (25% minor sample VAF) and 800 ng of one sample and 200 ng of another sample to generate an 80:20 mixed sample (10% minor sample VAF). Sequencing was performed as described for each mixed sample and for the two pure samples individually. Analysis was performed as described for the HapMap gold standard samples and mixes.
Results
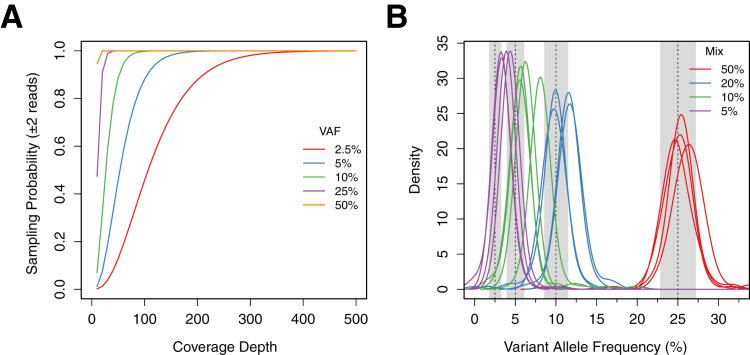
We used DNA extracted from HapMap cell lines to generate a dilution series of samples with defined mixture proportions to study the performance of common NGS analysis tools for detecting low-frequency variants. DNA from each of four HapMap samples (NA18484, NA18507, NA18872, and NA19127) was mixed with a fifth, ethnically distinct sample (NA17989) at proportions of 50%, 25%, 10%, and 5% by mass for a total of 16 mixed samples (Table 1). DNA, 1 μg, which is equivalent to approximately 3 × 105 haploid genome copies, for each mixed sample and the five individual samples was used as input for library construction and subsequent enrichment for coding and noncoding sequences for 26 cancer genes (Supplemental Table S2) and multiplex Illumina sequencing. The pooled sequencing experiments were designed to produce at least 1000× mean coverage of the targeted regions for each sample so that detection of low-frequency variants would not be limited by sampling. This high coverage requirement was predicted by simulations using binomial statistics, which showed that coverage depths as high as 200× predict only approximately 90% probability of observing two reads with a heterozygous variant present in the minor population at a mix proportion of 5% (expected VAF of 2.5%), the lowest in the present study (Figure 1A).
Figure 1.
A: Theoretical probability of sampling variant alleles of differing frequencies (25%, 10%, 5%, and 2.5%) two or more times versus coverage depth, based on binominal sampling statistics. B: Distribution of observed minor gold standard VAFs for all mixed samples. Each curve represents the distribution for a single sample with mixture proportions of 50%, 20%, 10%, and 5%, which were expected to have VAFs of 25%, 10%, 5%, and 2.5%, respectively. Gray bars show the middle 95% for each expected distribution.
Sequencing Results and Gold Standard Variant Identification
Sequencing resulted in a mean of 21 million reads (range, 14 million to 28 million) for each mixed and pure sample in the study (Table 1). Read quality and mapping statistics were examined and found to be similar for all sequenced samples. Coverage analysis revealed a mean coverage across the entire target region of 1606-fold for all samples (range, 1155 to 1969) and that a mean of 91% (range, 89% to 92%) and 81% (range, 75% to 84%) of the targeted positions in each sample had 500× and 1000× coverage, respectively. Complete sequencing and read mapping statistics are given in Supplemental Tables S4 and S5 for pure and mixed samples, respectively.
Two standard variant identification programs, GATK and SAMtools, were then used to identify all SNVs in the five pure samples to establish a set of gold standard SNVs for all subsequent analyses. These programs were used because their performance in identifying constitutional variants has been established in a large number of genomic studies.18,31,32 Variant identification was performed jointly on all samples, and discordant calls were masked in downstream analyses (see Materials and Methods). This resulted in 602 distinct gold standard SNVs across the five HapMap samples, with a mean of 267 per sample (range, 231 to 294). Of all identified SNVs, 97.7% (588 of 602) were present in dbSNP (version 137), and 100% of the genotypes that overlapped publically available, array-based HapMap genotypes for these samples were concordant; those variants that were not present in dbSNP were also included in the gold standard set, all of which seemed to be high-quality inherited variants on manual review. Variants were subsequently extracted for each sample used in the dilution series that were unique relative to NA17989 (which was used as the major sample in the dilution series) and heterozygous because these were expected to occur at VAFs that were half that of the mix proportion in the mixed samples (ie, 25%, 10%, 5%, and 2.5%). There were 427 of these minor gold standard variants across the four mixed samples (mean per sample, 107; range, 98 to 115), including 56 coding region variants (mean per sample, 14; range, 8 to 21), which were used to evaluate the performance of variant identification programs for detecting low-frequency variants.
In addition to these gold standard SNVs, we also identified a small number of insertions and deletions in the HapMap samples to assess the performance of the software tools for detection of indel variants. Because no concordant indels were identified via both GATK and SAMtools in the gold standard analysis, we reviewed potential indel calls made by either program and selected a subset that were not in microsatellites or long homopolymer runs for validation via Sanger sequencing. This resulted in five distinct Sanger-validated indel calls (three deletions and two insertions) of 1 to 6 bp (Supplemental Figures S2, S3, S4, S5, and S6) that were suitable for analysis in the mixed samples; none occurred in coding regions, and several were in regions adjacent to low-complexity sequences.
Observed VAFs
Before testing the low-frequency variant detection performance, we examined the observed frequencies of the minor gold standard variants to determine whether any allele bias occurred during targeted sequencing and to verify the VAFs in the mixes. This demonstrated that heterozygous minor gold standard variant VAFs generally fell within the expected range, although SNVs in the 10% mix had a mean VAF of 6.8% rather than the expected 5%, and the mean VAF of the 5% mix was 4.2% rather than 2.5% (Table 1 and Figure 1B). Inasmuch as mixing to make the most dilute samples involved DNA volumes as low as 1 μL, this discrepancy was likely due to inaccurate DNA quantitation and/or pipetting errors. However, despite these differences, the overall concordance between observed and expected VAFs in the mixed samples demonstrated that targeted hybridization capture did not bias enrichment against non-reference alleles in the sample, even though the capture reagents were designed from the human reference sequence and did not account for non-reference alleles present in the HapMap samples. In addition, this analysis verified that the lower boundary of VAFs in our dilution series was <5%.
