Abstract
The CD8+ T cell response to the immunodominant DbNP366 epitope has been analyzed sequentially to determine the prevalence and persistence of different T cell antigen receptor (TCR)Vβ8.3 clonotypes after primary and secondary influenza virus challenge. Based on the length and amino acid sequences of the complementarity-determining region 3 of TCRβ (CDR3β) loop and associated Jβ usage, the same dominant TCRβ signatures were found in the blood, the spleen, and the site of virus-induced pathology in the infected respiratory tract. Longitudinal analysis demonstrated that TCRβ prominent in the antigen-driven phase of response persisted into memory and were again expanded after secondary challenge. A proportion of these high-frequency TCRβ expressed “public” CDR3β sequences that were detected in every mouse sampled, whereas others were found more than once but were not invariably present. Analysis of N-region nucleotide diversity established that as many as 10 different nucleic acid sequences (maximum of four “nucleotypes” in any one mouse) could encode a single public TCRβ amino acid sequence. Conversely, whereas some of the unique, “private” TCRβ achieved a substantial clone size, they were always specified by a single nucleotype. Although there is a strong stochastic element in this response, the public TCRβ seem to represent a “best fit” for this immunodominant epitope, are selected preferentially from the naive TCR repertoire, and assume even greater prominence after secondary challenge.
Keywords: CD8+ T cells, T cell receptor repertoire, influenza A virus
Epitope-determined T cell-receptor (TCR) repertoires are selected from a pool of naïve precursors estimated to range in size from ≈107 and ≈108 distinct TCRαβ elements (1, 2). Antigen specific CD8+ T cell responses are sometimes biased toward usage of a particular TCRVα (3-5), but are more commonly determined by the TCRVβ profile (5-15). Most of the specificity of the TCRβ interaction is associated with the third complementarity-determining region (CDR3β) loop, which is generally positioned over the center of the antigenic peptide bound inside the groove of the MHC class I molecule (16, 17). Defining patterns of TCR CDR3β usage, either by determining amino acid sequences or by the spectratyping approach that measures CDR3β length and Jβ usage (18-20), provides a mechanism for following defined T cell populations through the course of an immune response into long-term memory (15, 21, 22).
Longitudinal single-cell analysis of CDR3β profiles in the CD8+Vβ7+ response to an influenza virus acid polymerase peptide (PA224-233) presented by the H-2Db MHC class I glycoprotein (DbPA224) provided evidence of substantial diversity in the absence of a single “public” TCRβ signature that could be detected in every mouse (15). A much earlier and more limited study with hybridoma cell lines identified what looked to be a more public response by CD8+Vβ8.3+ T cells specific for the DbNP336 inf luenza virus nucleoprotein (NP336-374) epitope (9). There was no consistent pattern of TCRVα usage by these CD8+Vβ8.3+ T cell hybridomas. This work explores this CD8+Vβ8.3+DbNP336+ repertoire further by using contemporary tetramer staining, single-cell sorting and PCR-based technology (15). The results provide a stark contrast in clonal diversity for these CD8+Vβ7+DbPA224+ and CD8+Vβ8.3+DbNP336+ T cell populations that expand equally and give rise to comparably sized memory T cell populations after primary influenza infection but differ greatly in numbers (CD8+Vβ8.3+DbNP336+ ≥ 10 × CD8+Vβ7+DbPA224+) subsequent to secondary virus challenge (23-25).
Materials and Methods
Mice and Viral Infection. Female C57BL/6J (B6, H2b) mice were bred at the University of Melbourne. Some were anaesthetized at 6 weeks of age by isofluorane inhalation and infected intranasally (i.n.) with 104 plaque forming units (pfu) of the HKx31 (H3N2) influenza A virus (26) in 30 μl of PBS. Memory mice for secondary challenge experiments were injected i.p. at least 6 weeks previously with 1.5 × 107 pfu of the PR8 (H1N1) influenza A virus (27, 28). Both virus stocks were grown in the allantoic cavity of 10-d embryonated hen's eggs and stored in aliquots at -80°C. Virus titers were determined as pfu on monolayers of Madin-Darby canine kidney cells.
Tissue Sampling and Cell Preparation. Mice were anaesthetized i.p with 3 mg of ketamine and 0.6 mg of Xylazil (Parnell Laboratories, Alexandria, Australia) and exsanguinated from the axillary artery; total body perfusion was performed with PBS/heparin. Lymphocytes recovered by bronchoalveolar lavage (BAL) of the infected lung were incubated on plastic Petri dishes for 1 h at 37°C to remove macrophages (27). Spleens were disrupted and enriched for CD8+ T cells by using goat anti-mouse IgG and IgM antibodies (Jackson ImmunoResearch). The longitudinal analysis of TCR repertoire used mice bled by means of the retroorbital sinus. The blood (50-100 μl) was collected into 50 μl of 10 units/ml heparin in PBS, followed by lysis of the RBCs with 10 ml of ammonium Tris chloride buffer at 37°C for 5 min (25).
Tetramer and TCRVβ Staining of CD8+ T Cells. The DbNP366-specific CD8+ T cells were identified by using tetrameric complexes (28) of the influenza virus H-2Db MHC class I glycoprotein and the NP366-374 (ASNENMETM) peptide (29) conjugated to streptavidin-phycoerythrin (Molecular Probes). Lymphocytes were stained with the DbNP366-phycoerythrin tetramer for 60 min at room temperature, followed by two washes in sort buffer (0.1% BSA in PBS), then anti-CD8α-allophycocyanin (for sorting) or anti-CD8α-PerCP Cy5.5 (for phenotyping) and anti-Vβ8.3-FITC (Pharmingen) for 30 min on ice, followed by two further washes. The stained lymphocytes were resuspended in sort buffer (2 × 107 cells per ml) and transferred to polypropylene tubes (BD Biosciences). Spleen or peripheral blood T cells from naïve, uninfected mice were stained with anti-CD8α-phycoerythrin and anti-Vβ8.3-FITC.
