Background: Ras proteins are critical regulators of cellular growth and are differentially modified by ubiquitination.
Results: Chemical ubiquitination and immunoprecipitation assays demonstrate that monoubiquitination causes sustained H-Ras activation in the absence of oncogenic mutations.
Conclusion: The mechanism by which H-Ras is activated by monoubiquitination is both isoform-specific and site-specific.
Significance: Monoubiquitination adds a new level of regulation and complexity to isoform-specific Ras signaling.
Keywords: GTPase, Oncogene, Post-translational Modification, Protein Chemical Modification, Ras, Signal Transduction, Ubiquitination, Monoubiquitination
Abstract
Ras GTPases are signaling switches that control critical cellular processes including gene expression, differentiation, and apoptosis. The major Ras isoforms (K, H, and N) contain a conserved core GTPase domain, but have distinct biological functions. Among the three Ras isoforms there are clear differences in post-translational regulation, which contribute to differences in localization and signaling output. Modification by ubiquitination was recently reported to activate Ras signaling in cells, but the mechanisms of activation are not well understood. Here, we show that H-Ras is activated by monoubiquitination and that ubiquitination at Lys-117 accelerates intrinsic nucleotide exchange, thereby promoting GTP loading. This mechanism of Ras activation is distinct from K-Ras monoubiquitination at Lys-147, which leads to impaired regulator-mediated GTP hydrolysis. These findings reveal that different Ras isoforms are monoubiquitinated at distinct sites, with distinct mechanisms of action, but with a common ability to chronically activate the protein in the absence of a receptor signal or oncogenic mutation.
Introduction
The small GTPase Ras is a signaling switch that regulates cellular growth and differentiation (1). In accord with its essential role in regulating cell growth, point mutations in Ras are common in cancer; about 30% of all human tumors contain an activating Ras mutation (2). To control the activation of essential pathways involved in cell proliferation, differentiation, and apoptosis, Ras binds and hydrolyzes GTP. Ras is active when it is GTP-bound and becomes inactive when GTP is hydrolyzed to GDP (3, 4). On its own, Ras is a poor enzyme and requires regulators to respond to cell signals on an appropriate timescale. The primary regulators of Ras cycling are guanine nucleotide exchange factors (GEFs),4 which increase the rate of GDP dissociation and GTP loading, leading to Ras activation (5, 6), and GTPase-activating proteins (GAPs), which down-regulate activated Ras by increasing the rate of GTP hydrolysis (4, 7).
There are three isoforms of Ras: K-Ras (two splice variants, K-Ras4A and K-Ras4B), H-Ras, and N-Ras. The core guanine nucleotide binding domain is highly conserved between these isoforms (>90% identical in the first 168 amino acids). However, despite the high sequence identity, Ras isoforms play distinct roles in regulating downstream signaling pathways. In particular, the three proteins exhibit differences in their localization in vivo, which alters access to regulatory proteins and downstream effectors (8–10). H-Ras and N-Ras are localized at both the plasma membrane and the Golgi, whereas K-Ras is predominately at the plasma membrane (11). Differences in Ras localization and activity are driven by differences in post-translational modifications (12–19). For example, differential lipidation of the Ras isoforms within their variable C-terminal domain is essential for proper membrane targeting and localization, which is in turn necessary for proper signaling (15, 20–24). Another post-translational modification, phosphorylation, has been shown to modulate K-Ras signaling output by changing its localization from the plasma membrane to the mitochondria and Golgi (25). Both lipid and phosphoryl modifications are dynamically regulated, which in turn dynamically controls Ras localization and activity (25).
Aside from lipidation and phosphorylation, there is mounting evidence that ubiquitination also contributes to the differentiation between Ras isoforms (26). Ubiquitination of an as yet unidentified site in H-Ras promotes its association with the endosome (26, 27). This localization change leads to altered effector interactions and signaling output (25). We and others have shown that monoubiquitination of Lys-147 in K-Ras does not appear to affect protein localization, but rather acts as a reversible trigger for signal initiation (28, 29). Monoubiquitination of K-Ras at this position impedes GAP-mediated GTP hydrolysis and promotes association with select downstream effectors (28, 29). This mechanism of K-Ras activation appears specific to ubiquitination at Lys-147 because modification of other lysines does not confer a GAP defect (29). Thus, H-Ras and K-Ras display differential regulation by ubiquitination.
In addition to Lys-147, Sasaki et al. (28) have identified Lys-117 as a site of ubiquitination in H-Ras and Lys-104 as a secondary site of ubiquitination in K-Ras. Thus, we postulated that different sites of H- and K-Ras ubiquitination have distinct consequences for Ras activity. Here, we show that monoubiquitination at Lys-104 does not affect nucleotide exchange or hydrolysis properties of Ras, whereas modification at Lys-117 promotes Ras activation. However, in this case, the mechanism of activation is unique. Although monoubiquitination of K-Ras at Lys-147 inhibits GAP-mediated hydrolysis, monoubiquitination of H-Ras at Lys-117 greatly enhances GTP-GDP exchange. These findings reveal that K-Ras and H-Ras can each be activated by monoubiquitination, but that activation occurs via distinct molecular mechanisms.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
All Ras constructs were generated with a C118S mutation to eliminate a native solvent exposed cysteine, as described previously (29, 30). The Ras domains (1–166) of H-RasC118S, H-RasC118S/K104C, and H-RasC118S/K117C were expressed and purified as described previously (29). The standard Ras protocol was revised for purification of H-RasC118S/K117C by the addition of 10% glycerol to all buffers to accommodate for the instability of this variant.