Low-Frequency SNV Identification
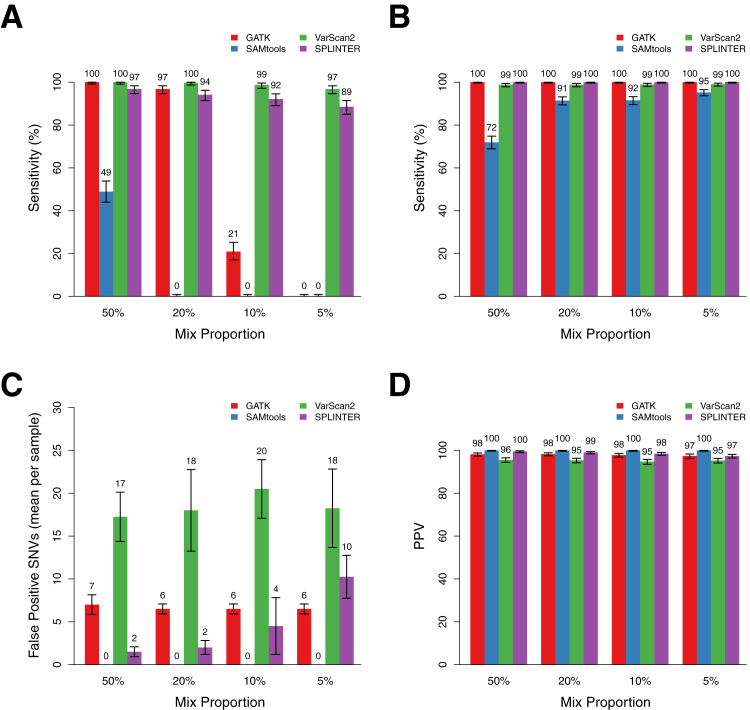
We used four publicly available NGS software tools to identify SNVs in the mixed samples. The programs we tested included the variant identification functions of popular tools SAMtools and GATK, which use bayesian genotype-calling algorithms, the somatic mutation program VarScan2, which uses a set of heuristic read and base quality filters, and SPLINTER, a low-frequency variant caller originally designed to detect rare alleles in pooled samples using custom run-specific error models based on large deviation theory.18,19,25,27 These programs were executed as described (see Materials and Methods) to obtain passing SNV calls (nonpassing variants using default filters were excluded) with a coverage depth of at least 100× across the entire target region; a mean of 97.6% of targeted positions met this coverage gold across all mixed samples (range, 97.3% to 98.2%). In most cases we used the default parameters for each program, which provided the best low-frequency variant performance with an acceptable specificity (although the best performance was variable, as we describe in Effect of Coverage on SNV Sensitivity and Specificity). However, we found changing some parameters to be critical and used optimized parameters for some programs (see Materials and Methods). Variant calling using these methods resulted in identification of between 212 and 442 total SNVs across all samples, depending on the particular mix proportion and program (Supplemental Table S5). We examined the sensitivity for each program across a range of variant frequencies by determining the proportions of minor gold standard variants that were successfully detected in the mixed samples; to prevent false negative results because of incomplete sampling, this analysis considered only positions in which observed coverage was ≥100×. Comparison of these sensitivities revealed substantial differences in performance across the programs tested. For example, SAMtools detected only 49% [201 of 411; 95% confidence interval (CI), 44% to 54%) of the minor gold standard variants in the 50% mixes (25.8% mean observed VAF), and few variants were detected at higher dilutions (Figure 2A). The other programs performed substantially better. GATK detected 97% (396 of 409; 95% CI, 94% to 98%) of the minor gold standard variants at the 20% dilution (11.2% mean observed VAF), although there was a substantial drop off at lower VAFs (Figure 2A). Indeed, this program detected 100% of variants with observed VAFs >10% and detected no variants with observed VAFs <7% regardless of coverage depth. VarScan2 and SPLINTER showed the best performance and detected at least 89% of the minor gold standard variants in each group of mixed samples. Sensitivity was dependent on VAF, as expected; however, these programs were still able to detect, respectively, 97% (VarScan2: 396 of 409; 95% CI, 95% to 98%) and 89% (SPLINTER: 363 of 409; 95% CI, 84% to 91%) of minor gold standard variants in the 5% mixed samples (4.2% mean observed VAF). Similar results were observed using only positions confirmed using array-based HapMap genotypes for all mixed samples (Supplemental Figure S7), and no differences in the minor gold standard variant calls were identified in one dilution series after remapping of the reads with a different program, BWA (data not shown). In most cases, minor gold standard variants that were not detected by these programs had lower coverage than those that were detected. For example, 17 of the 19 variants missed by VarScan2 occurred at positions with <200× coverage. Variants missed by SPLINTER had more variable coverage but frequently occurred at low coverage positions or had VAFs such that the variant allele was present in <25 reads in the sample.
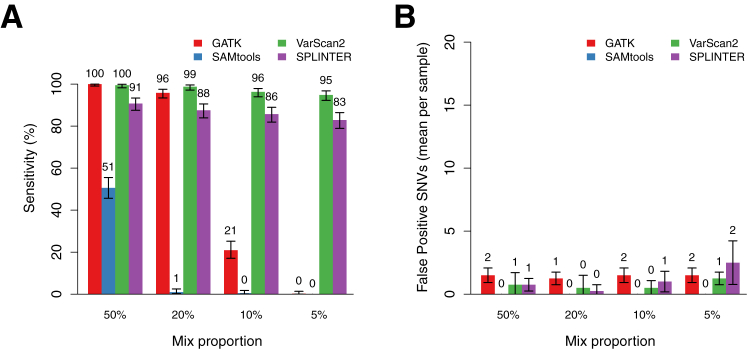
Figure 2.
Performance of GATK, SAMtools, VarScan2, and SPLINTER for detecting low-frequency variants in mixed samples at positions with coverage ≥100×. A: Sensitivity for detecting all heterozygous minor gold standard variants in samples with mix proportions of 50%, 20%, 10%, and 5% (mean observed gold standard VAFs, 25.5%, 11.2%, 6.8%, and 4.2%, respectively). Sensitivities (True positive/True positive + False negative) are point estimates based on detection of all minor gold standard variants at positions with ≥100× coverage in each set of mixed samples (n = 409, 406, 409, and 411, respectively). Error bars show the 95% binomial CI for each point estimate. B: Sensitivity for detecting homozygous and heterozygous gold standard variants in the major sample, which have estimated VAFs of >25%. Error bars show the 95% binomial CI for each point estimate. C: Mean number of false positive SNV calls per sample made by each program at the indicated mix proportion across the entire target region, encompassing coding and noncoding sequence of 26 genes (306,336 bp). Indel calls were excluded, as were positions with low coverage or discordant calls in the gold standard variant analysis (see Materials and Methods). Error bars show the SD across all samples with the indicated mix proportion (n = 4 for each mix proportion). D: PPV (True positive/True positive + False positive) for SNV calls by each program across the mix proportions. Error bars show the 95% binomial CI for each point estimate.