Isolation of Single CD8+ T Cells, RT-PCR, and Sequencing. Lymphocytes were isolated with a MoFlo sorter (Cytomation, Fort Collins, CO) fitted with a Cyclone single-cell deposition unit (15). Single immune (CD8+Vβ8.3+DbNP336+) or naïve CD8+Vβ8.3+ T cells were sorted directly into a 96-well PCR plate (Eppendorf) containing 5 μl of cDNA reaction mix. Negative controls were interspersed between the samples (1 in 10), and 50-80 cells were sorted per plate. The cDNA mix contained 0.25 μl of Sensiscript reverse transcriptase, 1× cDNA buffer, 0.5 mM dNTP (all from Qiagen), 0.125 μg of oligo dT (15) (Promega), 100 μg/ml gelatin (Roche), 100 μg/ml tRNA (Roche), 20 units of RNAsin (Invitrogen), and 0.1% Triton X-100 (Sigma). After sorting, plates were incubated at 37°C for 90 min for cDNA synthesis, followed by 5 min at 95°C to stop reverse transcriptase activity, and stored at -80°C. The Vβ8.3+ transcripts were amplified by nested PCR by using 2 μl of cDNA for a 25-μl amplification reaction. The first-round PCR was performed with 1.5 units of Taq polymerase (Invitrogen)/1.5 mM MgCl2/0.2 mM dNTP (Invitrogen)/10 pmol each of the external sense primer Vβ8.3-5′ (5′-AGCCCTAGAAACAAGGTGAC-3′) and the external antisense primer Cβa (5′-CCAGAAGGTAGCAGAGACCC-3′). A 2-μl aliquot of the first-round PCR was used as a template for the nested PCR with the internal sense Vβ8.3 primer (5′-ACCAGAACAACGCAAGAAGAC-3′) and the antisense primer Cβb (5′-CTTGGGTGGAGTCACATTTCTC-3′). The PCR conditions were 95°C for 5 min followed by 33 cycles of 95°C for 30 sec, 57°C for 30 sec, and 72°C for 1 min followed by 1 cycle of 95°C for 1 min, 57°C for 1 min, and 72°C for 7 min. The PCR products were resolved on a 2% agarose gel, purified with the MiniElute PCR purification kit (Qiagen), and sequenced with 20 pmol of the Vβ8.3 sense primer by using Big Dye 3.1 (Applied Biosystems). Sequencing products were purified by using Dye Ex 2.1 columns (Qiagen) and sequenced on an Applied Biosystems Prism 3700 sequence analyzer. The Vβ7+ transcripts were amplified and sequenced as described in ref. 15.
Results
The basic aim of these experiments is to define the nature of an antigen-specific CD8+ T cell repertoire, both within and between individuals responding to a primary or secondary virus challenge and in persistent immune memory (28, 30). The B6 mice were either immunologically naïve (primary response) or had been infected i.p. with the PR8 (H1N1) virus at least 8 weeks before i.n. challenge with the HKx31 (H3N2) influenza A virus (secondary response). The PR8 and HKx31 viruses share the same internal components, including the viral nucleoprotein and viral acid polymerase, but the surface influenza virus hemag-glutinin and viral neuraminidase glycoproteins are different, and there is no confounding effect of crossneutralization in these prime/boost studies (28, 30).
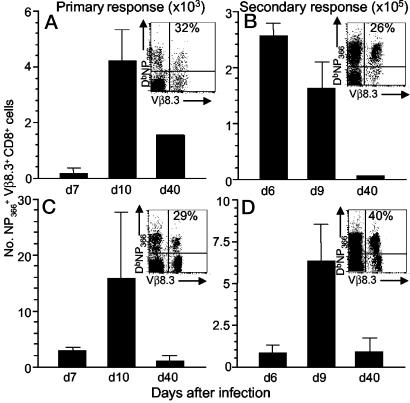
Quantification of the Vβ8.3+CD8+DbNP366-Specific T Cell Response. Typical flow cytometry profiles for CD8α+ T cells stained simultaneously with a mAb to Vβ8.3 and the DbNP366 tetramer are shown for BAL (Fig. 1 A and B) and spleen (Fig. 1 C and D) populations recovered following i.n. challenge with the HKx31 virus. The B6 mice were either immunologically naïve (primary response, Fig. 1 A and C), or had been infected i.p. at least 8 weeks previously with the PR8 virus (secondary response, Fig. 1 B and D). The range of ≈25-50% prevalence for the Vβ8.3+ component within these CD8+DbNP366+ sets (Fig. 1) is in accord with previous findings (9).
Fig. 1.
Kinetics of primary and secondary CD8+ Vβ8.3+DbNP366+ responses. Naïve or PR8-primed B6 mice were infected i.n. with the HKx31 virus. Lymphocytes were isolated from B6 mice at 7, 10, or 40 d of the primary response, or at 6, 9, or 40 d after secondary challenge. Enriched CD8+ T cells were stained with the DbNP366-phycoerythrin tetramer, anti-Vβ8.3-FITC, and anti-CD8-PerCP Cy5.5 mAbs and analyzed by flow cytometry. The fluorescence-activated cell sorter panels shown on the figures are for day 10 primary (A and C) and day 9 secondary (B and D) responses. The cell counts shown here were calculated from the percentage of cells staining and the numbers recovered in the BAL and spleen populations.