Catalytic domains of the GEF, Sos (Soscat), and GAP, p120GAP (GAP-334), were purified as described previously (29, 31). Full-length ubiquitinG76C was also purified as described previously (29).
Chemical Ubiquitination
The chemical ligation strategy used to link H-RasK104C and H-RasK117C to ubiquitinG76C was performed essentially as described in Baker et al. (29), except that 10% glycerol was added to buffers containing the RasK117C variant.
Thermal Stability of Ras
The fast quantitative cysteine reactivity (fQCR) method (32) was employed to measure changes in Ras thermal stability. Briefly, 2 μm RasK104C, mUbRasK104, RasK117C, or mUbRasK117 was incubated with 1 mm 4-fluoro-7-aminosulfonylbenzofurazan (AnaSpec) at pH 7.0 in the presence of 20 μm GTPγS and 2 mm MgCl2 in a gradient thermocycler (Biometra TProfessional thermocycler) at the desired temperature for 3 min. Reactions were quenched by the addition of 0.02 n HCl (final concentration). Fluorescence intensity was measured on a PHERAstar plate reader (BMG Labtech) with excitation and emission band-pass filters of 400 and 500 nm. The highest fluorescence for each unfolding curve was normalized to one after base lines were corrected for differences in background fluorescence. Data were fit using GraphPad Prism (GraphPad Software; San Diego, CA) to determine the melting temperature (Tm) for each construct.
Nucleotide Dissociation Assay
For the RasK104C and mUbRasK104 variants, the N-methylanthraniloyl (MANT)-GDP nucleotide dissociation assay was performed as described previously (29). The assay was adapted for RasK117C and mUbRasK117 as follows. Ras protein was exchanged into MANT nucleotide exchange buffer (50 mm Tris, pH 8.0, 50 mm NaCl, and 5 mm MgCl2). MANT-GDP (0.5 μm) was added to 1 μm of protein in the cuvette. The Ras protein was determined to be fully loaded when the fluorescence of the spectra no longer increased, ∼500 s. Unlabeled GDP (final concentration of 100 μm) was added to initiate dissociation. Soscat was mixed with H-Ras at a 1:1 molar ratio. MANT-GDP dissociation was measured as a change in fluorescence intensity over time (excitation: 360 nm, emission: 440 nm) (LS50B PerkinElmer luminescence spectrometer). Fluorescence data were fit in GraphPad Prism to a one-phase exponential decay curve to determine rates of nucleotide release.
Single Turnover Nucleotide Hydrolysis
Single turnover nucleotide hydrolysis assays were performed as described in Baker et al. (29). The ratio of fluorescence emission was measured at 480 and 530 nm with an excitation of 435 nm on a SpectraMax M5 (Molecular Devices). Fluorescence ratios were converted to phosphate concentrations using a standard curve. The measured rates of phosphate release over time were fit in GraphPad Prism to a one-phase exponential association curve.
Multiple Turnover Nucleotide Hydrolysis
Due to limitations associated with instability of RasK117C and mUbRasK117, GTP hydrolysis was measured in the presence of excess GTP (multiple turnover). Ras was exchanged (20-fold dilution and concentrated three times) into phosphate-free buffer with 5 mm EDTA. For intrinsic hydrolysis, 20 μm Ras was mixed with 60 μm GTP, and hydrolysis was initiated by the addition of magnesium. For GAP-mediated hydrolysis, GAP-334, at concentrations ranging from 0.0625 to 8 μm, was added to the reaction. The phosphate-binding protein Flippi 5U (Addgene) was used to detect inorganic phosphate released upon GTP hydrolysis (33). Fluorescence emission was measured as a ratio at 480 and 530 nm and an excitation of 435 nm (SpectraMax M5, Molecular Devices). Fluorescence ratios were converted to a phosphate concentration using a standard curve. Data from the first 10 min of hydrolysis were fit in GraphPad Prism to a linear regression and the resulting rates were plotted as a function of GAP concentration.
Mapping GEF and GAP Binding Sites on Ras
The pHinder algorithm was used to calculate a surface between Ras and the catalytic domains of Sos and p120GAP. Information on the triangulation used to define the surface has been described elsewhere (34). Residues were considered to be part of the interaction surface if they were less than 6 Å apart. Interaction surfaces were determined from the crystal structures of Ras with Sos (Protein Data Bank (PDB) 1BKD) and Ras with p120GAP (PDB 1WQ1).