We also assessed the performance of each program for detecting the minor gold standard variants present in coding regions and splice sites of the 26 targeted genes, which included 20% of the total targeted sequence of 61,221 bp (Supplemental Figure S8). There were a total of 56 coding variants among the four mixed samples in each dilution set, with a mean per sample of 14 (range, 8 to 21). Similar to the results for the entire target region, SAMtools demonstrated the poorest performance, detecting only 64% of the coding region variants in the 50% mix sample (36 of 56; 95% CI, 50% to 77%). GATK detected all of the coding region variants in the 50% mix and all but one variant in the 20% mixed sample (sensitivity 98%; 95% CI, 90.4% to 99.9%), but few variants in the more dilute samples. VarScan2 missed only one coding variant with a VAF of 5.4% that occurred at a position with relatively low coverage (111×) in a 5% mixed sample (sensitivity 98%; 95% CI, 90.4% to 99.9%), whereas SPLINTER achieved sensitivity between 96% and 98% across the four dilution sets, with the missed variants occurring at positions with coverage <600×.
Sensitivity was also evaluated for the gold standard variants present in the major HapMap sample in the dilution series to compare the performance of the programs for variants present at higher frequencies. The major gold standard variants in this analysis had expected VAFs of at least 25%, reflecting either heterozygous or homozygous variants diluted by a maximum of 50% with the minor HapMap sample. These sensitivities showed minor differences between the programs, with GATK and VarScan2 exhibiting the best performance. However, with the exception of SAMtools in the 50% mixed sample, all programs detected at least 91% of the variants, and performance was similar across all dilutions (Figure 2B).
Specificity and PPV
To determine whether there was a tradeoff between sensitivity and specificity across the variant identification programs, we identified variants in each mixed sample that were not present among the gold standard variants in either HapMap sample in the mix and were therefore likely to be false positive calls. Although some of these variants could be true positive variants that were missed in our gold standard analysis, we attempted to minimize this by excluding all positions that were low coverage or ambiguous in the gold standard analysis; we believe this approach provides a reasonable estimate of the specificity and PPV in interpretable regions of our target. This analysis revealed that GATK and SPLINTER made few false positive calls in any sample (GATK: mean, 6.6; range, 6 to 8; SPLINTER: mean, 4.6; range, 1 to 13) and that SAMtools made no false positive calls (Figure 2C). VarScan2 was the clear outlier, with a mean of 18.5 false positive calls per sample (range, 13 to 25). We calculated the PPV, ie, the proportion of called variants that were real, for each program by dividing the total number of gold standard variants identified (including both major and minor samples in each mixture) by the total number of variants called. This statistic ranged from 94% to 100% across the entire dataset, with VarScan2 having the lowest mean PPV, 95% (Figure 2D).
The vast majority of false positive SNV calls from all four programs occurred in noncoding regions with low complexity. Indeed, only one false positive call was made in a coding region by any program, which was a variant with VAF of 1% in a single sample that was called by VarScan2 (Supplemental Figure S9). There were no false positive calls in coding regions produced by the other three programs.
Indel Detection Sensitivity
We used five Sanger-validated indels identified in two of the HapMap samples to conduct a limited study of indel detection sensitivity of the four programs in the mixed samples. For this analysis, we considered any indel variant of the correct type (insertion or deletion) called within a 5-bp window of the validated indel position as correctly identified because the precise indel position in a sequence alignment can be arbitrary. Using this approach, we found that SAMtools detected none of the validated indels, even in the 50% mixed sample (Supplemental Table S6). SPLINTER detected only one of the indels, although it was detected at all mix proportions. GATK detected none of the indels using default parameters; however, after some optimization (see Materials and Methods), it detected all five indels in the 50% and 20% mixed samples, three of five in the 10% mixed sample and one of five in the 5% mixed sample. In contrast, VarScan2 detected all five indels at all mix proportions. Inasmuch as the sample size in this analysis was small, the results are likely influenced by factors such as indel length and sequence context of the specific variants in the set. Nonetheless, these results provide some preliminary indications of indel detection of these programs and suggest that VarScan2 may be an acceptable option for detecting low-frequency indel variants. However, we observed that VarScan2 called a large number of indels in noncoding regions (mean of approximately 54 per case), many of which were likely to be false positive on manual review. Although these spurious calls did not occur in coding regions, and, thus, would be unlikely to affect clinical interpretations, this observation suggests that using VarScan2 for indel detection would substantially increase the total number of variant calls.
Effect of Coverage on SNV Sensitivity and Specificity
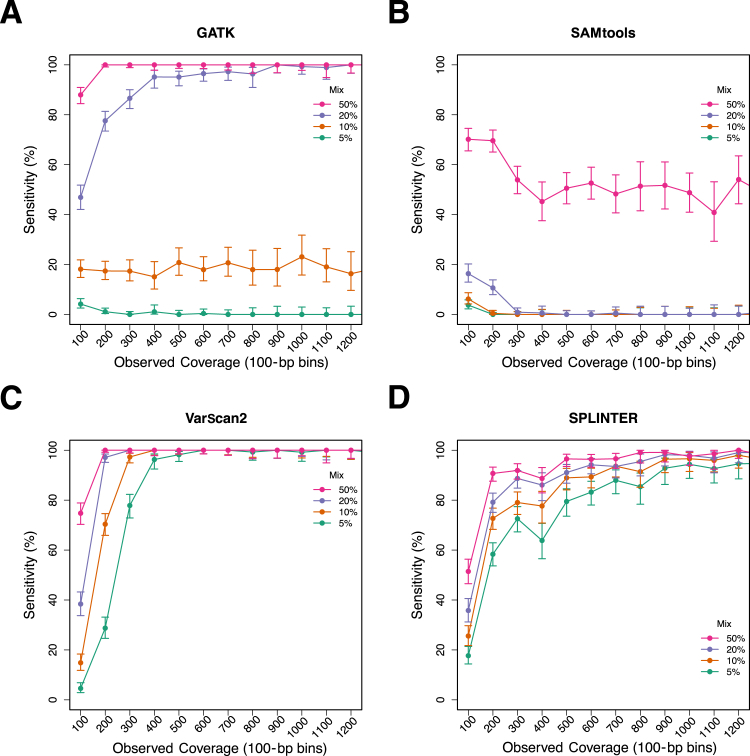
Because coverage depth is likely a critical variable in detecting low-frequency variants using these programs, we used a down-sampling approach to determine the minimum coverage required to confidently identify the minor gold standard variants in the mixed samples. Sequencing reads from the mixed samples were randomly sampled to achieve expected mean coverage depths for the entire target region, ranging from 100× to 1500×, and then the four programs were used to identify variants in these down-sampled read sets. The observed coverage depth at each minor gold standard variant position was also recorded, which allowed for analysis of variant detection with respect to the actual coverage at each variant position.