Earlier studies showed that the DbNP366-specific response dominates influenza virus-specific CD8+ T cell numbers after secondary (but not primary) exposure of B6 mice to the HKx31 virus (23, 25). The expansion of the Vβ8.3+ subset within the CD8+ DbNP366+ population parallels the expected profile after secondary challenge (Fig. 1 B and D). The recall CD8+ Vβ8.3+DbNP366+ response in the BAL (Fig. 1B) peaked earlier at a level that was more than 100-fold higher than that found after primary infection (Fig. 1 A). The differential between the primary and secondary CD8+ Vβ8.3+DbNP366+ response for maximum counts in the spleen was ≈30×. The CD8+ Vβ8.3+DbNP366+ T cell numbers in the spleen then progressively declined to day 40 to give greatly diminished memory T cell population (Fig. 1 C and D).
Thus, the overall pattern is of a very dynamic response with dramatic changes in virus-specific CD8+Vβ8.3+DbNP366+ T cell numbers from the effector phase to the memory phase (Fig. 1). Furthermore, both the magnitude and kinetics of T cell invasion into the virus-infected lung are greatly enhanced after secondary challenge (Fig. 1B). These dramatic changes reflect successive cycles of massive clonal expansion and editing (25). How stable is the profile of TCR repertoire usage throughout the course of this highly dynamic process?
Primary CD8+Vβ8.3+DbNP366+ TCRβ Profiles Between Individual Mice. Immune CD8+Vβ8.3+DbNP366+ T cells recovered from the BAL and spleen of naïve B6 mice infected i.n. with the HKx31 virus were sorted to give one cell per well and the CDR3β mRNA was expanded by RT-PCR with ≈80% efficiency to give the product used for sequencing. The Vβ8.3 CDR3β lengths were designated (31) from the analysis of amino acid sequences, whereas Jβ-element usage was determined after comparison with known genomic sequences (32, 33). The TCRβ identity was thus based on Jβ usage and the length and amino acid sequence of the CDR3β loop (15).
Two dominant TCRβ were present in the BAL and spleen of all three mice (Table 1, M1 to M3, first two lines) that were sampled on day 8 of the primary CD8+DbNP366-specific response. Furthermore, although they were less prominent, these same CDR3β signatures were found at significant prevalence in memory spleen populations recovered from different individuals infected more than 200 days previously (Table 1, M4 and M5). Such high-frequency TCRβ thus represent public amino acid sequences that are selected repeatedly in the CD8+Vβ8.3+DbNP366+ T cell response. Furthermore, the same pattern of Jβ2.2 usage and 9-aa CDR3β length was found for 13 other TCRβ recovered from these five mice, with 5 of 13 being detected in more than one individual (Table 1). In all, a total of 24 TCRβ were detected (n = 389 sequences). The amino acid at position 1 was S, K, G, or R for both the public and “private” TCRβ, and the CDR3β length ranged from 8 to 10 aa (Table 1).
Table 1. Acute phase and memory CDR3β repertoires for different individuals.
| TCRβ prevalence, %
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Acute (day 8)
|
Memory (day 200+)
|
|||||||||
| M1*
|
M2*
|
M3*
|
M4*
|
M5*
|
||||||
| CDR3β region | Jβ | Length, aa | Spl | BAL | Spl | BAL | Spl | BAL | Spl | Spl |
| SGGGNTGQL | 2.2 | 9 | 61 | 27 | 31 | 64 | 89 | 50 | 23.0 | 21.2 |
| SGGANTGQL | 2.2 | 9 | 7 | 36 | 54 | 21 | 6 | 44 | 4.1 | 1.5 |
| RGGANTGQL | 2.2 | 9 | — | 3 | — | 5 | — | 33 | 1.4 | 1.5 |
| RGGSNTGQL | 2.2 | 9 | — | — | 2 | 2 | — | — | 51.4 | 3.0 |
| SGGSNTGQL | 2.2 | 9 | — | — | 3 | — | — | — | — | 57.6 |
| SGGARTGQL | 2.2 | 9 | — | — | — | — | — | — | 20.3 | — |
| KGGSNTGQL | 2.2 | 9 | 13 | 27 | — | — | — | — | — | — |
| RGGGRTGQL | 2.2 | 9 | 3 | — | 6 | — | — | — | — | — |
| RGGGNTGQL | 2.2 | 9 | — | — | — | — | — | — | — | 1.5 |
| KGGQNTGQL | 2.2 | 9 | 2 | — | — | — | — | — | — | — |
| SGGGKHRQL | 2.2 | 9 | 2 | — | — | — | — | — | — | — |
| GGGANTGQL | 2.2 | 9 | — | 3 | — | — | — | — | — | — |
| GGGENTGQL | 2.2 | 9 | — | — | 2 | — | — | — | — | — |
| RGGNTGQL | 2.2 | 8 | — | — | 2 | — | — | — | — | — |
| SGGNTGQL | 2.2 | 8 | 2 | — | — | — | — | — | — | — |
| SARTANTEV | 1.1 | 9 | 10 | — | — | — | — | — | — | — |
| SARTGNTEV | 1.1 | 9 | 2 | — | — | — | — | — | — | — |
| RGAATTEV | 1.1 | 8 | — | — | — | 2 | — | — | — | — |
| SVAATTEV | 1.1 | 8 | — | — | — | 2 | — | — | — | — |
| SDWQGRGNTL | 1.3 | 10 | — | — | — | — | — | — | — | 13.6 |
| SARDGNYAEQ | 2.1 | 10 | — | — | — | — | — | 3 | — | — |
| SDLAGNAEQ | 2.1 | 9 | — | 3 | — | — | — | — | — | — |
| RDRESAETL | 2.3 | 9 | — | — | — | 2 | — | — | — | — |
| SDWGRDEQ | 2.6 | 8 | — | — | — | — | 6 | — | — | — |
| Total NP366 specific sequences | 61 | 33 | 61 | 42 | 18 | 34 | 74 | 66 | ||
Lymphocytes were obtained from either from BAL or spleens (Spl) of individual B6 mice at 8 d (or from spleens at 200 and 240 d) after i.n. infection with the HK×31 virus, then processed for single cell CDR3β analysis as described in Material and Methods. NP, viral nucleoprotein.