Assays in Cell Lysate
Ras activation was measured as described previously (28). The constructs FLAG-His-H-Ras or the mutant FLAG-His-H-RasK117N were co-expressed with HA-ubiquitin in HEK293T cells. The cells were prepared and lysed as described previously (29). For assays conducted in the presence of either Soscat or p120GAP, the catalytic domains were purified from bacterially produced protein and incubated with the cell lysate for 20 min at room temperature. The effect of the regulators on the population of active Ras or mUbRas was assessed by immunoblotting using GST-Raf-RBD (Ras binding domain) to detect active Ras. For the Soscat assay, 25 μm of GTPγS was added during the incubation. Samples were prepared from SDS-PAGE gels (Invitrogen, NuPAGE), and Ras was detected by varying exposure times.
RESULTS
Monoubiquitination at Lys-117 Activates H-Ras by Increasing Intrinsic Exchange
It was recently shown that H-Ras can be monoubiquitinated at Lys-117 (28). Lysine 117 is part of the conserved NKXD motif that contributes to nucleotide affinity and specificity by forming interactions with the guanine nucleotide base and ribose (Fig. 1A) (35). Conservative mutations at Lys-117 (K117R and K117N) are present in human cancers and developmental disorders and lead to Ras activation by reducing nucleotide binding affinity, resulting in increased rates of nucleotide exchange (36–38). Given these observations, we postulated that ubiquitination of Lys-117 is likely to disrupt interactions between the lysine and the guanine nucleotide base and activate Ras by a mechanism similar to mutation at the same site.
FIGURE 1.
Monoubiquitination at lysine 117 activates H-Ras. A, graphic highlighting interactions formed between the NKXD motif of Ras (GDP) and the guanine nucleotide base (PDB 1CRR). Nucleotide is color-coded by element (nitrogen in blue, oxygen in red, and carbon in green), and Lys-117 of Ras is shown in purple. B, formation of disulfide bond between ubiquitinG76C and RasK117C under nonreducing (NR) and reducing (R) conditions. The product of the reaction contains mUbRas, Ras, ubiquitin-ubiquitin dimer (Ub-Ub), and free ubiquitin (Ub). C, the thermal stability of Ras (black squares throughout), RasK117C (purple triangles throughout), and mUbRasK117 (blue circles throughout) measured by fQCR. The data were normalized using the maximum fluorescence intensity. Results are the mean ± S.E. (n = 4). D, intrinsic nucleotide release (closed symbols) for Ras, RasK117C, and mUbRasK117 bound to MANT-GDP. The rate of GDP dissociation was determined by monitoring the decrease in MANT-GDP fluorescence emission over time following the addition of unlabeled GDP. Nucleotide dissociation was also measured in the presence of a 1:1 molar ratio of Ras to Soscat (open symbols). Data were fit to an exponential dissociation curve, and the results are the mean ± S.E. (n = 4). E, multi-turnover hydrolysis for Ras, RasK117C, and mUbRasK117. GTP hydrolysis was measured by monitoring the change in fluorescence of the phosphate-binding protein, Flippi, upon binding to free phosphate. Data were converted to a phosphate concentration using a standard curve. Phosphate release was measured for 20 μm Ras in the presence of GAP concentrations ranging from 0.0625 to 8 μm. Results are the mean ± S.E. (n = 4). Data were fit to a one site binding model in GraphPad Prism.
We chemically ubiquitinated Ras at position 117 (mUbRasK117) using a cysteine ligation approach reported by us previously (Fig. 1B) (29). Mutation at a nearby site in the conserved NKXD motif, Asp-119, reduces protein stability and increases the rate of nucleotide exchange (39). We therefore investigated whether perturbations at Lys-117 could also alter protein stability. We measured the thermal stability of Ras using the fQCR assay (32). This assay measures protein unfolding as a function of temperature and chemical labeling of exposed cysteines. Because ubiquitin does not contain native cysteines, fQCR can be used to specifically determine whether ubiquitination affects Ras thermal stability. We found that mUbRasK117 retains the thermal stability observed for unmodified Ras (Fig. 1C) (Tm = 52.5 ± 0.5 °C). However, ubiquitination at Lys-117 is distinct from a RasK117C mutation, which leads to a 10° decrease in the thermal stability of Ras (RasK117C Tm = 43.8 ± 0.9 °C). Relative to unmodified Ras, RasK117C and mUbRasK117C exhibited less cooperative unfolding transitions (Fig. 1C) and increased rates of nucleotide dissociation (Fig. 1D). We attribute this correlation to loss of a key interaction (Lys-117) that impairs nucleotide binding, resulting in reduced cooperativity of the unfolding transition (40). Although either ubiquitination or mutation of Ras at position 117 alters the lysine side chain, which is crucial for forming interactions with the guanine nucleotide, these distinct alterations differentially affect the thermal stability of Ras.