The results of this analysis are summarized in Figure 3, which shows the sensitivity for minor gold standard variants as a function of the minimum observed coverage at variant positions for all four variant detection programs stratified into coverage bins of 100. The trends in sensitivity reflect what was observed in the full coverage data: SAMtools demonstrated poor sensitivity when compared with the other three programs, and VarScan2 and SPLINTER exhibited the best performance. As expected, sensitivity was decreased at lower coverage, with the most precipitous decline occurring at coverage depths <300×. GATK showed a more gradual decline in sensitivity in the 20% mixed sample (11.2% mean observed VAF) at coverages <800, but detected >90% of the minor gold standard variants at coverage depths between 400 and 800; at coverage depths <400, sensitivity decreased substantially. Compared with the other programs, SPLINTER also showed a gradual decline in sensitivity with coverage, with optimal sensitivity occurring at higher coverages (>800×). In contrast, the sensitivity of VarScan2 remained at >95% even for the 5% mix (4.2% mean observed VAF) at coverages of ≥400× before decreasing at lower coverage depths.
Figure 3.
Sensitivity of GATK, SAMtools, VarScan2, and SPLINTER for low-frequency variants as a function of observed coverage at variant positions. Sequencing reads from mixed samples were randomly sampled (see Materials and Methods) to obtain datasets with estimated mean coverage depths of 1500, 1250, 1000, 750, 500, 400, 200, and 100 across the entire target region for each of the mixed samples. The observed coverage depths were determined for all minor gold standard variants, and variant detection was performed using each of the four programs. Panels show the overall sensitivity for all variants from each mixed sample in the observed coverage in bins of 100 (eg, the 100 bin contains all gold standard variants with coverage depths between 0 and 100) for GATK (A), SAMtools (B), VarScan2 (C), and SPLINTER (D). Error bars show the 95% binomial CI for each point estimate.
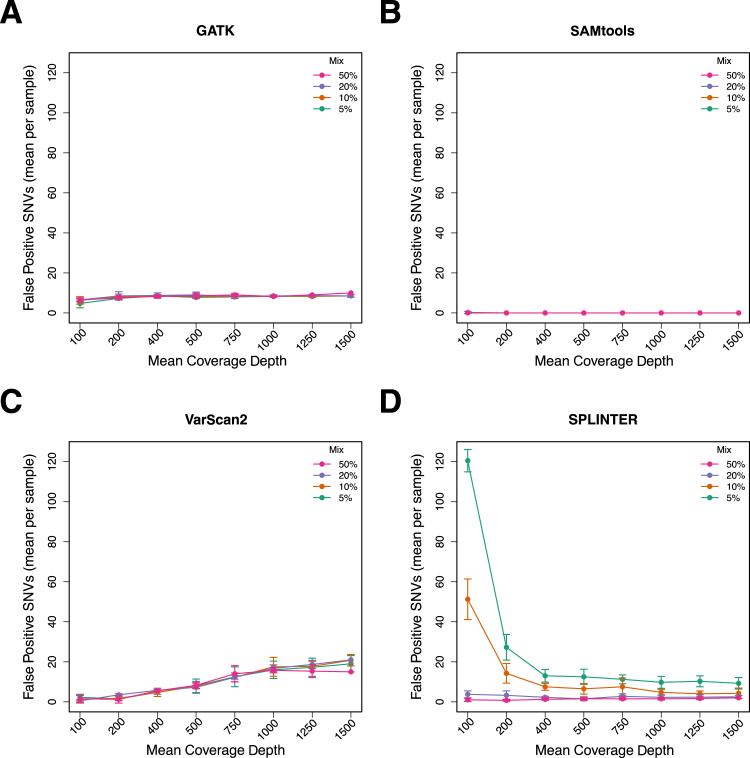
We also examined the specificity of each program in the down-sampled data to determine the effect of varying coverage levels on false positive calls (Figure 4). This revealed that VarScan2 called more false positives as the coverage increased; there were a mean of 1.4 false positives per sample in data with coverage of 100× compared with a mean of 18.5 per sample in the complete dataset (mean coverage, approximately 1500×). This was unexpected because higher coverage is generally associated with more confident base calls. GATK also called more false positives at higher coverage levels; however, the difference was small (100× coverage, mean of six false positive calls; 1500× coverage, mean of 8.6 false positive calls). In contrast, SPLINTER called a large number of false positives at low coverage levels (eg, >100 for the 100× coverage dataset), compared with fewer than 20 for the datasets with higher coverage. Similar to the complete dataset, SAMtools called virtually no false positives at any coverage level and mix proportion.
Figure 4.
False positives (mean per sample) called by GATK, SAMtools, VarScan2, and SPLINTER as a function of mean target coverage and mix proportion across the entire target region encompassing coding and noncoding sequences of 26 genes (306,336 bp). Variant SNV calls in down-sampled data that were not among the gold standard SNVs were called as false positive, after excluding indel calls and positions that were low coverage (<50) or discordant in the gold standard analysis. The number of false positive calls for GATK (A), SAMtools (B), VarScan2 (C), and SPLINTER (D) for each down-sampled coverage level and the mix proportion indicated by the legend. Error bars show the SD for each coverage level and mix proportion.
Manual review of selected false positive calls revealed that some of the variant positions themselves were of high quality and thus passed variant quality filters; however, the reads containing the variants had multiple discrepancies, which suggested that they were either incorrectly mapped or of variable quality. This observation provided some explanation for the greater number of false positives called by VarScan2 in datasets with higher coverage, because the greater number of reads could result in more instances of incorrect mapping or low-quality reads that generate false positive calls. To determine whether additional filtering to remove multiple-discrepancy and low-quality reads could reduce the number of false positives and improve specificity, we created a set of high-quality filtered reads from each mixed sample, comprised of reads with only high-quality bases (PHRED quality ≥20), a minimum mapping quality score of 20, and fewer than four total discrepancies per read. This removed 2 million to 9 million reads from the mixed samples (mean, 5.3 million) and had a relatively small effect on mean target coverage (mean prefiltered coverage, 1620; postfiltered coverage, 1387). Variant identification in this set of filtered reads using the same set of parameters as in our initial sensitivity analysis reduced the sensitivity of GATK and VarScan2 for minor gold standard variants by 1% to 3% and of SPLINTER by up to 6% (Figure 5A) because of reduced coverage at some positions. However, the number of false positive calls made by these programs was reduced substantially. In the filtered read set there was a mean of 0.8 (range, 0 to 2) false positive SNVs per sample for all programs (Figure 5B), compared with 6.7 (range, 6 to 8) false positives for GATK, 18.5 (range, 13 to 25) for VarScan2, and 5.1 (range, 1 to 12) for SPLINTER, using all reads (Figure 2C). This suggested that a prefiltering approach could be used to improve the specificity, in particular for VarScan2.