Individual mice tested.
Sequential Analysis Within Individuals. A further five mice were primed i.p. with the PR8 (H1N1) virus, then exposed i.n. to the HKx31 (H3N2) virus at least 90 d later. The peripheral blood lymphocyte compartment was sampled acutely on day 8 after the initial PR8 infection and again (day 30) in the early stage of memory (primary response). All mice were then exsanguinated on day 8 (days 98-148) after the HKx31 challenge and peripheral blood lymphocyte, spleen, and BAL were analyzed (secondary response). The results for one individual are shown in Table 2. A total of 31 different TCRβ were found in the 1,053 sequences that were analyzed, with 22 of these TCRβ being unique to individual mice (only in one of five mice). Two of the prominent TCRβ (SGGGNTGQL and SGGANTGQL; Table 2) were present in every individual (Table 3) sampled in this series and the previous one (Table 1).
Table 2. Sequential CDR3β analysis within an individual mouse.
| TCRβ prevalence, %
|
|||||||
|---|---|---|---|---|---|---|---|
| Blood
|
Spleen
|
BAL
|
|||||
| CDR3β region* | Jβ | Length, aa | day 8 (1°) | day 30 (1°) | day 138 (2°) | day 138 (2°) | day 138 (2°) |
| SGGSNTGQL | 2.2 | 9 | 54 | 22 | 56 | 29 | 65 |
| SGGGNTGQL | 2.2 | 9 | 18 | 22 | 13 | 6 | 6 |
| SGGANTGQL | 2.2 | 9 | 14 | 22 | 13 | 41 | 23 |
| KGGGNTGQL | 2.2 | 9 | 9 | — | 6 | 6 | — |
| KGGSNTGQL | 2.2 | 9 | — | — | — | 3 | — |
| RGGSNTGQL | 2.2 | 9 | — | — | 6 | 12 | — |
| RGGGNTGQL | 2.2 | 9 | 5 | — | — | 3 | — |
| RGGANTGQL | 2.2 | 9 | — | 7 | — | — | — |
| KAGGNTGQL | 2.2 | 9 | — | — | 4 | — | — |
| GGGSNTGQL | 2.2 | 9 | — | — | 2 | — | — |
| SGGARTGQL | 2.2 | 9 | — | — | — | — | 6 |
| SDRGRDTGQL | 2.5 | 10 | — | 7 | — | — | — |
| SDGTDY | 1.2 | 6 | — | 7 | — | — | — |
| SGGADTGQL | 1.3 | 9 | — | 7 | — | — | — |
| SDAGGRDEQ | 2.6 | 9 | — | 7 | — | — | — |
| Total NP366 specific sequences | 22 | 14 | 48 | 34 | 35 | ||
The primary TCRβV8.3 repertoire (1°) was analyzed from single CD8+Vβ8.3+DbNP366+ cells recovered from peripheral blood on day 8 and day 30 after i.p. challenge with the PR8 virus. The sequences for the secondary TCRβV8.3 repertoire (2°) were obtained from peripheral blood, BAL, and spleens on day 8 after subsequent i.n. infection with HKx31 virus (day 130 after the primary PR8 challenge).
The data are for mouse M9.
Table 3. Public and repeated CDR3β sequences that were found in at least three of eight mice.
| TCRβ prevalence, %
|
||||
|---|---|---|---|---|
| CDR3β region | Jβ | Length, aa | Primary | Secondary |
| SGGANTGQL | 2.2 | 9 | 19.1 (8) | 38.3 (5) |
| SGGGNTGQL | 2.2 | 9 | 36.2 (8) | 6.8 (5) |
| SGGSNTGQL | 2.2 | 9 | 15.9 (5) | 19.3 (5) |
| RGGSNTGQL | 2.2 | 9 | 8.5 (5) | 1.4 (2) |
| KGGGNTGQL | 2.2 | 9 | 1.6 (2) | 15.0 (3) |
| RGGANTGQL | 2.2 | 9 | 1.6 (7) | — |
| RGGGNTGQL | 2.2 | 9 | 0.6 (3) | 11.5 (3) |
| No. of NP366 specific sequences (mice) | 503 (8) | 828 (5) | ||
A total of eight mice (n = 503) were sampled after HKx31 or PR8 primary infection, and five mice (n = 828) were sampled after secondary challenge. Values in parentheses correspond to the number of mice in which a particular sequence was found. Samples of the primary data are shown in Tables 1, 2, and 4.