We next measured the effect of monoubiquitination on the rate of guanine nucleotide dissociation. We find that the intrinsic rate of nucleotide dissociation was 70-fold faster for mUbRasK117 as compared with unmodified Ras (8.3 ± 0.3 × 10−3 s−1) (Fig. 1D). This finding is consistent with the 100-fold increased rate of nucleotide release observed when Ras is mutated (RasK117C) at this same position (11.0 ± 0.4 × 10−1 s−1) (Fig. 1D). We also observed a small increase in GDP dissociation for mUbRasK117 in the presence of the GEF, Soscat, indicating that the nucleotide release can be further stimulated by GEFs. Even in the absence of GEF-mediated activation, the increased rate of intrinsic nucleotide dissociation should be sufficient to promote GTP exchange and mUbRasK117 activation in vivo (41).
We then measured intrinsic and GAP-mediated GTP hydrolysis using a multi-turnover GTP hydrolysis assay, as described previously (41). When Ras is monoubiquitinated at Lys-117, the rate of intrinsic GTP hydrolysis is similar (within 2-fold) to that of unmodified Ras (Fig. 1E). Furthermore, mUbRasK117 retains sensitivity to p120GAP (GAP-334), albeit with a reduced binding affinity. As seen in Fig. 1E, at 8 μm GAP-334 (5:2 ratio of Ras to GAP-334), the rate of GTP hydrolysis is not affected by monoubiquitination (0.024 ± 0.003 and 0.023 ± 0.002 s−1 for Ras and mUbRasK117, respectively). Therefore, mUbRasK117 is still sensitive to GAP-mediated down-regulation. Similar results were observed for both RasK117C (Fig. 1E) and RasK117R variants (41). Taken together, these data demonstrate that monoubiquitination of H-Ras at Lys-117 causes activation by enhancing the rate of nucleotide exchange.
Ras Monoubiquitinated at Lys-104 Retains the Activity of Unmodified Ras
Mass spectrometry characterization of K-Ras and H-Ras in HEK293T cells identified two major sites of ubiquitination, Lys-147 (K-Ras and H-Ras) and Lys-117 (H-Ras only) (28). Our data indicate that these modifications result in Ras activation, but act through distinct mechanisms. An additional minor site of ubiquitination was observed at position Lys-104 in K-Ras (28). Although Lys-147 and Lys-117 are in a conserved nucleotide binding motif, Lys-104 is located in helix 3 adjacent to the GEF binding interface (Fig. 2A). Substitutions at this position (K104Q) have been proposed to alter GEF-mediated dissociation by disrupting an electrostatic interaction that the Lys-104 side chain forms with the α2 helix. Moreover, acetylation at this position has recently been shown to impair GEF-mediated regulation of Ras (42). Therefore, we considered whether monoubiquitination at Lys-104 could also alter K-Ras activity.
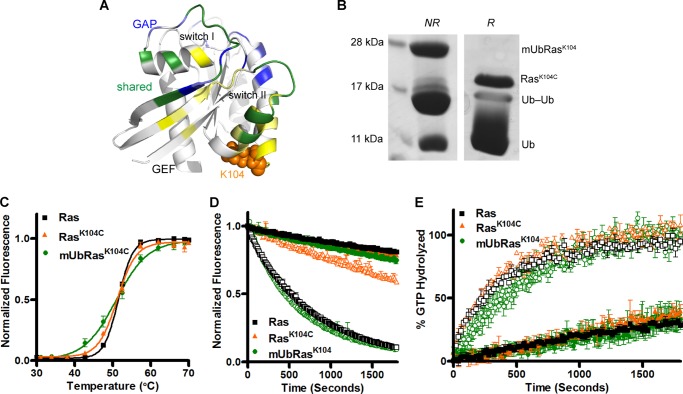
FIGURE 2.
Ras monoubiquitinated at lysine 104 retains the activity of unmodified Ras. A, structure of Ras-GDP (PDB 1CRR) with the Lys-104 side chain highlighted in orange. Backbone residues in yellow make contact with the GEF Sos (PDB 1BKD), backbone residues in blue make contact with the GAP p120GAP (PDB 1WQ1), and backbone residues in green are contacts common for both the GEF and the GAP. B, formation of disulfide bond between ubiquitinG76C and RasK104C under nonreducing (NR) and reducing (R) conditions. The product of the reaction contains mUbRas, Ras, ubiquitin-ubiquitin dimer (Ub-Ub), and free ubiquitin (Ub). C, the thermal stability of Ras (black squares throughout), RasK104C (orange triangles throughout), and mUbRasK104 (green circles throughout) measured by fQCR. The data were normalized using the maximum fluorescence intensity. Results are the mean ± S.E. (n = 4). D, intrinsic nucleotide dissociation (closed symbols) for Ras, RasK104C, and mUbRasK104 loaded with MANT-GDP. The rate of GDP dissociation was determined by monitoring the decrease in MANT-GDP fluorescence emission over time following the addition of unlabeled GDP. Nucleotide dissociation was also measured in the presence of a 1:1 molar ratio of Ras to Soscat (open symbols). Data were fit to an exponential dissociation curve, and the results are the mean ± S.E. (n = 4). E, single-turnover GTP hydrolysis for Ras, RasK104C, and mUbRasK104 in the absence (closed symbols) and presence (open symbols) of GAP-334 (1:500 GAP:Ras). Hydrolysis was initiated by the addition of Mg2+ and monitored by the change in fluorescence of the protein, Flippi, upon binding free phosphate. Data were converted to a phosphate concentration using a standard curve. Results are the mean ± S.E. (n = 6).