Figure 5.
Sensitivity and false positive calls made using only filtered reads compared with using all reads. Filtered reads included only those with a mapping quality >20, a minimum base quality of 20 for all bases, and no more than four discrepancies. A: Sensitivity for minor gold standard SNVs across all four programs after filtering of low-quality and multiple-discrepancy reads. Error bars show the 95% binomial CI for each point estimate. B: False positive SNV calls (mean per sample) using high-quality reads compared with all reads. Error bars show the SD across all samples at each mix proportion (n = 4).
Low-Frequency Variant Detection in Clinical Lung Adenocarcinoma FFPE Samples
We next used the four programs to identify variants in NGS data from 15 lung adenocarcinoma specimens to assess low-frequency variant detection performance in clinical NGS data. These samples were obtained from diagnostic FFPE tissue and were sequenced using the same targeted NGS platform that was used to sequence the HapMap dilution samples.28 Sequencing results were similar to the HapMap samples, with mean target coverage of 1100×, and 79% of target positions had coverage of at least 500× (Supplemental Table S7). Variant identification was performed in the same manner as described above with a few exceptions. Because no gold standard variants existed for these samples, we restricted our analysis to variants known to be highly recurrent in cancer samples on the basis of their presence in the COSMIC mutation database.33 Variants were identified in coding sequences; then only changes present in COSMIC were compared between the variant detection programs. In addition, we required the variants to be present at a frequency of at least 2% because our dilution series was based on a minimum VAF of 2.5%.
Approximately 30 total coding region variants were identified in the 130 kb of coding sequence in each sample (range, 22 to 45), about 3 (range, 0 to 7) of which were nonsynonymous SNVs that were not known polymorphisms present in dbSNP (version 137), and all four programs called a similar number of coding region and nonsynonymous variants in each case (Supplemental Table S8). In three cases, no variants were identified that were present in the COSMIC database, whereas in the remaining 12 cases, 15 recurrent cancer-associated variants (1 to 3 per sample) in five genes (KRAS, NRAS, TP53, EGFR, and BRAF) were detected by at least one of the four programs. Most of the variants, 11 of 15 (73%), had VAFs ranging between 24.6% and 86.7% and were detected by all four programs (Table 2). However, four variants (two KRAS codon 12, one in TP53, and another in BRAF) were present at lower frequencies (2.2% to 22.4%) and were called by only a subset of the programs. The detection of these mirrored the performance observed in the HapMap samples, with SAMtools missing all four low-frequency variants and only VarScan2 detecting a TP53 variant present at 2.2%, whereas GATK and SPLINTER detected three of the four low-frequency variants.
Table 2.
Detection of Recurrent Somatic Mutations in 15 FFPE Lung Adenocarcinoma Samples
| Case | Gene | Variant | Amino acid change | Coverage | VAF, % | SAMtools | GATK | VarScan2 | SPLINTER |
|---|---|---|---|---|---|---|---|---|---|
| 1 | KRAS | c.G35T | p.G12V | 3610 | 53.8 | + | + | + | + |
| 2 | No recurrent SNVs detected | – | – | – | – | ||||
| 3 | EGFR | c.T2573G | p.L858R | 3055 | 25.7 | + | + | + | + |
| 4 | NRAS | c.A182T | p.Q61L | 1875 | 67.8 | + | + | + | + |
| 5 | No recurrent SNVs detected | – | – | – | – | ||||
| 6 | KRAS | c.G35C | p.G12A | 2128 | 60.9 | + | + | + | + |
| TP53 | c.A840T | p.R280S | 730 | 42.5 | + | + | + | + | |
| TP53 | c.G734A | p.G245D | 407 | 2.2 | ND | ND | + | ND | |
| 7 | No recurrent SNVs detected | – | – | – | – | ||||
| 8 | KRAS | c.G35T | p.G12V | 1505 | 20.7 | ND | + | + | + |
| 9 | TP53 | c.C380T | p.S127F | 985 | 54.6 | + | + | + | + |
| 10 | TP53 | c.G517A | p.V173 mol/L | 791 | 38.7 | + | + | + | + |
| 11 | KRAS | c.G35T | p.G12V | 3430 | 54.8 | + | + | + | + |
| 12 | TP53 | c.C331A | p.L111 mol/L | 906 | 24.6 | + | + | + | + |
| BRAF | c.C897A | p.I300V | 1238 | 14 | ND | + | + | + | |
| 13 | KRAS | c.G35T | p.G12V | 1098 | 22.4 | ND | + | + | + |
| 14 | TP53 | c.G733T | p.G245C | 548 | 27 | + | + | + | + |
| 15 | TP53 | c.G818T | p.R273L | 1389 | 86.7 | + | + | + | + |
ND, not detected; SNVs, single nucleotide variants; VAF, variant allele fraction; +, positive; –, negative.
To further assess the ability of these programs to detect low-frequency variants in FFPE samples, we used DNA from two clinical specimens to generate samples with 50% and 20% mix proportions (25% and 10% expected minor sample VAF, respectively) and conducted an analysis similar to the HapMap mixes using 45 SNPs identified in each pure sample. The observed VAFs in these mixed samples were 16% and 7% for the 50% and 20% mixes, respectively, likely due to different DNA qualities and somatic copy number changes in the tumors. Variant detection performance using these samples was largely consistent with that observed in the HapMap mixes. SAMtools detected <15% of the variants in both mixes, and GATK detected 93% (42 of 45) of the variants in the 50% mix and 7% (3 of 45) in the 20% mix. VarScan2 detected 100% of the variants in the 50% mix and 93% (42 of 45) in the 20% mix; SPLINTER detected 80% (36 of 45) in both samples, with most missed sites due to low coverage. Overall, the results from both of these mixed samples and the pure clinical specimens supported those from the HapMap samples and showed that performance was similar using data from FFPE specimens. Detection performance in the pure clinical samples demonstrated that using SAMtools for somatic mutation detection may miss potentially relevant mutations because of low variant frequency.