The public TCRβ found at high frequencies (Table 3) in all of the mice tested during the primary or recall response were characterized by the usage of Jβ2.2, a 9-aa CDR3β loop, and the N-Dβ-N region SGGG or SGGA (Table 3). The remaining five sequences that were recovered from at least 3 of 10 mice (Table 3) also displayed the Jβ2.2, 9-aa profile. One “consensus” TRCRβ (SGGANTGQL) that was invariably selected in the CD8+Vβ8.3+DbNP366+ response (Table 3) was also found once in CD8+Vβ8.3+ T cells recovered from naïve, uninfected mice (total of 361 sequences analyzed; data not shown). Clearly, many more naïve TCRβ signatures would need to be examined before any more precise definition of the preimmune TCRβ repertoire can be made.
The 9 of 31 TCRβ that were found in more than one of the five sequentially sampled mice dominated the CD8+Vβ8.3+DbNP366+ response. These “repeat” CDR3β profiles were expressed on 80.4% ± 20.1% of the CD8+Vβ8.3+DbNP366+ T cells analyzed on day 8, 76.6% ± 22.8% on day 30, and 93.4% ± 9.0% on day 148. The perception that the recall response narrows to a more consensus profile is reinforced by the fact that, given the limited amount of blood that can be obtained from mice in survival experiments, many fewer T cells were analyzed on day 8 and day 30 (total 137) after the initial PR8 infection than from the peripheral blood lymphocyte, BAL, and spleen (total 916) following the HKx31 challenge.
Despite this >6-fold difference in sample size, some TCRβ were detected only in the blood following the initial exposure to PR8 and did not reemerge in the recall response. Most (12 of 18) of the TCRβ sequences, were private and were found in one of five of these sequentially sampled mice and were also absent from the five that were analyzed at a single time point (Table 1). All 12 lacked the characteristic 9-aa length and Jβ2S2 profile (Tables 1 and 3). A further 4 of 11 TCRβ detected only after secondary challenge also showed this private profile, but these could easily have been missed in the more limited analysis of the primary response.
Nucleotide Diversity Within Particular CDR3β Amino Acid Signatures. The variation permissible in the third nucleotide (”wobble”) encoding a particular amino acid in a TCRβ sequence allows a more precise definition of clonality. Evidence of considerable N-region nucleotide diversity was found within each of the public TCRβ that were repeated in this series of five mice (Tables 4 and 5). Although it was not necessarily the case that such public TCRβ were numerically dominant in any individual response (Tables 1, 2, and 4), the extent of this nucleotype diversity was significantly greater (P > 0.001) than for those that were present in <5 but >2 mice (Table 5). Furthermore, the private TCRβ that were found only once were always progeny of a single nucleotype (Table 5).
Table 4. Nucleotide and amino acid diversity profiles for a sequentially sampled mouse.
| Nucleotide prevalence, %
|
|||||
|---|---|---|---|---|---|
| Blood
|
Spleen
|
BAL
|
|||
| CDR3β region* | Day 8 (1°) | Day 30 (1°) | Day 148 (2°) | Day 148 (2°) | Day 148 (2°) |
| Public | |||||
| SGGANTGQL | 12.1 | 20.0 | 27.1 | 25.6 | 20.5 |
| AGTGGGGGGGCAAA | (9.1) | (10.0) | (20.3) | (17.2) | (8.7) |
| AGTGGGGGGGCTAA | (3.0) | (5.0) | (4.3) | (6.9) | (5.9) |
| AGTGGTGGGGCAAA | — | (5.0) | (2.5) | (1.5) | (5.9) |
| SGGGNTGQL | 18.2 | 10.0 | 4.9 | 3.4 | 8.2 |
| AGTGGGGGGGGAAA | (9.1) | (10.0) | — | (1.8) | (4.1) |
| AGTGGGGGAGGAAA | (3.1) | — | (1.6) | (0.8) | (1.6) |
| AGTGGGGGGGGGAA | — | — | — | — | (0.8) |
| AGTGGGGGGGGCAA | (6.0) | — | (3.3) | (0.8) | (1.6) |
| Repeated | |||||
| SGGSNTGQL | 21.2 | 70.0 | 9.8 | 7.7 | 9.0 |
| AGTGGGGGCTCAAA | (19.9) | (10.0) | (6.6) | (3.4) | (5.8) |
| AGTGGGGGGTCAAA | (1.3) | (60.0) | (3.2) | (4.3) | (3.2) |
| RGGGNTGQL | 3.0 | — | 19.7 | 20.5 | 14.8 |
| AGAGGAGGGGGGAA | (3.0) | — | (2.5) | (9.4) | (8.2) |
| AGGGGTGGGGGGAA | — | — | (17.2) | (11.1) | (6.6) |
| RGGSNTGQL | 3.0 | — | 0.8 | — | 3.3 |
| KGGGNTGQL | 18.2 | — | 32.8 | 32.5 | 28.7 |
| SDAANTEV | — | — | 0.8 | 1.7 | 3.3 |
| Private | |||||
| SGGGRSGQL | — | — | 0.8 | 0.9 | — |
| SDDRGRDQDTQ | 18.2 | — | — | 0.9 | — |
| SGGGTTGQL | — | — | — | — | 0.8 |
| KAGGNTGQL | — | — | 3.3 | 6.8 | 10.7 |
| KGGANTGQL | — | — | — | — | 0.8 |
| SEGAETL | 3.0 | — | — | — | — |
| SEQRLGRYEQ | 3.0 | — | — | — | — |
Nucleotide diversity for public CDR3β (Table 2) that were found in all of the mice, repeated CDR3β signatures recovered from at least two different individuals, or unique private CDRβ signatures. Nucleotide sequences are not provided when the TCRβ represented the progeny of a single clonotype. 1°, primary response; 2°, secondary response.
The data are for mouse M10.