To determine whether monoubiquitination at position 104 alters Ras activity, we chemically monoubiquitinated the protein at Lys-104 (mUbRasK104) (Fig. 2B) and measured thermal stability as well as intrinsic and regulator-mediated enzymatic activity. As seen in Fig. 2C, the thermal stability of mUbRasK104 is similar to that of Ras (Tm = 50.8 ± 0.4 °C), consistent with mutation at this same position. We next measured intrinsic and GEF-mediated rates of GDP dissociation using Soscat. This analysis showed that nucleotide release was unaffected by monoubiquitination or mutation at position 104 (Fig. 2D). However, we did not observe a change in Soscat-mediated dissociation for mUbRasK104 (Fig. 2D), although the RasK104C substitution decreased Soscat-mediated activation by 5-fold, consistent with previous studies (42). Neither monoubiquitination nor mutation at position 104 altered intrinsic or GAP-mediated single-turnover GTP hydrolysis (Fig. 2E). Together, these data demonstrate that Lys-104 on Ras can be monoubiquitinated without altering intrinsic or regulator-mediated activity. Moreover, although K-Ras can be ubiquitinated at two distinct lysines in vivo (Lys-147 and Lys-104), only the more prevalent site of ubiquitination, Lys-147, leads to a change in the active state of the protein.
H-Ras Is Activated and Primarily Modified at Lysine 117 in HEK293T Cells
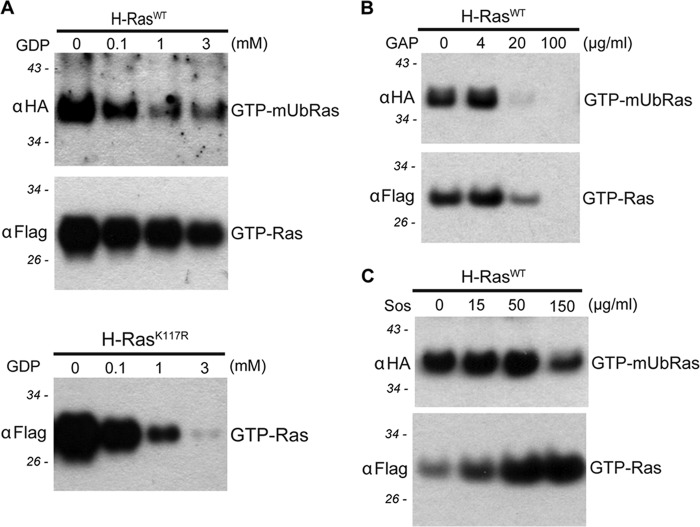
Our mechanistic studies indicate that monoubiquitination of H-Ras at Lys-117 enhances guanine nucleotide dissociation, leading to increased GTP loading and protein activation. We previously showed that the mechanism of K-Ras activation determined in vitro, using a chemical ubiquitination approach, recapitulated the GAP defect observed for mUbRasK147 in cell lysate assays (29). In the original study by Sasaki et al. (28), H-Ras was found to be ubiquitinated at both Lys-117 and Lys-147 in HEK293T cells. Because ubiquitination at Lys-117 and Lys-147 activates Ras by distinct mechanisms, we exploited this difference to determine whether modification at one or the other lysines predominates in HEK293T cells. Using Ras binding domain pulldown assays that specifically detect GTP-bound Ras, we measured intrinsic and regulator-mediated nucleotide exchange and hydrolysis. Monoubiquitinated Ras accounts for ∼1% of the total Ras protein and was detected by blotting for HA-ubiquitin. As seen in Fig. 3A, as the concentration of exogenously added GDP increases, the population of GTP-bound mUbRas decreases more than unmodified GTP-bound H-Ras. The increased loss of activated mUbRas relative to unmodified Ras indicates that nucleotide exchange is faster when H-Ras is monoubiquitinated. This result is similar to what is observed in the RasK117R mutant (Fig. 3A), which has previously been shown to possesses an 80-fold faster rate of intrinsic nucleotide dissociation relative to unmodified Ras, similar to the 70-fold increased rate of nucleotide exchange observed for mUbRasK117 (41). We also determined whether the population of monoubiquitinated H-Ras immunoprecipitated from HEK293T cell lysates was sensitive to Sos-mediated regulation. When H-Ras is monoubiquitinated, the population of active protein does not change significantly in the presence of recombinant GEF (Fig. 3B), indicating that mUbRas is not further activated by Soscat. Ras and mUbRas were equally responsive to the addition of recombinant GAP-334, suggesting that Ras had been modified at a site other than Lys-147 in these cells (Fig. 3C). Based on these findings, we propose that cellular H-Ras is predominately monoubiquitinated at Lys-117 in HEK293T cells and that monoubiquitination at this site activates the protein through a mechanism distinct from that of K-Ras. Although monoubiquitinated K-Ras is uncoupled from the GTPase-activating protein, monoubiquitinated H-Ras exhibits an increase in intrinsic nucleotide exchange activity.