Discussion
Next-generation sequencing in the clinical laboratory has become a practical alternative to conventional molecular techniques for detecting mutations. Testing for somatic mutations in cancer is a natural application for NGS because it enables multiplex testing of numerous cancer genes simultaneously, and deep sequencing can potentially detect low-frequency somatic mutations inherent in cancer specimens because of contamination with nontumor cells and tumor heterogeneity. Although the high throughput of NGS platforms and the digital nature of the resulting data make detection of low-frequency variants technically possible, the relatively high error rate of NGS platforms necessitates methods that are able to differentiate true variants from background noise.
In the present study, we assessed the ability of commonly used NGS analysis programs to detect low-frequency variants in high-coverage (>1000×) targeted NGS data using synthetic laboratory-derived mixtures of HapMap samples. Sequencing of these mixed samples showed no evidence for bias against non-reference alleles in targeted hybridization capture NGS data, and analysis of variant calls at known gold standard variant positions obtained by independent sequencing of the pure HapMap samples revealed substantial variability in sensitivity for low-frequency variants across the programs evaluated. We found that the variant identification function of the popular and widely used SAMtools program is particularly insensitive to low-frequency variants and would be poorly suited for a bioinformatics pipeline for de novo detection of somatic mutations in cancer specimens. The GATK variant caller demonstrated good sensitivity (97%) for variants with VAFs of approximately 10% but was unable to detect variants present at lower frequencies. VarScan2 and SPLINTER demonstrated the best performance and detected >89% of variant VAFs as low as approximately 4% on the basis of observed VAFs in our mixed samples, with even higher sensitivity in coding regions. We also conducted a limited analysis of indel detection performance using a small number of Sanger-validated indel variants and found that only VarScan2 and SPLINTER were able to detect any indels below 25%. However, our set of validated indels included only five distinct variants, all of which were noncoding, and thus these results may not be generalizable. As expected, detection of SNVs depended on coverage, with most missed calls occurring at positions with relatively low coverage (<200×) and the most robust detection occurring at a minimum coverage of approximately 400×. Because coverage in targeted NGS data can be quite variable, even higher mean target coverage would be required to ensure that this coverage level is achieved at all critical positions in an NGS assay. Although higher coverage improved sensitivity, it was associated with more false positive variant calls, in particular with VarScan2, although specificity could be improved by filtering sequencing reads to eliminate those with low quality and questionable alignments. SPLINTER exhibited the opposite trend, with a large number of false positive calls in the low-coverage data (<200×) and few at higher coverage levels. It is important to note, however, that the PPV of all callers for coding region variants was high. In these data, we observed only one coding region false positive call, a variant with a VAF of 1% made by VarScan2, which suggests that clinically relevant false positives are infrequent in high-coverage targeted NGS data. Finally, we used these programs to detect variants in a set of targeted sequence data from lung adenocarcinoma samples and found similar performance and results. Several variants were identified at canonical loci (eg, KRAS codon 12) that were at low frequency and thus were detected by only the best-performing software evaluated in the present study.
These results provide some guidance to molecular diagnostic laboratories as they design and implement NGS-based assays for somatic mutations in samples for either clinical testing or research studies. We tested two of the most commonly used open source NGS analysis tools, SAMtools and GATK, which were not designed for somatic mutation detection in NGS data but nonetheless might be the initial choice for laboratories implementing a test for cancer-associated somatic mutations because of their availability and widespread use as reported in the literature. The general conclusions of the present study with respect to these particular programs would likely be discovered early in the validation process of a clinical test; however, the range of mix proportions and the coverage analysis we performed should provide laboratories with more detailed performance characteristics for the common analysis tools we evaluated when applied to deep coverage capture enrichment NGS data targeted to any locus. Because our study also included DNA from FFPE samples, we believe our observations can be generalized to routine clinical testing that typically involves this specimen type. In addition, we believe our mixing study design, using well-characterized HapMap samples, is an example of the type of experiment that could be implemented for initial test validation and for routine quality assurance/quality control in the clinical laboratory.
In the present study we also compared the performance of the common general purpose NGS software tools SAMtools and GATK with that of VarScan2 and SPLINTER, the latter of which is a specialized program that requires spiked-in control sequences to establish run-dependent error models. The sensitivity of this program at low VAFs (approximately 4%) was higher than that of GATK and SAMtools and was similar to that of VarScan2, while maintaining higher specificity. However, the algorithm used by SPLINTER requires high coverage depth for acceptable performance, which was demonstrated in our study by the lower sensitivity and greater number of false positive calls at lower coverage (eg, <200×). Because SPLINTER relies on spike-in control samples to calibrate internal error models, control reagents must also be included in the assay design. Control sequences are already part of the standard NGS workflow (eg, the phiX sequencing control), and these can double as controls for SPLINTER without additional cost or inputs. Our data suggest substantial benefit of using SPLINTER rather than SAMtools or GATK; however, its performance is similar to that of VarScan2; eg, the sensitivity of VarScan2 seems to be at least as good as, if not better than, SPLINTER, and VarScan2 made only one false positive call in a coding region across all cases in our dataset, which occurred at a VAF of 1%, the practical lower limit of the sequencing platform. Although we found that VarScan2 produces a relatively large number of likely false positive calls in noncoding regions, this can be reduced substantially by using a prefiltering step to remove reads with a high number of discrepancies. In our experiments, removing reads with more than four discrepancies reduced the mean number of likely false positive SNVs from approximately 20 in our target region to less than one. This approach has been used by other groups and thus may be a general method for increasing the specificity of NGS platforms.34
Although the present study explored some variables that affect low-frequency variant sensitivity such as coverage depth, other important variables were not examined. For example, we used 1 μg of input DNA for library preparation, which is equivalent to approximately 300,000 genome copies; thus detection of low-frequency variants was not limited because of low library complexity. Low input DNA amounts could result in diminished sensitivity because of sampling statistics, even at deep coverage levels. In addition, we tested a single version of four variant identification programs, although others have been developed. These may perform differently, and future versions of the programs we tested that incorporate improvements to the calling algorithms or additional parameters could also change the performance. However, any software that uses a bayesian algorithm designed for constitutional variants, and there are many in this category, would be expected to have limited sensitivity because such methods anticipate only homozygous and heterozygous variants.35–37 Another limitation of the present study is that there were few gold standard insertion or deletion variants. The few Sanger-validated indels that were available for analysis offer some information about the detection performance of the programs we tested, which was largely similar to the performance for SNVs. Further, we have previously found that other programs have good sensitivity for larger indel events such as FLT3 insertions, which suggests that NGS approaches can be used to identify low-frequency indel variants in some cases.38 Inasmuch as the present study was not designed to fully assess indel detection performance, this will need to be addressed in future studies and/or validation experiments performed by individual laboratories using gold standard indel variants obtained using other methods.