Table 5. Nucleotide diversity in CDR3β amino acid sequences for mice analyzed longitudinally.
| Mouse* | Public (five of five mice) | Repeated (less than five and at least two mice) | Private (one moue) |
|---|---|---|---|
| M10 | 3-4 | 1-2 | 1 |
| M9 | 2-3 | 1-2 | 1 |
| M8 | 2-3 | 1-2 | 1 |
| M7 | 1-3 | 1-2 | 1 |
| M6 | 1-3 | 1-2 | 1 |
| Mean† ± SD | 2.5 ± 0.97 | 1.4 ± 0.5 | 1 ± 0 |
| Total in five mice | 7-10 | 1-4 | 1 |
The extent of nucleotide diversity within public, repeated, and private TCRβ signatures is shown for five mice that were sampled sequentially through the primary and recall response (Tables 2 and 4).
More detailed data for mice M9 and M10 are shown in Tables 2 and 4, respectively. At least one private TCRβ in these mice accounted for >10% of the total analysed.
The mean values are all significantly different (P > 0.001).
Looking at all five mice (Table 5), we found a total of seven different nucleotypes encoding SGGANTGQL. Similarly, the overall SGGGNTGQL response was comprised of 10 clonotypes. Furthermore, although the most frequently repeated nucleotype predominated throughout for the sequentially sampled mouse shown in Table 4, this was not necessarily the case for the other individuals analyzed in this way (data not shown). However, all of the 7 SGGANTGQL clones and 5 of 10 SGGGNTGQL clones found after primary PR8 infection were present again in the recall response, establishing that the majority produced progeny T cells that enter the memory pool.
Discussion
The present CDR3β analysis of CD8+Vβ8.3+DbNP366+ T cells, together with our prior dissection of the CD8+Vβ7.1+DbPA224+ repertoire (15), provide systematic, longitudinal studies of primary and secondary responses to different epitopes within the same virus. The characteristics in common are that the likelihood of persistence into memory reflects the clonal burst size (34) achieved during the antigen-driven acute response and that the clonotypic profile of T cell memory shows substantial stability in the long term (22). Also, although TCRβ that are prominent after primary infection continue to be detected after secondary challenge, stochastic events (35) clearly contribute to the recall response. More evidence for the emergence of “novel” TCRβ after secondary antigen stimulation was found for the CD8+Vβ7.1+DbPA224+ than the CD8+Vβ8.3+DbNP366+ response, although all such analyses probably underestimate the diversity of memory (36) represented by unique, private clonotypes that are present at very low frequencies (15).
Interestingly, although no public TCRβ (37) were found for the CD8+ Vβ7.1+DbPA224+ response (15), such sequences are a prominent feature of the CD8+ Vβ8.3+DbNP366+ repertoire. These repeat CD8+Vβ8.3+DbNP366+ TCRβ are characterized by the almost exclusive usage of Jβ2.2 and a 9-aa CDR3β, whereas the CDR3β loop of DbPA244-specific T cells is generally shorter (6 aa) and has a more diverse Jβ (1.1, 1.4, and 1.6) profile. The likely reasons for these differences rest in the molecular characteristics of the CDR3β-DbNP366 and DbPA224 interactions (5, 14). The structure of DbNP366 is known (38), and the solution of DbPA224 may be imminent (J. Rossjohn, personal communication), but there are as yet no cocrystals with representative TCR for either of these epitopes. We have not attempted to analyze TCRα expression in these experiments, but the spectrum of TCRα usage in the CD8+Vβ8.3+DbNP366+ response shows considerable variability (ref. 9 and K.K., unpublished data), and it seems likely that diversity is largely a function of the CDR3β profile.
Although the secondary CD8+ T cell response to DbNP366 response is much larger than that to DbPA224 (23, 25, 39), an accumulating body of evidence indicates that the TCR-DbPA224 interaction is of higher avidity (40, 41). The correlation of greater diversity and higher avidity suggests the possibility that the development of an appropriate binding event between a selected TCR CDR3β and the immunogenic epitope is much more readily achieved for DbPA224 than for DbNP366. The level of diversity may reflect the selection of dominant characteristics important for specificity. The CD8+ Vβ8.3+DbNP366+ T cells clearly “prefer” to use a limited set of public TCRβ and draw these from a spectrum of related nucleotide sequences. The use of the same amino acid sequences encoded by different gene rearrangements is evidence for strong antigen selection for these public TCRβ. This implies that optimal recognition of the DbNP366 complex requires a combination of structural factors which are only found within the specific TCRVβ8.3/CDR3β/Jβ2.2 combination (5, 36). Although large clone sizes can be achieved by unique private CDR3β that bind DbNP366, these are only ever specified by a single nucleotide sequence and tend to be less prominent after secondary challenge.
There is also the possibility that higher avidity T cells may be preferentially deleted during the resolution phase of the host response. If this possibility were to bias profiles of TCR retention by, for example, selectively diminishing the prevalence of public CD8+Vβ7.1+DbPA224+ TCR (42), we might expect to see obvious changes in repertoire between the initial stage of antigen-driven clonal expansion and the development of established T cell memory. However, no such effects have been found for primary CD8+Vβ8.3+DbNP366+ or CD8+Vβ7.1+DbPA224+ responses. In addition, although there is evidence of preferential tumor necrosis factor-α-mediated loss of the CD8+Vβ7.1+DbPA224+ T cells during the acute response, this is limited to the small minority of highly activated, tumor necrosis factor-receptor positive T cells recovered from the virus-infected lung (41). The TCRβ profiles are essentially equivalent for both the CD8+Vβ8.3+DbNP366+ and CD8+Vβ7.1+DbPA224+ BAL and spleen populations, indicating that this differential editing process (if active) does not modify the repertoire.