FIGURE 3.
Assays in cell lysate indicate H-Ras is primarily modified at lysine 117. A, immunoblotting of GTP-bound H-Ras, GTP-bound mUbRas, and GTP-bound H-RasK117R from cell extracts in the presence of the indicated concentrations of GDP. Anti-FLAG and anti-HA antibodies reveal the relative fraction of total Ras and mUbRas, respectively, for all assays. Molecular masses are shown in italics for all panels. B, immunoblotting of GTP-bound Ras and GTP-bound mUbRas from cell extracts in the presence of the indicated concentrations of recombinant p120GAP, GAP-334. C, immunoblotting of GTP-bound Ras and GTP-bound mUbRas in cell extract in the presence of increasing concentrations of recombinant Soscat and 25 μm GTPγS.
DISCUSSION
We have shown previously that monoubiquitination of K-Ras at Lys-147 promotes protein activation by rendering the protein insensitive to GAP-mediated GTP hydrolysis (29). This same modification does not alter the intrinsic nucleotide exchange and GTP hydrolysis activities of Ras, and it has only a modest effect on GEF-mediated nucleotide exchange. Here, we characterize two additional sites of monoubiquitination, one acting on K-Ras at Lys-104 and the other acting on H-Ras at Lys-117. We find that in vitro, monoubiquitination at Lys-117 uniquely up-regulates H-Ras activity by promoting exchange of GDP for GTP. Furthermore, our cell lysate assays indicate that ubiquitination of H-Ras at Lys-117 predominates in HEK293T cells because monoubiquitinated H-Ras was sensitive to GAP-mediated regulation but displayed an increased rate of intrinsic nucleotide exchange. Activation of Ras by sustained GTP binding (monoubiquitination of K-Ras at Lys-147) may have distinct cellular outcomes from that of constitutive GTP cycling (monoubiquitination of H-Ras at Lys-117). In support of this premise, disease-related Ras mutations that are activated through increased exchange have different phenotypes than those that cause inhibition of GAP-mediated hydrolysis (43).
In contrast to monoubiquitination of Ras at Lys-147 and Lys-117, monoubiquitination of Lys-104 does not affect Ras activity in vitro. The absence of any functional difference is striking, particularly because monoubiquitination at a nearby site (Lys-117) was sufficient to fully activate the protein. Nevertheless, the identification of a monoubiquitination site on Ras that does not alter activity is consistent with previous data obtained in CHOK-1 cells, where trafficking to the endosome rather than direct H-Ras activation was the consequence of monoubiquitination (26). Together, these data suggest that site-specific ubiquitination may occur in both an isoform-specific and a cell line-specific manner to regulate Ras localization and signaling. Left unresolved is how the differences in ubiquitination are controlled, although they are likely to depend on differences in E3 ubiquitin ligase and deubiquitinating enzyme expression or localization (28).
It was recently shown that Lys-104 is also a site of acetylation in K-Ras in HEK293T cells (42). Molecular dynamics simulations suggest that acetylation at Lys-104 alters switch II due to a perturbation in electrostatic interaction in the α2 helix that populates the Ras switch region in a disordered conformation, which is less favorable for Ras-GEF interaction (42). Acetylation of K-Ras at Lys-104 conferred a defect in GEF-mediated exchange, resulting in its accumulation in the inactive GDP-bound state both in vitro and in cells (42). Given that acetylation and monoubiquitination are likely to be mutually exclusive, there may be a role for Lys-104 ubiquitination in protecting Ras from acetylation, or vice versa.
Ras proteins are well known to be activated by growth factor receptors and by oncogenic mutations. Although the structure and sequence of the three Ras isoforms are highly conserved, they play distinct roles in signaling and cell regulation in vivo. Differences in Ras localization and activity are driven by differences in post-translational modifications (12–19). Our analysis has revealed two distinct mechanisms of Ras activation through the same post-translational modification. Furthermore, our biochemical analysis of site-specific Ras ubiquitination revealed that ubiquitination alters Ras through mechanisms distinct from mutations at the same positions. Assuming that these events are regulated independently, our findings reveal a potential new mechanism for dictating functional hierarchy and conferring isoform-dependent differences in Ras activation and signaling.
This work was supported, in whole or in part, by National Institutes of Health Grants R01-GM106227A (to S. L. C.) and R01-GM101560 (to H. G. D.). This work was also supported by University Cancer Research Fund (UCRF) innovative grants (to S. L. C.) and by the University of Cincinnati DNS/MERF (to A. T. S.).