The lowest VAFs explored using the mixed samples in the present study ranged between 2% and 4%, which we believe is a reasonable lower limit of detection for tumor sequencing from diagnostic tissue samples. Although the clinical importance of detecting such low-frequency variants is unclear, we note that conventional targeted methods for detecting canonical mutations at a single position, such as FLT3 D835 and KRAS codon 12 mutations, are often fairly sensitive and can detect variants down to allele burdens of 3% to 5%.39,40 These tests have been the basis for clinical assays and numerous clinical studies and are routinely used to guide clinical management of patients with cancer.41–44 The present study demonstrates that standard NGS-based tests are at least capable of achieving similar sensitivity. Somatic mutations certainly exist at frequencies lower than those we explored, in particular in the setting of minimal residual disease testing, which often requires accurate detection of variants at frequencies substantially <1%. Although it is possible that the best-performing variant identification tools may have reasonable sensitivity below the lowest VAF examined in this study, it is likely that current error rates of standard targeted NGS make such platforms poorly suited for reliable discovery of variants much below approximately 2% without compromising specificity. In practice, we believe a low-tech approach involving manual review of base counts at specific hot spot loci with high a priori likelihood of somatic mutation (eg, KRAS codon 12, FLT3 codon 835, and BRAF codon 600) provides a means for sensitive detection of critical variants without resulting in a large number of false positive calls that could occur if a high-sensitivity approach is broadly applied across the targeted genomic regions. Ultimately, it is likely that recent advances in NGS methods that use molecular tags to increase sequencing accuracy will replace these ad hoc methods and enable reliable detection of low-frequency variants for cancer testing and minimal residual disease assays.20,21,45
Acknowledgments
We thank Karen Seibert for assisting with the project; Maggie O’Guin of WU-GPS for technical assistance; Haley Abel for critiquing the manuscript; and the Genome Technology Access Center at the Department of Genetics, Washington University School of Medicine, for assistance with genomic analysis.
Footnotes
Supported by the Departments of Pathology and Genetics, Washington University, St. Louis, Missouri. The Genome Technology Access Center at the Department of Genetics, Washington University School of Medicine, is partially supported by NCI Cancer Center Support grant P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA grant UL1RR024992 from the National Center for Research Resources, a component of the National Institutes of Health, and the National Institutes of Health Roadmap for Medical Research.
Supplemental Data
Approximate tumor cellularity as determined by visual estimate by a study pathologist (E.J.D.) for FFPE lung adenocarcinomas used in analysis. Original magnification ×20.
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Deletion of A in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Insertion of AA in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Insertion of CC in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Insertion of TG in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Deletion of CTCTTC in case NA18507 (arrow).
Sensitivity of minor gold standard variants confirmed by array-based genotype calls from undiluted samples.
Sensitivity and PPV for coding regions only. Methods and calculations are identical to those in Figure 2, A and C, except applied only to the protein coding regions and adjacent splice sites (±10 bp) for all targeted genes.
Read alignments showing the single false positive coding region variant call, made by VarScan2. The VAF of this variant is 1%.
References
- 1.Druker B.J., Talpaz M., Resta D.J., Peng B., Buchdunger E., Ford J.M., Lydon N.B., Kantarjian H., Capdeville R., Ohno-Jones S., Sawyers C.L. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 2.Kohl T.M., Schnittger S., Ellwart J.W., Hiddemann W., Spiekermann K. KIT exon 8 mutations associated with core-binding factor (CBF)-acute myeloid leukemia (AML) cause hyperactivation of the receptor in response to stem cell factor. Blood. 2005;105:3319–3321. doi: 10.1182/blood-2004-06-2068. [DOI] [PubMed] [Google Scholar]
- 3.Kottaridis P.D., Gale R.E., Frew M.E., Harrison G., Langabeer S.E., Belton A.A., Walker H., Wheatley K., Bowen D.T., Burnett A.K., Goldstone A.H., Linch D.C. The presence of a FLT3 internal tandem duplication in patients with acute myeloid leukemia (AML) adds important prognostic information to cytogenetic risk group and response to the first cycle of chemotherapy: analysis of 854 patients from the United Kingdom Medical Research Council AML 10 and 12 trials. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 4.Ley T.J., Ding L., Walter M.J., McLellan M.D., Lamprecht T., Larson D.E. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lièvre A., Bachet J.B., Le Corre D., Boige V., Landi B., Emile J.F., Côté J.F., Tomasic G., Penna C., Ducreux M., Rougier P., Penault-Llorca F., Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 6.Miki Y., Swensen J., Shattuck-Eidens D., Futreal P.A., Harshman K., Tavtigian S., Liu Q., Cochran C., Bennett L.M., Ding W. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 7.Schnittger S., Schoch C., Kern W., Mecucci C., Tschulik C., Martelli M.F., Haferlach T., Hiddemann W., Falini B. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106:3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 8.Yan H., Parsons D.W., Jin G., McLendon R., Rasheed B.A., Yuan W., Kos I., Batinic-Haberle I., Jones S., Riggins G.J., Friedman H., Friedman A., Reardon D., Herndon J., Kinzler K.W., Velculescu V.E., Vogelstein B., Bigner D.D. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncavage E.J., Abel H.J., Szankasi P., Kelley T.W., Pfeifer J.D. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Mod Pathol. 2012;25:795–804. doi: 10.1038/modpathol.2012.29. [DOI] [PubMed] [Google Scholar]