Analysis of antigen-presenting cells recovered directly ex vivo from infected mice indicates that, although DbNP366 can be detected on epithelial cells, macrophages, and dendritic cells, DbPA224 is found only on dendritic cells (43). A formal possibility is that the much wider distribution of DbNP366 on suboptimal, “nonprofessional” antigen-presenting cells skews the response to use of the “best fit” (44, 45), public TCRβ. Studies with viruses modified by reverse genetic technology to change the spectrum of epitope presentation may provide further information. Hopefully, experiments such as these will help to develop a better understanding of the nature of immunodominance (24, 46) in virus-specific CD8+ T cell responses and the role that TCR repertoire usage and diversity plays in such processes.
Acknowledgments
We thank Dr. Nicole La Gruta for critical review and discussion, Dina Stockwell for technical assistance, and Phyllis Halliday for assistance with the manuscript. This work was supported by a Burnet Award to P.C.D.; a Peter Doherty Postdoctoral Fellowship to K.K. (both from the Australian National Health and Medical Research Council); Science, Technology, and Innovation funds from the Government of Victoria, Australia; United States Public Health Service Grant AI29579; and funds from American Lebanese Syrian Associated Charities at St. Jude Children's Research Hospital.
Abbreviations: TCR, T cell receptor; CDR3β, complementarity-determining region 3 of the TCRβ; DbNP366, epitope comprised of the nucleoprotein NP366-374 peptide bound to MHC class I glycoprotein H-2Db; DbPA224, complex of H-2Db and virus acid polymerase peptide PA224-233; i.n., intranasally; BAL, bronchoalveolar lavage.
References
- 1.Arstila, T. P., Casrouge, A., Baron, V., Even, J., Kanellopoulos, J. & Kourilsky, P. (1999) Science 286, 958-961. [DOI] [PubMed] [Google Scholar]
- 2.Casrouge, A., Beaudoing, E., Dalle, S., Pannetier, C., Kanellopoulos, J. & Kourilsky, P. (2000) J. Immunol. 164, 5782-5787. [DOI] [PubMed] [Google Scholar]
- 3.Davis, M. M., McHeyzer-Williams, M. & Chien, Y. H. (1995) Ann. N.Y. Acad. Sci. 756, 1-11. [DOI] [PubMed] [Google Scholar]
- 4.Mikszta, J. A., McHeyzer-Williams, L. J. & McHeyzer-Williams, M. G. (1999) J. Immunol. 163, 5978-5988. [PubMed] [Google Scholar]
- 5.Kjer-Nielsen, L., Clements, C. S., Purcell, A. W., Brooks, A. G., Whisstock, J. C., Burrows, S. R., McCluskey, J. & Rossjohn, J. (2003) Immunity 18, 53-64. [DOI] [PubMed] [Google Scholar]
- 6.Acha-Orbea, H., Mitchell, D. J., Timmermann, L., Wraith, D. C., Tausch, G. S., Waldor, M. K., Zamvil, S. S., McDevitt, H. O. & Steinman, L. (1988) Cell 54, 263-273. [DOI] [PubMed] [Google Scholar]
- 7.Urban, J. L., Kumar, V., Kono, D. H., Gomez, C., Horvath, S. J., Clayton, J., Ando, D. G., Sercarz, E. E. & Hood, L. (1988) Cell 54, 577-592. [DOI] [PubMed] [Google Scholar]
- 8.Yanagi, Y., Maekawa, R., Cook, T., Kanagawa, O. & Oldstone, M. B. (1990) J. Virol. 64, 5919-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deckhut, A. M., Allan, W., McMickle, A., Eichelberger, M., Blackman, M. A., Doherty, P. C. & Woodland, D. L. (1993) J. Immunol. 151, 2658-2666. [PubMed] [Google Scholar]
- 10.Imarai, M., Goyarts, E. C., van Bleek, G. M. & Nathenson, S. G. (1995) Cell Immunol. 160, 33-42. [DOI] [PubMed] [Google Scholar]
- 11.Prevost-Blondel, A., Chassin, D., Zeliszewski, D., Dorval, I., Sterkers, G., Pannetier, C. & Guillet, J. G. (1995) J. Virol. 69, 8046-8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maryanski, J. L., Jongeneel, C. V., Bucher, P., Casanova, J. L. & Walker, P. R. (1996) Immunity 4, 47-55. [DOI] [PubMed] [Google Scholar]
- 13.Wallace, M. E., Bryden, M., Cose, S. C., Coles, R. M., Schumacher, T. N., Brooks, A. & Carbone, F. R. (2000) Immunity 12, 547-556. [DOI] [PubMed] [Google Scholar]
- 14.Stewart-Jones, G. B., McMichael, A. J., Bell, J. I., Stuart, D. I. & Jones, E. Y. (2003) Nat. Immunol. 4, 657-663. [DOI] [PubMed] [Google Scholar]
- 15.Turner, S. J., Diaz, G., Cross, R. & Doherty, P. C. (2003) Immunity 18, 549-559. [DOI] [PubMed] [Google Scholar]
- 16.Garboczi, D. N., Ghosh, P., Utz, U., Fan, Q. R., Biddison, W. E. & Wiley, D. C. (1996) Nature 384, 134-141. [DOI] [PubMed] [Google Scholar]
- 17.Garboczi, D. N. & Biddison, W. E. (1999) Immunity 10, 1-7. [DOI] [PubMed] [Google Scholar]