- GEF
- guanine nucleotide exchange factor
- GAP
- GTPase-activating protein
- fQCR
- fast quantitative cysteine reactivity
- mUbRas
- monoubiquitinated Ras
- MANT
- N-methylanthraniloyl
- Ub
- ubiquitin
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate.
REFERENCES
- 1. Herrmann C. (2003) Ras-effector interactions: After one decade. Curr. Opin. Struct. Biol. 13, 122–129 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A., Siegel R., Ward E., Hao Y., Xu J., Thun M. J. (2009) Cancer statistics, 2009. CA Cancer J. Clin. 59, 225–249 [DOI] [PubMed] [Google Scholar]
- 3. Campbell S. L., Khosravi-Far R., Rossman K. L., Clark G. J., Der C. J. (1998) Increasing complexity of Ras signaling. Oncogene 17, 1395–1413 [DOI] [PubMed] [Google Scholar]
- 4. Geyer M., Wittinghofer A. (1997) GEFs, GAPs, GDIs and effectors: Taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins. Curr. Opin. Struct. Biol. 7, 786–792 [DOI] [PubMed] [Google Scholar]
- 5. Karnoub A. E., Symons M., Campbell S. L., Der C. J. (2004) Molecular basis for Rho GTPase signaling specificity. Breast Cancer Res. Treat. 84, 61–71 [DOI] [PubMed] [Google Scholar]
- 6. Sprang S. (2001) GEFs: Master regulators of G-protein activation. Trends Biochem. Sci. 26, 266–267 [DOI] [PubMed] [Google Scholar]
- 7. Sprang S. R. (1997) G proteins, effectors and GAPs: Structure and mechanism. Curr. Opin. Struct. Biol. 7, 849–856 [DOI] [PubMed] [Google Scholar]
- 8. Daniels M. A., Teixeiro E., Gill J., Hausmann B., Roubaty D., Holmberg K., Werlen G., Holländer G. A., Gascoigne N. R. J., Palmer E. (2006) Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444, 724–729 [DOI] [PubMed] [Google Scholar]
- 9. Omerovic J., Laude A. J., Prior I. A. (2007) Ras proteins: Paradigms for compartmentalised and isoform-specific signalling. Cell. Mol. Life Sci. 64, 2575–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolfman A. (2001) Ras isoform-specific signaling: Location, location, location. Sci STKE 2001, pe2. [DOI] [PubMed] [Google Scholar]
- 11. Hancock J. F. (2003) Ras proteins: Different signals from different locations. Nat. Rev. Mol. Cell. Biol. 4, 373–384 [DOI] [PubMed] [Google Scholar]
- 12. Pfleger C. M. (2011) Ubiquitin on Ras: Warden or partner in crime? Sci. Signal. 4, pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tidyman W. E., Rauen K. A. (2009) The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 19, 230–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Omerovic J., Prior I. A. (2009) Compartmentalized signalling: Ras proteins and signalling nanoclusters. FEBS J. 276, 1817–1825 [DOI] [PubMed] [Google Scholar]
- 15. Ahearn I. M., Haigis K., Bar-Sagi D., Philips M. R. (2012) Regulating the regulator: Post-translational modification of RAS. Nat. Rev. Mol. Cell. Biol. 13, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. (1989) All Ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57, 1167–1177 [DOI] [PubMed] [Google Scholar]
- 17. Laude A. J., Prior I. A. (2008) Palmitoylation and localisation of RAS isoforms are modulated by the hypervariable linker domain. J. Cell Sci. 121, 421–427 [DOI] [PubMed] [Google Scholar]
- 18. Choy E., Chiu V. K., Silletti J., Feoktistov M., Morimoto T., Michaelson D., Ivanov I. E., Philips M. R. (1999) Endomembrane trafficking of Ras: The CAAX motif targets proteins to the ER and Golgi. Cell 98, 69–80 [DOI] [PubMed] [Google Scholar]
- 19. Apolloni A., Prior I. A., Lindsay M., Parton R. G., Hancock J. F. (2000) H-Ras but not K-Ras traffics to the plasma membrane through the exocytic pathway. Mol. Cell. Biol. 20, 2475–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prior I. A., Muncke C., Parton R. G., Hancock J. F. (2003) Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 160, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plowman S. J., Muncke C., Parton R. G., Hancock J. F. (2005) H-Ras, K-Ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 102, 15500–15505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prior I. A., Harding A., Yan J., Sluimer J., Parton R. G., Hancock J. F. (2001) GTP-dependent segregation of H-Ras from lipid rafts is required for biological activity. Nat. Cell Biol. 3, 368–375 [DOI] [PubMed] [Google Scholar]