- 10.Lipson D., Capelletti M., Yelensky R., Otto G., Parker A., Jarosz M., Curran J.A., Balasubramanian S., Bloom T., Brennan K.W., Donahue A., Downing S.R., Frampton G.M., Garcia L., Juhn F., Mitchell K.C., White E., White J., Zwirko Z., Peretz T., Nechushtan H., Soussan-Gutman L., Kim J., Sasaki H., Kim H.R., Park S.I., Ercan D., Sheehan C.E., Ross J.S., Cronin M.T., Jänne P.A., Stephens P.J. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pritchard C.C., Smith C., Salipante S.J., Lee M.K., Thornton A.M., Nord A.S., Gulden C., Kupfer S.S., Swisher E.M., Bennett R.L., Novetsky A.P., Jarvik G.P., Olopade O.I., Goodfellow P.J., King M.C., Tait J.F., Walsh T. ColoSeq provides comprehensive lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis M.J., Ding L., Shen D., Luo J., Suman V.J., Wallis J.W. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P., Varela I., Phillimore B., Begum S., McDonald N.Q., Butler A., Jones D., Raine K., Latimer C., Santos C.R., Nohadani M., Eklund A.C., Spencer-Dene B., Clark G., Pickering L., Stamp G., Gore M., Szallasi Z., Downward J., Futreal P.A., Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing [published correction appears in N Engl J Med 2012, 367:976] N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nik-Zainal S., Alexandrov L.B., Wedge D.C., Van Loo P., Greenman C.D., Raine K., Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yachida S., Jones S., Bozic I., Antal T., Leary R., Fu B., Kamiyama M., Hruban R.H., Eshleman J.R., Nowak M.A., Velculescu V.E., Kinzler K.W., Vogelstein B., Iacobuzio-Donahue C.A. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Welch J.S., Ley T.J., Link D.C., Miller C.A., Larson D.E., Koboldt D.C. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weir B.A., Woo M.S., Getz G., Perner S., Ding L., Beroukhim R. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiatt J.B., Pritchard C.C., Salipante S.J., O’Roak B.J., Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc Natl Acad Sci U S A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flaherty P., Natsoulis G., Muralidharan O., Winters M., Buenrostro J., Bell J., Brown S., Holodniy M., Zhang N., Ji H.P. Ultrasensitive detection of rare mutations using next-generation targeted resequencing. Nucleic Acids Res. 2012;40:e2. doi: 10.1093/nar/gkr861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstung M., Beisel C., Rechsteiner M., Wild P., Schraml P., Moch H., Beerenwinkel N. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat Commun. 2012;3:811. doi: 10.1038/ncomms1814. [DOI] [PubMed] [Google Scholar]
- 24.Harismendy O., Schwab R.B., Bao L., Olson J., Rozenzhak S., Kotsopoulos S.K., Pond S., Crain B., Chee M.S., Messer K., Link D.R., Frazer K.A. Detection of low prevalence somatic mutations in solid tumors with ultra-deep targeted sequencing. Genome Biol. 2011;12:R124. doi: 10.1186/gb-2011-12-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M., Stoneking M. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol. 2012;13:R34. doi: 10.1186/gb-2012-13-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vallania F.L., Druley T.E., Ramos E., Wang J., Borecki I., Province M., Mitra R.D. High-throughput discovery of rare insertions and deletions in large cohorts. Genome Res. 2010;20:1711–1718. doi: 10.1101/gr.109157.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spencer D.H., Sehn J.K., Abel H.J., Watson M.A., Pfeifer J.D., Duncavage E.J. Comparison of clinical targeted next-generation sequence data from formalin-fixed and fresh-frozen tissue specimens. J Mol Diagn. 2013;15:623–633. doi: 10.1016/j.jmoldx.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clark M.J., Chen R., Lam H.Y., Karczewski K.J., Chen R., Euskirchen G., Butte A.J., Snyder M. Performance comparison of exome DNA sequencing technologies. Nature Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K., Munson J., Hiatt J.B., Turner E.H., Levy R., O’Day D.R., Krumm N., Coe B.P., Martin B.K., Borenstein E., Nickerson D.A., Mefford H.C., Doherty D., Akey J.M., Bernier R., Eichler E.E., Shendure J. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., Teague J.W., Campbell P.J., Stratton M.R., Futreal P.A. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39(Database issue):D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. ArXiv e-prints. 2012:1207:3907. [Google Scholar]
- 36.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li R., Li Y., Fang X., Yang H., Wang J., Kristiansen K., Wang J. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19:1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spencer D.H., Abel H.J., Lockwood C.M., Payton J.E., Szankasi P., Kelley T.W., Kulkarni S., Pfeifer J.D., Duncavage E.J. Detection of FLT3 internal tandem duplication in targeted, short-read-length, next-generation sequencing data. J Mol Diagn. 2013;15:81–93. doi: 10.1016/j.jmoldx.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Ogino S., Kawasaki T., Brahmandam M., Yan L., Cantor M., Namgyal C., Mino-Kenudson M., Lauwers G.Y., Loda M., Fuchs C.S. Sensitive sequencing method for KRAS mutation detection by Pyrosequencing. J Mol Diagn. 2005;7:413–421. doi: 10.1016/S1525-1578(10)60571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy K.M., Levis M., Hafez M.J., Geiger T., Cooper L.C., Smith B.D., Small D., Berg K.D. Detection of FLT3 internal tandem duplication and D835 mutations by a multiplex polymerase chain reaction and capillary electrophoresis assay. J Mol Diagn. 2003;5:96–102. doi: 10.1016/S1525-1578(10)60458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pratz K.W., Sato T., Murphy K.M., Stine A., Rajkhowa T., Levis M. FLT3-mutant allelic burden and clinical status are predictive of response to FLT3 inhibitors in AML. Blood. 2010;115:1425–1432. doi: 10.1182/blood-2009-09-242859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogino S., Meyerhardt J.A., Cantor M., Brahmandam M., Clark J.W., Namgyal C., Kawasaki T., Kinsella K., Michelini A.L., Enzinger P.C., Kulke M.H., Ryan D.P., Loda M., Fuchs C.S. Molecular alterations in tumors and response to combination chemotherapy with gefitinib for advanced colorectal cancer. Clin Cancer Res. 2005;11:6650–6656. doi: 10.1158/1078-0432.CCR-05-0738. [DOI] [PubMed] [Google Scholar]
- 43.Meshinchi S., Alonzo T.A., Stirewalt D.L., Zwaan M., Zimmerman M., Reinhardt D., Kaspers G.J., Heerema N.A., Gerbing R., Lange B.J., Radich J.P. Clinical implications of FLT3 mutations in pediatric AML. Blood. 2006;108:3654–3661. doi: 10.1182/blood-2006-03-009233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao X., Lochhead P., Nishihara R., Morikawa T., Kuchiba A., Yamauchi M., Imamura Y., Qian Z.R., Baba Y., Shima K., Sun R., Nosho K., Meyerhardt J.A., Giovannucci E., Fuchs C.S., Chan A.T., Ogino S. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Approximate tumor cellularity as determined by visual estimate by a study pathologist (E.J.D.) for FFPE lung adenocarcinomas used in analysis. Original magnification ×20.
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Deletion of A in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Insertion of AA in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Insertion of CC in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Insertion of TG in case NA18484 (arrow).
Sanger validation traces from indel sensitivity comparison. Top panel: NA17989 normal reference. Bottom panel: Deletion of CTCTTC in case NA18507 (arrow).
Sensitivity of minor gold standard variants confirmed by array-based genotype calls from undiluted samples.
Sensitivity and PPV for coding regions only. Methods and calculations are identical to those in Figure 2, A and C, except applied only to the protein coding regions and adjacent splice sites (±10 bp) for all targeted genes.
Read alignments showing the single false positive coding region variant call, made by VarScan2. The VAF of this variant is 1%.