- 18.Pannetier, C., Cochet, M., Darche, S., Casrouge, A., Zoller, M. & Kourilsky, P. (1993) Proc. Natl. Acad. Sci. USA 90, 4319-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sourdive, D. J., Murali-Krishna, K., Altman, J. D., Zajac, A. J., Whitmire, J. K., Pannetier, C., Kourilsky, P., Evavold, B., Sette, A. & Ahmed, R. (1998) J. Exp. Med. 188, 71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, M. Y. & Welsh, R. M. (1998) J. Exp. Med. 188, 1993-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McHeyzer-Williams, M. G. & Davis, M. M. (1995) Science 268, 106-111. [DOI] [PubMed] [Google Scholar]
- 22.Walker, P. R., Wilson, A., Bucher, P. & Maryanski, J. L. (1996) Int. Immunol. 8, 1131-1138. [DOI] [PubMed] [Google Scholar]
- 23.Belz, G. T., Xie, W., Altman, J. D. & Doherty, P. C. (2000) J. Virol. 74, 3486-3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belz, G. T., Stevenson, P. G. & Doherty, P. C. (2000) J. Immunol. 165, 2404-2409. [DOI] [PubMed] [Google Scholar]
- 25.Marshall, D. R., Turner, S. J., Belz, G. T., Wingo, S., Andreansky, S., Sangster, M. Y., Riberdy, J. M., Liu, T., Tan, M. & Doherty, P. C. (2001) Proc. Natl. Acad. Sci. USA 98, 6313-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilbourne, E. D. (1969) Bull. W. H. O. 41, 643-645. [PMC free article] [PubMed] [Google Scholar]
- 27.Allan, W., Tabi, Z., Cleary, A. & Doherty, P. C. (1990) J. Immunol. 144, 3980-3986. [PubMed] [Google Scholar]
- 28.Flynn, K. J., Belz, G. T., Altman, J. D., Ahmed, R., Woodland, D. L. & Doherty, P. C. (1998) Immunity 8, 683-691. [DOI] [PubMed] [Google Scholar]
- 29.Townsend, A. R., Rothbard, J., Gotch, F. M., Bahadur, G., Wraith, D. & McMichael, A. J. (1986) Cell 44, 959-968. [DOI] [PubMed] [Google Scholar]
- 30.Flynn, K. J., Riberdy, J. M., Christensen, J. P., Altman, J. D. & Doherty, P. C. (1999) Proc. Natl. Acad. Sci. USA 96, 8597-8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chothia, C., Boswell, D. R. & Lesk, A. M. (1988) EMBO J. 7, 3745-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gascoigne, N. R., Chien, Y., Becker, D. M., Kavaler, J. & Davis, M. M. (1984) Nature 310, 387-391. [DOI] [PubMed] [Google Scholar]
- 33.Malissen, M., Minard, K., Mjolsness, S., Kronenberg, M., Goverman, J., Hunkapiller, T., Prystowsky, M. B., Yoshikai, Y., Fitch, F., Mak, T. W., et al. (1984) Cell 37, 1101-1110. [DOI] [PubMed] [Google Scholar]
- 34.Hou, S., Hyland, L., Ryan, K. W., Portner, A. & Doherty, P. C. (1994) Nature 369, 652-654. [DOI] [PubMed] [Google Scholar]
- 35.Attuil, V., Bucher, P., Rossi, M., Mutin, M. & Maryanski, J. L. (2000) Proc. Natl. Acad. Sci. USA 97, 8473-8478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pewe, L. L., Netland, J. M., Heard, S. B. & Perlman, S. (2004) J. Immunol., 172, 3151-3156. [DOI] [PubMed] [Google Scholar]
- 37.Levraud, J. P., Pannetier, C., Langlade-Demoyen, P., Brichard, V. & Kourilsky, P. (1996) J. Exp. Med. 183, 439-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young, A. C., Zhang, W., Sacchettini, J. C. & Nathenson, S. G. (1994) Cell 76, 39-50. [DOI] [PubMed] [Google Scholar]
- 39.Webby, R. J., Andreansky, S., Stambas, J., Rehg, J. E., Webster, R. G., Doherty, P. C. & Turner, S. J. (2003) Proc. Natl. Acad. Sci. USA 100, 7235-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.La Gruta, N. L., Turner, S. J. & Doherty, P. C. (2004) J. Immunol., in press.
- 41.Turner, S. J., La Gruta, N. L., Stambas, J., Diaz, G. & Doherty, P. C. (2004) Proc. Natl. Acad. Sci. USA 101, 3545-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cibotti, R., Cabaniols, J. P., Pannetier, C., Delarbre, C., Vergnon, I., Kanellopoulos, J. M. & Kourilsky, P. (1994) J. Exp. Med. 180, 861-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crowe, S. R., Turner, S. J., Miller, S. C., Roberts, A. D., Rappolo, R. A., Doherty, P. C., Ely, K. H. & Woodland, D. L. (2003) J. Exp. Med. 198, 399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Argaet, V. P., Schmidt, C. W., Burrows, S. R., Silins, S. L., Kurilla, M. G., Doolan, D. L., Suhrbier, A., Moss, D. J., Kieff, E., Suclley, T. B., et al. (1994) J. Exp. Med. 180, 2335-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McHeyzer-Williams, L. J., Panus, J. F., Mikszta, J. A. & McHeyzer-Williams, M. G. (1999) J. Exp. Med. 189, 1823-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yewdell, J. W. & Bennink, J. R. (1999) Annu. Rev. Immunol. 17, 51-88. [DOI] [PubMed] [Google Scholar]