- 23. Roy S., Plowman S., Rotblat B., Prior I. A., Muncke C., Grainger S., Parton R. G., Henis Y. I., Kloog Y., Hancock J. F. (2005) Individual palmitoyl residues serve distinct roles in H-Ras trafficking, microlocalization, and signaling. Mol. Cell. Biol. 25, 6722–6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Niv H., Gutman O., Kloog Y., Henis Y. I. (2002) Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 157, 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castellano E., Santos E. (2011) Functional Specificity of Ras isoforms: So similar but so different. Genes Cancer 2, 216–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jura N., Scotto-Lavino E., Sobczyk A., Bar-Sagi D. (2006) Differential modification of Ras proteins by ubiquitination. Mol. Cell 21, 679–687 [DOI] [PubMed] [Google Scholar]
- 27. Colicelli J. (2010) Signal transduction: RABGEF1 fingers RAS for ubiquitination. Curr. Biol. 20, R630–R632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sasaki A. T., Carracedo A., Locasale J. W., Anastasiou D., Takeuchi K., Kahoud E. R., Haviv S., Asara J. M., Pandolfi P. P., Cantley L. C. (2011) Ubiquitination of K-Ras enhances activation and facilitates binding to select downstream effectors. Sci. Signal. 4, ra13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baker R., Lewis S. M., Sasaki A. T., Wilkerson E. M., Locasale J. W., Cantley L. C., Kuhlman B., Dohlman H. G., Campbell S. L. (2013) Site-specific monoubiquitination activates Ras by impeding GTPase-activating protein function. Nat. Struct. Mol. Biol. 20, 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mott H. R., Carpenter J. W., Campbell S. L. (1997) Structural and functional analysis of a mutant Ras protein that is insensitive to nitric oxide activation. Biochemistry 36, 3640–3644 [DOI] [PubMed] [Google Scholar]
- 31. Sondermann H., Soisson S. M., Boykevisch S., Yang S. S., Bar-Sagi D., Kuriyan J. (2004) Structural analysis of autoinhibition in the Ras activator Son of Sevenless. Cell 119, 393–405 [DOI] [PubMed] [Google Scholar]
- 32. Isom D. G., Marguet P. R., Oas T. G., Hellinga H. W. (2011) A miniaturized technique for assessing protein thermodynamics and function using fast determination of quantitative cysteine reactivity. Proteins 79, 1034–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gu H., Lalonde S., Okumoto S., Looger L. L., Scharff-Poulsen A. M., Grossman A. R., Kossmann J., Jakobsen I., Frommer W. B. (2006) A novel analytical method for in vivo phosphate tracking, FEBS Lett. 580, 5885–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Isom D. G., Sridharan V., Baker R., Clement S. T., Smalley D. M., Dohlman H. G. (2013) Protons as second messenger regulators of G protein signaling. Mol. Cell 51, 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vetter I. R., Wittinghofer A. (2001) The guanine nucleotide-binding switch in three dimensions. Science 294, 1299–1304 [DOI] [PubMed] [Google Scholar]
- 36. Schubbert S., Bollag G., Lyubynska N., Nguyen H., Kratz C. P., Zenker M., Niemeyer C. M., Molven A., Shannon K. (2007) Biochemical and functional characterization of germ line KRAS mutations. Mol. Cell. Biol. 27, 7765–7770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Janakiraman M., Vakiani E., Zeng Z., Pratilas C. A., Taylor B. S., Chitale D., Halilovic E., Wilson M., Huberman K., Ricarte Filho J. C., Persaud Y., Levine D. A., Fagin J. A., Jhanwar S. C., Mariadason J. M., Lash A., Ladanyi M., Saltz L. B., Heguy A., Paty P. B., Solit D. B. (2010) Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res. 70, 5901–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K. V., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 39. Schmidt G., Lenzen C., Simon I., Deuter R., Cool R. H., Goody R. S., Wittinghofer A. (1996) Biochemical and biological consequences of changing the specificity of p21Ras from guanosine to xanthosine nucleotides. Oncogene 12, 87–96 [PubMed] [Google Scholar]
- 40. Zhang J., Matthews C. R. (1998) Ligand binding is the principal determinant of stability for the p21H-ras protein. Biochemistry 37, 14881–14890 [DOI] [PubMed] [Google Scholar]
- 41. Denayer E., Parret A., Chmara M., Schubbert S., Vogels A., Devriendt K., Frijns J.-P., Rybin V., de Ravel T. J., Shannon K., Cools J., Scheffzek K., Legius E. (2008) Mutation analysis in Costello syndrome: Functional and structural characterization of the HRAS p.Lys117Arg mutation. Hum. Mutat. 29, 232–239 [DOI] [PubMed] [Google Scholar]
- 42. Yang M. H., Nickerson S., Kim E. T., Liot C., Laurent G., Spang R., Philips M. R., Shan Y., Shaw D. E., Bar-Sagi D., Haigis M. C., Haigis K. M. (2012) Regulation of RAS oncogenicity by acetylation. Proc. Natl. Acad. Sci. U.S.A. 109, 10843–10848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gremer L., Merbitz-Zahradnik T., Dvorsky R., Cirstea I. C., Kratz C. P., Zenker M., Wittinghofer A., Ahmadian M. R. (2011) Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 32, 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]